SYNOPSIS
Sss1p, an essential component of the heterotrimeric Sec61 complex in the endoplasmic reticulum (ER) #, is a tail-anchored protein whose precise mechanism of action is largely unknown. Tail-anchored proteins are involved in many cellular processes and are characterized by a single transmembrane sequence (TMS) at or near the carboxyl-terminus. The Sec61 complex is the molecular machine through which secretory and membrane proteins translocate into and across the ER membrane. To understand the function of the tail-anchor of Sss1p, we introduced mutations into the tail-anchor sequence and analyzed the resulting yeast phenotypes. Point mutations in the C-terminal hydrophobic core of the tail-anchor of Sss1p were identified that allowed Sss1p assembly into Sec61 complexes but resulted in diminished growth, defects in co- and post-translational translocation, diminished ribosome binding to Sec61 complexes, reduced stability of both heterotrimeric Sec61 and heptameric Sec complexes, and a complete breakdown of ER structure. The underlying defect caused by the mutations involves loss of a stabilizing function of the Sss1p tail-anchor sequence for both the heterotrimeric Sec61 and the heptameric Sec complexes. These data indicate that by stabilizing multiprotein membrane complexes, the hydrophobic core of a tail-anchor sequence can be more than a simple membrane anchor.
Keywords: tail-anchor proteins, translocon, endoplasmic reticulum, membrane protein complexes, Sec61, yeast
INTRODUCTION
Transport across the ER is a vital, first committed step in the biogenesis of secretory and integral membrane proteins. These proteins are targeted to the ER either co-translationally as ribosome-bound nascent chains, or in a post-translational manner, as fully synthesized polypeptides.
The translocon is the molecular machine that forms the channel in the ER through which proteins are conducted. The core of the translocon is comprised of a heterotrimeric membrane complex called the Sec61 complex [1–5]. The α and γ subunits of this complex (Sec61p and Sss1p in yeast, and SecY and SecE in eubacteria) show significant sequence conservation and are essential for cell viability. In contrast, the β subunits (Sbh1p/SecG) are dispensable and are similar in archaea and eukaryotes, but show little homology to the corresponding subunit in eubacteria.
In yeast, the Sec61 complex is required for both co- and post-translational translocation. During co-translational translocation, the Sec61 complex, the ER membrane protein Sec63p, and the ER lumenal chaperone, Kar2p (the yeast homologue of BiP) are necessary for efficient protein import [6–9]. In reconstituted systems, the co-translational translocation of proteins synthesized from membrane-docked ribosomes requires only the Sec61 complex [1]. However, post–translational translocation requires the Sec61 complex as well as another multi-protein complex that contains Sec62p, Sec63p, Sec71p, and Sec72p. The resulting heptameric complex is known as the Sec complex [10, 11]. The Sec complex consists of both essential (Sec62p and Sec63p) and non-essential (Sec71p and Sec72p) proteins [10, 12, 13]. In addition, Kar2p is required for post-translational transport; it binds to the J domain of Sec63p and facilitates translocation of the emerging polypeptide through successive rounds of binding that are driven by ATP hydrolysis [14, 15]. Proteins utilizing the post-translational pathway are maintained in a translocation competent conformation by molecular chaperones, which also play a role in targeting them to the heptameric Sec complex [16, 17].
The γ–subunit of the Sec61complex, or Sss1p in the yeast Saccharomyces cerevisiae, is a 9 kDa tail-anchored protein. Depletion of Sss1p leads to a severe defect in co- and post-translational translocation [18]. Tail-anchored proteins, such as Sss1p, are inserted into membranes post-translationally via a carboxyl-terminal hydrophobic region [19–21]. It is not completely clear how distinct tail-anchor sequences favour the association of a protein with one intracellular membrane versus another, even though the machinery that inserts tail-anchor proteins into the ER membrane has been better defined in recent years [22–27].
A number of models have been proposed to define why Sss1p is essential and why protein depletion leads to profound translocation defects. Sss1p may help to maintain the membrane permeability barrier by acting as a place holder for signal peptides within the Sec61 complex [28], it may facilitate the oligomerization of the heterotrimeric Sec61 complex [29], and/or it may clamp together the two halves of the Sec61α subunit [30]. The cytoplasmic region of Sss1p has also been shown to interact with oligosaccharyl-transferase [31, 32], suggesting that the protein helps assemble other components required for nascent secretory protein biogenesis. The hydrophobic core of tail-anchor sequences are generally considered to have limited function(s) other than positioning the protein at the correct subcellular membrane location. Consistent with this view, point mutations of the conserved glycines in the hydrophobic core of Sss1p were tolerated but deletion of the 12 residues C-terminal of the hydrophobic sequence disrupted the translocation function of the Sec61 complex [33]. The molecular cause of the functional defect in translocons with large deletions in Sss1p remains unclear because these mutant Sss1 proteins still targeted to and interacted with Sec61p [33].
To determine the molecular mechanism for which the tail-anchor sequence of Sss1p is important, we introduced specific mutations into this motif. Point mutations in conserved sequences in both the transmembrane and flanking regions were analyzed phenotypically and by biochemical methods. The impact of single point mutations on cellular physiology was profound, including complete alteration of the ER structure and cessation of cell division. Data from this genetic analysis were then complemented by biochemical analyses of isolated Sec61 complexes and the larger Sec complexes that revealed the molecular defects in specific mutant proteins. Our collective results suggest that the specific sequence of the hydrophobic core of the tail-anchor of Sss1p is critical for stabilizing the Sec61 complex, and therefore Sec complexes. Surprisingly, Sss1 proteins with mutations in this region still assemble into heterotrimeric Sec61 complexes and heptameric Sec complexes, yet the oligomers are less stable and exhibit translocation defects that lead to profound defects in ER morphology. Even small perturbations such as extending the length of the hydrophobic core by a single residue or transposing two adjacent residues were sufficient to abolish translocon function. We conclude that residues at the C-terminal end of the Sss1p tail-anchor sequence are critical for translocon architectural integrity and function and that translocon function places severe constraints on the stability of the assembled complexes.
EXPERIMENTAL
Plasmids
A low-copy yeast centromere plasmid (YCp) vector with a HIS3 marker (pRS313 [34]) was used to express wild-type (WT) and mutant Sss1p proteins. All mutant coding sequences were initially constructed in psPUTK (Stratagene, La Jolla, California) or a derivative thereof and later sub-cloned into a pRS313 derivative containing the endogenous promoter and 3’UTR of Sss1p. All plasmid sequences are available upon request (also see Supplemental Information).
Viability Assays and Immunoblotting
Saccharomyces cerevisiae haploid strain FKY173 (MATα sss1:: URA3 leu2-3,-112 ade2-1 ura3-1 his3-11,-15 trp1-1 can1-100 (pFKp106 [PGAL10-CYC1-SSS1-ADE2]) was a gift from François Képès (GENOPOLE Evry - Ile de France) [18]. Strains expressing WT and mutants of Sss1p were created by transforming FKY173 with the plasmids described above and grown as described in the Supplemental Material. Viability assays were performed as follows: Yeast strains expressing Sss1p mutants were grown in 0.67% synthetic drop-out medium (supplemented with 2% galactose and 0.2% amino acids) without adenine and histidine overnight in a rotating incubator at 30°C. A total of 1 OD unit (measured at 600nm) of cell culture was centrifuged and resuspended in 1ml of distilled water. From this cell suspension, four ten-fold serial dilutions were prepared. A total of 5µl from each of the five cell suspensions was then spotted onto selective solid media plates (containing 2% dextrose as the carbon source) and incubated at 30°C for 2 to 3 days.
Protein samples were separated by SDS-PAGE and transferred to nitrocellulose. Immunodetection performed using antibodies described in the supplemental methods and in [7] were visualized using the Western Lightning Chemiluminescence Reagent Plus Kit (PerkinElmer Life Sciences, Boston, MA). The decorated blots were exposed to Bioflex Econofilm (Clonex Corporation, Interscience, Markam, ON) and quantified with ImageQuant 5.0 Software (Molecular Dynamics, Inc. Sunnyvale, CA). Data were subjected to statistical analysis using the Student’s t-test.
Purification of Post-Nuclear Membranes
Yeast strains were grown in selective liquid media containing dextrose at 30°C for 12 h from a starting density measured at OD 600nm of 0.1–0.15 to a final density of between 0.5 and 2.0. Cells were centrifuged at 5000 rpm (2700g, avg) in a Beckman JA-10 rotor for 5 min, washed once with 200–300ml of distilled water and re-centrifuged as before. Cells were suspended in 7ml/g (wet weight) of 100mM Tris-Cl, pH 8.4, and 10mM DTT and pre-incubated at 30°C in an orbital shaker at 150 rpm for 10 min. Cells were centrifuged at 5000g for 5 min in an Eppendorf model 5084 centrifuge, resuspended in 7ml/g (wet weight) of Zymolase Buffer (10mM Tris-Cl, pH 7.4, 0.5% dextrose, 0.7M Sorbitol, and 1mM DTT in 0.75X YP medium), and incubated with Zymolase-20T (Seikagaku Corporation, Japan) at 5mg/g (wet weight) of cells for 60–90 min at 30°C in an orbital shaker at 150 rpm. Spheroplasts were centrifuged at 1000g for 10 min, resuspended in 2 ml/g of ice cold SKEEM Buffer (5mM MES-KOH, pH 5.5, 1M Sorbitol, 0.5M EDTA, 1mM KCl, 0.1% ethanol, 1mM Phenylmethylsulfonyl fluoride (PMSF), (Sigma-Aldrich, Canada), 5X Complete Protease Inhibitor Cocktail (CPIC), (Roche Diagnostics, Laval, QC), and 1mM iodoacetamide (Sigma-Aldrich, Canada)), transferred to a pre-chilled stainless steel handheld homogenizer and disrupted with 30 strokes. Homogenized spheroplasts were centrifuged for 10 min at 1000g at 4°C in a Beckman JA25.5 rotor (Beckman Instruments, Palo Alto, CA) to pellet intact cells and nuclei. This last step was repeated and the resulting supernatant was centrifuged at 35000 rpm (110,000g, avg) in a Beckman Type Ti50.2 rotor for 1 h at 4°C. Pellets containing post-nuclear membranes were re-suspended in 20mM Tris-Cl, pH 7.4, 5mM magnesium acetate, 250mM sucrose, and 1mM PMSF and stored at a concentration of 100–200 A280 units/ml at −80°C.
Ribosome-Associated Membrane Protein Assay
The ribosome-associated membrane protein assay was performed as follows: 40 equivalents (1equivalent=1ul of a solution of 50 A280 units/ml) of post-nuclear membranes were solubilized in a final volume of 400µl of TAG-M buffer (20mM Tris-Cl, pH7.4, 500mM aminocaproic acid (Sigma-Aldrich, Canada), 5mM magnesium acetate, 10% (v/v) glycerol, 1% Triton X-100 (BDH Inc., Toronto, ON), 1XCPIC (EDTA-free), and 1mM PMSF), and homogenized in a 2 ml Dounce, Pestle “B”, (Kontes, Vineland, NJ) for 20 strokes. The sample was then incubated on ice for 15 min; 20µl was removed for total products, and the remaining sample loaded onto a 10 to 30% linear sucrose gradient and centrifuged at 41,000 rpm (207,000g, avg) for 5 h in a Beckman SW41Ti rotor at 4°C. Linear gradients were prepared as described [35] with the following modifications: 3.35ml of 10, 20 and 30% w/v sucrose solutions in TAG-M buffer were layered in a centrifuge tube and allowed to diffuse overnight at room temperature. Twelve equal fractions were collected, and the proteins were precipitated with a final concentration of 15–20% trichloroacetic acid, subjected to electrophoresis by SDS-PAGE and transferred to nitrocellulose. Immunoblots were decorated with antibodies against Sec61p, Sss1p and L3 (see above). Fractions containing the majority of L3 protein (5–10 inclusive) were quantified and expressed as a percent of the total protein in all fractions excluding fraction 13, which consisted primarily of aggregated protein.
Sucrose Density Gradient Separation of Digitonin-Soluble Sec Complexes
Digitonin-soluble Sec complexes were prepared by solubilizing 50 equivalents of post-nuclear membranes in 100µl of 50mM HEPES-KOH, pH 7.5, 500mM potassium acetate, 2.5% Digitonin (Sigma-Aldrich, Canada), 10mM EDTA, 5mM β-mercaptoethanol, 1mM PMSF, and 5XCPIC. Next, the mixture was homogenized with 20 strokes in a 2 ml Dounce, Pestle “B”, and incubating on ice for 15 min. After removal of 10µl of the sample for a pre-spin control, the remaining sample was layered onto a 25–35% linear sucrose density gradient and centrifuged for 16 h at 50,000 rpm (166,000g, avg) in a Beckman TLS-55 rotor at 4°C. Thirteen fractions of 158µl each were collected and processed as described above except that immunoblots were also probed for Sec63p, Sec62p and Sec72p. Linear gradients were prepared as described above using 1ml each of 25% and 35% w/v sucrose solutions prepared in 50mM HEPES-KOH, pH 7.5, 500mM potassium acetate, 1mM EDTA, 0.1% digitonin, 5mM β-mercaptoethanol, 1mM PMSF, and 1XCPIC, and allowed to form for 2 h at room temperature. Triton-soluble Sec61 complexes and Sec62p/Sec63p/Sec71p/Sec72p complexes were also resolved in this manner by substituting 1% Triton X-100 for 2.5% digitonin in the solubilization buffer, and 0.1% Triton X-100 for 0.1% digitonin in the linear gradients.
Sucrose Density Gradient Separation of Triton-Soluble Sec61 Complexes
Triton–soluble Sec61 complexes were prepared and assayed as follows: 50 equivalents of post-nuclear membranes were solubilized in 100µl of 50mM HEPES-KOH, pH 7.5, 500mM potassium acetate, 1% Triton X-100, 10mM EDTA, 5mM β-mercaptoethanol, 1mM PMSF, and 5XCPIC, agitated on a Vortex mixer at full speed for 15 sec and incubated on ice for 15 min. A total of 10µl was removed for a pre-spin sample and the remainder was layered onto a 0 to 15% linear sucrose density gradient and centrifuged for 16 h at 50,000 rpm (166,000g, avg) in a Beckman TLS-55 rotor at 4°C. Fractions were collected and processed as described above. Linear gradients were prepared as described above using 500µl each of 0, 5, 10, and 15% w/v sucrose solutions prepared in 50mM HEPES-KOH, pH 7.5, 500mM potassium acetate, 1mM EDTA, 0.1% Triton X-100, 5mM β-mercaptoethanol, 1mM PMSF, and 1XCPIC.
Gel Filtration Assay
Post-nuclear membranes from yeast strains expressing myc-tagged Sss1pWT or Sss1pYG and from strains expressing Sss1pWT, Sss1pTMSa and Sss1pCTSa were first salt extracted at a concentration of 0.050 A280 units /µl in 25mM HEPES-KOH, pH 7.5, 500mM potassium acetate, 10mM EDTA, 1mM iodoacetamide, and 1XCPIC for 30 min on ice and then centrifuged at 60,000 rpm (150,000g) for 10 min in a Beckman TLA120.2 rotor. Membranes totaling 200–800 equivalents were resuspended in 400µl of 50mM HEPES-KOH, pH 7.5, 500mM potassium acetate, 2.5% digitonin, 10% glycerol, 10mM EDTA, 5mM β-mercaptoethanol, 1mM iodoacetamide, and 5XCPIC, processed in a 2 ml Dounce, Pestle “B” for 20 strokes, and allowed to incubate on ice for 30 min. The sample was pre-cleared at 72,000 rpm (225,000g) for 15 min in a Beckman TLA120.2 rotor to remove insoluble complexes. A total of 300µl of the cleared sample was loaded directly onto a Superdex 200 HR 10/30 column (or Superose 6 HR 10/30 column for WT, CTSa and TMSa samples) (Amersham Pharmacia Biotech, GE Healthcare Life Sciences, Canada) pre-equilibrated in 50mM HEPES-KOH, pH 7.5, 500mM potassium acetate, 0.1% digitonin, 1mM EDTA, 5mM β-mercaptoethanol, and 1XCPIC and run at 0.2ml/min on an ÄKTA FPLC System (Amersham Pharmacia Biotech, GE Healthcare Life Sciences, Canada) at 4°C. Fifty 0.5ml fractions were collected, and the proteins were precipitated with trichloroacetic acid, separated by SDS-PAGE, and transferred to nitrocellulose. Immunoblots were decorated with anti-myc antibodies and bands were quantified as described above.
Concanavalin A Binding Assay
A total of 100 equivalents of post-nuclear membranes were recovered by centrifugation at 50,000 rpm for 5 min at 4°C in a Beckman TLA100 rotor, re-suspended in 400µl of 25mM HEPES-KOH, pH 7.5, 500mM potassium acetate, 50mM EDTA, and 1XCPIC, and incubated on ice for 15 min. Membranes were again recovered by centrifugation at 50,000 rpm (100,000g, avg) for 5 min at 4°C in a Beckman TLA120.2 rotor and this step was repeated two more times. After the final wash, membranes were re-suspended in 200µl of 50mM HEPES-KOH, pH 7.5, 400mM potassium acetate, 2.5% digitonin or 1% Triton X-100, 10% (v/v) glycerol, 5mM β-mercaptoethanol, 1mM PMSF, and 1XCPIC, homogenized in a 2 ml Dounce, Pestle “B” for 20 strokes, and placed on ice for 15 min. A 150µl aliquot of the sample was pre-cleared by centrifugation at 65,000 rpm (165,000g, avg) for 10 min at 4°C in a Beckman TLA100 rotor to remove insoluble components, and 100µl of the resulting supernatant was added to 450µl of ConA Buffer (50mM HEPES-KOH, pH 7.5, 293mM potassium acetate, 0.5mM MgCl2, 0.5mM MnCl2, 0.5mM CaCl2, 5mM β-mercaptoethanol, 10% (v/v) glycerol, 1XCPIC, 1mM PMSF) and 50µl packed beads of Concanavalin A Sepharose-4B (ConA) (Sigma-Aldrich, Canada) equilibrated in ConA Buffer. The mixture was incubated for 1 h at 4°C on an end-over-end rotator after which the beads were recovered by centrifugation at 2500g for 2.5 min in a microcentrifuge. The supernatant (containing unbound Sec61 complexes) was transferred to a fresh tube and unbound proteins were precipitated with trichloroacetic acid. The recovered beads were washed three times in 1ml of ConA Buffer plus 1mM PMSF and resuspended in 300µl of 50mM HEPES-KOH, pH 7.5, 500mM NaCl, 1% Triton X-100, 10% (v/v) glycerol, 0.5mM MgCl2, 0.5mM MnCl2, 0.5mM CaCl2, 5mM β-mercaptoethanol, and 1mM PMSF, and incubated for 30 min on an end-over-end rotator at 4°C. After centrifugation at 2500g for 2.5 min in a microcentrifuge, the supernatant (containing bound Sec61 complexes) was precipitated with trichloroacetic acid and the beads (containing bound Sec complexes) were washed two times in ConA Buffer plus 1mM PMSF and once in ConA Buffer containing 100mM NaCl (instead of potassium acetate) plus 1mM PMSF. Precipitated samples and beads were resuspended in 30µl SDS-PAGE Loading Buffer, and analyzed by SDS-PAGE and immunoblotting with the indicated antibodies.
RESULTS
Yeast viability is compromised by the expression of Sss1p with mutations in the CTS and TMS
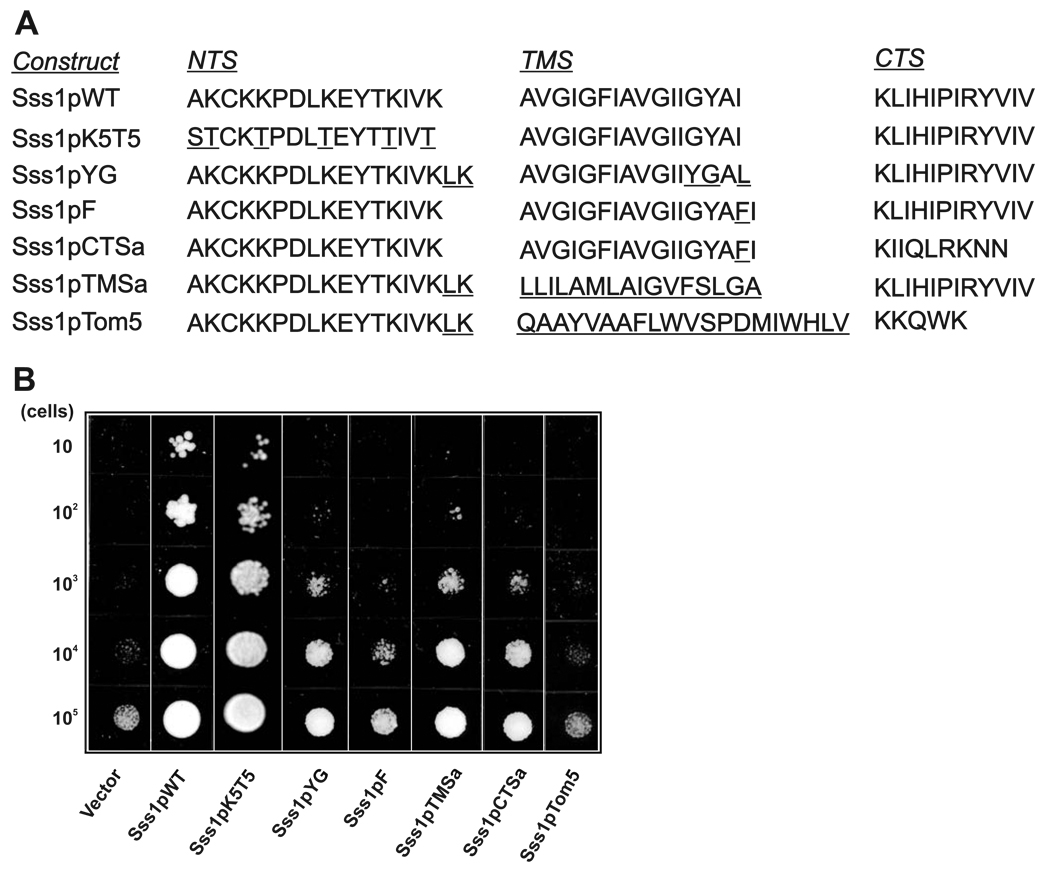
Mutants of Sss1p were constructed with deletions or substitutions in the three sub-regions of the tail-anchor (NTS, TMS, and CTS) in order to determine the effect on cell viability (Figure 1A). Each Sss1p variant was expressed under the control of the SSS1 promoter and transformed into yeast strain FKY173, which lacks a chromosomal copy of the SSS1 gene. The viability of this strain is maintained by galactose-inducible expression of WT Sss1p; therefore, after transformation with an engineered Sss1p expression vector and growth on glucose only the mutant Sss1p is expressed. Consequently, cell viability reflects the function of the mutant.
Figure 1. Mutations in the TMS and CTS of the tail-anchor of Sss1p result in growth defects in yeast.
(A) Amino acid sequence of the tail-anchor of WT and mutant Sss1p proteins. The tail-anchor located at the extreme carboxyl-terminus of the protein was subdivided into three regions: the amino-terminal sequence (NTS), the transmembrane sequence (TMS), and the carboxyl-terminal sequence (CTS). The name and sequence of the tail-anchor of each construct are indicated. Amino acid changes from WT are underlined. (B) 10-fold serial dilutions of exponentially growing cultures of the FKY173 strain carrying an empty plasmid (vector), or plasmids expressing WT Sss1p or tail-anchor mutants of Sss1p described in (A), were spotted onto solid minimal media plates (minus adenine and histidine) supplemented with 0.2% amino acids and 2% glucose, and incubated at 30°C for 3 days.
The NTS of Sss1p is composed of an amphipathic helix with 5 lysines on one face. Strains expressing Sss1p with all 5 of the lysines mutated to threonine (Sss1pK5T5) showed no change in viability compared to the WT (Sss1pWT) strain (Figure 1B). In contrast, mutations in both the TMS and CTS resulted in a marked decrease in growth. Inversion of two amino acids (glycine and tyrosine) at the carboxyl end of the TMS (Sss1pYG) dramatically inhibited yeast growth. The Sss1pYG mutant includes three other conservative mutations that in control experiments were shown not to affect growth (data not shown). Comparison of the growth of Sss1pYG with Sss1pTMSa and Sss1pCTSa revealed that inversion of the GY with a YG had the same effect on viability as complete substitution of the TMS or CTS, respectively (Figure 1A, B). Remarkably, insertion of a single phenylalanine at the carboxyl end of the TMS (Sss1pF) resulted in even more severe growth defects similar to substitution of the entire tail-anchor region with one that normally targets to mitochondria (Sss1pTom5; Figure1A, B). Although yeast expressing Sss1pYG or Sss1pF failed to grow, the strains are not dead even 24 hours after shutting off expression of Sss1p as induction of the WT gene with galactose completely rescued cell growth (not shown).
Substitution of the entire CTS (Sss1pCTSa) or the transmembrane region (Sss1pTMSa) did not prevent protein integration into the ER membrane (Supplemental Figure 1). Because some mutated TA sequences can target proteins for dislocation and degradation (55), we also examined the amount of each of the mutant proteins in the strains. In fact, the mutants were expressed at slightly higher levels than the WT proteins. Additional control experiments for selected constructs demonstrated that the effects reported here were not related to the expression level of the different mutants (see below).
Mutations in the tail-anchor of Sss1p result in translocation defects
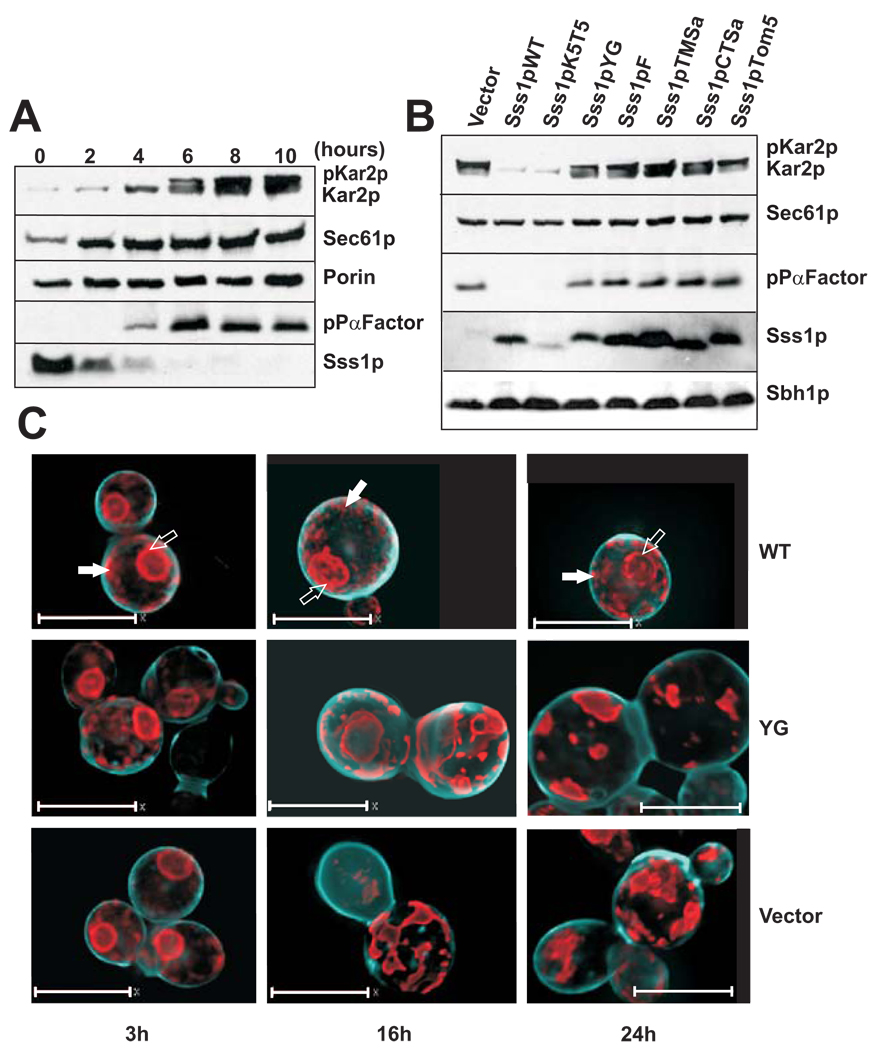
It was reported previously that depletion of Sss1p or deletion of the last 12 amino acids of the protein results in the accumulation of secretory proteins in the cytoplasm of yeast [18]. We obtained similar results in strain FKY173 after the cells had been switched to glucose-containing media (Figure 2A). Sss1p was no longer detectable by immunoblotting at 6 h and pre-Kar2p (pKar2p) and pre-Pro-alpha factor (pPαFactor) began to accumulate at 6 to 8 h. Both Kar2p and pPαFactor are processed from precursor to mature form by signal peptidase. Accumulation could result from faulty signal cleavage and retention of the pKar2p and pPαFactor in the ER, and/or from a defect in translocation resulting in accumulation of the precursors in the cytosol. Our results strongly suggest that there are translocation defects, as precursors begin to accumulate before there is substantial reorganization of the ER. Moreover, there is a corresponding defect in ribosome binding to Sec61 (see below) which would result in the synthesis of membrane and secreted proteins in the cytoplasm. There was a slight increase in the expression level of Sec61p over this time course, whereas the expression level of mitochondrial porin (voltage dependent anion channel or VDAC) remained constant. Sec61p induction may reflect an attempt by the cells to compensate for reduced translocation, or it may reflect induction of the unfolded protein response [36]. Consistent with this second hypothesis, Kar2 levels also increased significantly over the same time course.
Figure 2. Depletion of WT Sss1p and expression of mutant tail-anchor proteins result in translocation defects and altered ER morphology.
(A) Depletion of WT Sss1p and accumulation of pre-Kar2p (pKar2p) and prepro-alpha-factor (pPαFactor) was determined by immunoblot analysis of whole cell lysates from an equal number of cell equivalents of yeast strain FKY173 grown in minimal media (minus adenine and histidine) supplemented with 0.2% amino acids and 2% glucose at 30°C. Yeast cells were grown for 10 hours and samples taken every two hours. Immunoblots were probed with antibodies to Kar2p, Sec61p, Porin, prepro-alpha-factor, and Sss1p and the relevant bands are identified to the right of the panels. (B) Expression levels of Sss1p and accumulation of pre-Kar2p and prepro-alpha-factor in yeast strain FKY173 transformed with an empty plasmid (vector) or with plasmids expressing mutant Sss1p proteins. Immunoblot analysis was performed on whole cell lysates of yeast cells grown for 8 hours at 30°C as described in (A). Expression of Sec61p and Sbh1p is also shown. (C) Deconvolved widefield microscopy images of yeast strain FKY173 containing an empty plasmid (vector), or expressing wild-type (WT) and Sss1pYG (YG) proteins, and co-transformed with a plasmid expressing the ER marker SRβSA-mRFP (SRβ signal-anchor fused to mRFP. Time points at 3, ,16 and 24 hours of growth in glucose are shown. The cell wall (cyan) was stained blue with Calcofluor White. Scale bar: 7 µm.
In order to determine whether the tail-anchor variants could repair the translocation defect, the FKY173 strain was transformed with either an empty plasmid or plasmids expressing the WT or one of the Sss1p tail-anchor mutants. The resultant strains were then grown in glucose for 8 h in order to repress the expression of WT Sss1p. We discovered that cells expressing each of the tail-anchor mutants, except Sss1pK5T5, accumulated both pKar2p and pPαFactor. To rule out that low expression of the Sss1pK5T5 mutant was responsible for the lack of accumulation of pre-proteins, myc-tagged Sss1pK5T5 was expressed from a high copy vector in the same strain. In these cells expression of myc-tagged Sss1pK5T5 was higher than expression of Sss1pWT and roughly equal to that of the other mutants. There were no growth defects and minimal accumulation of pre-proteins (Supplemental Figure 2A–C). It has been shown previously that expression of myc-tagged WT Sss1p has no effect on yeast cell growth [31]. Under these conditions, Sec61p and Sbh1p levels remained relatively constant (Figure 2B), suggesting that translocation defects do not arise from a loss of the other components of the Sec61 complex. Interestingly, most of the Sss1p variants that were unable to repair the translocation defect were present at significantly higher levels than the WT and K5T5 proteins, indicating that the mutants are unable to compensate for the loss of Sss1p even when over-expressed. In addition, because pPαFactor translocates post-translationally, whereas pKar2p can translocate in a co- and post-translational manner [37, 38], these data suggest that translocation defects occur in both of these pathways (also see below).
The expression of tail-anchor mutants of Sss1p alters ER morphology
Efficient trafficking between the ER and Golgi complex and ribosome binding to the Sec61 complex are important to maintain ER structure [39]. To determine if depletion of Sss1p and expression of tail-anchor mutants affected ER morphology, yeast strain FKY173 transformed with an empty vector or transformed with vectors expressing Sss1pWT or Sss1pYG were co-transformed with a vector expressing two fluorescent reporters: GFP fused to the tail-anchor of yeast cytochrome b5 (GFP-Cyb5TA), and the signal anchor of the β-subunit of the canine SRP receptor fused to the N-terminus of mRFP (SA-mRFP). Both proteins are targeted to the ER in WT yeast (Supplemental Figure 1B) but localization of GFP-Cyb5TA does not require a functional Sec61p based translocon‡. Thus, the distribution of these proteins reports on ER structural changes, which may occur upon Sss1p depletion or mutation. We observed that depletion of Sss1p resulted in considerable changes to both the peri-nuclear as well as the peripheral ER structure. Nevertheless the SA-mRFP and the GFP-Cyb5TA reporters continued to colocalize (compare Figure 2C and supplemental Figure 3A) and cofractionate with an ER marker (supplemental Figure 2B), demonstrating that the alteration was due to a change in ER morphology. At 3 h the expression of Sss1pYG had little effect on the ER structure, since at least 6 h of expression under the GAL promoter is required to shut down the expression of WT Sss1p (Figure 2C, “YG 3h”). At 16 and 24 h, when only the mutant Sss1p proteins are present, radical structural changes are evident in both the peri-nuclear and peripheral ER. At these later time points, both of these compartments coalesce into very dense structures in strains containing both the YG mutant and the empty vector (Figure 2C, compare “YG” and “Vector” at 16 and 24 h) but not the WT. . Time lapse imaging revealed that unlike yeast expressing Sss1pWT, cells expressing Sss1pYG were unable to bud and by 21 h the ER had appeared to collapse. (Supplemental Figures 3B–C).
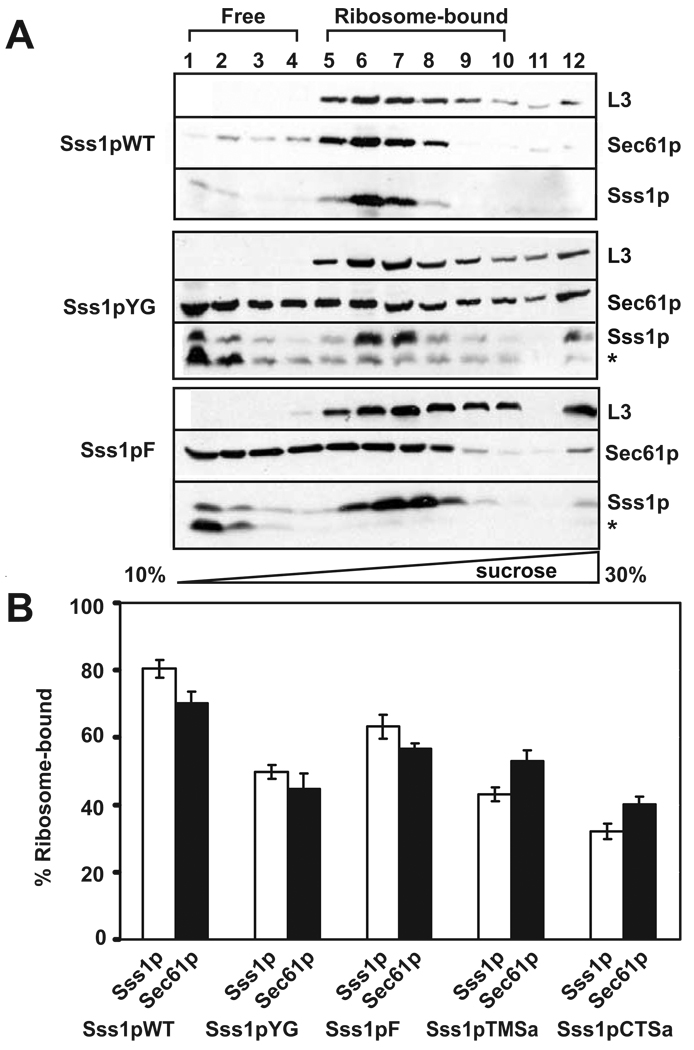
Tail-anchor mutations decrease ribosome binding to the Sec61 complex
Binding of heterotrimeric Sec61 complexes to the ribosome-nascent chain complex is required for co-translational translocation in eukaryotes [40]. Therefore, we used a quantitative assay to determine for selected Sss1p tail-anchor mutants if the defect in co-translational translocation was due to impaired ribosome binding to Sec61 complexes. To this end, membranes from strains expressing WT and the tail-anchor mutants were solubilized in a low ionic strength buffer in order to preserve ribosome interaction. The lysate was then resolved by sucrose density gradient centrifugation to determine the degree of Sec61 complex-ribosome binding. Complexes separated by this method are almost completely soluble (Supplemental Figure 4A); a relatively minor amount of each protein is present at the bottom (last fraction) of the gradient. Immunoblot analysis of the gradient samples showed that both Sec61p and Sss1p co-fractionated with the yeast 60S ribosomal protein L3 in membranes expressing Sss1pWT (Figure 3A, lanes 5–10). In gradient samples from membranes expressing Sss1pYG and Sss1pF, however, a considerable amount of Sec61p and Sss1p failed to co-fractionate with the L3 peak fractions but instead fractionated at the top of the gradient, indicating reduced but not abolished binding of Sec61 complexes to ribosomes. A similar effect was seen for the TMSa and CTSa mutants (Supplemental Figure 4B). In a previous study, substitution of the entire tail-anchor sequence of Sss1p with that of Ubc6 resulted in translocation defects, yet pre-proteins crosslinked to Sec61p [33]. The partial defect in ribosome binding observed here is consistent with this result and suggests that the defect underlying both observations involves the structure and/or function of the Sec61 complex.
Figure 3. Sec61 complexes isolated from strains expressing mutant Sss1p proteins are defective for ribosome binding.
(A) Post-nuclear membranes isolated from yeast strains expressing wild-type Sss1p, Sss1pYG or Sss1pF were solubilized in a low ionic strength buffer to preserve the interaction with the ribosomes. Complexes resolved by sucrose gradient centrifugation in the same buffer containing a linear gradient of 10 to 30% sucrose were precipitated, separated by SDS-PAGE, and identified by immunoblotting with antibodies to the yeast 60S ribosomal protein L3, Sec61p and Sss1p. The sedimentation position of the ribosomes is indicated. * denotes a degradation product of Sss1p. (B) The amount of Sss1p (white bars) and Sec61p (black bars) that co-fractionated with the L3 peak fractions was quantified as a percent of the total (excluding fraction 13) for multiple experiments (described in (A)) for the indicated Sss1p mutants. Averages of 6 independent experiments are shown for all constructs except Sss1pYG (7) and Sss1pCTSa (5). All data are statistically significantly different from the wild type (p < 0.005)
Gradient samples from Sss1pYG, Sss1pF, Sss1pTMSa and Sss1pCTSa also showed partial proteolysis of Sss1p, particularly for Sss1p that did not co-migrate with ribosomes (Figure 3A and Supplemental Figure 4B–C, a faster migrating form of Sss1p is denoted with an asterisk). Degradation appears to occur during gradient centrifugation, as the amount of this product is minimal upon solubilization (Supplemental Figure 4C). These data are consistent with studies indicating that the Sec61 complex is more susceptible to proteolysis when unprotected by bound ribosomes [41]. Consistent with this interpretation, Sss1pWT shows no susceptibility to degradation. Quantitative analysis of ribosome binding for the Sss1pYG, Sss1pF, Sss1pTMSa and Sss1pCTSa mutants indicated that binding of Sec61 complexes to ribosomes was compromised by 20–50% relative to the WT control (Figure 3B).
Heptameric Sec complexes containing tail-anchor mutants remain intact but are unstable
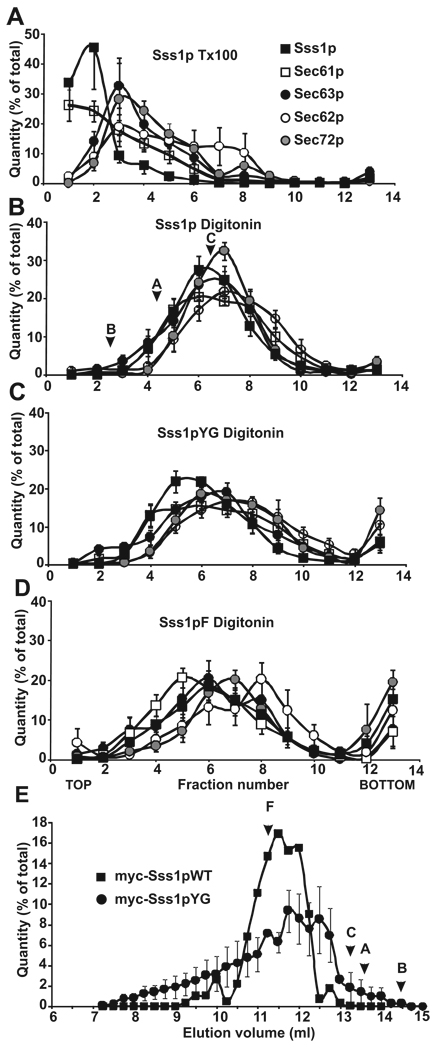
The heptameric Sec complex, consisting of the heterotrimeric Sec61 complex, and the tetrameric Sec62p-Sec63p-Sec71p-Sec72p complex, is required for post-translational translocation in yeast [10, 42]. These complexes can function without binding ribosomes. It has been shown that solubilization of yeast microsomes in digitonin preserves the interaction between these sub-complexes, whereas they are dissociated after solubilization in Triton X-100 [15]. To investigate whether the post-translational translocation defect in strains expressing the Sss1p tail-anchor mutants resulted from impaired assembly of the heptameric Sec complex into its sub-complexes, membranes from strains expressing WT and tail-anchor mutants were solubilized in digitonin or Triton X-100 and centrifuged through a 25% to 35% sucrose density gradient. Immunoblots were probed for Sss1p and Sec61p, which form part of the Sec61 complex, and for Sec62p, Sec63p and Sec72p. The amount of each product was quantified, expressed as percent of total, and the results were plotted against the fraction number to generate gradient profiles for each protein. In control samples solubilized with Triton X-100, we found that profiles of complexes derived from yeast expressing Sss1pWT resulted in two major peaks. The first peak (Figure 4A, Fraction 2), contained Sss1p and Sec61p, and migrated at the size expected for the heterotrimeric Sec61 complex (~70 kDa). The second peak, enriched for Sec62p, Sec63p and Sec72p, is consistent with the size of the Sec62p-Sec63p-Sec71p-Sec72p sub-complex (~160 kDa) (Figure 4A, Fraction 3). This second peak also contained a significant fraction of the total amount of Sec61p.
Figure 4. Sec complexes containing tail-anchor mutants are intact but altered when assayed by sucrose density gradient centrifugation.
Post-nuclear membranes derived from strains expressing Sss1pWT, Sss1pYG, or Sss1pF were solubilized in 1% Triton X-100 (A) or 2.5% digitonin (B–D) and complexes were resolved by sucrose gradient centrifugation (linear gradient of 25–35% sucrose). Gradients were divided into 13 fractions, acid precipitated, and analyzed by SDS-PAGE and immunoblotting with antibodies to Sec63p (black circles), Sec62p (white circles), Sec72p (grey circles), Sec61p (white squares), and Sss1p (black squares). Protein bands in each fraction were quantified, expressed as a percent of the total, and plotted by fraction number. The sedimentation positions of molecular weight standards run in parallel (digitonin only) are marked: B, Bovine Serum Albumin (67 kDa), A, Aldolase (158 kDa), C, Catalase (232 kDa). (E) Post-nuclear membranes derived from yeast expressing myc- tagged Sss1pWT (black squares) and myc-tagged Sss1pYG (black circles) were salt extracted, solubilized in 2.5% digitonin buffer, and Sec complexes were resolved by separation on a Sephadex 200 FPLC column. Samples eluting from 7 to 15 ml were collected in 0.5ml fractions, precipitated, and analyzed as in (A). Immunoblots were probed with antibodies to myc, quantified, expressed as a percent of the total, and plotted by elution volume. Elution positions of molecular weight standards are marked: F, Ferritin (440 kDa); C, Catalase (232 kDa); A, Aldolase (158 kDa); B, BSA (67 kDa). Panels A–D represent averages of 6 experiments.
As anticipated, WT complexes solubilized in digitonin fractionated as one major peak at approximately 230 kDa consistent with a heptameric complex (Figure 4B, Fraction 7). This peak contained all of the examined members of the Sec complex. These results confirm previous observations suggesting that the integrity of the Sec complex is preserved in digitonin but not in Triton X-100. Digitonin-solubilized complexes isolated from yeast expressing the mutant proteins Sss1pYG and Sss1pF resulted in a distribution similar to the larger Sec complexes but was more diffuse (Figure 4C, D). Moreover, individual proteins peaked in different fractions (Figure 4C, D), and a large portion of some of the proteins was found at the bottom of the gradient (compare fraction 13 in Figures 4B–D; Supplemental Figure 5). Gradient profiles for complexes derived from yeast expressing Sss1pTMSa, Sss1pCTSa, and Sss1pTOM5 tail-anchor mutants showed similar results to that of the Sss1pYG and Sss1pF mutants (Supplemental Figure 5). The heterogeneous nature of these samples on density gradients suggests that the compositions of the Sec complexes solubilized in digitonin were not uniform, consistent with either impaired assembly or reduced stability of complexes containing the Sss1p mutants.
The oligomeric state of digitonin-soluble Sec complexes was also investigated by gel filtration chromatography using myc-tagged versions of Sss1pWT and Sss1pYG (Figure 4E). Similar to the results obtained with sucrose density gradients, WT complexes eluted primarily as a single peak while complexes from the YG mutant (Figure 4E) and the other mutants, except for Sss1pK5T5 (Supplemental Figure 6A), had a much broader size distribution. Not surprisingly, the elution profile for the K5T5 mutant was indistinguishable from that of Sss1pWT. To directly compare the sizes of complexes eluting from the gel filtration column for each mutant, the Stokes radii at 50% peak height (at the upper and lower ends of elution) were calculated for the Sss1pK5T5, Sss1pTMSa, Sss1pCTSa, and Sss1pTOM5 mutants (Supplemental Figure 6B). The results reveal that complexes containing mutant Sss1 proteins had a greater variation in size and shape than those formed from WT protein and that the Sec complexes containing Sss1p mutants were less soluble in the extraction buffer (Supplemental Figure 6C).
There is a discrepancy between the molecular weights of the major Sss1p containing complexes estimated by the sucrose gradient and gel filtration analyses: 200 kDa for density gradient data versus 400 kDa for gel filtration data. This may be due to differences in the protein composition of the complexes, or the effects of osmolytes on detergent binding since the density gradient contained increasing concentrations of sucrose, while the gel filtration buffer contained 10% glycerol. Another explanation for the difference is the possible existence of dimers of Sec61 heterotrimers that may have been stable during gel filtration but that dissociated during density gradient centrifugation. Consistent with this interpretation there are several studies indicating that the Sec61 complex can form oligomers of the Sec61 heterotrimer [43–45]. Nonetheless, the results of both techniques provide evidence that detergent-soluble heptameric Sec complexes derived from yeast expressing Sss1p tail-anchor mutants are more heterogeneous in mass and shape than those derived from yeast expressing WT Sss1p. We propose that the amino acid sequence of the tail-anchor impacts the final assembly of the Sec61 heterotrimer into Sec complexes.
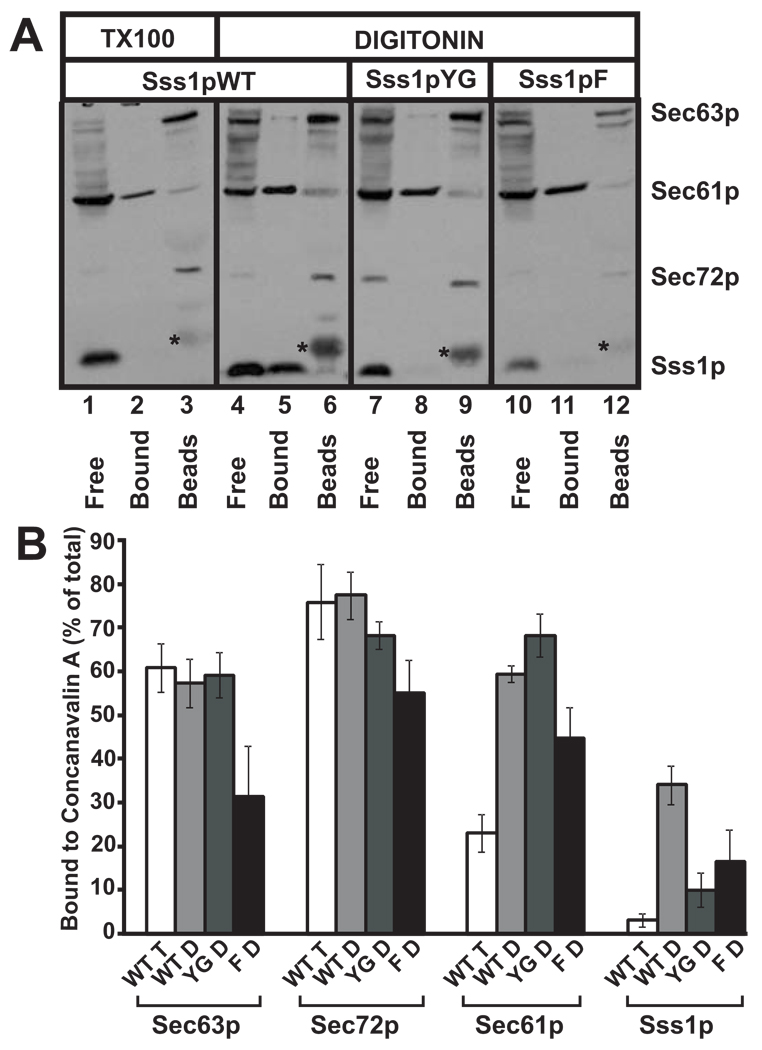
The stability of Sec complexes containing mutant tail-anchor proteins was further investigated by affinity separation with ConA. In WT membranes, some of the Sec61 complexes form part of the heptameric Sec complex, which remains intact in digitonin and binds to ConA due to the presence of oligosaccharides on the Sec71p component [10, 11]. In agreement with published data [46], when WT, salt-extracted, post-nuclear membranes were solubilized in digitonin and incubated with Con A Sepharose, approximately half of the Sec61 complexes (represented by Sec61p and Sss1p) were found in the supernatant fraction (Figure 5A, lane 4, “Free”). Washing the ConA beads with a high salt buffer disrupted the interaction between the Sec61 complex and the Sec62p-Sec63p-Sec71p-Sec72p sub-complex. As a result, the Sec61 complexes were released into the high salt wash (Figure 5A, lane 5, “Bound”). This fraction of Sec61p represents the Sec61 complexes that were part of the heptameric Sec complex. In contrast, Sec63p and Sec72p form part of the Sec62p-Sec63p-Sec71p-Sec72p complex and remained bound to ConA even in high salt (Figure 5A, lane 6). In Triton X-100, most of the Sec61 complex dissociates from the Sec62p-Sec63p-Sec7p-Sec72p sub-complex, leaving the latter bound to ConA. This results in only a very small amount of Sec61p in the salt wash (Figure 5A, lane 2). Therefore, exposure to Triton X-100 mimics the expected result if mutation of Sss1p destabilizes the Sec complex.
Figure 5. Digitonin-soluble Sec complexes prepared from membranes from mutant Sss1p strains are unstable when assayed by binding to Concanavalin A Sepharose.
(A) Post-nuclear membranes derived from yeast expressing Sss1pWT, Sss1pYG, and Sss1pF were salt extracted, washed several times to remove EDTA, solubilized in either 2.5% digitonin or 1% Triton X-100 (wild type only), and pre-cleared by centrifugation to remove insoluble components. Soluble Sec complexes were bound to Concanavalin A Sepharose (Con A beads) which were separated from free Sec61 complexes by low speed centrifugation. Bound Sec61 complexes forming part of the Sec complex were then eluted from the Con A beads with 1% Triton X-100 and high salt. Samples were either acid precipitated or had loading buffer added (Con A beads) and were analyzed by SDS-PAGE and immunoblotting with antibodies to Sec63p, Sec61p, Sec72p, and Sss1p. Free, fraction of detergent-soluble proteins not bound to Concanavalin-A; Bound, proteins bound to Concanavalin-A and eluted with 1% Triton X-100 and 500mM NaCl; Beads, proteins still bound to Concanavalin-A after elution with 1% Triton and 500mM NaCl; * contaminant eluted from beads. (B) Experiments performed as described in (A) were quantified and expressed as percent bound to Con A for each protein indicated. WT T, Sss1pWT-derived post-nuclear membranes solubilized in 1% Triton X-100; WT D, YG D, F D, digitonin-solubilized post-nuclear membranes derived from strains expressing Sss1pWT, Sss1pYG, and Sss1pF, respectively.
Based on the data presented above, we used the digitonin-solubilization/ConA assay to determine the stability of the interaction between Sec61 complexes and the Sec62p-Sec63p-Sec71p-Sec72p sub-complex in strains expressing the Sss1p mutants. As expected, digitonin-solubilized complexes from strains expressing the Sss1pTMSa, Sss1pCTSa and Sss1pTom5 mutants showed reduced solubility compared to complexes from strains expressing Sss1pWT (Supplemental Figure 7A). To our surprise, ConA binding experiments using complexes containing the Sss1pYG and Sss1pF mutant proteins produced results in which Sec61p was present in the salt wash similar to the WT control, but Sss1p was detected in the unbound fractions. This result suggests that some fraction of Sec61p remains bound to the Sec62p-Sec63p-Sec71p-Sec72p complex in digitonin (Figure 5A; compare amounts of Sec61p in lanes 8 and 11 to lane 2). However, the Sss1p mutants are no longer part of the Sec complexes under these conditions (Figure 5A; compare amounts of Sss1p in lanes 8 and 11 to lane 5). Quantification of the results of multiple experiments showed that the amount of Sec61p bound to ConA in digitonin lysates was significantly reduced for the Sss1pF, Sss1pTMSa, Sss1pCTSa, and Sss1pTOM5 mutants but not for the YG mutant (Figure 5B; Supplemental Figure 7B). These results indicate that Sec complexes containing these mutant Sss1ps are less stable than WT complexes under the assay conditions. Moreover, it appears that the mutant Sss1ps are not as stably bound within the Sec complex as Sec61p and that the presence of the mutant Sss1p protein affects the stable association of the Sec61 complex with the Sec62p-Sec63p-Sec71p-Sec72p complex. We also noticed that complexes isolated from yeast expressing Sss1pF contained substantially less Sec63p (Figure 5A–B), which is also consistent with Sec complex destabilization and may explain why the effect of the F insertion on cell growth is more severe than the YG mutation.
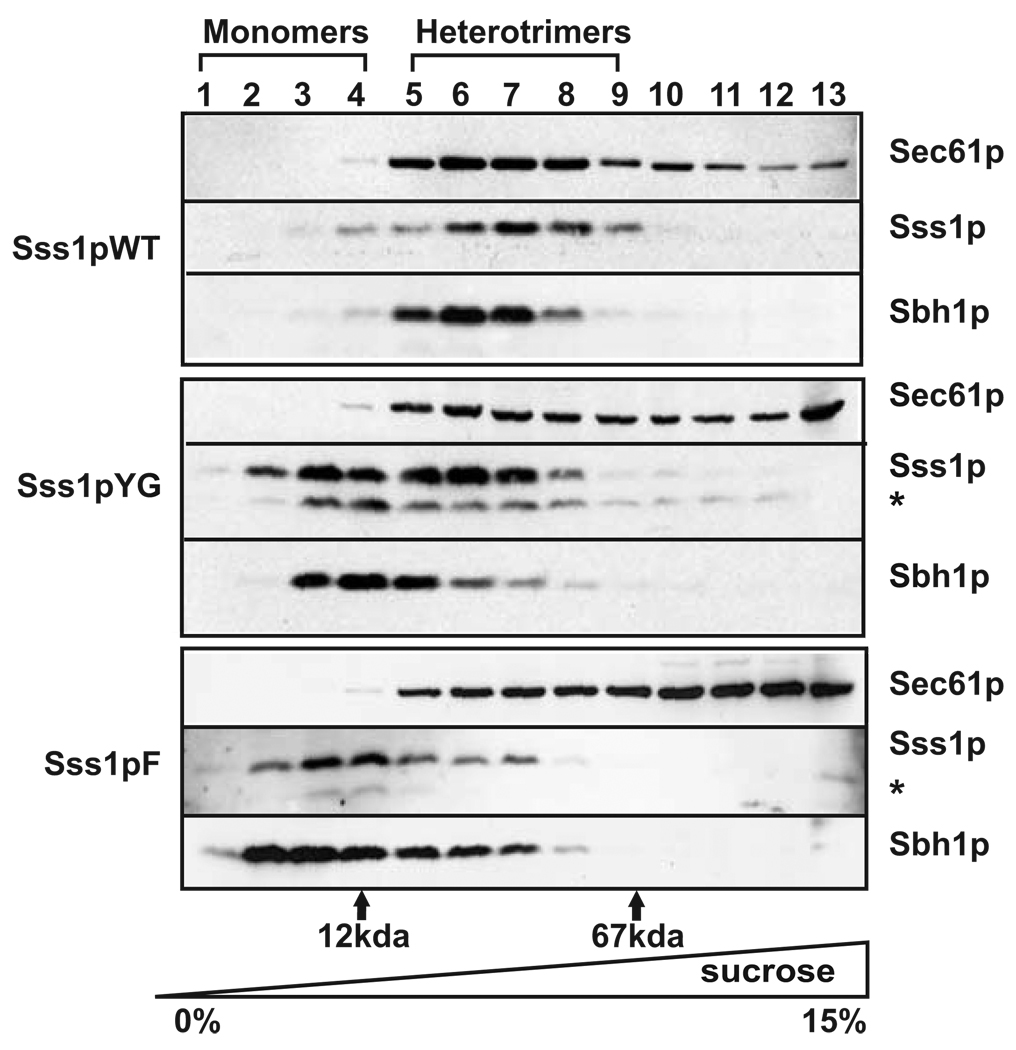
Tail-anchor mutants of Sss1p form unstable heterotrimeric Sec61 complexes
To determine if the reduced binding of the Sss1p mutants to the Sec complexes was due to an altered composition or conformation of the heterotrimeric Sec61 complex, membranes were solubilized in Triton X-100 and resolved by centrifugation through a 0–15% linear sucrose density gradient. After centrifugation, gradient samples were probed for each of the three components of the Sec61 complex: Sec61p, Sss1p, and Sbh1p. Unlike heptameric Sec complexes, heterotrimeric Sec61 complexes are stable in Triton X-100 [15]. Thus, as expected, the three proteins co-migrated when the complex prepared from WT membranes was examined (Figure 6, lanes 5–9). In samples derived from yeast expressing either the Sss1pYG or Sss1pF mutant, both Sss1p and Sbh1p exhibited decreased sedimentation and were found near the top of the gradient while Sec61p sedimented toward the bottom (Figure 6, lanes 2–4), suggesting protein mis-folding and aggregation. Similar results were obtained for complexes derived from strains expressing the Sss1pTMSa, Sss1pCTSa, and Sss1pTOM5 mutants (Supplemental Figure 8A). Quantification of both Sec61p and Sss1p in all of the mutant strains exhibiting translocation defects showed significant disruption of heterotrimers by Triton X-100 (Supplemental Figure 8C). Together these results indicate that heterotrimeric Sec61 complexes are less stable when they contain mutant Sss1p proteins, and are consistent with the proteolytic processing of the Sss1p mutants (denoted by the asterisk in Figure 6; Supplemental Figure 8A,). Because the Sss1p mutants were intact when they were first solubilized (Supplemental Figure 8B), proteolysis must occur during gradient centrifugation.
Figure 6. Tail-anchor mutants of Sss1p form Sec61 complexes unstable in Triton X-100 when assayed by low percentage sucrose density gradient centrifugation.
Post-nuclear membranes derived from yeast strains expressing Sss1pWT, Sss1pYG, or Sss1pF were solubilized in 1% Triton X-100 and Sec61 complexes were resolved by sucrose gradient centrifugation through a linear 0–15% sucrose gradient. Gradient samples were divided into 13 fractions, acid precipitated, and analyzed by SDS-PAGE and immunoblotting with antibodies to Sec61p, Sss1p, and Sbh1p. The positions of molecular weight standards run in parallel experiments are shown: Cytochrome c, 12 kDa; BSA, 67 kDa.
DISCUSSION
Our data indicate that subtle mutations in the tail-anchor sequence of Sss1p affect the integrity and function of the translocon. More generally, these results indicate that even the hydrophobic core of tail-anchor sequences can impact the assembly and stability of integral membrane protein partners. This result is noteworthy because most attention has been paid to the role of the tail-anchor sequence in protein targeting [20, 47, 48]. Although the mutations we created did not reduce the hydrophobicity of the TM core of Sss1p and therefore did not alter ER targeting, the stability of the Sec and Sec61 complexes was still compromised. Moreover, this change in stability results in a severe defect in the translocation function of the Sec61 complex and possibly other defects that lead to complete reorganization of ER structure (Figure 2C & Supplemental Figure 3). This strongly suggests that the primary sequence of this region is involved in complex integrity.
It is surprising that a small change to the primary sequence of the transmembrane domain of Sss1p could have such a dramatic effect on ER function. With the exception of binding sites within membrane transporters, most transmembrane segments tend to be relatively insensitive to modest changes in primary sequence: The main role of these segments is to anchor the protein within the lipid bilayer. This is especially true when one considers the tail-anchor protein family. As a result, membrane spanning segments are viewed as requiring a specific length and hydrophobicity, but are often considered rather generic in terms of primary amino acid sequence [49]. Our data instead suggest that there can be very strict sequence requirements for the hydrophobic sequence of a tail-anchored protein. In contrast, even non-conservative mutations of the sequence N-terminal to the transmembrane domain, which is soluble, had no effect on translocon function.
Studies of the prokaryotic SecYEG/SecA complex reveal interesting interpretations of the data presented here. Mutagenic studies of the SecE protein, which is the bacterial homologue of Sss1p, support the hypothesis that the membrane domain of Sss1p may have functions beyond that of a simple membrane anchor. Although the E. coli protein contains three transmembrane domains while archaea and eukaryotes have only one, the most conservation between them lies in the third transmembrane domain and its flanking regions. However, even in this region the consensus sequence for the SecE region includes a 28-residue hydrophobic region containing only a single charged residue. In Eukaryotes the hydrophobic core of Sss1p is only 21 residues flanked by hydrophilic sequences (Supplementary Figure 8A–C) Deletion analysis of E. coli Sec E shows that only the third transmembrane domain and a portion of the conserved second cytoplasmic region are necessary for its function during protein transport [50]. PrlG mutations that are located in the third transmembrane domain of SecE and are close to the mutations made in this study have been shown to weaken the interactions between the SecY, SecE and SecG subunits. This loosened association enhances translocation rates and is thought to increase the insertion of SecA due to increased conformational flexibility of the SecYEG complex [51].
Sequence alignments of Sec61γ from a number of different species show that the G residue mutated in the YG variant is absolutely conserved (Figure 7). Comparison of the sequence of Sss1p with that of SecE proteins from eubacteria and archaea (Figure 7 and Supplemental Figure 9B) shows that the GY residues mutated in this study are close to the positions of two of the four prlG mutations in SecE. However substitution of the conserved glycine residues with leucine in the same region of Sss1p (including the G in the YG substitution here) had no effect on Sss1p function [33]. In contrast, addition of the F to Sss1p, which had an even more dramatic effect on Sec61 complex stability than the YG inversion, is at a non-conserved position that is frequently an F in other organisms. Moreover, the number of residues within this region of the tail-anchor is variable. Although changes to the primary sequence of the transmembrane domain of SecE have been proposed to change the positions of the α- and γ-subunits with respect to one another, ultimately altering the stability and function of the entire complex [51, 52], mutations in the TMS of yeast Sss1p serve to disrupt translocation rather than enhance it, as is the case in eubacteria. This may be due to the differences between the SecYEG and Sec61 complexes. Crystal structures of SecA-bound SecYEG complexes superimposed onto ribosome nascent chain-bound Sec61 complexes show differing conformations; neither lateral gate opening nor plug movement was observed when SecA was bound. Also, comparison of the crystal structures of the two complexes shows that SecE is less structured than Sss1p and not as tightly associated with the complex [53]. These observations suggest that, although homologous, Sss1p and SecE play unique roles in their interactions with the translocon.
Figure 7. Sec61-γ is highly conserved.
Alignments of the γ- subunit from Homo sapiens (1), Arabidopsis thaliana (2), Saccharomyces cerevisiae (3), Methanocaldococcus jannaschii (4) and SecE from Escherichia coli K12 (substrain W3110) (5) made with the EMBL-EBI ClustalW2 program. The GY residues mutated to YG (black solid arrows) and the F insertion (grey solid arrow) in Sss1p (3) are indicated. PrlG mutations in SecE (5) are indicated by open arrows. Identical (*), conserved (:), and semi-conserved (.) residues are indicated.
Nevertheless, there is evidence from both eukaryotic and bacterial studies that the transmembrane region of the Sec61 γ-subunit is in close proximity to several membrane helices of the Sec61 α-subunit and the γ-subunit is required for complex stability. A representation of the yeast Sec61 complex [54], as modeled on the crystal structure of the M. jannaschii SecYEβ translocon [30], shows that the transmembrane helix of Sss1p spans the membrane plane at an ~35° angle [30], and one side appears to be in contact with Sec61p membrane helices. Studies in yeast showed that strains expressing combinations of Sec61p polypeptides that were unable to form stable complexes were partially rescued by over expression of Sss1p, providing evidence that Sss1p is required for complex stability [29]. Perhaps the close proximity of the membrane helix of the γ-subunit to various regions of the α-subunit confers complex stability and thus constrains amino acid composition, rendering the protein sensitive to mutations within this region.
In our study, substitution of the entire transmembrane domain of Sss1p results in unstable Sec61 and Sec complexes; however, these strains are not significantly more defective than strains expressing Sss1pYG or Sss1pF. This suggests that the C-terminal end of the hydrophobic sequence in Sss1p plays a role in the proper assembly of these complexes. Transmembrane helix rotational angle calculations [55] for the mutant proteins, Sss1pYG and Sss1pF, predict an increase in the α-helical angle when compared to the predicted angle of the WT helix. Analysis of the crystal structure of the yeast Sec61 channel [53] suggests that the region of Sss1p containing these highly conserved glycines may be in close proximity to the plug. Molecular dynamics simulations of SecYEβ indicate that the translocon is a highly dynamic structure, accommodating both secretory proteins, which requires plug movement, as well as transmembrane proteins, which requires opening of the lateral gates [56]. Therefore, it is somewhat surprising that substitution of GY with YG or the introduction of an F some distance away would have such a profound effect on Sec complex assembly. It may be that as a partner of Sec61p, Sss1p contributes to translocon dynamics. Thus, small changes in amino-acid sequence may have profound effects by interfering with the required dynamics of the complexes. We speculate that small flexible residues allow the appropriate “fit” between Sss1p and the neighboring helices of Sec61p. As the smallest side group, glycine is often found where main chains closely approach each other; it is more flexible than other residues, contributing to parts of the protein that need to move or act as hinges [57]. It may also be that these residues contribute to an Sss1p interaction not seen in prokaryotes that requires the appropriate positioning of the charged region C-terminal of the hydrophobic region sequence. Consistent with this view, this charged region is not conserved in SecE proteins. Clearly, high-resolution structural data that display protein-protein contacts within this region will be particularly telling.
Supplementary Material
Acknowledgements
This work was supported by a grant from the Canadian Institute of Health Research (FRN 10490) to D.W.A., who also holds a Canada Research Chair in Membrane Biogenesis, and by grant GM75061 from the National Institutes of Health to J.L.B. The authors also thank Vicki Pierre for providing the data in Supplemental Figure 1B and acknowledge the thesis students N. Usmani, J.M. Tkach and M. Campbell whose efforts contributed to the genesis of this project.
Footnotes
Manuscript in preparation, Henderson M.P. et. al.
Abbreviations: Endoplasmic Reticulum (ER), wild-type (WT), carboxyl-terminal sequence (CTS), amino-terminal sequence (NTS), transmembrane sequence (TMS), Concanavalin A (Con A).
REFERENCES
- 1.Gorlich D, Rapoport TA. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- 2.Flower AM. The SecY translocation complex: convergence of genetics and structure. Trends Microbiol. 2007;15:203–210. doi: 10.1016/j.tim.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 4.Robson A, Collinson I. The structure of the Sec complex and the problem of protein translocation. EMBO Rep. 2006;7:1099–1103. doi: 10.1038/sj.embor.7400832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stephenson K. Sec-dependent protein translocation across biological membranes: evolutionary conservation of an essential protein transport pathway (review) Mol Membr Biol. 2005;22:17–28. doi: 10.1080/09687860500063308. [DOI] [PubMed] [Google Scholar]
- 6.Brodsky JL, Goeckeler J, Schekman R. BiP and Sec63p are required for both co- and posttranslational protein translocation into the yeast endoplasmic reticulum. Proc Natl Acad Sci U S A. 1995;92:9643–9646. doi: 10.1073/pnas.92.21.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brodsky JL, Schekman R. A Sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J Cell Biol. 1993;123:1355–1363. doi: 10.1083/jcb.123.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jermy AJ, Willer M, Davis E, Wilkinson BM, Stirling CJ. The Brl domain in Sec63p is required for assembly of functional endoplasmic reticulum translocons. J Biol Chem. 2006;281:7899–7906. doi: 10.1074/jbc.M511402200. [DOI] [PubMed] [Google Scholar]
- 9.Young BP, Craven RA, Reid PJ, Willer M, Stirling CJ. Sec63p and Kar2p are required for the translocation of SRP-dependent precursors into the yeast endoplasmic reticulum in vivo. Embo J. 2001;20:262–271. doi: 10.1093/emboj/20.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deshaies RJ, Sanders SL, Feldheim DA, Schekman R. Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature. 1991;349:806–808. doi: 10.1038/349806a0. [DOI] [PubMed] [Google Scholar]
- 11.Panzner S, Dreier L, Hartmann E, Kostka S, Rapoport TA. Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell. 1995;81:561–570. doi: 10.1016/0092-8674(95)90077-2. [DOI] [PubMed] [Google Scholar]
- 12.Deshaies RJ, Schekman R. SEC62 encodes a putative membrane protein required for protein translocation into the yeast endoplasmic reticulum. J Cell Biol. 1989;109:2653–2664. doi: 10.1083/jcb.109.6.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rothblatt JA, Deshaies RJ, Sanders SL, Daum G, Schekman R. Multiple genes are required for proper insertion of secretory proteins into the endoplasmic reticulum in yeast. J Cell Biol. 1989;109:2641–2652. doi: 10.1083/jcb.109.6.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liebermeister W, Rapoport TA, Heinrich R. Ratcheting in post-translational protein translocation: a mathematical model. J Mol Biol. 2001;305:643–656. doi: 10.1006/jmbi.2000.4302. [DOI] [PubMed] [Google Scholar]
- 15.Matlack KE, Plath K, Misselwitz B, Rapoport TA. Protein transport by purified yeast Sec complex and Kar2p without membranes. Science. 1997;277:938–941. doi: 10.1126/science.277.5328.938. [DOI] [PubMed] [Google Scholar]
- 16.McClellan AJ, Brodsky JL. Mutation of the ATP-binding pocket of SSA1 indicates that a functional interaction between Ssa1p and Ydj1p is required for post-translational translocation into the yeast endoplasmic reticulum. Genetics. 2000;156:501–512. doi: 10.1093/genetics/156.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becker J, Walter W, Yan W, Craig EA. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Molecular and cellular biology. 1996;16:4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esnault Y, Blondel MO, Deshaies RJ, Scheckman R, Kepes F. The yeast SSS1 gene is essential for secretory protein translocation and encodes a conserved protein of the endoplasmic reticulum. Embo J. 1993;12:4083–4093. doi: 10.1002/j.1460-2075.1993.tb06092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mandon EC, Gilmore R. The tail end of membrane insertion. Cell. 2007;128:1031–1032. doi: 10.1016/j.cell.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Rabu C, Schmid V, Schwappach B, High S. Biogenesis of tail-anchored proteins: the beginning for the end? Journal of cell science. 2009;122:3605–3612. doi: 10.1242/jcs.041210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yabal M, Brambillasca S, Soffientini P, Pedrazzini E, Borgese N, Makarow M. Translocation of the C terminus of a tail-anchored protein across the endoplasmic reticulum membrane in yeast mutants defective in signal peptide-driven translocation. J Biol Chem. 2003;278:3489–3496. doi: 10.1074/jbc.M210253200. [DOI] [PubMed] [Google Scholar]
- 22.Rabu C, Wipf P, Brodsky JL, High S. A precursor-specific role for Hsp40/Hsc70 during tail-anchored protein integration at the endoplasmic reticulum. J Biol Chem. 2008;283:27504–27513. doi: 10.1074/jbc.M804591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuldiner M, Metz J, Schmid V, Denic V, Rakwalska M, Schmitt HD, Schwappach B, Weissman JS. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stefanovic S, Hegde RS. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128:1147–1159. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 25.Wang F, Brown EC, Mak G, Zhuang J, Denic V. A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol Cell. 40:159–171. doi: 10.1016/j.molcel.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mariappan M, Li X, Stefanovic S, Sharma A, Mateja A, Keenan RJ, Hegde RS. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature. 466:1120–1124. doi: 10.1038/nature09296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brodsky JL. The special delivery of a tail-anchored protein: why it pays to use a dedicated courier. Mol Cell. 40:5–7. doi: 10.1016/j.molcel.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plath K, Mothes W, Wilkinson BM, Stirling CJ, Rapoport TA. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 1998;94:795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- 29.Wilkinson BM, Esnault Y, Craven RA, Skiba F, Fieschi J, K'Epes F, Stirling CJ. Molecular architecture of the ER translocase probed by chemical crosslinking of Sss1p to complementary fragments of Sec61p. Embo J. 1997;16:4549–4559. doi: 10.1093/emboj/16.15.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van den Berg B, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 31.Chavan M, Yan A, Lennarz WJ. Subunits of the translocon interact with components of the oligosaccharyl transferase complex. J Biol Chem. 2005;280:22917–22924. doi: 10.1074/jbc.M502858200. [DOI] [PubMed] [Google Scholar]
- 32.Scheper W, Thaminy S, Kais S, Stagljar I, Romisch K. Coordination of N-glycosylation and protein translocation across the endoplasmic reticulum membrane by Sss1 protein. J Biol Chem. 2003;278:37998–38003. doi: 10.1074/jbc.M300176200. [DOI] [PubMed] [Google Scholar]
- 33.Wilkinson BM, Brownsword JK, Mousley CJ, Stirling CJ. Sss1p is required to complete protein translocon activation. J Biol Chem. 285:32671–32677. doi: 10.1074/jbc.M110.128256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanese N. Small-scale density gradient sedimentation to separate and analyze multiprotein complexes. Methods. 1997;12:224–234. doi: 10.1006/meth.1997.0475. [DOI] [PubMed] [Google Scholar]
- 36.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 37.Rothblatt JA, Webb JR, Ammerer G, Meyer DI. Secretion in yeast: structural features influencing the post-translational translocation of prepro-alpha-factor in vitro. Embo J. 1987;6:3455–3463. doi: 10.1002/j.1460-2075.1987.tb02669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng DT, Brown JD, Walter P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J Cell Biol. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prinz WA, Grzyb L, Veenhuis M, Kahan JA, Silver PA, Rapoport TA. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Biol. 2000;150:461–474. doi: 10.1083/jcb.150.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalies KU, Gorlich D, Rapoport TA. Binding of ribosomes to the rough endoplasmic reticulum mediated by the Sec61p-complex. J Cell Biol. 1994;126:925–934. doi: 10.1083/jcb.126.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murphy EC, 3rd, Zheng T, Nicchitta CV. Identification of a novel stage of ribosome/nascent chain association with the endoplasmic reticulum membrane. J Cell Biol. 1997;136:1213–1226. doi: 10.1083/jcb.136.6.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finke K, Plath K, Panzner S, Prehn S, Rapoport TA, Hartmann E, Sommer T. A second trimeric complex containing homologs of the Sec61p complex functions in protein transport across the ER membrane of S. cerevisiae. Embo J. 1996;15:1482–1494. [PMC free article] [PubMed] [Google Scholar]
- 43.Hanein D, Matlack KE, Jungnickel B, Plath K, Kalies KU, Miller KR, Rapoport TA, Akey CW. Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell. 1996;87:721–732. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- 44.Kalies KU, Stokes V, Hartmann E. A single Sec61-complex functions as a protein-conducting channel. Biochim Biophys Acta. 2008;1783:2375–2383. doi: 10.1016/j.bbamcr.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 45.Mitra K, Frank J, Driessen A. Co- and post-translational translocation through the protein-conducting channel: analogous mechanisms at work? Nat Struct Mol Biol. 2006;13:957–964. doi: 10.1038/nsmb1166. [DOI] [PubMed] [Google Scholar]
- 46.Pilon M, Romisch K, Quach D, Schekman R. Sec61p serves multiple roles in secretory precursor binding and translocation into the endoplasmic reticulum membrane. Mol Biol Cell. 1998;9:3455–3473. doi: 10.1091/mbc.9.12.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borgese N, Colombo S, Pedrazzini E. The tale of tail-anchored proteins: coming from the cytosol and looking for a membrane. J Cell Biol. 2003;161:1013–1019. doi: 10.1083/jcb.200303069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gilmore R. Protein translocation across the endoplasmic reticulum: a tunnel with toll booths at entry and exit. Cell. 1993;75:589–592. doi: 10.1016/0092-8674(93)90476-7. [DOI] [PubMed] [Google Scholar]
- 49.Chen H, Kendall DA. Artificial transmembrane segments. Requirements for stop transfer and polypeptide orientation. J Biol Chem. 1995;270:14115–14122. doi: 10.1074/jbc.270.23.14115. [DOI] [PubMed] [Google Scholar]
- 50.Murphy CK, Beckwith J. Residues essential for the function of SecE, a membrane component of the Escherichia coli secretion apparatus, are located in a conserved cytoplasmic region. Proc Natl Acad Sci U S A. 1994;91:2557–2561. doi: 10.1073/pnas.91.7.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duong F, Wickner W. The PrlA and PrlG phenotypes are caused by a loosened association among the translocase SecYEG subunits. Embo J. 1999;18:3263–3270. doi: 10.1093/emboj/18.12.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pohlschroder M, Murphy C, Beckwith J. In vivo analyses of interactions between SecE and SecY, core components of the Escherichia coli protein translocation machinery. J Biol Chem. 1996;271:19908–19914. doi: 10.1074/jbc.271.33.19908. [DOI] [PubMed] [Google Scholar]
- 53.Becker T, Bhushan S, Jarasch A, Armache JP, Funes S, Jossinet F, Gumbart J, Mielke T, Berninghausen O, Schulten K, Westhof E, Gilmore R, Mandon EC, Beckmann R. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science. 2009;326:1369–1373. doi: 10.1126/science.1178535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Junne T, Schwede T, Goder V, Spiess M. The plug domain of yeast Sec61p is important for efficient protein translocation, but is not essential for cell viability. Mol Biol Cell. 2006;17:4063–4068. doi: 10.1091/mbc.E06-03-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dastmalchi S, Beheshti S, Morris MB, Church WB. Prediction of rotational orientation of transmembrane helical segments of integral membrane proteins using new environment-based propensities for amino acids derived from structural analyses. Febs J. 2007;274:2653–2660. doi: 10.1111/j.1742-4658.2007.05800.x. [DOI] [PubMed] [Google Scholar]
- 56.Gumbart J, Schulten K. Structural determinants of lateral gate opening in the protein translocon. Biochemistry. 2007;46:11147–11157. doi: 10.1021/bi700835d. [DOI] [PubMed] [Google Scholar]
- 57.Richardson JS. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.