Abstract
Cryo-electron tomography and subtomogram averaging are utilized to determine that the bacteriophage ϕ12, a member of the Cystoviridae family, contains surface complexes that are: toroidal in shape; composed of six globular domains with six-fold symmetry; and have a discrete density connecting them to the virus membrane-envelope surface. The lack of a hexameric spike in a reassortant of ϕ12 demonstrates that the gene for the hexameric spike is located in ϕ12’s medium length genome segment, likely to the P3 open reading frames which are the proteins involved in viral-host cell attachment. Based on this, and on protein mass estimates derived from the obtained averaged structure, it is suggested that each of the globular domains is most likely composed of a total of four copies of P3a and/or P3c proteins. Our findings may have implications in the study of the evolution of the cystovirus species in regard to their host specificity.
Keywords: Cystovirus, Cryo-electron tomography, ϕ12 six-fold symmetry, Subtomogram averaging, ϕ12 surface proteins
Introduction
The bacteriophage ϕ12 is a member of the family Cystoviridae (ϕ6 to ϕ14), a unique group of lipid-containing, membrane-enveloped bacteriophages that infect strains of the plant pathogen Pseudomonas syringae pv. phaseolicola. This group of viruses contain a segmented double-stranded RNA genome (Mindich et al., 1999; Semancik et al., 1973), divided into three segments named according to their size - small (S), medium (M), and large (L). Members of Cystoviridae have been very useful research models for elucidating replication mechanisms of several RNA viruses. Its RNA packaging and replicative mechanism with a multi-shell structure are analogous to members of the Reoviridae family (Mindich, 2004). Species of both families package their mRNA as precursors to the double-stranded RNA genomic segments. In addition, the RNA-directed RNA polymerase is structurally and mechanistically related to the comparable enzyme of the flaviviruses and it has been used to study de novo initiation of viral RNA synthesis (Butcher et al., 2001).
Within the cystoviruses, all species share a very similar genetic organization, and encode a comparable set of proteins (Mindich et al., 1999). The virion architecture and the genomic organization are conserved across the classified species. Nevertheless, there is considerable variability in the amino acid sequences of analogous proteins from individual species thus producing a ready-made mutant library (Gottlieb et al., 2002a; Gottlieb et al., 2002b; McGraw et al., 1986). ϕ6 is the best characterized member of Cystoviridae (Vidaver et al., 1973) and represents a distant relative of ϕ8, ϕ12 and ϕ13 (Gottlieb et al., 2002a; Gottlieb et al., 2002b; Hoogstraten, 2000; Qiao, 2000). Infection of P. syringae pv. phaseolicola by ϕ6 is initiated by binding of the P3 protein complex to the host cell type IV pili. Although ϕ8, ϕ12 and ϕ13 also rely on P3 open reading frames (ORFs) for infection, they attach to a truncated lipopolysaccharide O chain in the host cell outer membrane - the rough lipopolysaccharide (rlps) (Mindich et al., 1999), utilizing a complex of at least two proteins encoded by P3 (Gottlieb et al., 2002a; Gottlieb et al., 2002b; Hoogstraten, 2000; Qiao, 2000). Unlike ϕ6, which can only infect Pseudomonads, ϕ8, ϕ12 and ϕ13 can also infect other Gram-negative hosts such as Escherichia coli and Salmonella typhimurium through the rlps (Mindich et al., 1999). These differences illustrate how the outer virion layer, which mediates host interactions, has diverged to assist recognition and entry into a variety of host cells and thus determines the host range (Poranen et al., 2005). On the other hand, the inner-most virion layers are similar at the structural level, indicating conserved mechanisms of genome packaging, replication and transcription.
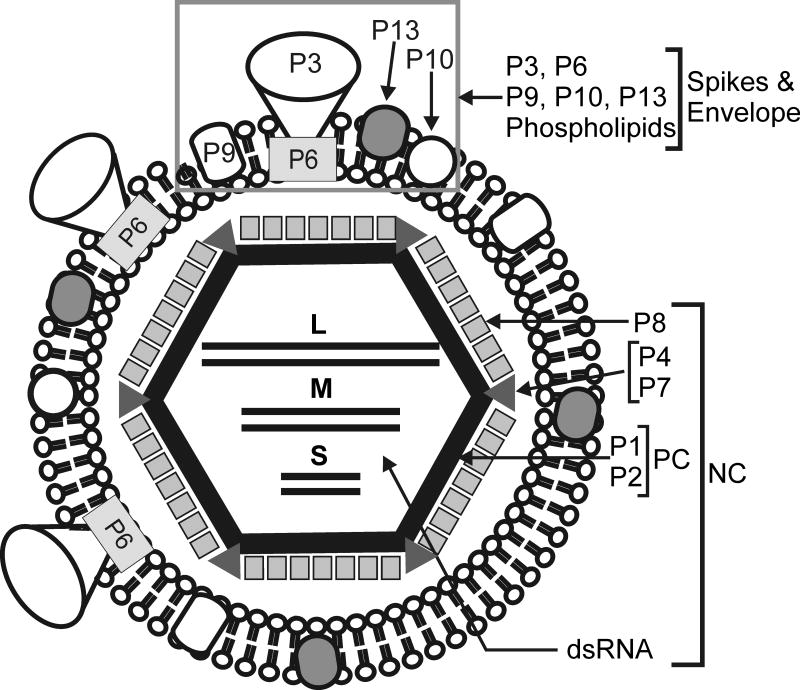
A schematic depiction of the structure of ϕ12 is shown in Fig. 1. The virus is composed of three concentric layers. The innermost layer in ϕ12 has a dodecahedral shape (black hexagon in Fig. 1) and is composed of only two protein species, P1 and P2 (Wei et al., 2009). This dodecahedron is referred to as the polymerase complex (PC) and represents the viral core that encases the genomic material. In ϕ6, the PC is composed of P1, P2, P4 and P7 (Bamford and Mindich, 1980; Mindich and Davidoff-Abelson, 1980), but in ϕ12, P4 and transcription factor, P7, are less tightly associated with the PC (Wei et al., 2009). The next layer is formed by P8 which forms trimers that are arranged on an incomplete T = 13 icosahedral lattice. It is plausible that P7 occupies type II holes in this P8 lattice and hexameric P4 turrets (grey triangles in Fig. 1) extend from the PC at the five-fold axes (Wei et al., 2009). These first two layers, together, form the nucleocapsid (NC) (Etten et al., 1976; Hantula and Bamford, 1988). Interestingly, an equivalent P8 layer is absent in species ϕ8 suggesting that ϕ12 has a structural state that is intermediate between that of ϕ6 and ϕ8.
Fig. 1.
ϕ12 schematic. Schematic depiction of the structure of ϕ12 showing its multilayered architecture with the general location of proteins indicated.
The third, outermost, layer of ϕ12 is a lipid bilayer (Mindich et al., 1999). More is known about the bilayer of ϕ6, which contains phospholipids from the host plasma membrane (Laurinavicius et al., 2004), and four virally encoded integral membrane proteins, P6, P9, P10, and P13 (Etten et al., 1976; Gottlieb et al., 1988; Sinclair et al., 1975); P3 forms an external spike that binds the host receptor and is anchored to the bilayer by P6 (Stitt and Mindich, 1983a, b). Although the ϕ12 envelope also originates from the host cell membrane (Gottlieb et al., 1988) its surface proteins differ substantially from ϕ6, reflecting the different host-cell attachment mechanisms i.e. recognition of rlps by ϕ12 vs. pili by ϕ6. Nevertheless, in both cases, P6 appears to anchor the P3 complex to the membrane and to mediate membrane fusion, thus releasing the NC into the periplasmic space. P12 is a nonstructural protein that mediates membrane acquisition during assembly of the ϕ6 viral particle and presumably does the same for ϕ12 (Johnson and Mindich, 1994; Sinclair et al., 1975). At the genomic level, P3 encodes a single polypeptide in ϕ6 (Gottlieb et al., 1988), whereas in ϕ12, P3 encodes at least two proteins, P3a and P3c (Gottlieb et al., 2002a; Gottlieb et al., 2002b). The presence of two ORFs within P3 is also observed in ϕ8 and ϕ13, which like ϕ12 recognize rlps as a host-cell receptor (Hoogstraten, 2000; Mindich et al., 1999; Qiao, 2000). This observation suggests that the oligomeric P3 complex extending from the viral surface uniquely encodes host cell specificity.
Cryo-electron microscopy has previously been used to reveal the organization of proteins making up the NC of ϕ6 as well as the docking of the hexameric ATPase at its five-fold vertices (Butcher et al., 1997). These studies relied on the icosahedral symmetry innate to the NC, either for averaging of symmetry-related subunits or for orientation of the PC at the vertex. Surface proteins involved in ϕ12 host recognition and fusion, do not conform to the icosahedral symmetry of the NC and therefore their shape and distribution on the surface of the virus must be studied by cryo-electron tomography. In an earlier study (Hu et al., 2008), cryo-electron tomography of ϕ12 revealed two discrete shells corresponding to the NC and the membranous envelope. Periodic connections were seen between the NC and the inner surface of the envelope that were postulated to maintain the NC’s centralized position (Hu et al., 2008). The viral envelope’s outer surface is decorated with two types of protruding densities: elongated structures that are closely associated with the membrane surface and distinctive toroidal elements that are set further away from this surface (Hu et al., 2008). In this regard the surface of ϕ12 differs considerably from that of ϕ6, which only has a closer set of densities (Kenney et al., 1992). The averaged structure of the toroidal element, at 4.6 nm resolution, was approximately 15 nm in diameter and contained a central hole of approximately 3.5 nm diameter (Hu et al., 2008). In the current study, we extend the resolution of this toroidal complex, demonstrate its six-fold symmetry, and reveal discrete densities connecting it to the virus envelope surface. Furthermore, we used a genetic reassortant of ϕ12 to assign unambiguously the gene of this hexameric surface element to the viral RNA M segment and to the P3 ORFs and unambiguously demonstrate its role in host specificity (Gottlieb et al., 2002b). This detailed analysis of the structure and function of the ϕ12 surface-protein has implications for the evolution of the cystovirus species in regard to their host specificity and replication fitness.
Results and Discussion
Comparison of P3 proteins of ϕ12 to ϕ2954 and ϕ2996
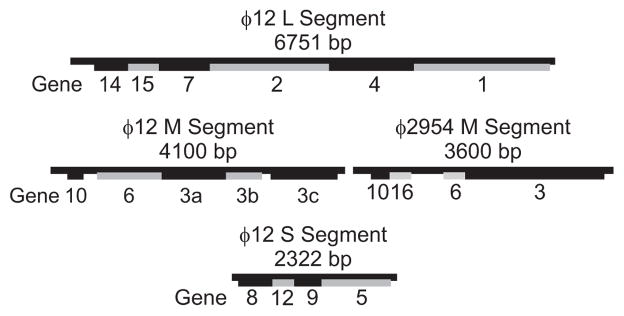
Bacteriophage ϕ2954 is a member of Cystoviridae with a genomic organization similar to ϕ12 (Qiao et al., 2010). However, the host specificity of ϕ2954 indicated that it recognized pili, rather than the usual rlps. Given the similarity in the genome, the M genomic segment could be acquired by ϕ12, thus switching its host recognition to pili. We sought to use this phenomenon to demonstrate that the M genome segment of ϕ12 encodes the toroidal element previously seen on the surface of ϕ12 and that this element is responsible for host recognition. To do this, ϕ12 M segment was substituted by the M segment of ϕ2954 by gene segment pick up from a host cell carrying plasmid pLM3497 (Qiao et al., 2010). The resulting reassortant virus, dubbed ϕ2996, contained the L and S segments from ϕ12 and the M segment of ϕ2954. A genetic map using data from Gottlieb (Gottlieb et al., 2002a; Gottlieb et al., 2002b) and Qiao (Qiao et al., 2010) is shown in Fig. 2.
Fig. 2.
Genetic maps. Genetic map showing the L, M and S segments of ϕ12 and the M segment of ϕ2954. The genome of the reassortant ϕ2996 contains the L and S segments of ϕ12 and the M segment of ϕ2954.
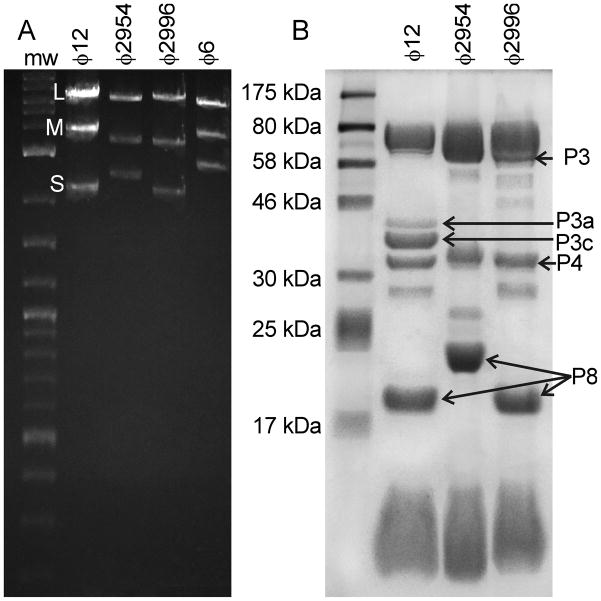
Agarose gel electrophoresis confirms that the M dsRNA segment of ϕ12 has been substituted with the M segment of ϕ2954 in the ϕ2996 reassortant (Fig. 3A) as reported by (Qiao et al., 2010). Similarity, SDS-PAGE analysis shows that the single P3 protein of ϕ2954 has replaced the P3a and P3c proteins of ϕ12 in reassortant ϕ2996 (Fig. 3B). All proteins in ϕ12 that are encoded by the S and L segments are also found in ϕ2996 (Gottlieb et al., 2002a; Gottlieb et al., 2002b). Because the P3 surface element is thought to confer host binding specificity, the reassortant ϕ2996 can be expected, like ϕ2954, to recognize pili instead of rlps and infect P. syringae pv. phaseolicola strain HB10Y (HB10Y) (Qiao et al., 2010).
Fig. 3.
Agarose gels and SDS-PAGE analysis illustrates the substitution of the M segment to produce ϕ2996. A) Agarose gels of genomic RNA show that the M segment of the ϕ2996 reassortant is the same as that of ϕ2954. B) SDS-PAGE of the proteins show that the P3 surface protein band of ϕ2996 runs at the same mw as that of ϕ2954, whereas all the other bands are comparable to ϕ12.
Other cystoviruses with P3 structure similar to ϕ12 may exist. ϕ13 which also binds to the rlps may share structural similarities in P3. It has been shown by sequence alignment that ϕ12 proteins P3a, P3b, P3c, and P6 have considerable identities to the corresponding proteins of ϕ13 (Gottlieb et al., 2002b).
Host range is determined by the M genome segment
To test host recognition, four kinds of cystoviruses – ϕ6, ϕ2954, ϕ2996 and ϕ12 – were plated on two different strains of P. syringae pv. phaseolicola – LM2509 and HB10Y. HB10Y is the wild-type strain containing smooth lps and the type IV pili recognized by ϕ6 and ϕ2954, whereas strain LM2509 lacks the specific type IV pili but has rlps recognized by ϕ12 (Mindich et al., 1999). Pili-specific species such as ϕ6, ϕ2954 and ϕ2996 can infect HB10Y but not LM2509. On the other hand, ϕ12 displays the opposite range. The fact that the reassortant ϕ2996 only infects HB10Y is consistent with the swapping of its M genomic segment containing the P3 host recognition complex on its surface. Results are summarized in Table 1.
Table 1.
Ability of ϕ6, ϕ12, ϕ2954 and ϕ2996 to infect HB10Y (pili) and LM2509 (no pili).
| Ability to infect pili and non-pili Pseudomonads
| ||||
|---|---|---|---|---|
| Phage | P3a,c | P3 | HB10Y | LM2509 |
|
| ||||
| ϕ6 | − | + | + | − |
| ϕ2954 | − | + | + | − |
| ϕ2996 | − | + | + | − |
| ϕ12 | + | − | − | + |
The ϕ12 surface complex is toroidal in shape
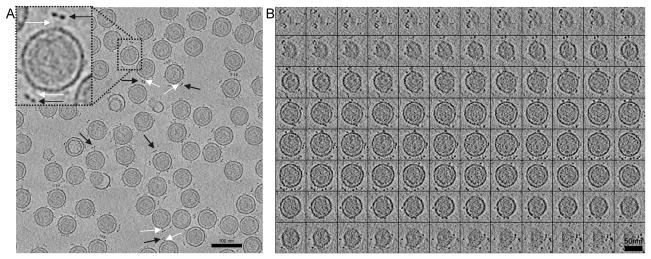
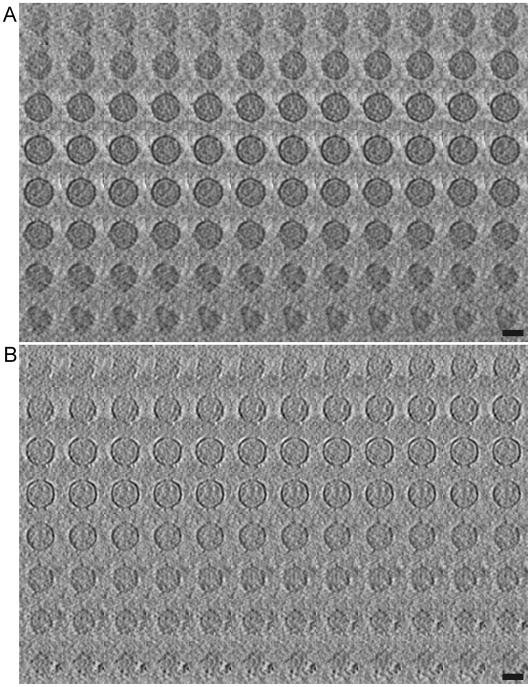
We next sought to correlate host specificity with the structural features on the surface of the virus as seem in electron tomograms. Fig. 4A shows a 7 nm thick tomographic slice from a field of ϕ12 viruses. This image shows that these viruses are quite homogeneous in size and shape with an envelope diameter of approximately 70 nm. Distinct densities are evident around the surface of the viruses, the majority of which appear as bi-lobed structures when viewed in a single slice (black arrows in Fig. 4A). Connections between these bi-lobed densities and the membrane are also visible in some cases (white arrows). Fig. 4B contains a complete set of 96 slices, 0.7 nm thick, through an individual virus, which is representative of 158 viruses observed in three tomograms. This series of images reveal several toroidal surface elements set ~13 nm away from the viral surface. In addition, a second set of surface densities are visible immediately adjacent to the membrane envelope, as previously reported by Hu et al (Hu et al., 2008).
Fig. 4.
Slices of ϕ12 tomogram. A) A 7 nm thick xy-slice in the z-axis of one tomogram showing ~ 60 virions. A few of the visible protruding toroidal complexes are indicated by black arrows and the connectors to the virion surface by white arrows. B) Gallery view of 0.7 nm-thick xy-slices along the z-axis of one single virion tomogram sliced from top to bottom. Scale bar is A) 100 nm and B) 50 nm.
The ϕ12 toroidal surface complex is encoded on the M segment
In order to confirm our hypothesis that the toroidal surface element corresponds to the P3 protein complex and is responsible for host recognition and binding, electron tomograms of ϕ2954 and the reassortant ϕ2996 were collected. This hypothesis is based on the observation that ϕ6, which recognizes pili instead of rlps, lacks a toroidal surface element and has a completely different organization of the P3 ORFs which are expected to confer host specificity. Although ϕ6 has many other genomic differences that could account for its lack of a toroidal surface element, the reassortant ϕ2996 differs from ϕ12 only in the M genomic segment which encodes the P3 complex.
Five tomograms each of ϕ2954 and ϕ2996 were acquired and provided a total of 117 ϕ2954 and 115 ϕ2996 intact virions for examination. No evidence of any toroidal surface structures was found in any of these virions, confirming that ϕ2954 and ϕ2996 lack a toroidal surface element. The panels in Figs. 5A and B show a series of 0.7 nm thick slices through typical ϕ2954 and ϕ2996 virions, respectively. Close examination of the ϕ2954 in Fig. 5A show single-lobed structures next to the virion surface, similar to the second set of densities seen on ϕ12. The ϕ2996 surface (Fig. 6B) has more single-lobed surface protrusions than ϕ2954, but none of these protrusions resemble the toroidal complex found in ϕ12 (Fig. 5). This observation strongly suggests that the toroidal complex is encoded by the M segment, most likely by gene 3 which gives rise to the P3 protein. Given the different host specificity shown in Fig. 3, this observation also supports the conclusion that the toroidal complex is responsible for binding to rlps. Proteins P10 and P6 of ϕ2954 are also present on the M segment that was acquired by ϕ2996. Qiao et al. reported that P10 and P6 have little amino acid sequence similarity to the corresponding proteins of ϕ12 (Qiao et al., 2010), leaving open the possibility that P10 and P6 might also contribute to the toroid conformation seen exclusively in ϕ12. However, P10 and P6 both appear to be integral membrane proteins in both viruses and would therefore be expected to lie within the membrane envelop and not contribute to the external surface features.
Fig. 5.
Individual virion tomogram slices. Gallery views of 0.7 nm-thick tomogram slices of individual A) ϕ2954 and B) ϕ2996 virions. Scale bars are 50 nm.
Fig. 6.
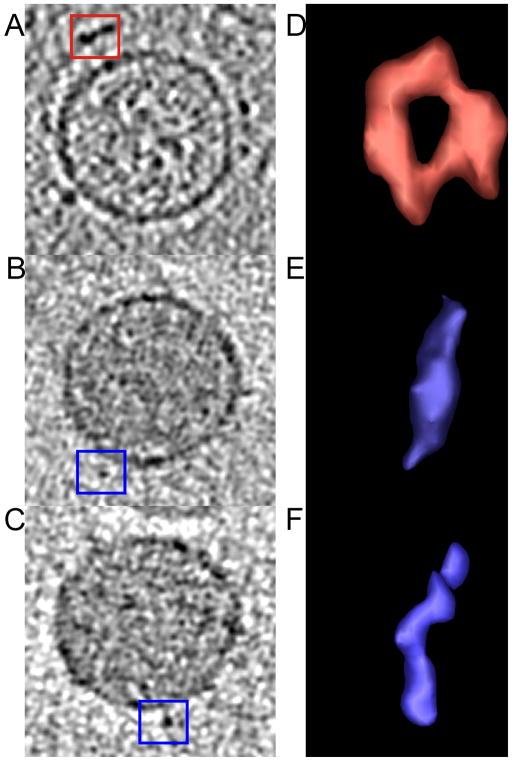
Representative surface elements decorating the membrane. Regions selected for isosurface rendering of surface decorations of A) ϕ12, B) ϕ2954 and C) ϕ2996 are indicated by the boxed areas in the central slices shown. The isosurface representations are shown in D) ϕ12, E) ϕ2954, and F) ϕ2996.
The membrane proximal surface structures seen on ϕ2954 and ϕ2996 also appear on the surface of ϕ12 surface (Hu et al., 2008) (Figs. 5 and 6A). This finding strongly implies that this elongated membrane proximal structure is not encoded by the M segment and could be another envelope protein, such as P9 encoded by the S segment. The isosurface renderings for representative surface densities decorating the membranes of individual ϕ12, ϕ2954 and ϕ2996 virions were created. The regions used to create the renderings are indicated by the boxed regions in the central xy-slices shown in Figs. 6A, B, and C for ϕ12, ϕ2954 and ϕ2996, respectively. The isosurface representations are shown in Figs. 6D, E and F for ϕ12, ϕ2954 and ϕ2996, respectively. In Figs. 6D–F the image plane is tangential to the virion (i.e. the virion radius is normal to the plane of the Figure). Fig. 6D reveals that the ϕ12 surface density is roughly toroidal in shape with six distinct globular densities composing the toroid. In contrast to the ϕ12 virus, in which two types of surface elements, toroids (Fig. 6D) and elongated spikes located closer to the virus surface, can be seen (Hu et al., 2008), ϕ2954 has only one type of surface structure attached to the membrane. It is elongated and exhibits none of the features that are characteristic of a hexameric toroid. Similarly, ϕ2996 has the same type of elongated surface protrusion as ϕ2996 and does not present any toroidal-shaped structures protruding from the membrane. That ϕ12 loses the toroidal element when it acquires the ϕ2954 M genome segment to become the reassortant ϕ2996 is evidence that the M genome segment is responsible for this feature. That this physical alteration in the viral surface elements coincides with a change in host range indicates that the toroidal complex has a significant role in viral attachment to the rlps rather than retractable type IV pili.
Subtomogram averaging of the toroidal surface complex
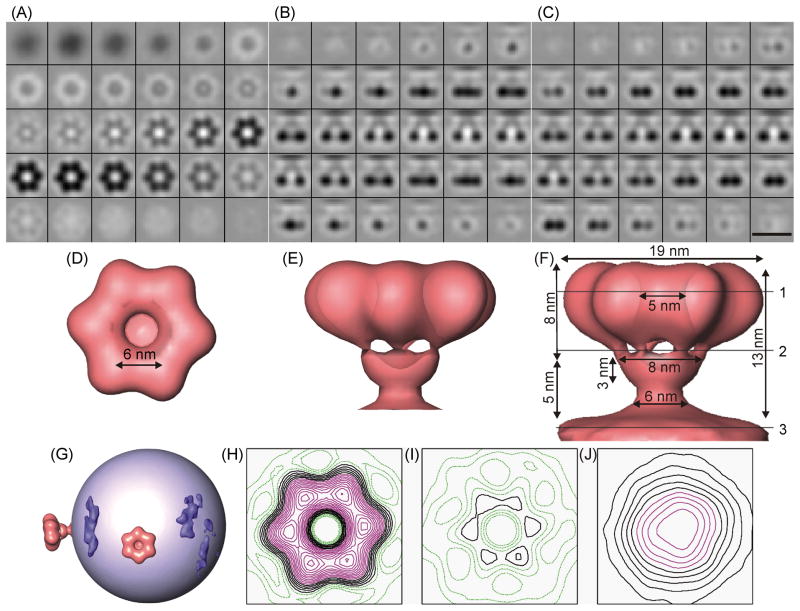
In order to obtain a higher resolution structure for the toroidal surface element of ϕ12, we aligned and averaged a total of 744 copies of this feature from 158 virions. On average, about five of this kind of surface protrusion can be found per virion with a random distribution around the perimeter of the membrane. Each toroidal structure was extracted from the original tomogram in 32 × 32 × 32 voxel boxes (22 × 22 × 22 nm3) and subjected to an iterative procedure of alignment and averaging (see Materials and Methods Section) to obtain the final map. Fig. 7 shows the resulting structure. A connection between the toroid and membrane envelope is visible near the center of the image stack on Figs. 7B and C and the isosurface renderings in Figs. 7E and F. A distinct six-fold symmetry is apparent in Fig. 7A and D, even though this symmetry was not applied during the averaging process as can be appreciated in the slight deviations from six-fold symmetry that are apparent in the contour plots (Fig. 7H–J). The sizes of toroid features are indicated in Fig. 7F. Notable features are the funnel-like stem which is capped by six globular densities with a center-to-center distance of 6 nm that confer the toroidal shape and with an internal hole ~5 nm in diameter. Fig. 7G is a schematic representation of one virion showing the membrane envelope surrounded by two toroidal structures and other elongated surface structures (not averaged) in dark blue.
Fig. 7.
Structure of the toroidal complex. A) Sections 0.7 nm thick cut perpendicular the radial vector connecting the toroid to the center of the virus. The top-most sections are closer to the membrane envelope. B) Sections cut parallel to this radial vector. The membrane envelope is at the top of each section and connecting density is visible in the center of the image stack. C) Sections cut parallel to the radial vector, but perpendicular to B. Scale bar is 20 nm. D–F) Isosurface representations of the averaged spike in the same orientations as in A–C. G) Schematic representation of a representative ϕ12 virus in which the membrane is represented as a light blue sphere, with two toroidal spikes (red) and several elongated elements (dark blue) visible on the surface. H–J) Contour plots of averaged toroid at cuts 1–3 shown in F, respectively.
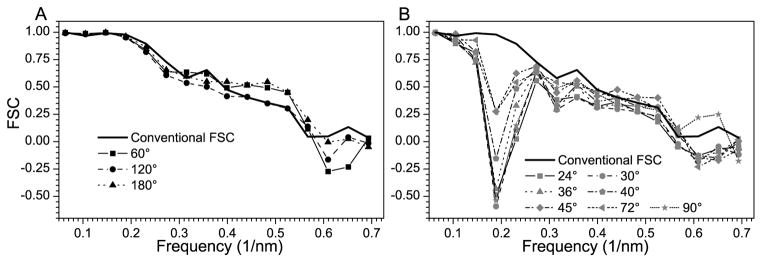
In order to characterize the six-fold symmetry apparent in the averaged structure, a correlation analysis was performed, in which the structure was compared after rotation through various angles about the radial axis (i.e, the axis connecting the toroid to the membrane envelope). Objects with six-fold symmetry should be invariant when rotated in multiples of 60°; therefore after rotation of 60°, 120°, or 180°, the Fourier Shell Correlation (FSC) should be comparable to that obtained by the conventional FSC (i.e., splitting the data into two arbitrary half data sets). FSC plots shown in Fig. 8A indicate that this is indeed the case. The slightly higher FSC for rotated structures compared to the conventional FSC at higher spatial frequencies probably reflects a lower signal-to-noise ratio from the half data sets used for the conventional FSC. Fig. 8B shows the results for rotations of 24°, 30°, 36°, 40°, 45°, 72° and 90°, which are not consistent with six-fold symmetry and which display very poor correlations. We estimate the resolution of the final averaged toroidal complex to be 2.6 nm based on the conventional FSC plot and a threshold of 0.5 (Fig. 8).
Fig. 8.
Resolution and symmetry assessment. Conventional FSC (solid line) and FSC curves resulting from correlating the averaged structure with versions of itself rotated different angles about the Z-axis: A) FSC for 60°, 120°, and 180° rotations; B) FSC for 24°, 30°, 36°, 40°, 45°, 72° and 90° rotations.
Each of the six globular domain of the toroidal spike is most likely composed of three copies of P3a and one copy of P3c
We have demonstrated that the toroidal surface element is composed of the M genome segment. The major products are P3, which is responsible for receptor binding, and P6, which is a very hydrophobic protein that likely resides in the membrane and anchors P3 to the virion surface. Thus, the six globular domains of the toroidal surface element in ϕ12 are almost certainly composed of P3. We used the averaged structure together with SDS-PAGE to estimate the oligomeric composition of the toroid. Volume boundaries were chosen by using a single density threshold such that the toroid maintained its connection to the membrane. Contour plots at Z-positions of the toroid (indicated in Fig. 7F) corresponding to the six-fold domain, stalk, and viral membrane are shown in Figs. 7H-J, respectively.
The mass of the corresponding volume was calculated using a protein density of 1.35 gm/cm3 (Fischer et al., 2004). Thus, each globular domain (1.8×102 nm3) is estimated to be 146 kDa. Given molecular weights for P3a and P3c of 41.3 kDa and 39.5 kDa, respectively (Gottlieb et al., 2002b), it is a plausible estimated that each domain consists of a total of four copies of P3a and/or P3c (total mass of ~160 kDa). The SDS-PAGE analysis (Fig. 3B) indicates P3c is more prevalent than P3a, suggesting an oligomer composed of three copies of P3c and one copy of P3a.
Materials and Methods
Preparation of pure virus
Bacteriophages used in this study are described in Gottlieb et. al (Gottlieb et al., 2002a; Gottlieb et al., 2002b), Mindich et. al (Mindich et al., 1999), Qiao et. al (Qiao et al., 2010). ϕ12 were plated into soft agar with a culture of LM2333 that had been grown overnight. A total of 20–40 plate lysates were incubated overnight at room temperature. The top layer of agar, which contained the ϕ12 bacteriophage, was then collected and the cell debris and agar were removed by centrifugation in a Sorvall SS-34 rotor at 15,000 rpm for 15 min at 4°C. Phage was collected by centrifugation of the supernatant in a Beckman TI270 rotor at 33,000 rpm for 1 hr at 4°C. The resulting pellet was resuspended in 1 ml of buffer P (pH 7.5, 20 mM Tris-HCL, 150 mM NaCl, 0.5 mM CaCl2, and 1 mM MgCl2). The bacteriophage sample was next layered on a 10–30% sucrose gradient and centrifuged at 23,000 rpm for 1 hr at 23°C using a Beckman SW 50.1 rotor. The band of bacteriophage was detected by light scattering and collected by needle puncture. After pelleting and resuspension in buffer P, final purification of the bacteriophage was accomplished by equilibrium centrifugation in a 40–60% sucrose gradient using a Beckman SW 50.1 rotor at 23,000 rpm, overnight at 4°C. The purified bacteriophage band was located as above and was finally centrifuged in a Beckman TI270 rotor at 33,000 rpm for 2 hr at 4°C and resuspended in 100 μl ACN buffer (pH 7.5, 10 mM KPO4, 1 mM MgSO4, 200 mM NaCl, and 0.5 mM CaCl2).
Reassortant ϕ2996 was constructed by transcript pick-up from plasmid pLM3497 (Qiao et al., 2010). This plasmid carries a cDNA copy of the M genomic segment of ϕ2954. The M segment of ϕ12 was replaced with the comparable segment of the pili receptive ϕ2954. Selection for the reassortant virus, ϕ2996, was made on HB10Y. ϕ2954 was grown on plate lysates as described above.
Host bacteria preparation
HB10Y is the host of phage ϕ6 and was utilized as a phenotypic screen based on its non-infectivity by ϕ12. P. syringae pv. phaseolicola strain LM2333 is a mutant of HB10Y which is productively infected by ϕ12 (Mindich et al., 1999). The media used to grow the P. syringae pv. phaseolicola strain host cells was Luria-Bertani.
Cryo-electron tomography
Suspensions of ϕ12, ϕ2954 and ϕ2996 were placed onto glow-discharged, perforated Quantifoil grids and plunge-frozen in liquid ethane. Images were recorded either on a Tecnai F20 electron microscope (FEI, Inc, Hillsboro, OR) operating at 200kV or on a JEOL JEM3200FSC electron microscope (JEOL Ltd, Tokyo, Japan) operating at 300kV with an in-column energy filter. Images were collected on both microscopes at a set magnification of 25k and 8 um underfocus. Images recorded on the Tecnai were collected on a 4k × 4k CCD camera (TVIPS) set to 2× binning, giving an effective pixel size of 7 Å. Images recorded on the JEOL were collected on a 4k × 4k CCD camera (Gatan Inc., Pleasanton, CA), also set to 2× binning, giving an effective pixel size of 8.8 Å. Tilt series on both microscopes were recorded using the SerialEM program (Mastronarde, 2005) at specimen angles between ±65° with 2° steps according to the Saxton scheme (Saxton et al., 1984). The total specimen dose was limited to 60–80 electrons/Å2 for each complete tilt series.
Image processing
Tomographic image reconstruction was performed using eTomo software suite (Mastronarde, 1997). Images without fiducials were aligned by cross correlation, and then refined using the centers of virus particles as virtual markers. For better visualization, tomograms were denoised by nonlinear anisotropic diffusion (Frangakis and Hegerl, 2001) using the SPIDER (Frank et al., 1996) image processing software.
Subtomogram averaging
If species occurring in identical copies within one or many tomograms can be brought into precise register (that is, aligned to a common frame by determining the Euler angles and spatial shifts in each case), it has been shown that by averaging them, their weak individual signals can be amplified. The resulting average can be interpreted at a higher resolution than the original tomogram. Many successful examples exist in the literature, like the case of nuclear pore complex (Beck et al., 2004); the unit-cell of the viral protein lattice (Briggs et al., 2009); the ribosome (Ortiz et al., 2010); and other envelope spikes (Bostina et al., 2007; Förster et al., 2005; Winkler et al., 2009), to name some of them.
Here, we use the method described by Förster and Hegerl (Forster and Hegerl, 2007) consisting of iteratively optimizing a scoring function based on cross-correlation and accounting for the “missing-wedge effect”. This function adopts its maximum value when all particles are in register. This procedure does not require the input of an arbitrary starting model.
Toroidal-shaped spikes protruding away from the viral membrane were manually located in the tomograms and extracted out in subvolumes of 32 × 32 × 32 voxels. By assuming that the spike is approximately oriented in a direction perpendicular to the viral membrane and taking advantage of the fact that the virus is approximately spherical, the Ψ and θ Eulerian angles for each manually selected spike could be initially estimated. The polar angle ϕ was initially chosen randomly for each spike. The extracted subtomograms were rotated according to these angles and an initial reference was created by averaging them. This average was used as a starting model for further alignment (with six degrees of freedom, considering translation and rotation), followed by averaging. The procedure is iterated until convergence is achieved. The resolution of the final structure was estimated by FSC.
All subtomogram processing was performed using MATLAB (Mathworks, Natick, MA, USA), equipped with the TOM (Nickell et al., 2005) and AV3 (Förster et al., 2005) software packages. Visualization and representations were generated using IMOD (University of Colorado, Boulder CO) (Mastronarde, 1997) and Amira (Visage Imaging, San Diego, CA) together with the EM Package (Pruggnaller et al., 2008).
Conclusions
The protein structure protruding 13 nm from the membrane of the cystovirus ϕ12 has been shown by cryo-electron tomography and subtomogram averaging to be a 19 nm diameter toroid with six-fold symmetry that is attached to the virus envelope via a 5 nm connecting density. The 2.6 nm resolution, obtained by subtomogram averaging, of the current structure improves upon the previous imaging of this viral element which first defined its general shape and position (Hu et al., 2008). Furthermore, we provide evidence that the toroid is encoded by the M segment dsRNA genome; that each of the hexameric domains most likely consists of one copy of P3a and three copies of P3c; and that the toroid is primarily responsible for recognizing rlps on the surface of the host bacteria (Gottlieb et al., 2002b).
Our findings have important implications in regard to the mechanisms of viral evolution and their adaptation to additional host cells. The receptor binding protein, P3 is the viral component usually altered in response to host change. Future work will focus on the structure of P3 and its relationship to host cell binding in different species and strains of cystoviruses such as ϕ13.
Acknowledgments
This work is support in part by the National Institute of General Medical Science - grant #SC1 GM092781-01 and the Research Centers in Minority Institutions (NIH/NCRR/RCMI) CCNY/Grant G12-RR03060. The electron microscopy facilities at the New York Structural Biology Center are supported by grant C000087 from the New York State Foundation for Science, Technology and Innovation (NYSTAR) and NIH grant S10 RR017291. ALM is recipient of a postdoctoral fellowship from the Spanish Ministry of Science. We wish to thank Dr. Leonard Mindich of the Public Health Research Institute of UMDNJ, Newark NJ, for the generous gift of ϕ2954 and ϕ2996.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bamford DH, Mindich L. Electron microscopy of cells infected with nonsense mutants of bacteriophage ϕ6. Virology. 1980;107:222–228. doi: 10.1016/0042-6822(80)90287-1. [DOI] [PubMed] [Google Scholar]
- Beck M, Forster F, Ecke M, Plitzko JM, Melchior F, Gerisch G, Baumeister W, Medalia O. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306:1387–1390. doi: 10.1126/science.1104808. [DOI] [PubMed] [Google Scholar]
- Bostina M, Bubeck D, Schwartz C, Nicastro D, Filman DJ, Hogle JM. Single particle cryoelectron tomography characterization of the structure and structural variability of poliovirus-receptor-membrane complex at 30 A resolution. J Struct Biol. 2007;160:200–210. doi: 10.1016/j.jsb.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs JA, Riches JD, Glass B, Bartonova V, Zanetti G, Krausslich HG. Structure and assembly of immature HIV. Proc Natl Acad Sci U S A. 2009;106:11090–11095. doi: 10.1073/pnas.0903535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher SJ, Dokland T, Ojala PM, Bamford DH, Fuller SD. Intermediates in the assembly pathway of the double-stranded RNA virus ϕ6. Embo J. 1997;16:4477–4487. doi: 10.1093/emboj/16.14.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI. A mechanism for initiating RNA-dependent RNA polymerization. Nature. 2001;410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- Etten JV, Lane L, Gonzalez C, Partridge J, Vidaver A. Comparative properties of bacteriophage ϕ6 and ϕ6 nucleocapsid. J Virol. 1976;18:652–658. doi: 10.1128/jvi.18.2.652-658.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer H, Polikarpov I, Craievich AF. Average protein density is a molecular-weight-dependent function. Protein Sci. 2004;13:2825–2828. doi: 10.1110/ps.04688204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster F, Hegerl R. Structure determination in situ by averaging of tomograms. Methods Cell Biol. 2007;79:741–767. doi: 10.1016/S0091-679X(06)79029-X. [DOI] [PubMed] [Google Scholar]
- Förster F, Medalia O, Zauberman N, Baumeister W, Fass D. Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc Natl Acad Sci U S A. 2005;102:4729–4734. doi: 10.1073/pnas.0409178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangakis AS, Hegerl R. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J Struct Biol. 2001;135:239–250. doi: 10.1006/jsbi.2001.4406. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Gottlieb P, Metzger S, Romantschuk M, Carton J, Strassman J, Bamford DH, Kalkkinen N, Mindich L. Nucleotide sequence of the middle dsRNA segment of bacteriophage phi6: placement of the genes of membrane-associated proteins. Virology. 1988;163:183–190. doi: 10.1016/0042-6822(88)90245-0. [DOI] [PubMed] [Google Scholar]
- Gottlieb P, Potgieter C, Wei H, Toporovsky I. Characterization of ϕ12, a bacteriophage related to ϕ6: nucleotide sequence of the large double-stranded RNA. Virology. 2002a;295:266–271. doi: 10.1006/viro.2002.1436. [DOI] [PubMed] [Google Scholar]
- Gottlieb P, Wei H, Potgieter C, Toporovsky I. Characterization of ϕ12, a bacteriophage related to ϕ6: nucleotide sequence of the small and middle double-stranded RNA. Virology. 2002b;293:118–124. doi: 10.1006/viro.2001.1288. [DOI] [PubMed] [Google Scholar]
- Hantula J, Bamford DH. Chemical crosslinking of bacteriophage phi 6 nucleocapsid proteins. Virology. 1988;165:482–488. doi: 10.1016/0042-6822(88)90592-2. [DOI] [PubMed] [Google Scholar]
- Hoogstraten D, Qiao X, Sun Y, Hu A, Onodera S, Mindich L. Characterization of ϕ8, a bacteriophage containing three double-stranded RNA genomic segments and distantly related to ϕ6. Virology. 2000;272:218–224. doi: 10.1006/viro.2000.0374. [DOI] [PubMed] [Google Scholar]
- Hu GB, Wei H, Rice WJ, Stokes DL, Gottlieb P. Electron cryo-tomographic structure of cystovirus ϕ12. Virology. 2008;372:1–9. doi: 10.1016/j.virol.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MD, 3rd, Mindich L. Plasmid-directed assembly of the lipid-containing membrane of bacteriophage phi 6. J Bacteriol. 1994;176:4124–4132. doi: 10.1128/jb.176.13.4124-4132.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney JM, Hantula J, Fuller SD, Mindich L, Ojala PM, Bamford DH. Bacteriophage phi 6 envelope elucidated by chemical cross-linking, immunodetection, and cryoelectron microscopy. Virology. 1992;190:635–644. doi: 10.1016/0042-6822(92)90901-z. [DOI] [PubMed] [Google Scholar]
- Laurinavicius S, Kakela R, Bamford DH, Somerharju P. The origin of phospholipids of the enveloped bacteriophage phi6. Virology. 2004;326:182–190. doi: 10.1016/j.virol.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- McGraw T, Mindich L, Frangione B. Nucleotide sequence of the small double-stranded RNA segment of bacteriophage phi 6: novel mechanism of natural translational control. J Virol. 1986;58:142–151. doi: 10.1128/jvi.58.1.142-151.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindich L. Packaging, replication and recombination of the segmented genome of bacteriophage Phi6 and its relatives. Virus Res. 2004;101:83–92. doi: 10.1016/j.virusres.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Mindich L, Davidoff-Abelson R. The characterization of a 120 S particle formed during phi 6 infection. Virology. 1980;103:386–391. doi: 10.1016/0042-6822(80)90197-x. [DOI] [PubMed] [Google Scholar]
- Mindich L, Qiao X, Qiao J, Onodera S, Romantschuk M, Hoogstraten D. Isolation of additional bacteriophages with genomes of segmented double-stranded RNA. J Bacteriol. 1999;181:4505–4508. doi: 10.1128/jb.181.15.4505-4508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickell S, Forster F, Linaroudis A, Net WD, Beck F, Hegerl R, Baumeister W, Plitzko JM. TOM software toolbox: acquisition and analysis for electron tomography. J Struct Biol. 2005;149:227–234. doi: 10.1016/j.jsb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Ortiz JO, Brandt F, Matias VR, Sennels L, Rappsilber J, Scheres SH, Eibauer M, Hartl FU, Baumeister W. Structure of hibernating ribosomes studied by cryoelectron tomography in vitro and in situ. J Cell Biol. 2010;190:613–621. doi: 10.1083/jcb.201005007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poranen MM, Tuma R, Bamford DH. Assembly of double-stranded RNA bacteriophages. Adv Virus Res. 2005;64:15–43. doi: 10.1016/S0065-3527(05)64002-X. [DOI] [PubMed] [Google Scholar]
- Pruggnaller S, Mayr M, Frangakis AS. A visualization and segmentation toolbox for electron microscopy. J Struct Biol. 2008;164:161–165. doi: 10.1016/j.jsb.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Qiao X, Qiao J, Onodera S, Mindich L. Characterization of ϕ13, a bacteriophage related to ϕ6 and containing three dsRNA genomic segments. Virology. 2000;275:218–224. doi: 10.1006/viro.2000.0501. [DOI] [PubMed] [Google Scholar]
- Qiao X, Sun Y, Qiao J, Di Sanzo F, Mindich L. Characterization of ϕ2954, a newly isolated bacteriophage containing three dsRNA genomic segments. BMC microbiology. 2010;10:55. doi: 10.1186/1471-2180-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton WO, Baumeister W, Hahn M. Three-dimensional reconstruction of imperfect two-dimensional crystals. Ultramicroscopy. 1984;13:57–70. doi: 10.1016/0304-3991(84)90057-3. [DOI] [PubMed] [Google Scholar]
- Semancik JS, Vidaver AK, Van Etten JL. Characterization of segmented double-helical RNA from bacteriophage phi6. J Mol Biol. 1973;78:617–625. doi: 10.1016/0022-2836(73)90283-0. [DOI] [PubMed] [Google Scholar]
- Sinclair JF, Tzagoloff A, Levine D, Mindich L. Proteins of bacteriophage phi6. J Virol. 1975;16:685–695. doi: 10.1128/jvi.16.3.685-695.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt BL, Mindich L. Morphogenesis of bacteriophage phi 6: a presumptive viral membrane precursor. Virology. 1983a;127:446–458. doi: 10.1016/0042-6822(83)90157-5. [DOI] [PubMed] [Google Scholar]
- Stitt BL, Mindich L. The structure of bacteriophage phi 6: protease digestion of phi 6 virions. Virology. 1983b;127:459–462. doi: 10.1016/0042-6822(83)90158-7. [DOI] [PubMed] [Google Scholar]
- Vidaver AK, Koski RK, Van Etten JL. Bacteriophage phi6: a Lipid-Containing Virus of Pseudomonas phaseolicola. J Virol. 1973;11:799–805. doi: 10.1128/jvi.11.5.799-805.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Cheng RH, Berrimen J, Rice WJ, Stokes DL, Katz A, Morgan DG, Gottlieb P. Three-dimensional Structure of the Enveloped Bacteriophage ϕ12: An incomplete T=13 lattice is superposed on an enclosed T=1 shell. PLoS One. 2009;4:e6850. doi: 10.1371/journal.pone.0006850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler H, Zhu P, Liu J, Ye F, Roux KH, Taylor KA. Tomographic subvolume alignment and subvolume classification applied to myosin V and SIV envelope spikes. J Struct Biol. 2009;165:64–77. doi: 10.1016/j.jsb.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]