Abstract
Golgi phosphoprotein 2 (GP73) is a type II Golgi protein, which was found on examination of the fucosylated proteome as a potential tumor marker for hepatocellular carcinoma (HCC). The serum levels of both total and fucosylated GP73 were increased in the sera of patients with HCC. Fucosylation is one of the most important oligosaccharide modifications involved in cancer and is catalyzed by α1,6-fucosyltransferase (Fut8). In the present study, we investigated the effect of Fut8 overexpression on GP73 production in the human hepatoma cell line Hep3B. The Fut8 expression vector was transfected into Hep3B cells and the expression of GP73 was investigated by Western blotting and real-time PCR. Overexpression of Fut8 dramatically enhanced the expression of GP73 at the transcriptional level. Surprisingly, this effect was not dependent on cellular fucosylation. Overexpression of a mutant Fut8, which was unable to be localized to the Golgi, did not induce GP73 production, suggesting that the localization of Fut8 in the Golgi apparatus was important for the increase in GP73 expression. This is the first demonstration of GP73 regulation through overexpression of a glycosyltransferase, which may lead to Golgi stress.
Keywords: Fut8, GP73, HCC, fucosylation, oligosaccharide
Introduction
Golgi phosphoprotein 2 (GP73) is a type II Golgi protein of unknown function that is up-regulated on viral infection (1). Expression of GP73 in the normal liver is quite low, but increases with liver damage, especially that with viral hepatitis (2–4). The mechanism underlying up-regulation of GP73 remains unknown but may be due to increases in cytokine responses as well as the viral infection itself. Kladney et al reported that GP73 increased in response to interferon γ and was inhibited by tumor necrosis factor α (2). Based on this basic information, several groups developed an enzyme-linked immunosorbent assay for GP73 and measured the serum levels of GP73 in patients with liver disease (5–7). As a result, the serum levels of GP73 were found to be increased in chronic liver diseases such as chronic hepatitis and liver cirrhosis, and further enhanced in hepatocellular carcinomas (HCCs). GP73 was identified as a cancer biomarker in the sera of woodchucks bearing HCC through glyco-proteomic analyses (8). This study indicated that GP73 appeared to be enhanced in the core fucosylated proteome. Indeed, while the GP73 level is increased in sera of patients with HCC, the fucosylation of GP73 is more specifically increased in HCC as compared to in chronic liver diseases (8,9).
Fucosylation is one of the most important oligosaccharide modifications involved in cancer and inflammation, and is regulated through complicated mechanisms (10). Increases in core fucosylated α-fetoprotein (AFP-L3) are used as both diagnostic and prognostic markers for HCC, and α1,6-fucosyltransferase (Fut8) is a key enzyme for producing AFP-L3 (11). GP73 is fucosylated specifically in HCC and undergoes the same type of fucosylation as AFP-L3. Recently, it was reported that the oligosaccharide structure of GP73 in HCC was analyzed in detail (9). Modification of α1,6-fucosylation (core fucosylation) through Fut8 manipulation leads to dramatic changes in the phenotypes of cancer cells or animals (12,13). The mechanisms underlying these alterations are dependent on direct or indirect pathways of changes in cellular fucosylation.
In the present study, the effect of fucosylation on GP73 production was investigated by overexpression of Fut8 in a human hepatoma cell line, Hep3B. We found that expression of GP73 markedly increased on Fut8 overexpression. Surprisingly, this increase was independent of cellular fucosylation, but dependent on overexpression of Fut8 in the Golgi apparatus. A novel concept is that Golgi stress might induce the production of GP73 in hepatoma cells. Furthermore, this phenomenon was investigated in human HCC tissues.
Materials and methods
Establishment of an expression vector and stable transfectant of the Fut8 gene
Chinese hamster Fut8 cDNA was modified with a single nucleotide change of G to A at position 512 resulting in a serine-to-asparagine substitution at amino acid position 171, and subcloned into pcDNA3.1 (+) (Invitrogen, Carlsbad, CA) (16). An expression vector of wild-type Chinese hamster Fut8 was also established, using pcDNA3.1 (+). These constructs were transfected into Hep3B cells using FuGENE® 6 Reagent (Roche, Basel, Switzerland) according to the manufacturer's protocol. Selection was performed by the addition of G418 disulfate (Nacalai Tesque, Kyoto, Japan).
Cell culture and transfection
The human hepatoma cell line, Hep3B, was provided by the ATCC (American Type Culture Collection, Manassas, USA). The cells were cultured in D-MEM (Sigma, St. Louis, MO) supplemented with 100 U/ml penicillin, 100 µg/ml streptomycin and 10% fetal calf serum. Transfectants of fucosylation-related genes such as Fut8 and GDP-L-fucose transporter (GDP-Fuc Tr) were established in our previous studies (14,15).
Immunoblot analysis
Hep3B cells were quickly harvested from a 10 cm dish in ice-cold PBS. After precipitation by centrifugation at 2000 rpm for 5 min at 4°C, they were resuspended in TNE buffer [10 mM Tris-HCl (pH 7.8), 1% NP40, 1 mM EDTA, 0.15 M NaCl] including a protease inhibitor cocktail (Roche), and then placed on ice for 30 min to allow solubilization. Samples were then centrifuged at 15,000 rpm for 15 min at 4°C and the supernatants were collected as cell lysates. The conditioned media of Hep3B cells were collected under subconfluent conditions in serum-free D-MEM for 24 h. Samples were quantitated using a Bicinchoninic acid kit (BCA kit, Pierce, Rockford, IL). Cell lysates were subjected to 8% SDS-PAGE under reducing conditions and then transferred to a nitrocellulose membrane (GE Healthcare BioSciences, Chalfont St. Giles, UK). After blocking with phosphate-buffered saline (PBS) containing 5% skim milk for 1 h at room temperature (RT), the membrane was incubated overnight at 4°C with ~1/2,000–1/5,000 diluted primary antibodies, anti-GP73 antibodies (8), anti-β-actin antibodies (Cell Signaling, Beverly, MA), and anti-Fut8 antibodies (15C6; Fujirebio Inc., Tokyo, Japan). After washing the membrane three times with Tris-buffered saline containing 0.05% Tween-20 (TBST) (pH 7.4), it was incubated for 1 h at RT with 1/2,000 diluted secondary antibodies, horseradish peroxidase-conjugated anti rabbit-IgG (Promega, Madison, WI and Cell Signaling) or anti-mouse IgG (Promega). It was again washed three times and developed with an enhanced chemiluminescence system, Immobilon Western (Millipore Corp., Billerica, MA), according to the manufacturer's protocol.
RNA extraction and real-time RT-PCR
One milliliter of Tri zol (Invitrogen) was added to Hep3B cells after removing the conditioned medium and washing with PBS, and then the cells were collected with a scraper. After keeping the cells in 2 ml micro-tubes for 15 min at RT, 200 µl of chloroform was added, followed by vortexing for 15 sec and then standing for 15 min at RT. The tubes were centrifuged at 15,000 rpm at 4°C for 15 min. The upper layers were out into new tubes and the same volume of 2-propanol was added, followed by vortexing and standing for 15 min. The tubes were then centrifuged at 15,000 rpm at 4°C for 10 min. After ethanol precipitation, the precipitates were dissolved in 50 µl of diethylpyrocarbonate (DEPC)-treated H2O. The concentrations of all RNA samples were determined spectrophotometrically and the samples were stored at −80°C until used. RNA (500 µg) containing 1X M-MLV buffer, 0.5 mM dNTP mixture, 50 pmol Random 6 mers, 50 U M-MLV RTase and 10 U RNase inhibitor was adjusted to a volume of 10 µl with RNase-free distilled H2O followed by reverse-transcription for 15 min at 42°C and 2 min at 95°C (Takara Bio, Shiga, Japan). Each PCR product was then adjusted to 25 µl solution containing 1X SYBR Premix Ex Taq, 0.2 µM forward and reverse primers. Real-time PCR analysis was carried out using a Chromo 4 Real-Time Detector (Bio-Rad Laboratories, Inc., Richmond, CA). The primers for GP73 and hypoxanthine guanine phosphoribosyl transferase (HPRT) used in this study are presented in Table I. HPRT was used as an internal control. The results were normalized as relative values using HPRT as a reference to compare the expression of GP73 mRNA.
Table I.
Sequence of primers for real-time PCR analysis.
| mRNA | Sequence | |
|---|---|---|
| GP73 | F | 5'-TTGGTAACAGCAAGTCCCAGACA-3' |
| GP73 | R | 5'-ACCACCTGGATCTCATTGGTTTC-3' |
| HPRT | F | 5'-TGACACTGGCAAAACAATGCA-3' |
| HPRT | R | 5'-GGTCCTTTTCACCAGCAAGCT-3' |
Lectin blot analysis
Duplicate samples were subjected to 8% SDS-PAGE under reducing conditions. One gel was subjected to Coomassie Brilliant Blue (CBB) R-250 staining and another was transferred to a nitrocellurose membrane for Western blot analysis, using Aleuria aurantia lectin (AAL). AAL interacts with fucosylated oligosaccharides (17). After blocking with PBS containing 3% BSA overnight at 4°C, the membrane was incubated in 1/5,000 diluted biotinylated AAL (Seikagaku Corp., Tokyo, Japan) for 30 min at RT. It was then washed three times with TBST and incubated with 1/2,500 diluted avidin-peroxidse conjugates (ABC kits, Vector Res. Corp., Burlingame, CA) for 30 min at RT. The membrane was again washed three times with TBST and then developed with an Immobilon Western (Millipore).
Immunocytochemistry
Cells were prepared in a 35 mm collagen-coated glass dish bottom. The cells were fixed in 4% paraformaldehyde/PBS at RT for 20 min, permeabilized with 0.25% Triton X-100 in PBS for 5 min, washed twice with PBS, and then incubated with blocking buffer (1% bovine serum albumin in PBS) at RT for 30 min. The fixed cells were then incubated with 1/2,000 diluted anti-Fut8 antibody at RT for 20 min, and washed three times in PBS for 2 min. Alexa Fluor® 594 donkey anti-mouse IgG (H+L) (Invitrogen) was used as the secondary antibody at a dilution of 1/500 for 1 h at RT. After washing 3 times with PBS, NBD-C6-Ceramide (Invitrogen) was used to label the Golgi complex according to the manufacturer's protocol. The dishes were mounted with Prolong® Gold Antifade Reagent (Invitrogen). Confocal microscopy was performed using a laser scanning microscope (model LSM 5 PASCAL; Carl Zeiss Micoimaging, Inc., Germany) with a 63×1.4 Plan-Apochromat oil immersion lens. Images were processed using Adobe Photoshop CS4 software (Adobe Systems, Inc., CA).
Results
Levels of GP73 expression in Hep3B cells and Fut8-transfectants
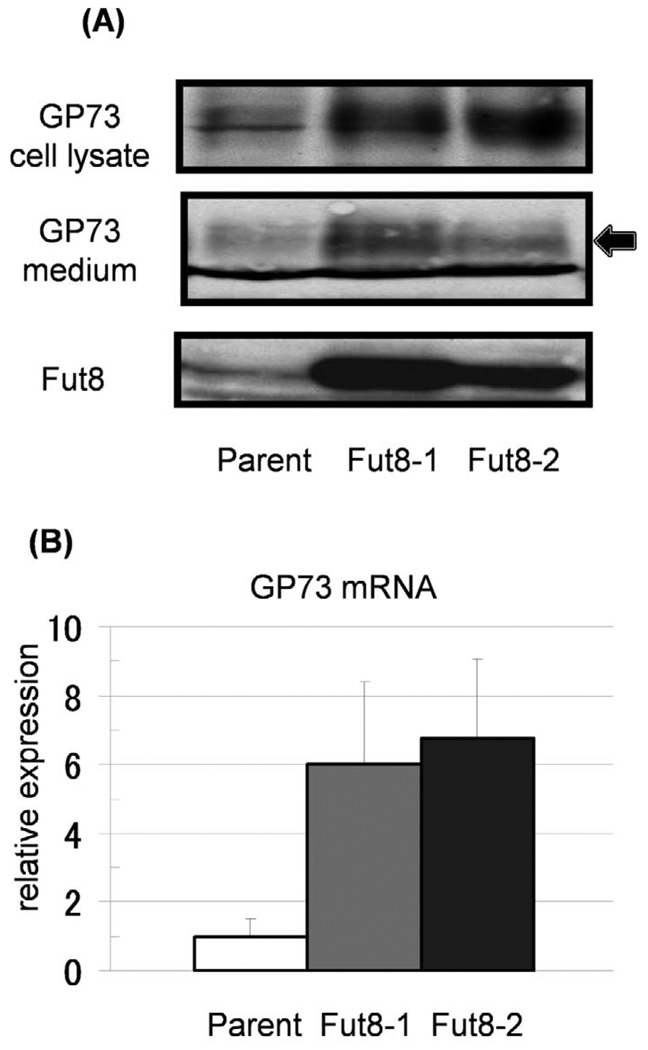
As alteration of gene expression has been observed following the upregulation of certain glyco-genes (18), we investigated the expression of GP73 in Fut8-transfectants by Western blot analysis (Fig. 1A). Interestingly, the expression of total cellular GP73 proteins was dramatically increased in the two Fut8 transfectants of Hep3B cells, as compared within their parental cells. The GP73 level in the conditioned medium was also increased for the Fut8 transfectants, suggesting that increased GP73 with Fut8 transfection was cleaved in the Golgi apparatus and then secreted into the conditioned medium. This increase in GP73 protein was due to up-regulation of GP73 mRNA (Fig. 1B). The levels of GP73 mRNA in Fut8 transfectants were increased 6–7 times compared to in the parental cells.
Figure 1.
Increases in production and secretion of GP73 were observed in Hep3B cells transfected with the Fut8 gene. (A) Both cell lysates (15 µg/each lane) and conditioned media (15 µl of 10 times concentrated medium) were collected from parental Hep3B cells and independent Fut8 transfectants (2 clones). Expression of GP73 and Fut8 was investigated by immunoblot analysis. The arrow indicates GP73 and the lower bands are non-specific ones. (B) Expression of GP73 mRNA was analyzed by real-time RT-PCR. Bars indicate SD for individual. The GP73 mRNA levels in Fut8 transfectants were significantly higher than those in the Hep3B parental cells. Data are means for triplicate experiments.
Production of GP73 in Hep3B cells transfected with fucosylation-regulatory genes
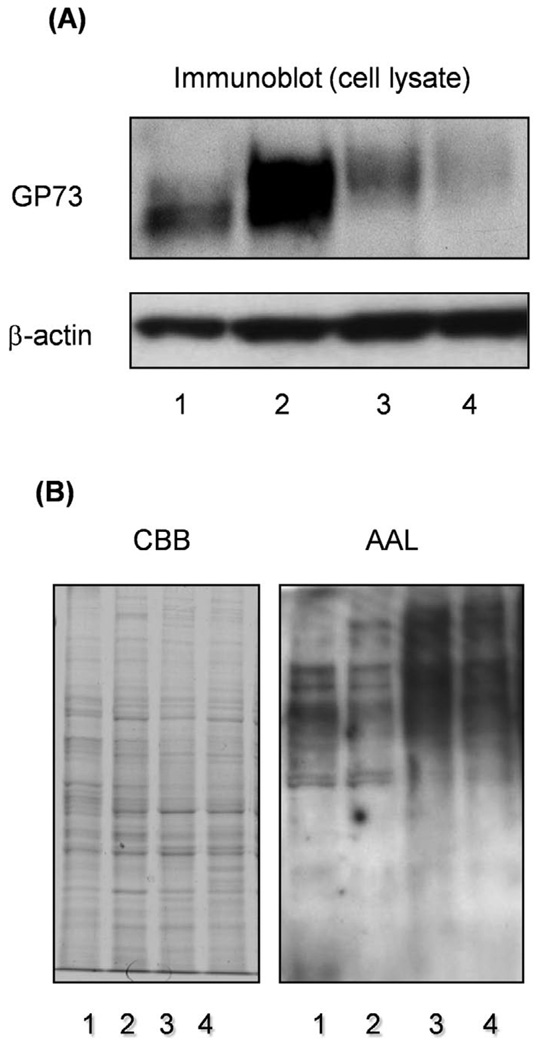
Fucosylation is regulated by several factors including a set of fucosyltransferases, GDP-L-fucose as the donor substrate, and GDP-Fuc Tr. Fucosyltransferases regulate linkage-specific fucosylation while the GDP-L-fucose level regulates total fucosylation, which is independent of the linkage. To determine which modes of fucosylation are important for increases in GP73 expression, immunoblot analysis of GP73 was performed, using Hep3B cells transfected with Fut8 or GDP-Fuc Tr. As shown in Fig. 2A, the up-regulation of GP73 was not observed in the transfectants of GDP-Fuc Tr, up-regulation of GP73 being only observed in the Fut8 transfectants. To investigate changes in the fucosylation level, we performed lectin blot analysis using AAL (Fig. 2B). Significant increases in total cellular fucosylation were observed in GDP-Fuc Tr transfectants. In the case of Fut8 transfectants, increases in cellular fucosylation were scarcely detected, high molecular bands being obtained, suggesting that enhancement of GP73 expression by Fut8 gene transfection was independent of cellular fucosylation.
Figure 2.
An increase in production of GP73 was not observed in Hep3B cells transfected with the GDP-Fuc Tr gene. Immunoblotting of GP73 was performed for Hep3B cells transfected with GDP-Fuc Tr. Lanes 1 and 2 are control and Fut8 transfectant, respectively. Lanes 3 and 4, independent clones of GDP-Fuc Tr transfectants. Lectin blotting with AAL was performed, using the same samples. Coomassie brilliant blue (CBB) staining indicated that equal amounts of protein were loaded in the lanes.
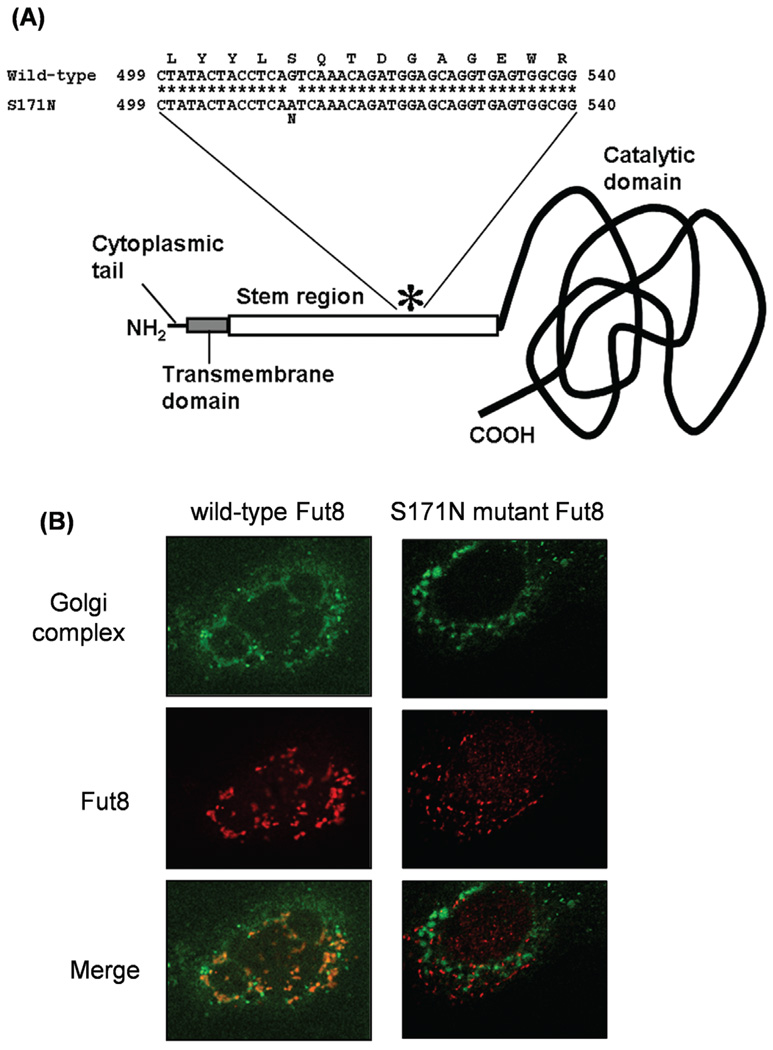
Intracellular localization of the S171N mutant Fut8
To further elucidate the functional role of Fut8 in GP73 expression, we constructed a vector carrying the Fut8 mutant gene bearing a point mutation at amino acid position 171 (S171N) in the stem region with aberrant intracellular localization (Fig. 3A). In vitro Fut8 activity was not detected in the extract of S171N mutant Fut8-overexpressing cells derived from CHO cells on the enzymatic activity assaying using a fluorescence-labeled sugar chain substrate (14). We have analyzed the intracellular localization of Fut8 in transfectants cells by confocal microscopy. Localization to the Golgi apparatus was investigated by co-staining using a Golgi-specific probe, NBD-C6-Ceramide, and anti-Fut8 monoclonal antibodies (Fig. 3B). While the immunofluorescence of wild-type Fut8 was confined to the Golgi apparatus, the S171N mutant signal was not co-localized to the Golgi structure but dispersed throughout the whole intracellular space. These results suggested that the S171N mutant has lost both Fut8 activity and the proper localization to the Golgi apparatus.
Figure 3.
Establishment of an expression vector for mutant Fut8. (A) Diagram of the Fut8 S171N mutant. The asterisk represents a serine-to-asparagine substitution at amino acid position 171. (B) Localization of Fut8 was observed in Hep3B cells transfected with the wild-type (left) or S171N mutant (right) hamster Fut8, on confocal microscopy.
The effect of Fut8 localization on GP73 expression
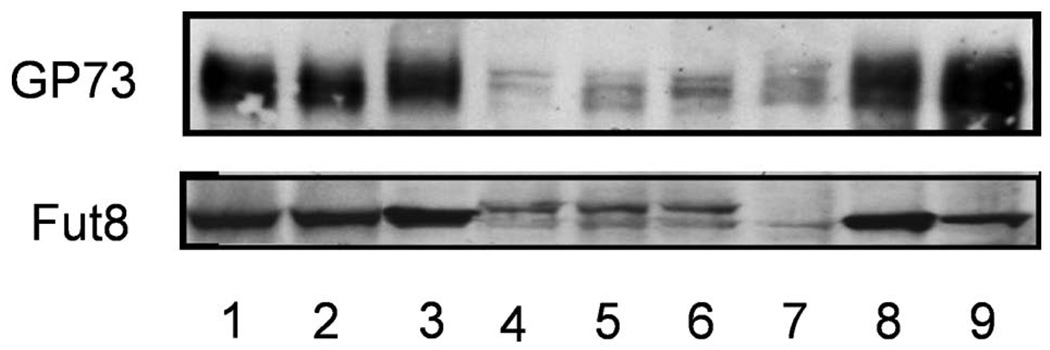
Since cellular fucosylation does not regulate GP73 expression directly, the effect of Fut8 localization in the Golgi on GP73 expression was investigated (Fig. 4). Western blot analysis showed 2 bands of Fut8 mutant proteins, suggesting that an oligosaccharide was attached to the N-linked oligosaccharide site on mutant Fut8, which resulted from the point mutation. When cell lysates were treated with N-glycanase followed by Western blot analysis, the upper band of Fut8 disappeared, suggesting that N-glycan was attached to this site of mutant Fut8 (data not shown). However, a similar sized wild-type Fut8 band was also observed for transfectants of Fut8 mutants. While the expression of GP73 was increased on wild-type hamster Fut8 overexpression in Hep3B cells, like in the Fut8 transfectants described in Fig. 1A, this increase was not observed in 3 independent transfectants of mutant Fut8. These results suggest that the localization of Fut8 in the Golgi apparatus is important for the enhancement of GP73 expression.
Figure 4.
No increase in production of GP73 was observed in Hep3B cells transfected with the Fut8 mutant. Western blot analysis of GP73 was performed for Hep3B cells transfected with the wild-type hamster Fut8 (lanes 1–3) and hamster Fut8 mutant (lanes 4–6). Lane 7, parental Hep3B cells, and lanes 8 and 9, the same Fut8 transfectants as in Fig. 1.
Discussion
The expression of GP73 dramatically increases with viral infection of the liver (1,2). However, the reason why the expression of GP73 is more enhanced in liver cirrhosis and HCC remains unknown. It is also unclear whether or not fucosylated GP73 is produced in the same manner as AFP-L3 and fucosylated haptoglobin (8,19–23). To address these issues, we investigated the effect of overexpression of Fut8 on the production of GP73. Surprisingly, the expression of GP73 is dramatically enhanced at the transcriptional level with overexpression of Fut8. But this effect was independent of cellular fucosylation (Fig. 2). Therefore, we think that overexpression of Fut8 proteins in the Golgi may be a key factor for up-regulation of GP73. One more interesting phenomenon is that the molecular weight of GP73 was slightly increased after transfection of Fut8 and GDP-Fuc Tr (Fig. 2), however the underlying mechanism remains unknown.
Fucosylation mutants have been found during the selection of Lens culinaris (LCA) resistant CHO cells. LCA-resistant cells show low levels of fucosylation on the cell surface. Most of these cells had mutations of the GMDS gene, which is one of the fucosylation regulatory genes (16). Loss of cellular fucosylation, especially in the case of the GMDS mutation, leads to a marked change in biological function (23). In the case of the Fut8 mutant, which has an amino acid alteration (S171N), the mutation also yields an LCA-resistant phenotype of the cells. This Fut8 mutant exhibited no enzymatic activity in vitro. A point mutation with an amino acid alteration in Fut8 yielded a potential site for N-glycans. As shown in Fig. 3A, there were 2 types of Fut8 mutants with/without N-glycans. Confocal microscopy showed that mutant Fut8 is localized outside of the Golgi apparatus. These data suggest that the amino acid sequence of Fut8 is important for the localization to the Golgi, which is independent of N-glycans. While overexpression of wild-type Fut8 induced enhancement of GP73 expression, that of mutant Fut8 had no effect, suggesting that the localization of the Fut8 protein is essential for the induction of GP73.
Since GP73 is a Golgi-resident protein, overexpression of Fut8 might lead to certain kinds of stress in the Golgi. The accumulation of misfolded proteins in the endoplasmic reticulum (ER) induces ER-stress. Our concept of Fut8-induced GP73 expression might suggest some level of Golgi stress. Viral infection induces stress in this organelle of the infected cells. Morphological analyses of the Golgi apparatus in these cells should be performed, to prove the novel concept, Golgi stress (24). Thus, induction of GP73 caused by an increase in Fut8 was observed in vivo on immunohistochemistry (data not shown). Further studies are required to clarify the biological function of GP73.
Acknowledgements
This study was supported by a grant for the New Energy and Industrial Technology Development Organization (NEDO) as a part of the developing technology project for implementing sugar chain functions in Japan, a Grant-in-Aid for Scientific Research (A) No. 21249038, from the Japan Society for the Promotion of Science, and the Global COE Program of Osaka University funded by the Ministry of Education, Culture, Sports, Science, and Technology of Japan. A.S.M. was funded in part by grant R01 CA120206-01 from the National Cancer Institute (NCI). Naoko Yamane-Ohnuki and Mitsuo Satoh are employees of Kyowa Hakko Kirin Co., Ltd.
Abbreviations
- Fut8
α1,6-fucosyltransferase
- GP73
Golgi phosphoprotein 2
- HCC
hepatocellular carcinoma
- LCA
Lens culinaris lectin
- AAL
Aleuria aurantia lectin
- AFP
α-fetoprotein
- AFP-L3
fucosylated-AFP
- GDP-Fuc Tr
GDP-L-fucose transporter
References
- 1.Kladney RD, Bulla GA, Guo L, et al. GP73, a novel Golgi-localized protein upregulated by viral infection. Gene. 2000;249:53–65. doi: 10.1016/S0378-1119(00)00136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kladney RD, Cui X, Bulla GA, Brunt EM, Fimmel CJ. Expression of GP73, a resident Golgi membrane protein, in viral and nonviral liver disease. Hepatology. 2002;35:1431–1440. doi: 10.1053/jhep.2002.32525. [DOI] [PubMed] [Google Scholar]
- 3.Iftikhar R, Kladney RD, Havlioglu N, et al. Disease- and cell-specific expression of GP73 in human liver disease. Am J Gastroenterol. 2004;99:1087–1095. doi: 10.1111/j.1572-0241.2004.30572.x. [DOI] [PubMed] [Google Scholar]
- 4.Marrero JA, Romano PR, Nikolaeva O, et al. GP73, a resident Golgi glycoprotein, is a novel serum marker for hepatocellular carcinomas. Hepatology. 2005;43:1007–1012. doi: 10.1016/j.jhep.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 5.Riener MO, Stenner F, Liewen H, et al. Golgi phosphoprotein 2 (GOLPH2) expression in liver tumors and its value as a serum marker in hepatocellular carcinomas. Hepatology. 2009;49:1602–1609. doi: 10.1002/hep.22843. [DOI] [PubMed] [Google Scholar]
- 6.Wang M, Long RE, Comunale MA, et al. Novel fucosylated biomarkers for the early detection of hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev. 2009;99:939–948. doi: 10.1158/1055-9965.EPI-08-0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu Y, Chen W, Zhao Y, Chen L, Peng T. Quantitative analysis of elevated serum Golgi protein-73 expression in patients with liver diseases. Ann Clin Biochem. 2009;46:38–43. doi: 10.1258/acb.2008.008088. [DOI] [PubMed] [Google Scholar]
- 8.Block TM, Comunale MA, Lowman M, et al. Use of targeted glycoproteomics to identify serum glycoproteins that correlate with liver cancer in woodchucks and humans. Proc Natl Acad Sci USA. 2005;102:779–784. doi: 10.1073/pnas.0408928102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Norton PA, Comunale MA, Krakover J, et al. N-linked glycosylation of the liver cancer biomarker GP73. J Cell Biochem. 2008;104:136–149. doi: 10.1002/jcb.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyoshi E, Moriwaki K, Nakagawa T. JB Minireview-Biological function of fucosylation in cancer biology. J Biochem. 2008;143:725–729. doi: 10.1093/jb/mvn011. [DOI] [PubMed] [Google Scholar]
- 11.Noda K, Miyoshi E, Uozumi N, et al. Gene expression of alpha 1–6 fucosyltransferase in human hepatoma tissues: a possible implication for increased fucosylation of alpha-fetoprotein. Hepatology. 1998;28:944–952. doi: 10.1002/hep.510280408. [DOI] [PubMed] [Google Scholar]
- 12.Miyoshi E, Noda K, Ko JH, et al. Overexpression of α1-6 fucosyl-transferase in hepatoma cells suppresses intrahepatic metastasis after splenic injection in athymic mice. Cancer Res. 1999;59:2237–2243. [PubMed] [Google Scholar]
- 13.Wang X, Inoue S, Gu J, et al. Dysregulation of TGF-β1 receptor activation leads to abnormal lung development and emphysema-like phenotype in core fucose-deficient mice. Proc Natl Acad Sci USA. 2005;102:15791–15796. doi: 10.1073/pnas.0507375102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noda K, Miyoshi E, Gu J, et al. Relationship between elevated FX expression and increased production of GDP-L-fucose, a common donor substrate for fucosylation in human hepatocellular carcinoma and hepatoma cell lines. Cancer Res. 2003;63:6282–6289. [PubMed] [Google Scholar]
- 15.Moriwaki K, Noda K, Nakagawa T, et al. A high expression of GDP-fucose transporter in hepatocellular carcinoma is a key factor for increases in fucosylation. Glycobiology. 2007;17:1311–1320. doi: 10.1093/glycob/cwm094. [DOI] [PubMed] [Google Scholar]
- 16.Yamane-Ohnuki N, Kinoshita S, Inoue-Urakubo M, et al. Establishment of FUT8 knockout Chinese hamster ovary cells: an ideal host cell line for producing completely defucosylated antibodies with enhanced antibody-dependent cellular cytotoxicity. Biotechnol Bioeng. 2004;87:614–622. doi: 10.1002/bit.20151. [DOI] [PubMed] [Google Scholar]
- 17.Yamashita K, Kochibe N, Ohkura T, Ueda I, Kobata A. Fractionation of L-fucose-containing oligosaccharides on immobilized Aleuria aurantia lectin. J Biol Chem. 1985;260:4688–4693. [PubMed] [Google Scholar]
- 18.Miyoshi E, Ihara Y, Hayashi N, Fusamoto H, Kamada T, Taniguchi N. Transfection of N-acetylglucosaminyltransferase III gene suppresses expression of hepatitis B virus in a human hepatoma cell line, HB611. J Biol Chem. 1995;270:28311–28315. doi: 10.1074/jbc.270.47.28311. [DOI] [PubMed] [Google Scholar]
- 19.Aoyagi Y, Suzuki Y, Igarashi K, et al. The usefulness of simultaneous determinations of glucosaminylation and fucosylation indices of alpha-fetoprotein in the differential diagnosis of neoplastic diseases of the liver. Cancer. 1991;67:2390–2394. doi: 10.1002/1097-0142(19910501)67:9<2390::aid-cncr2820670928>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 20.Taketa K, Endo Y, Sekiya C, et al. A collaborative study for the evaluation of lectin-reactive alpha-fetoproteins in early detection of hepatocellular carcinoma. Cancer Res. 1993;53:5419–5423. [PubMed] [Google Scholar]
- 21.Okuyama N, Ide Y, Nakano M, et al. Fucosylated haptoglobin is a novel marker for pancreatic cancer: a detailed analysis of the oligosaccharide structure and a possible mechanism for fucosylation. Int J Cancer. 2006;118:2803–2808. doi: 10.1002/ijc.21728. [DOI] [PubMed] [Google Scholar]
- 22.Miyoshi E, Nakano M. Fucosylated haptoglobin is a novel marker for pancreatic cancer: detailed analyses of oligosaccharide structures. Proteomics. 2008;8:3257–3262. doi: 10.1002/pmic.200800046. [DOI] [PubMed] [Google Scholar]
- 23.Moriwaki K, Noda K, Furukawa Y, et al. Deficiency of GMDS leads to escape from NK cell-mediated tumor surveillance through modulation of TRAIL signaling. Gastroenterology. 2009;137:188–198. doi: 10.1053/j.gastro.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida H. ER stress response, peroxisome proliferation, mitochondrial unfolded protein response and Golgi stress response. IUBMB Life. 2009;61:871–879. doi: 10.1002/iub.229. [DOI] [PubMed] [Google Scholar]