Abstract
Low-grade serous ovarian carcinoma is believed to arise from serous borderline ovarian tumors, yet the progression from serous borderline tumors to low-grade serous ovarian carcinoma remains poorly understood. The purpose of this study was to identify differentially expressed genes between the two groups. Expression profiles were generated from 6 human ovarian surface epithelia (HOSE), 8 serous borderline ovarian tumors (SBOT), 13 low-grade serous ovarian carcinomas (LG), and 24 high-grade serous ovarian carcinomas (HG). The anterior gradient homolog 3 (AGR3) gene was found to be highly upregulated in serous borderline ovarian tumors; this finding was validated by real-time quantitative RT-PCR, Western blotting, and immunohistochemistry. Anti-AGR3 immunohistochemistry was performed on an additional 56 LG and 103 HG tissues and the results were correlated with clinical data. Expression profiling determined that 1254 genes were differentially expressed (P < 0.005) between SBOT, LG and HG tumors. Serous borderline ovarian tumors exhibited robust positive staining for AGR3, with a lower percentage of tumor cells stained in LG and HG. Immunofluorescence staining indicated that AGR3 expression was limited to ciliated cells. Tumor samples with a high percentage (>10%) of AGR3 positively stained tumor cells were associated with improved longer median survival in both the LG (P = 0.013) and HG (P = 0.008) serous ovarian carcinoma groups. The progression of serous borderline ovarian tumors to low-grade serous ovarian carcinoma may involve the de-differentiation of ciliated cells. AGR3 could serve as a prognostic marker for survival in patients with low-grade and high-grade serous ovarian carcinomas.
Keywords: AGR3, BCMP11, ovarian cancer, biomarker, cilia
Ovarian cancer is the most lethal gynecologic cancer in the United States, causing approximately 13,850 deaths per year.7 Among the different histologic subtypes, serous ovarian carcinoma is the most common, yet its etiology remains unclear. Our previous work has demonstrated that the gene expression profiles of serous borderline ovarian tumors are more closely related to those of low-grade serous ovarian carcinoma than to those of high-grade serous ovarian carcinoma.2, 26 The concept that SBOT progresses to low-grade serous ovarian carcinoma (SOC), while high-grade SOC likely develops from an unidentified precursor, is also gaining acceptance. 2, 12, 19-21, 23, 24, 27 Nevertheless, the progression from serous borderline ovarian tumor to low-grade serous ovarian carcinoma remains poorly understood.
To better understand the progression from serous borderline ovarian tumor to low-grade serous ovarian carcinoma, and validate that high-grade and low-grade SOCs are molecularly distinct entities, we compared the gene expression profiles of these groups and identified a list of differentially expressed genes. This list included one particular gene of interest: the anterior gradient homolog 3 (AGR3) gene, which has been implicated in breast cancer. AGR3 (or breast cancer membrane protein 11 [BCMP11]) was originally identified as a membrane protein from breast cancer cell lines using a proteomic approach.1 AGR3 contains putative CXXS Tx-domain motifs and has been implicated in the growth and metastasis of hormone-responsive breast tumors. 1, 4 Here, we report for the first time the differential expression of AGR3 in ovarian cancer and evaluate its expression in serous ovarian cancer. We also correlate its expression with patient survival.
Materials and Methods
Tissue samples
Paraffin-embedded sections from patients with SBOT, advanced-stage LG serous ovarian carcinoma, and advanced-stage HG serous ovarian carcinoma were obtained from the archives of the Department of Pathology at The University of Texas MD Anderson Cancer Center (Houston, Texas). All cases were reviewed and classified as low-grade or high-grade by gynecologic pathologists (A.M. and M.T.D.) using histologic criteria described previously. 13, 14 Frozen tissues were obtained from the Multidisciplinary Gynecologic Cancer Translational Research Tissue Bank at MD Anderson Cancer Center, and all cases were validated by morphological review at the time of retrieval. All tissue specimens had been collected and archived previously under protocols approved by the institutional review board.
Gene expression profiling and data analysis
Expression profiles were extracted from 6 human ovarian surface epithelia (HOSE), 8 serous borderline ovarian tumors, 13 low-grade serous ovarian carcinomas (LG), and 24 high-grade serous ovarian carcinomas (HG), and profiles were then generated with commercial GeneChip Human Genome U133 Plus 2.0 Array (all from Affymetrix except where noted otherwise). To successfully generate a sufficient amount of labeled cRNA for microarray analysis from 25 ng total RNA, two rounds of amplification were necessary and were achieved with the Two-Cycle cDNA Synthesis Kit. For the first round of synthesis of double-stranded cDNA, total RNA was reverse transcribed using the oligo(dT)24-T7 (5′-GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG-3′) primer according to the manufacturer's instructions, followed by amplification with the MEGAscript T7 Kit (Ambion, Inc.). After cleanup of the cRNA with a GeneChip Sample Cleanup Module, second-round double-stranded cDNA was amplified using the Gene Chip IVT Labeling Kit. A 15.0-μg aliquot of labeled product was fragmented by heat and ion-mediated hydrolysis at 94°C for 35 min in 24 μL H2O and 6 μL of 5× fragmentation buffer. The fragmented cRNA was hybridized for 16 h at 45°C in a Hybridization Oven 640 to a GeneChip Human Genome U133 Plus 2.0 oligonucleotide array. Washing and staining the arrays with phycoerythrin-conjugated streptavidin (Molecular Probes) was completed in a GeneChip Fluidics Station 450. The arrays were then scanned using a confocal laser GeneChip Scanner 3000 and GeneChip operating software.
Real-time quantitative reverse-transcription polymerase chain reaction (RT-PCR)
Total RNA extraction (RNeasy Mini Kit, Qiagen) and cDNA synthesis (High-Capacity cDNA Archive Kit, Applied Biosystems) were done according to the manufacturer's instructions. Total RNA was extracted from 51 microdissected samples (6 HOSE, 8 SBOT, 13 LG, and 24 HG serous ovarian tumors). Real-time RT-PCR was performed twice in triplicate using TaqMan primer sets for AGR3 and the housekeeping gene cyclophilin (Applied Biosystems). All reactions were run in an 7300 Real Time PCR System (Applied Biosystems). The amount of change (as a percentage) was calculated with the ΔΔCt method as described previously.25
Immunohistochemistry
The AGR3 polyclonal antibody was raised in rabbits immunized with two specific peptides, CAQNEEIQEMAQNKFIMLNLMHET and CTYEPRDLPLLIENMKKALRLIQSEL, as described previously.1 Custom antibodies were obtained from 21st Century Biochemicals. Paraffin sections (4 μm thick) were cut from formalin-fixed, paraffin-embedded specimens and then placed on glass slides. Before staining, the paraffin-embedded tissue sections were incubated at 60°C overnight to improve adherence to the slide. At room temperature, the specimens were deparaffinized in xylene and rehydrated through 100%, 95%, and 75% ethanol and dH2O. Antigen retrieval was then performed in 1× Reveal (Biocare Medical) in a Decloaking Chamber (Biocare Medical) heated at 120°C for 15 min. The cooled slides were washed with dH2O and 1× Immunocare TBS (Biocare Medical), each for 5 min. Background SNIPER (Biocare Medical) was applied for 20 min to reduce nonspecific background staining, and TBS-rinsed sections were then incubated at a dilution of 1:250 in 1% fresh bovine serum albumin buffer for 120 min. To remove unbound antibodies, the slides were washed twice in TBS buffer for 10 min. Slides were then incubated with Mach3 rabbit IgG probe (Biocare Medical) and then Mach3 polymer alkaline-phosphatase (Biocare Medical); after each treatment, the slides were washed twice with TBS buffer for 10 min. Vulcan Fast Red (Biocare Medical) was applied for 10 min, and the slides were then rinsed with dH2O, counterstained with Vector Hematoxylin QS (Vector Laboratories), and rinsed again. The slides were fixed with Permount Mounting Medium (Fisher Scientific) and evaluated by light microscopy at 10× and 20×, with photography at 20× documenting staining intensity.
As the staining of the AGR3 polyclonal antibody was particularly robust, staining was classified according to percentages of cells staining positively (0, <10%, 10%–25%, 25%–50%, 50%–75%, and >75%). Intestinal and fallopian tube tissues served as positive controls, and 17 SBOT slides were stained. For the survival analysis, 159 slides were stained (56 LG and 103 HG specimens). As the percentage of AGR3-positive staining in HG serous ovarian carcinomas was very low, a subset of 30 HG slides containing specimens from patients at the survival extremes (<20 months, >90 months) was selected initially and stained. Because the initial correlation between positive staining and survival time was positive, an additional 73 slides from patients with various survival times, who were randomly selected, were examined. Slides were reviewed and divided into high (>10%) and low (<10%) staining intensity categories. Clinical data were collected on survival time (from diagnosis to death), International Federation of Gynecology and Obstetrics (FIGO) stage, and tumor cytoreduction status (defined as <2 cm of residual disease at the completion of tumor-reductive surgery).
Western blot analysis
Total protein lysates from 5 SBOT, 7 LG, and 6 HG tissues, which were different from the set of 51 tumor samples used for expression analysis, were extracted for Western blot analysis. Tumor tissues were homogenized and subjected to RIPA buffer to extract protein lysates. SDS-PAGE was performed using protein lysates (10 μg) on a 10% SDS-polyacrylamide gel; the protein lysates were then electroblotted to a polyvinylidene difluoride membrane and blocked in PBS-Tween (PBS-T; 50 mM potassium phosphate, pH 7.2, 150 mmol/L NaCl, and 0.5% Tween 20) with 5% nonfat dry milk powder for 1 h. Blots were incubated overnight with a 1:1,000 dilution of the primary antibody (anti-AGR3, 0.4 mg/mL) in 1× PBS-T with 5% bovine serum albumin, and washed three times in PBS-T buffer, followed with the horseradish peroxidase-conjugated secondary antibody (1:1,000; Amersham-GE Healthcare) diluted in PBS-T with nonfat dry milk powder for 1 h at room temperature. Amersham ECL Plus Detection Reagent (GE Healthcare) was used to visualize the protein bands.
AGR3 immunofluorescence double staining
Freshly cut frozen tissue specimens embedded in Tissue Tek OCT Compound (Fisher Scientific) were fixed initially in 4% paraformaldehyde (Sigma-Aldrich). Samples were then permeabilized using 0.5% Triton X-100 for 15 min and blocked in 5% normal goat serum for 30 min. The first primary antibody, rabbit anti-AGR3 immunoglobulin G at a 1:200 dilution (initial concentration 0.4 mg/mL), was incubated for 1.5 h at room temperature, followed by incubation with the secondary antibody for 1 h. Specimens were blocked again with 5% normal goat serum for 30 min and then were incubated with the second primary antibody, monoclonal mouse anti-β tubulin IV immunoglobulin G 1 at a 1:500 dilution (Sigma-Aldrich), at 4°C overnight; the specimens were then incubated with the secondary antibody for 1 h. The secondary antibodies used were Cy2-conjugated (green) AffiniPure Goat Anti-Mouse IgG, FCγ Subclass 1 Specific, at a 1:200 dilution (Jackson ImmunoResearch Laboratories) and Cy3-conjugated (red) AffiniPure Goat Anti-Rabbit IgG (H&L), at a 1:500 dilution (both from Jackson ImmunoResearch Laboratories). Tissue sections were mounted with 10 μL of Prolong Gold Antifade Reagent with DAPI (Molecular Probes, Invitrogen) and viewed with a Leica DMLA fluorescence microscope. Images were captured using Image-Pro Plus (version 6) (Media Cybernetics).
Statistical analysis
Statistical analyses were done using the statistical package SPSS 15.0 for Windows (SPSS, Inc.). A nonparametric Mann-Whitney test was used to assess the statistical significance of the differences in AGR3 mRNA expression between normal human ovarian surface epithelia cells and microdissected ovarian cancer tissues. P values < 0.05 were considered statistically significant. The Kaplan-Meier method was used to generate survival curves for patients with high- and low-staining tissues, and staining intensity was correlated with survival time (in months) for both low-grade and high-grade serous ovarian carcinomas. The Cox proportional hazards regression model was used to evaluate the relationship between survival time and staining intensity, FIGO stage, and cytoreduction status.
Results
Genes upregulated in serous borderline ovarian tumors/low-grade serous ovarian carcinoma compared with high-grade serous ovarian carcinoma
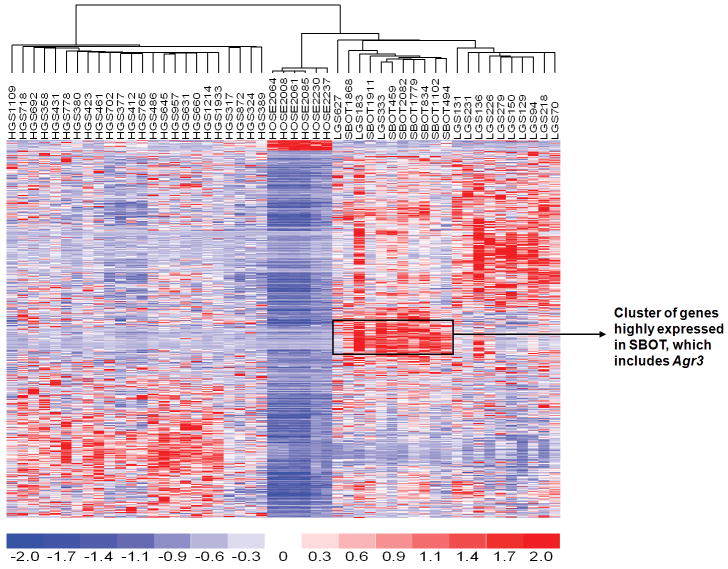
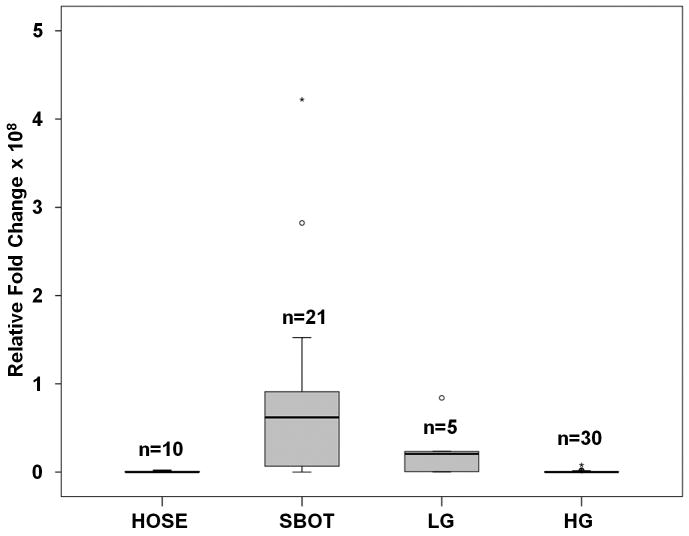
In comparing the gene expression profiles of all ovarian tumors with those of the human ovarian surface epithelia, 1254 genes were found to be differentially expressed (P < 0.005; Supplemental Table 1), which separated SBOT/LG from HG (Fig. 1) as determined by unsupervised clustering. In significance analysis of microarrays, 122 genes were found to be significantly upregulated in the SBOT/LG group compared to the HG group (Supplemental Table 2). The 20 most-commonly upregulated genes are listed in Table 1. Several of these genes—including DNAH9,5 RSPHA4,3 SNTN,10 SPATA18, 9 TPPP3,30 RSPH1,18 and TEKT1 11—are components of cilium/flagellum or are involved in microtubule-based cellular movement. The overexpression of AGR3 in serous borderline ovarian tumors and low-grade serous ovarian carcinoma samples was validated by RT-PCR as shown in Fig. 2. The relative expression of AGR3 in tumors was compared with that of the human ovarian surface epithelia samples. AGR3 was significantly upregulated in SBOT and LG (P < 0.001) but not in HG samples.
Figure 1.
Differentially expressed genes between 6 HOSE, 8 SBOT, 13 LG, and 24 HG tumors were identified by t-test with 50 permutations to estimate false discovery rate (FDR) using dChip2009. A list of genes with median 2.2% FDR was identified. Unsupervised analysis using this set of genes was able to separate the SBOT/LG from HG.
Table 1. The top 20 most frequently overexpressed genes in serous borderline ovarian tumors and low-grade serous ovarian carcinomas.
| Probe ID | Fold-change | Symbol | Entrez gene name | Biological function |
|---|---|---|---|---|
| 229331_at | 19.0 | SPATA18 | Spermatogenesis-associated 18 homolog | Component of flagellum |
| 220269_at | 18.9 | ZBBX | Zinc finger, B-box domain containing | Metal ion binding |
| 211996_s_at | 18.1 | NPIPL3 | Nuclear pore complex interacting protein-like 3 | Unknown |
| 240065_at | 16.4 | FAM81B | Family with sequence similarity 81, member B | Unknown |
| 233157_x_at | 16.3 | CCDC114 | Coiled-coil domain containing 114 | Unknown |
| 240857_at | 15.1 | DNAH9 | Dynein, axonemal, heavy chain 9 | Cell movement |
| 233071_at | 14.7 | RSPH4A | Radial spoke head 4 homolog A | Microbule-based movement |
| 220390_at | 14.3 | AGBL2 | ATP/GTP binding protein-like 2 | Metal ion binding |
| 239150_at | 13.8 | SNTN | Sentan, cilia apical structure protein | Cilium component |
| 229170_s_at | 13.3 | TTC18 | Tetratricopeptide repeat domain 18 | Protein binding |
| 223924_at | 11.8 | TTC25 | Tetratricopeptide repeat domain 25 | Protein binding |
| 1556158_at | 11.7 | FAM154B | Family with sequence similarity 154, member B | Unknown |
| 218876_at | 10.0 | TPPP3 | Tubulin polymerization-promoting protein family member 3 | Microtubule bundle formation |
| 231729_s_at | 9.4 | CAPS | Calcyphosine | Calcium binding protein |
| 228241_at | 9.2 | AGR3 | Anterior gradient homolog 3 | Unknown |
| 230093_at | 9.1 | RSPH1 | Radial spoke head 1 homolog | Component of flagellum |
| 212525_s_at | 8.3 | H2AFX | H2A histone family, member X | Histone binding |
| 227062_at | 7.9 | NEAT1 | Nuclear paraspeckle assembly transcript 1 | Unknown |
| 239216_at | 7.6 | TEKT1 | Tektin 1 | Microtubule cytoskeleton organization |
| 219332_at | 7.6 | MICALL2 | MICAL-like 2 | Actin filament bundle |
Figure 2.
Real-time RT-PCR validating overexpression of AGR3 in SBOT compared to LG, HG, and HOSE tumors. AGR3 was significantly up-regulated in SBOT and LG (p <0.001) but not in HG samples.
Immunohistochemistry of AGR3
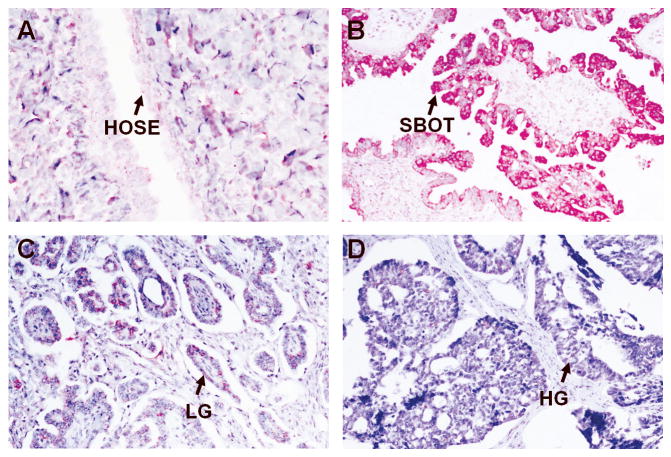
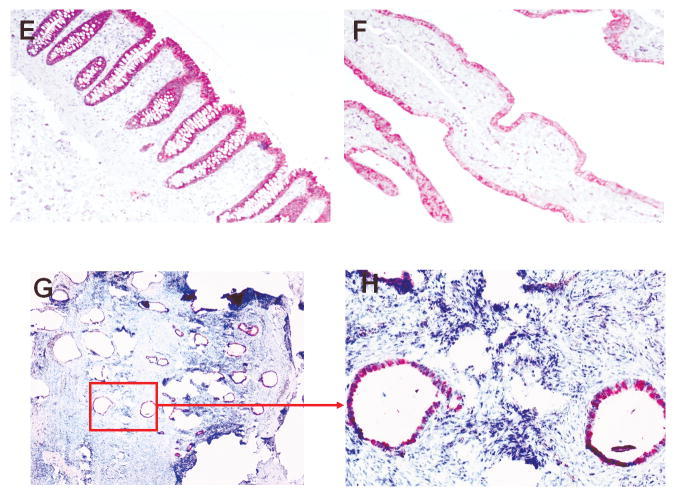
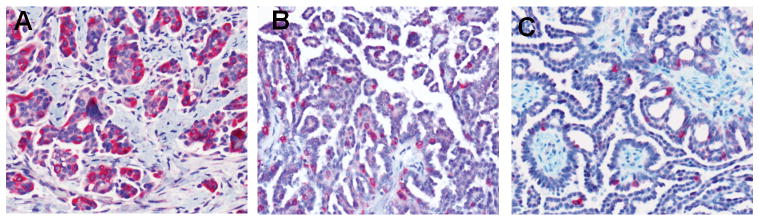
Table 2 summarizes the immunostaining of AGR3 on the paraffin-embedded tissue sections. Representative stains of various sections are shown in Fig. 3. Epithelial cells from normal ovaries were negative for AGR3 (Fig. 3A). All SBOT samples except one were positive for AGR3, with >50% of tumor cells staining (Fig. 3B), while 20% of the LG samples stained. Moreover, in the majority of LG cases, <10% of the tumor cells exhibited staining (Fig. 3C). High-grade serous ovarian carcinoma specimens also exhibited rare AGR3 staining (Fig. 3D). We also stained three ovarian samples with ciliated epithelial inclusion cysts, two normal intestinal epithelial cells, and three normal fallopian tubes (Fig. 3E-3H). All these samples also were positive for AGR3, with more than 50% of the epithelial cells exhibiting staining. Another 12 recurrent low-grade serous ovarian carcinoma samples from patients diagnosed initially with serous borderline ovarian tumors were stained; the initial SBOT samples all stained, but loss of staining occurred with progression to LG serous ovarian carcinoma (Fig. 4).
Table 2. Summary of immunohistochemistry of AGR3.
| Samples | Stained Positive n (%) |
Stained Negative* n (%) |
|---|---|---|
| Normal ovary (n = 4) | 0 (0) | 4 (100) |
| Serous borderline ovarian tumors (n = 16) | 16 (94) | 1 (6) |
| Serous borderline ovarian tumors with progression to low-grade serous ovarian carcinoma (n = 4) | 4 (80) | 1 (20) |
| Low-grade serous ovarian carcinoma (n = 56) | 11 (20) | 45 (80) |
| High-grade serous ovarian carcinoma (n = 103) | 20 (19) | 83 (81) |
Low-grade tumor samples that were called negative have 0-9% tumor cells with AGR3 positive staining as Fig. 4C. However, High-grade tumor samples that were called negative have no tumor cells with AGR3 positive staining. Tumor samples with 10% or more AGR3 positively stained tumor cells were called positive.
Figure 3.
Representative stains of various tissue sections for AGR3 expression. All figures were obtained at 100× unless otherwise noted. A. Normal ovarian surface epithelium (400×); B. Serous borderline ovarian tumor; C. Low grade OSC; D. High grade OSC; E. Intestinal epithelium; F. Fallopian tube; G/H. Ovarian endosalpingiosis.
Figure 4.
Examples of low-grade serous ovarian carcinomas with decreasing percentage of AGR3 positively stained tumor cells (from A to C). All images at 200×.
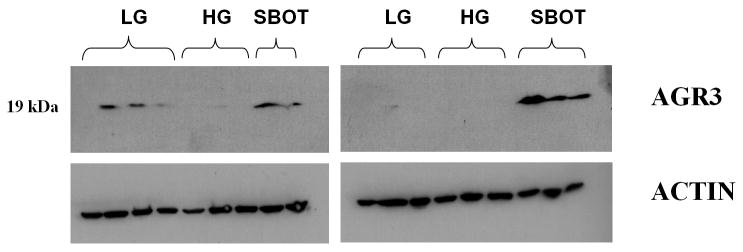
AGR3 protein was detected mainly in serous borderline ovarian tumors and low-grade serous ovarian carcinoma. In the Western blot analysis, all 5 SBOT demonstrated robust expression of AGR3 (19-kDa protein band), 2 of the 7 LG samples exhibited robust expression of AGR3, and none of the 6 HG samples expressed AGR3 (Fig. 5).
Figure 5.
Western blot of AGR3 protein expression in low-grade serous ovarian carcinoma, high-grade serous ovarian carcinoma, and serous borderline ovarian tumor.
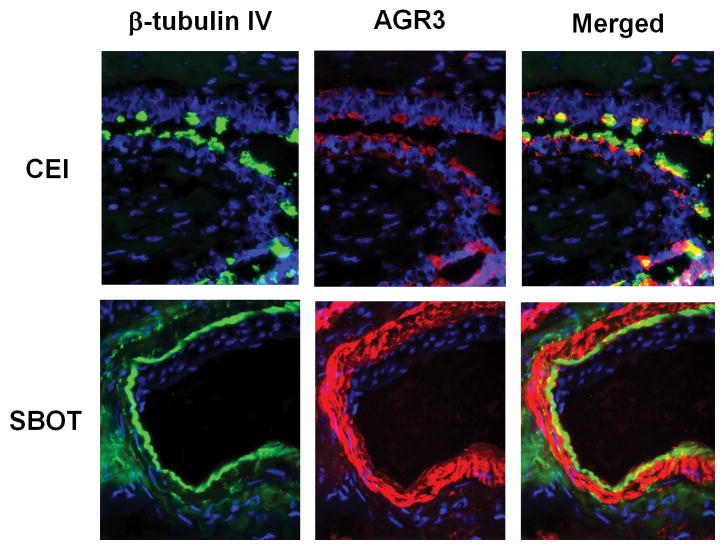
AGR3 was expressed in ciliated cells. Double immunofluorescence staining of ciliated epithelial inclusion cysts and SBOT revealed that AGR3 was expressed in ciliated cells. In ciliated epithelial inclusion cysts, only the ciliated cells expressed AGR3. Conversely, all tumor cells in the serous borderline ovarian tumors were positive for both β-tubulin IV (a cilia-specific protein marker) and AGR3 (Fig. 6).8
Figure 6.
Immunofluorescent stain of β-tubulin IV and AGR3 in cliliated epithelial inclusion cyst versus serous borderline ovarian tumor.
AGR3 expression was correlated with longer survival time
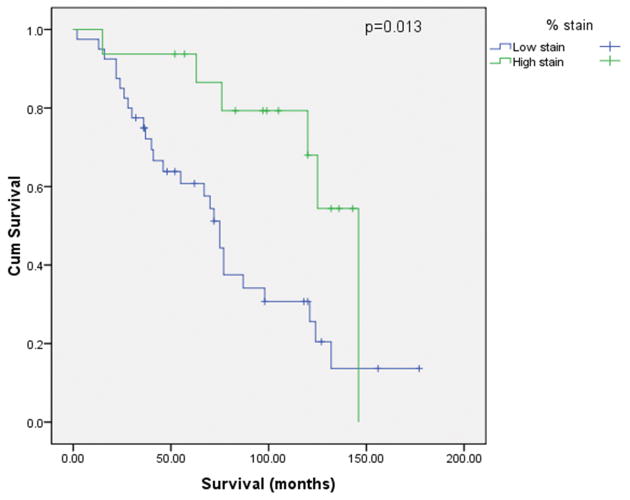
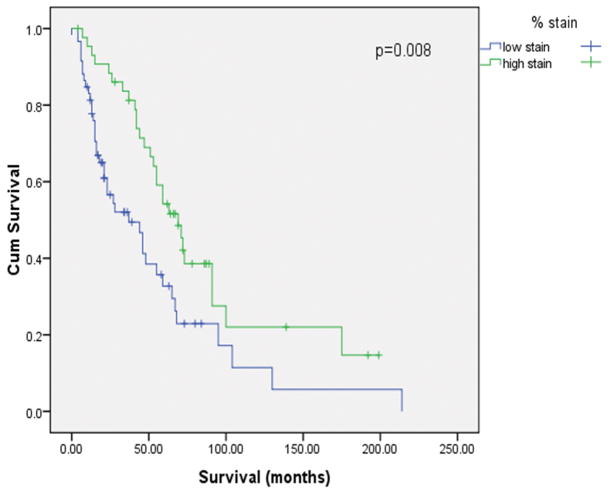
Survival analyses were performed for patients with low-grade and high-grade serous carcinomas separately. The Kaplan-Meier curves (Figs. 7A and 7B) indicated that AGR3 positive staining correlated significantly with a better survival in patients within the low-grade group (p = 0.013) or the high-grade group (p = 0.008). The median survival time for patients with low-grade serous ovarian carcinoma was 72 months (range: 2-177 months), compared with 39 months (range: 0-214 months) for those with high-grade serous ovarian carcinoma. In the low-grade group (median age, 52; range: 23-85), 79% of patients had stage III disease, and 18% of patients had stage IV disease at the time of diagnosis; 68% of patients underwent optimal cytoreductive surgery. In the high-grade group (median age, 61; range: 33-95), 93% of patients had stage III disease and 5% had stage IV disease at the time of diagnosis; 85% of patients underwent optimal cytoreductive surgery. Cox proportional hazards regression model analysis was further performed to test the significance of AGR3 staining after adjusted for age, stage and debulking status. The Cox regression result revealed that AGR3 positive staining was still a significant predictor of better survival for both the low-grade group (p = 0.029, HR = 3.42 with 95% CI of 1.13 to 10.36) and the high-grade group (p = 0.021, HR = 1.86 with 95% CI of 1.1 to 3.15).
Figure 7.
Kaplan-Meier survival curve according to high- and low-AGR3 staining in serous ovarian carcinoma paraffin sections. A. Low-grade; B. High-grade.
Discussion
In this study, we have identified a list of genes that are highly upregulated in serous borderline ovarian tumors and low-grade serous ovarian carcinomas, several of which are involved in cilia structure and/or microtubule-based cellular motility. It is generally accepted that cilia play a crucial role in normal development. Mice born lacking the cilia-encoding gene Kif3a die during embryogenesis,15 and human ciliopathies lead to a range of defects. It is now becoming clear that cilia—or rather the lack thereof—also contribute to tumorigenesis. Wong et al.28 demonstrated a loss of cilia during the development of basal cell carcinoma in mice and showed that this particular loss led to accelerated tumorigenesis.
While much remains unknown about their exact function and their role in various cancers, it is postulated that cilia may behave as tumor-suppressing organelles. In breast cancer cell lines, loss of primary cilia has been associated with tumor progression and the basal B subtype, which is analogous to triple-negative breast cancer.29 Other studies have demonstrated similar results; electron microscopy of ovarian tumors has revealed tubal cells with numerous microvilli projecting into the lumen and the presence of ciliated cells in both benign and borderline tumors, but not in malignant tumors. 22 Our co-immunofluorescent staining demonstrated similar results, with AGR3 being highly expressed in the ciliated cells of epithelial inclusion cysts and serous borderline ovarian tumors.
AGR3 has been described as a membrane protein in breast cancer cells and may have a role in tumorigenesis by interacting with metastasis-associated genes. 4 The overexpression of AGR3 in serous borderline ovarian tumors was also validated in our study using Gilks' microarray data,6 which are available at the Oncomine website.16 In our study, the overexpression of AGR3 in borderline serous tumors and low-grade serous ovarian carcinomas was further confirmed by real-time reverse-transcription polymerase chain reaction, Western blotting, and immunostaining. These findings are consistent with the pathologic observation that serous borderline ovarian tumors and low-grade serous ovarian carcinomas are well-differentiated tumors compared with high-grade serous ovarian carcinomas and further support the use of a two-tiered grading system.
In our study, we also found that AGR3 staining correlated with the level of tumor differentiation. A similar percentage of high- and low-grade cells stained for AGR3, which is likely attributable to our initial selection of the high-grade serous ovarian carcinoma subset according to survival extremes, with a disproportionate number of longer-surviving individuals in the high-grade group relative to those individuals randomly selected in the low-grade group. Thus, a higher-than-expected number of stained slides was observed in the high-grade serous ovarian carcinoma group.
Similar results were obtained with 12 recurrent low-grade serous ovarian carcinoma samples from patients who were initially diagnosed with serous borderline ovarian tumors, suggesting that during the progression of serous borderline ovarian tumor to low-grade serous ovarian carcinomas, the number of AGR3-positive cells decreases. Since many of the overexpressed genes in serous borderline ovarian tumors and low-grade serous ovarian carcinoma are involved in the cilia structure, the decrease in ciliated cells in low-grade serous ovarian carcinomas may suggest that the progression of serous borderline ovarian tumors to low-grade serous ovarian carcinomas involves the de-differentiation or loss of ciliated structures. Therefore, AGR3 may serve as a differentiation factor, and the loss of AGR3 is associated with progression of serous borderline ovarian tumors.
It is interesting that we also established a correlation between survival and the percentage of AGR3 positively stained tumor cells in tumor samples. Though uncommon in the low- and high-grade serous ovarian carcinoma subtypes,17 those patients with tumors expressing AGR3 survived significantly longer than those without AGR3 positively stained tumor cells. AGR3 thus may be a biomarker for serous borderline ovarian tumors and may indicate a better prognosis in patients who are newly diagnosed with ovarian cancer.
In conclusion, our findings support the concept that serous borderline ovarian tumors and low-grade serous ovarian carcinoma form a continuum, while high-grade serous ovarian carcinomas arise from a clinically and pathologically distinct entity. Moreover, they suggest that AGR3 serves as a marker of tumor differentiation, more favorable prognosis, and longer survival and is present in ciliated cells. These findings warrant further investigation into the role of cilia in tumorigenesis and the utility of AGR3 as a biomarker and potential therapeutic target.
Supplementary Material
Acknowledgments
We thank Joseph Celestino for retrieving the frozen samples, and Tri Nguyen for cutting the paraffin sections. We also thank Christina Yeung for optimizing the immunostaining conditions and Kristi M. Speights for editing the manuscript.
The authors declare that neither pharmaceutical nor industry support was provided for this work. Funding for this project was received from the National Institutes of Health, including The University of Texas MD Anderson Cancer Center Specialized Program of Research Excellence in Ovarian Cancer (P50 CA08369), grant R01CA133057, grant RC4CA156551, and MD Anderson's Cancer Center Support Grant (CA016672); the Hera Foundation; the Entertainment Industry Foundation; Marsha Rivkin Center; and the Blanton-Davis Ovarian Cancer Research Program. Authors ERK and CST are supported by NCI-DHHS-NIH T32 Training Grant (T32 CA101642). GTML is partly supported by the Molecular Biology Program, CUHK.
References
- 1.Adam PJ, Boyd R, Tyson KL, et al. Comprehensive proteomic analysis of breast cancer cell membranes reveals unique proteins with potential roles in clinical cancer. J Biol Chem. 2003;278:6482–6489. doi: 10.1074/jbc.M210184200. [DOI] [PubMed] [Google Scholar]
- 2.Bonome T, Lee JY, Park DC, et al. Expression profiling of serous low malignant potential, low-grade, and high-grade tumors of the ovary. Cancer Res. 2005;65:10602–10612. doi: 10.1158/0008-5472.CAN-05-2240. [DOI] [PubMed] [Google Scholar]
- 3.Castleman VH, Romio L, Chodhari R, et al. Mutations in radial spoke head protein genes RSPH9 and RSPH4A cause primary ciliary dyskinesia with central-microtubular-pair abnormalities. Am J Hum Genet. 2009;84:197–209. doi: 10.1016/j.ajhg.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fletcher GC, Patel S, Tyson K, et al. hAG-2 and hAG-3, human homologues of genes involved in differentiation, are associated with oestrogen receptor-positive breast tumours and interact with metastasis gene C4.4a and dystroglycan. Br J Cancer. 2003;88:579–585. doi: 10.1038/sj.bjc.6600740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fliegauf M, Olbrich H, Horvath J, et al. Mislocalization of DNAH5 and DNAH9 in respiratory cells from patients with primary ciliary dyskinesia. Am J Respir Crit Care Med. 2005;171:1343–1349. doi: 10.1164/rccm.200411-1583OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilks CB, Vanderhyden BC, Zhu S, et al. Distinction between serous tumors of low malignant potential and serous carcinomas based on global mRNA expression profiling. Gynecol Oncol. 2005;96:684–694. doi: 10.1016/j.ygyno.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 7.Jemal A, Siegel R, Xu J, et al. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 8.Jensen-Smith HC, Luduena RF, Hallworth R. Requirement for the betaI and betaIV tubulin isotypes in mammalian cilia. Cell Motil Cytoskeleton. 2003;55:213–220. doi: 10.1002/cm.10122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaneko T, Murayama E, Kurio H, et al. Characterization of Spetex-1, a new component of satellite fibrils associated with outer dense fibers in the middle piece of rodent sperm flagella. Mol Reprod Dev. 2010;77:363–372. doi: 10.1002/mrd.21154. [DOI] [PubMed] [Google Scholar]
- 10.Kubo A, Yuba-Kubo A, Tsukita S, et al. Sentan: a novel specific component of the apical structure of vertebrate motile cilia. Mol Biol Cell. 2008;19:5338–5346. doi: 10.1091/mbc.E08-07-0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larsson M, Norrander J, Graslund S, et al. The spatial and temporal expression of Tekt1, a mouse tektin C homologue, during spermatogenesis suggest that it is involved in the development of the sperm tail basal body and axoneme. Eur J Cell Biol. 2000;79:718–725. doi: 10.1078/0171-9335-00097. [DOI] [PubMed] [Google Scholar]
- 12.Longacre TA, McKenney JK, Tazelaar HD, et al. Ovarian serous tumors of low malignant potential (borderline tumors): outcome-based study of 276 patients with long-term (> or =5-year) follow-up. Am J Surg Pathol. 2005;29:707–723. doi: 10.1097/01.pas.0000164030.82810.db. [DOI] [PubMed] [Google Scholar]
- 13.Malpica A, Deavers MT, Lu K, et al. Grading ovarian serous carcinoma using a two-tier system. Am J Surg Pathol. 2004;28:496–504. doi: 10.1097/00000478-200404000-00009. [DOI] [PubMed] [Google Scholar]
- 14.Malpica A, Deavers MT, Tornos C, et al. Interobserver and intraobserver variability of a two-tier system for grading ovarian serous carcinoma. Am J Surg Pathol. 2007;31:1168–1174. doi: 10.1097/PAS.0b013e31803199b0. [DOI] [PubMed] [Google Scholar]
- 15.Marszalek JR, Ruiz-Lozano P, Roberts E, et al. Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc Natl Acad Sci U S A. 1999;96:5043–5048. doi: 10.1073/pnas.96.9.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sciallis AP, Aubry MC, Bell DA. Ciliated adenocarcinoma of the ovary with evidence of serous differentiation: report of a case. Int J Gynecol Pathol. 2009;28:447–452. doi: 10.1097/PGP.0b013e3181a0717f. [DOI] [PubMed] [Google Scholar]
- 18.Shetty J, Klotz KL, Wolkowicz MJ, et al. Radial spoke protein 44 (human meichroacidin) is an axonemal alloantigen of sperm and cilia. Gene. 2007;396:93–107. doi: 10.1016/j.gene.2007.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shih Ie M, Kurman RJ. Molecular pathogenesis of ovarian borderline tumors: new insights and old challenges. Clin Cancer Res. 2005;11:7273–7279. doi: 10.1158/1078-0432.CCR-05-0755. [DOI] [PubMed] [Google Scholar]
- 20.Shvartsman HS, Sun CC, Bodurka DC, et al. Comparison of the clinical behavior of newly diagnosed stages II-IV low-grade serous carcinoma of the ovary with that of serous ovarian tumors of low malignant potential that recur as low-grade serous carcinoma. Gynecol Oncol. 2007;105:625–629. doi: 10.1016/j.ygyno.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 21.Silva EG, Gershenson DM, Malpica A, et al. The recurrence and the overall survival rates of ovarian serous borderline neoplasms with noninvasive implants is time dependent. Am J Surg Pathol. 2006;30:1367–1371. doi: 10.1097/01.pas.0000213294.81154.95. [DOI] [PubMed] [Google Scholar]
- 22.Stenback F. Benign, borderline and malignant serous cystadenomas of the ovary. A transmission and scanning electron microscopical study. Pathol Res Pract. 1981;172:58–72. doi: 10.1016/S0344-0338(81)80123-9. [DOI] [PubMed] [Google Scholar]
- 23.Tung CS, Mok SC, Tsang YT, et al. PAX2 expression in low malignant potential ovarian tumors and low-grade ovarian serous carcinomas. Mod Pathol. 2009;22:1243–1250. doi: 10.1038/modpathol.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vang R, Shih Ie M, Kurman RJ. Ovarian low-grade and high-grade serous carcinoma: pathogenesis, clinicopathologic and molecular biologic features, and diagnostic problems. Adv Anat Pathol. 2009;16:267–282. doi: 10.1097/PAP.0b013e3181b4fffa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong KK, Chang YM, Tsang YT, et al. Expression analysis of juvenile pilocytic astrocytomas by oligonucleotide microarray reveals two potential subgroups. Cancer Res. 2005;65:76–84. [PubMed] [Google Scholar]
- 26.Wong KK, Gershenson D. The continuum of serous tumors of low malignant potential and low-grade serous carcinomas of the ovary. Dis Markers. 2007;23:377–387. doi: 10.1155/2007/204715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong KK, Tsang YT, Deavers MT, et al. BRAF mutation is rare in advanced-stage low-grade ovarian serous carcinomas. Am J Pathol. 2010;177:1611–1617. doi: 10.2353/ajpath.2010.100212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong SY, Seol AD, So PL, et al. Primary cilia can both mediate and suppress Hedgehog pathway-dependent tumorigenesis. Nat Med. 2009;15:1055–1061. doi: 10.1038/nm.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan K, Frolova N, Xie Y, et al. Primary cilia are decreased in breast cancer: analysis of a collection of human breast cancer cell lines and tissues. J Histochem Cytochem. 58:857–870. doi: 10.1369/jhc.2010.955856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou W, Li J, Wang X, et al. Stable knockdown of TPPP3 by RNA interference in Lewis lung carcinoma cell inhibits tumor growth and metastasis. Mol Cell Biochem. 2010 doi: 10.1007/s11010-010-0518-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.