Abstract
Compstatin family peptides are potent inhibitors of the complement system and promising drug candidates against diseases involving under-regulated complement activation. Compstatin is a 13-residue cyclized peptide that inhibits cleavage of complement protein C3, preventing downstream complement activation. We present three new compstatin variants, characterized by tryptophan replacement at positions 1 and/or 13. Peptide design was based on physicochemical reasoning and was inspired by earlier work which identified tryptophan substitutions at positions 1 and 13 in peptides with predicted C3c binding abilities (Bellows, M. L.; Fung, H. K.; Taylor, M. S.; Floudas, C. A.; López de Victoria, A.; Morikis, D. (2010) Biophys J 98: 2337–2346). The new variants preserve distinct polar and nonpolar surfaces of compstatin, but have altered local interaction capabilities with C3. All three peptides exhibited potent C3 binding by surface plasmon resonance (SPR) and potent complement inhibition by ELISAs. We also present ELISA data and detailed SPR kinetic data of three peptides from previous computational design.
Keywords: Structure-Based Drug Design, Peptide Design, Compstatin Family, Complement System, Complement System Inhibitors
Introduction
The compstatin family consists of peptides that bind to C3 and inhibit the activation of the complement system1,2 by hindering the cleavage of C3 to C3a and C3b.3 Controlled inhibition of the complement system is desirable in cases of several autoimmune and inflammatory diseases and pathological situations that involve its inappropriate activation.4–6 Compstatin was first discovered using a phage-displayed random peptide library for binding against C3b7 and subsequently optimized over several years using knowledge-based design and experimental and computational combinatorial methods (reviewed in Refs. 1, 8–13). The compstatin family peptides consist of 13 amino acids, 11 of which form a cyclic chain through a disulfide bridge with an additional two amino acids that are located outside the cyclic peptidic ring. The parent compstatin sequence is I[CVVQDWGHHRC]T (Peptide II), where brackets denote cyclization between Cys2 and Cys12 hereafter, and the most active sequence with natural amino acids is I[CVWQDWGAHRC]T (Peptide III or W4/A9). There are several benchmarks in the process of designing more potent peptides than parent compstatin. The design process involved basic understanding of physicochemical properties that underlie structure and inhibitory activity and the optimization of specific amino acids which resulted in enhanced inhibitory activity. The determination of the three-dimensional solution structure of compstatin by NMR, in combination with an alanine scan, established the first sequence-structure-activity relations.14 NMR and inhibitory activity studies of compstatin peptides with rationally-designed mutations, aimed at introducing local perturbations affecting structure and activity, were responsible for identifying the physicochemical properties that were important for structural stability and inhibitory activity.8,15 These studies proposed the sequence template X[CVXQDWGXXXC]X, which distinguished 7 amino acids indispensable for activity from 6 amino acids (noted by X) which were amenable to further optimization, with the latter maintaining their dominant parent physicochemical properties during optimization.8,15 Application of an integer linear optimization and deterministic global optimization computational method, using the solution structure of compstatin and the aforementioned sequence template, was responsible for pinpointing the need for an aromatic amino acid at position 4, such as Tyr and Trp, in order to optimize the activity of parent compstatin.16 This substitution led to the sequence of Peptide III, above, and opened the way for the inclusion of non-natural amino acids at position 4, which further increased the activity of compstatin family peptides.17 Other major breakthroughs involved the first study of compstatin dynamics using molecular dynamics simulations,18 the first pharmacophore study of compstatin family peptides using quasi-dynamic pharmacophore models,19 the most potent peptide with methylated Trp at position 4,20 and the crystallographic structure of the complex between C3c and the most potent compstatin peptide comprising of natural amino acids (Peptide III or W4/A9, above).21
In a recent study, we used a new de novo design computational framework to design new compstatin peptides with predicted binding abilities to C3c and expected inhibitory activities against complement activation.22 Three of the sequences of that study are studied in detail experimentally here (Peptides IV–VI). The computational framework was based on sequence selection, fold specificity, and approximate binding affinity calculations, using the crystallographic structure of the C3c-W4/A9 complex (Fig. 1). The computational studies revealed new key positions in the sequence of compstatin, within the optimizable amino acid positions of the sequence template mentioned above, which were predicted to greatly affect the binding affinity to C3c. Based on these new key features and molecular analysis of the physicochemical properties of compstatin family peptides, we have rationally designed three new sequences with predicted binding abilities to C3/C3c (Peptides VII–IX).
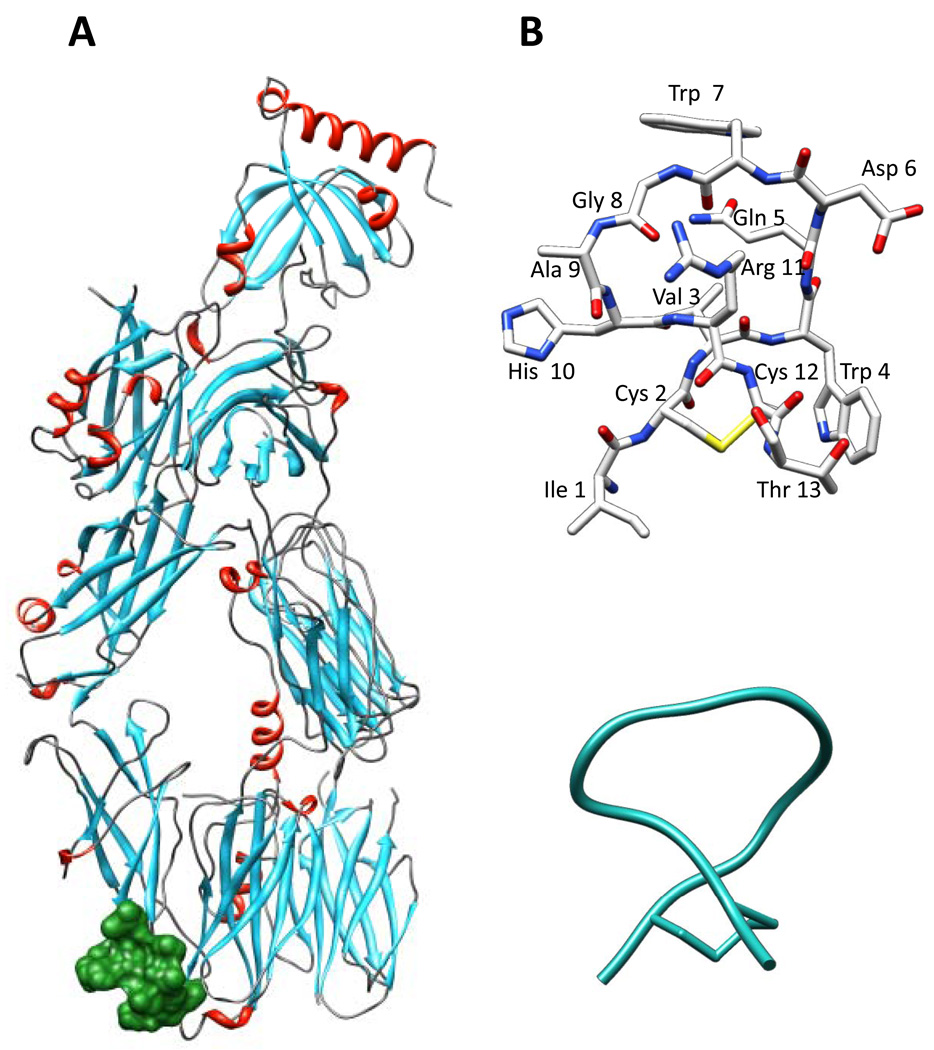
Figure 1.
Location of binding site. (A) Molecular model of the C3c:W4/A9 complex (PDB Code 2QKI21). C3c is colored by secondary structure: helices are in red, sheets are in cyan, and loops are in gray. W4/A9 (Peptide III) is shown in surface representation colored green. (B) Stick representation of Peptide III, depicting the peptide backbone and side chains. The color code is by atom type: grey for C, blue for N, red for O, and yellow for S. (C) ribbon representation of Peptide III, depicting the peptide backbone.
In this paper, we present experimental binding studies to C3, using SPR, and complement inhibitory activity studies, using ELISAs, for the three new rationally designed peptides (Peptides VII–IX). We also present SPR and ELISA data for three of the peptides designed computationally, whose sequences were previously reported (Peptides IV–VI).22 In the previous study we had reported KD values from SPR data for Peptides IV–VI, but not the detailed kinetics, which are presented here. The ELISA data for Peptides IV–VI are reported here for first time. We have also used two positive controls, the parent compstatin (Peptide II) and the most active peptide with natural amino acids, W4/A9 (Peptide III), and a negative control, linear compstatin with sequence IAVVQDWGHHRAT (Peptide I).
Herein, we use the terms Peptides I–X referring to their amino acid sequences (positions 1–13) irrespective of the presence, or not, of blocking groups at the termini or pegylation and biotinylation extensions at the C-terminus for the SPR experiments. The specifics of each sequence per experimental study is given in Tables (vide infra).
A main aspect in the design of efficacious compstatin analogs is the balance between hydrophobicity and polarity. Enhanced hydrophobicity at key sequence positions can provide enhanced binding, whereas increasing the polarity at key sequence positions can enhance solubility of the free peptides. Solubility is important for storage and delivery of biopharmaceuticals administered in solution. The newly designed active peptides presented here pave the way for new combinations of natural and non-natural amino acids, which will have improved balance between hydrophobicity and polarity.
Materials and Methods
Surface Plasmon Resonance (SPR)
Compstatin peptides were synthesized by Abgent Inc. (San Diego, CA). The peptide sequences and mass spectral analysis are shown in Table 1. Three peptides served as known controls. Two were positive controls, parent compstatin (Peptide II) and W4/A9 (Peptide III), and one was negative control, linear compstatin (Peptide I). At the carboxy-terminus, the peptides were pegylated with an 8-mer PEG (polyethylene glycol) block spacer, followed by a lysine, and biotinylated, followed by an NH2 block. Four peptides were acetylated at the amino-terminus (Peptides VII–X).
Table 1.
Amino acid sequences and mass spectral analysis for compstatin peptides used in SPR experiments.
| Peptide # |
Peptide Name | Amino Acid Sequencea | Mass Spectral Analysisb |
|---|---|---|---|
| I | Linear | I AVVQDWGHHRA T-(PEG)8-K-(biotin)-NH2 | 2267.06 |
| II | Parent | I[CVVQDWGHHRC]T-(PEG)8-K-(biotin)-NH2 | 2327.6 |
| III | W4/A9 | I[CVWQDWGAHRC]T-(PEG)8-K-(biotin)-NH2 | 2349.2 |
| IV | SQ027 | W[CVWQDWGTNRC]W-(PEG)8-K-(biotin)-NH2 | 2514.9 |
| V | SQ059 | D[CVWQDWGTNKC]W-(PEG)8-K-(biotin)-NH2 | 2414.93 |
| VI | SQ086 | Q[CVWQDWGQNQC]W-(PEG)8-K-(biotin)-NH2 | 2454.4 |
| VII | W4/A9/W13 | Ac-I[CVWQDWGAHRC]W-(PEG)8-K-(biotin)-NH2 | 2476.0 |
| VIII | W1/W4/A9 | Ac-W[CVWQDWGAHRC]T-(PEG)8-K-(biotin)-NH2 | 2464.0 |
| IX | W1/W4/A9/W13 | Ac-W[CVWQDWGAHRC]W-(PEG)8-K-(biotin)-NH2 | 2548.7 |
| X | W(i, i+3)/A9 | Ac-W[CVWQDWGAWRC]W-(PEG)8-K-(biotin)-NH2 | 2598.5 |
Brackets denote cyclization. Sequence differences of various compstatin peptides from parent compstatin are shown in bold characters.
Molecular masses were provided by Abgent Inc.
Binding of compstatin peptides to C3 was determined by SPR with Biacore X100 (GE healthcare, Piscataway, NJ) according to previous studies23–26 with modifications. The PEG spacer in the peptides increases mobility, solubility, and accessibility, and also decreases non-specific interactions. Streptavidin sensor chip SA was used to immobilize biotinylated compstatin peptides as the ligands. PBS containing 0.05% tween-20 was used as the running buffer and the assay was carried out at 25°C.
Human C3 was purchased from Complement Technology Inc. (Tyler, TX). Approximately 300 µL of C3 were placed in a Microcon MWCO 100 kD (Millipore, Billerica, MA) and centrifuged at 10000 rcf for 15 minutes at 4°C. After discarding the flow-through, 100 µL of running buffer was added to the C3 and the solution was centrifuged again. This process was repeated 3 times. C3 was recovered after the last run by adding 100 µL of running buffer at a time, for 5 times. Concentration was determined using the Beer-Lambert Law with an extinction coefficient of ε1% at 280 nm of 10.3 (g/100mL)−1 cm−1 (obtained from the Complement Technology Inc. report).
The compstatin peptides were immobilized onto the chip through their carboxy-terminus, as previously suggested.23 That study had shown that binding of compstatin to human C3 was not possible if the peptide was immobilized through the amino-terminus, thus revealing the importance of a free amino-terminus for the interaction.23 The cyclic compstatin family peptides were immobilized in the flow cell 2 (Fc2) and the linear negative control was immobilized in flow cell 1 (Fc1), both to a reading of 1000 response units (RU) with a flow rate of 10 µL/min. The binding was measured at 30 µL/min by adding several concentrations of human C3 (analyte) for 120 s and dissociation was monitored for 180 s, followed by regeneration with 10mM NaOH for 60 s. Repeats of each experiment were performed for 180 s of association and 240 s of dissociation, followed by regenerations with 10mM NaOH for 30 s and 0.05% SDS for 30 s. The solutions were placed in the Biacore sample rack in the following positions: (1) 0nM C3, (2) 25nM C3, (3) 50nM C3, (4) 100nM C3 (run twice), (5) 200nM C3, (6) 400nM C3, (7) 800nM C3, (8) 1600nM C3, (9) running buffer, (10) 10mM NaOH, (11) 0.05% SDS.
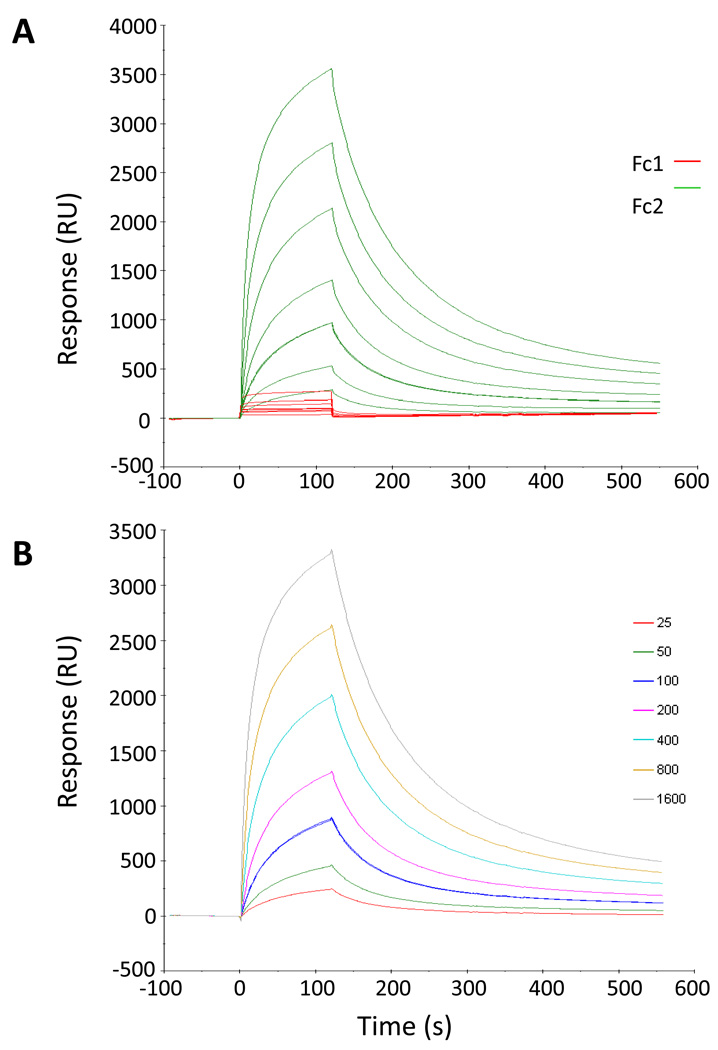
Figure 2 demonstrates the data quality, using Peptide VII as an example. Figure 2A shows data obtained for Fc1 and Fc2 before the sensorgram was constructed. The sensorgrams were adjusted in the x-axis to start at the injection of the first sample, and in the y-axis to start at the baseline. Figure 2B shows the sensorgram produced by the difference between Fc2 and Fc1. Kinetic parameters were obtained using the Biacore X100 Evaluation Software. Global fittings of the Biacore sensorgrams were performed using the 1:1 binding model (A + B → AB) and the two-state reaction model (A + B → AB → AB*), according to which 1:1 binding is followed by a conformational change that stabilizes the complex.27 Although data were collected at 0nM–1600nM C3 concentration, best fits were produced using the data in the 25nM–800nM or the 0nM–800nM range and the two-state model. The quality of the fits was assessed by visual inspection of the fitted data and their residuals and using the Rmax and χ2 values.
Figure 2.
Example of SPR sensorgram adjustment. (A) Inactive linear compstatin (Peptide I) used as a control in flow cell one (Fc1) is shown in red. Peptide VII in flow cell two (Fc2) is shown in green. (B) Adjusted sensorgram of Peptide VII by taking the difference Fc2-Fc1. The color code corresponds to C3 concentrations shown in the legend at the right.
Normal Human Serum (NHS) Kinetics
Normal human serum (NHS) was obtained from Complement Technology Inc. and was centrifuged for 2 min at 12000 rpm prior to its use. In the course of ELISA (enzyme-linked immunosorbent assays) inhibition experiments, we observed that assay parameters were dependent on reagents used. Specifically, the assays were sensitive to the lot of serum used. Prior to performing ELISAs, the optimal NHS incubation time was determined for each lot used, by performing assay kinetics. A 96-well plate was coated with a 40 µg/mL solution of lipopolysaccharides (LPS) from E. coli in PBS and left overnight at 4°C. The plate was washed with PBS-Tween (PBS-T), 5% milk was added, and the plate was incubated for an hour at room temperature. 92 µL of diluent solution (GVB, 5mM MgEGTA) and of 8 µL NHS was added to each well. In the 96-well plate, serum incubation times were varied between 15 and 120 min. After incubation, the wells were washed to remove unbound C3b. A 1:1000 dilution of Horseradish peroxidase-conjugated antihuman C3 antibody (C3-HRP) in PBS-T was added to the wells and incubated for an hour at room temperature. The wells were washed to remove unbound antibody. Addition of 1 mM ABTS-H2O2 causes a color change (green) that was measured spectrophotometrically at 415 nm after 10 min of incubation. A graph of absorbance versus incubation time was constructed to determine the proper incubation time, based on the highest time point in the linear segment of the curve.
Human C3b ELISAs
Several concentrations of compstatin peptides were tested using human C3b ELISAs to determine the concentration at which half of the C3 present is inhibited by the peptide. Compstatin peptides were synthesized by Abgent Inc. The peptides were acetylated at the N-terminus and amidated at the C-terminus, with the exception of Peptides IV and VI, which were only amidated at the C-terminus.
Compstatin peptides were dissolved in physiological strength PBS, with exception of Peptides VII and IX, which were dissolved in DMSO due to poor solubility. Peptide IV was initially dissolved in PBS, but subsequently lyophilized and dissolved in DMSO to improve solubility. The DMSO concentration used was 6%. Initial concentrations of compstatin peptides were calculated using the Beer-Lambert Law with an extinction coefficient of 5500 M−1 cm−1 for each Trp present in the sequence, at 280 nm.
A 96-well plate was coated with a 40 µg/mL solution of LPS from E. coli in PBS and left overnight at 4°C. The plate was washed with PBS-T, 5% milk was added, and the plate was incubated for an hour at room temperature. The plate was washed and placed on ice while the dilutions of the compstatin peptides were prepared.
Serial dilutions of the compstatin peptides were prepared in an empty 96-well plate, using a diluent solution of GVB and 5mM MgEGTA. The first column (8 wells) consisted of the highest concentration of the peptide, and ten serial (two-fold) dilutions were performed. A volume of 92 µL of each serial dilution was transferred to corresponding wells in the ELISA plate.
NHS was heat inactivated (Hi-NHS) in a 60°C water bath for 30 min, and used as the negative control for complement activation in each experiment. A volume of 8 µL NHS was added to each well, and the plate was incubated at room temperature for 45–120 min, depending on the serum sample. If the compstatin peptide was present at an inhibitory concentration, cleavage of C3 was prevented. After incubation, the wells were washed to remove unbound C3b. A 1:1000 dilution of C3-HRP in PBS-T was added to the wells and incubated for an hour at room temperature. C3-HRP binds to the C3b-LPS complex. The wells were washed to remove unbound antibody. Addition of 1 mM ABTS-H2O2 causes a color change (green) that is measured spectrophotometrically at 415 nm after 10 min of incubation. Percent C3b was plotted against peptide concentration and was fit using a logistic dose response curve with the software Prism (GraphPad, San Diego, CA) to determine IC50.
Computational Studies
Approximate binding affinities for Peptides II–IX were calculated using the second stage of the computational framework, described before.22 In brief, these calculations generate rotamerically-based ensembles of structures for the free peptide, the free target protein, and the peptide-protein complex. The energies, Ei, of each of the conformers in each of the ensembles are used to calculate the partition functions, q, in Equation 1, where the subscripts PL, P, and L refer to the protein-peptide complex, the free target protein, and the free peptide, respectively
| (1) |
Equation 2 is then used to calculate an approximate binding affinity,28 K*, for the peptide-protein complex
| (2) |
The more accurate the partition functions are, the more precise the binding affinity will be. The approximate binding affinities are used to rank the sequences of Peptides II–IX. Sequences that are ranked higher than the parent sequence are predicted to be better binders than the parent.
Results
Rational Design
The major finding of the previous computational study was the persistent observation of Trp at sequence position 13, whereas a variety of combinations of polar amino acids were observed at positions 9–11.22 Trp was occasionally observed at position 1; however, position 1 was dominated by polar amino acids.22 To assess the effect of a single Trp amino acid at position 13, we reasoned that we could use the sequence of the most active peptide to-date, comprised entirely of natural amino acids (Peptide III), with the substitution of Thr13 with Trp13 (Peptide VII). Since the two end amino acids are hanging outside the cyclization ring (positions 1 and 13) and thus having backbone which is conformationally less restrained than the rest of the peptide, we reasoned to insert Trp at position 1 only and at positions 1 and 13 simultaneously with the remaining sequence being that of Peptide III (Peptides VIII and IX). These substitutions enhance the hydrophobic character of the surface of compstatin formed by amino acids 1–4/12–13 at the linked termini, and thus contribute to the hydrophobic interactions which dominate binding to C3c.21 They also introduce additional interaction capabilities with C3/C3c in the form of possible hydrogen bonding through the Trp indole amide or pi-cation or pi-stacking interaction through the Trp benzene ring. At the same time the polar surface of the peptides at positions 5–6/8–11 is preserved. All three peptides (Peptides VII–IX) were found experimentally to be strong binders to C3 and inhibitors of complement activation (vide infra). Upon examination of the sequence of Peptide IX, we observed that four Trp amino acids were present at positions 1, 4, 7, and 13. By inserting another Trp at position 10, we could symmetrically arrange (in sequence) Trp amino acids at positions (i, i+3) (Peptide X). We reasoned that this arrangement could probably enhance the possibility of Trp ring stacking which could stabilize the peptide structure and perhaps binding to C3. However, this was not the case as Peptide X was found not to bind to C3 (vide infra). This finding suggests the need for a polar patch at positions 9–11, which was interrupted by the insertion of the benzene ring-containing Trp10.
Surface Plasmon Resonance (SPR) Studies
We have performed SPR studies to experimentally assess the binding abilities of Peptides II–X. Among them there are two positive controls, parent compstatin (Peptide II) and W4/A9, the most active compstatin peptide consisting of natural amino acids (Peptide III). We also present detailed SPR data for three top-ranking peptides from our previous computational study.22 Figure 3 shows the SPR sensorgrams for the two positive controls and the two of the top-rankers in the computational predictions. Figure 4 shows the SPR sensorgrams for the third top-ranker in the computational predictions and the three rationally-designed peptides, whose design was inspired by the computational data. Table 2 summarizes the dissociation constants, KD, for the binding Peptides II–IX. Maximum response units (Rmax) and χ2 values are also listed in Table 2 to assess the quality of the fits, in addition to visual inspection of Figs. 3 and 4. As shown in Table 2, the KD values are sensitive to the fitting model used. Although better fits were generated using the 2-state model (Figs. 3 and 4), we also include in Table 2 the parameters from fits using the 1:1 model for comparison (sensorgrams not shown). In general, better fits were generated using C3 concentrations up to and including 800nM. It is likely that at 1600nM C3 concentration aggregates may be forming, which change the protein conformation. This may cause rapid surface saturation, which can affect the binding kinetics. The fits were not very sensitive upon inclusion of a 0nM concentration or not. The 2-state model suggests conformational change of compstatin and/or C3 upon binding, which has been previously proposed using isothermal calorimetry data29 and by a comparison of the crystal structure of the C3c:W4/A9 complex21 and the solution structure of free parent compstatin.14
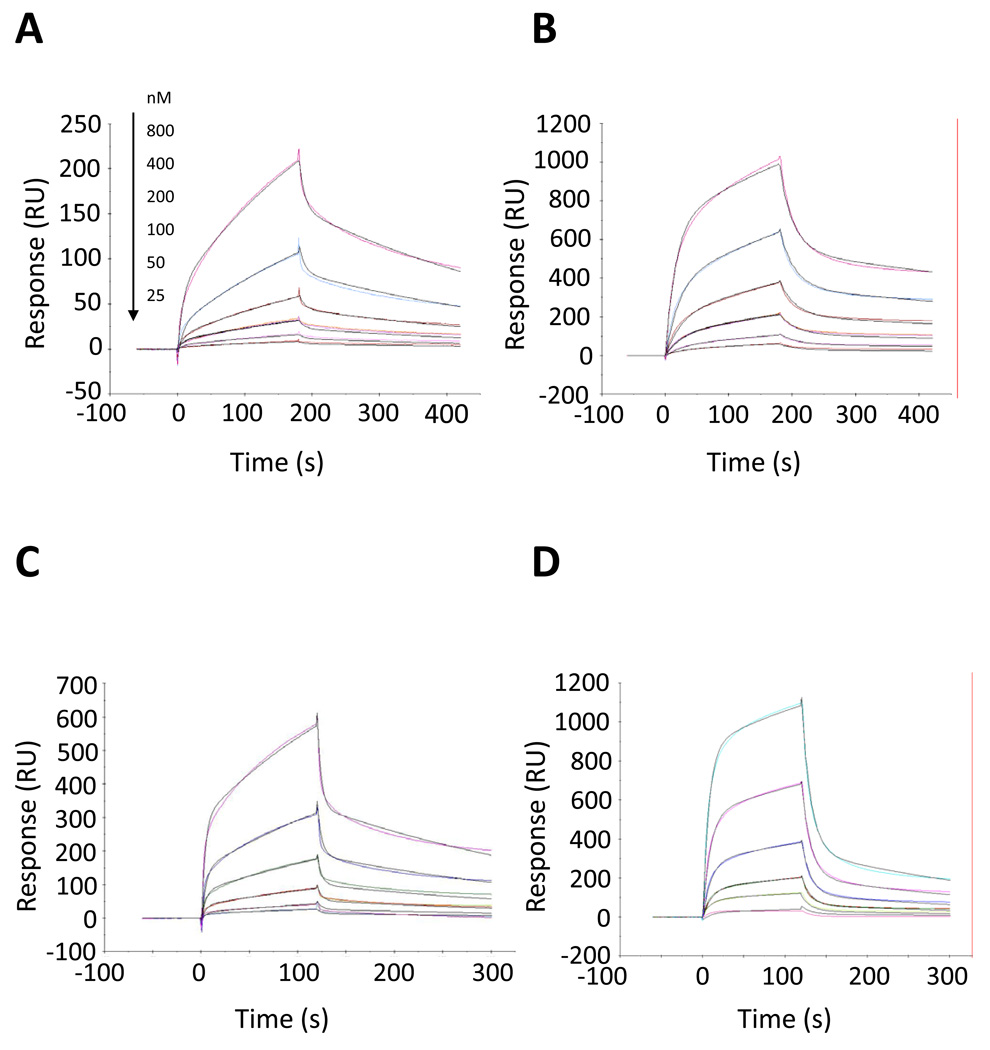
Figure 3.
Kinetic analysis of Peptides II–V to human C3. The C3 concentrations are 800, 400, 200, 100, 50, and 25nM (from top to bottom). Solid color lines correspond to the experimental data and solid black lines correspond to the fits. All sensorgrams were fitted to a two-state reaction model. The panels show: (A) Peptide II, (B) Peptide III, (C) Peptide IV, and (D) Peptide V.
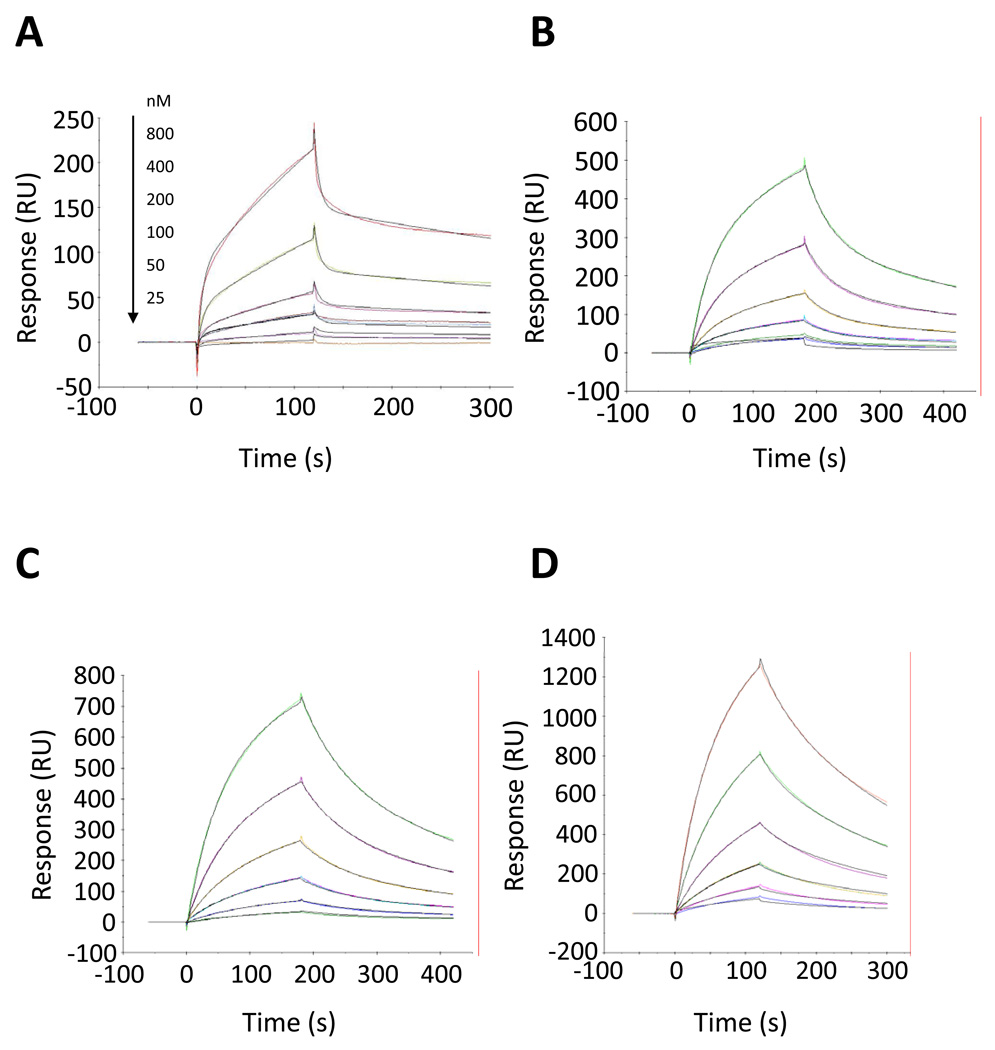
Figure 4.
Kinetic analysis of Peptides VI–IX to human C3. The C3 concentrations are 800, 400, 200, 100, 50, and 25nM (from top to bottom). Solid color lines correspond to the experimental data and solid black lines correspond to the fits. Solid color lines correspond to the experimental data and solid black lines correspond to the fits. All sensorgrams were fitted to a two-state reaction model. The panels show: (A) Peptide VI, (B) Peptide VII, (C) Peptide VIII, and (D) Peptide IX.
Table 2.
Kinetic and affinity parameters calculated for the interaction of active compstatin peptides with human C3.a
| Peptide # |
Peptide Name | KD (10−6 M) |
Rmax | Kon1 (105/M·s) |
Kon2 (10−2/s) |
Koff1 (10−2/s) |
Koff2 (10−2/s) |
χ2 | Model |
|---|---|---|---|---|---|---|---|---|---|
| II | Parent | 1.26 | 937.7 | 0.172 | 1.40 | 13.04 | 0.282 | 4.69 | 2-State Reaction |
| 0.81 | 507.6 | 0.036 | 0.294 | 12 | 1:1 Binding | ||||
| III | W4/A9 | 0.19 | 1781 | 0.558 | 0.52 | 6.402 | 0.106 | 78.1 | 2-State Reaction |
| 0.23 | 1000 | 0.129 | 0.296 | 813 | 1:1 Binding | ||||
| IV | SQ027 | 0.76 | 2781.5 | 0.561 | 0.36 | 8.73 | 0.337 | 52.27 | 2-State Reaction |
| 0.51 | 1086.2 | 0.412 | 2.111 | 1522.7 | 1:1 Binding | ||||
| V | SQ059 | 0.74 | 1150 | 1.051 | 1.29 | 74.30 | 1.289 | 9.21 | 2-State Reaction |
| 0.45 | 484.9 | 0.050 | 0.225 | 28.3 | 1:1 Binding | ||||
| VI | SQ086 | 0.99 | 2154 | 2.091 | 1.03 | 84.76 | 0.326 | 36 | 2-State Reaction |
| 0.50 | 799 | 0.085 | 0.418 | 181 | 1:1 Binding | ||||
| VII | W4/A9/W13 | 0.74 | 1397 | 0.100 | 0.55 | 2.184 | 0.282 | 23.8 | 2-State Reaction |
| 0.70 | 939.8 | 0.069 | 0.483 | 108 | 1:1 Binding | ||||
| VIII | W1/W4/A9 | 0.50 | 1516 | 0.108 | 0.48 | 1.146 | 0.425 | 6.83 | 2-State Reaction |
| 0.52 | 1297 | 0.087 | 0.456 | 61.4 | 1:1 Binding | ||||
| IX | W1/W4/A9/W13 | 0.38 | 2442 | 0.154 | 1.38 | 1.534 | 0.866 | 41.7 | 2-State Reaction |
| 0.38 | 2187 | 0.126 | 0.477 | 111.3 | 1:1 Binding |
The C3 concentration range used for the fits is 0–800nM.
The SPR data using the 2-state model suggest the following binding order (from stronger to weaker): Peptide III > Peptide IX > Peptide VIII > Peptide VII ~ Peptide V ~ Peptide IV > Peptide VI > Peptide II (Table 2). Small differences in binding order are seen in the SPR data using the 1:1 model: Peptide III > Peptide IX > Peptide V > Peptide VI ~ Peptide IV ~ Peptide VIII > Peptide VII > Peptide II (Table 2). Although the fits using the 2-state model were better (see Materials and Methods), the results from 1:1 model produced comparable KD values but with different kinetics. We report data extracted from both models to demonstrate the sensitivity of the obtained KD values and their effect on binding order. The KD values for the two positive controls, Peptides II and III, are in agreement with recently reported data.30 Overall, the SPR data are indicative of strong binding for the newly designed peptides (Peptides IV–IX).
The complexity of the binding kinetics is reflected in the variation of the curve shape and maximum binding response values in the plots of Figs. 3 and 4. Based on the previous NMR data of free peptides and preliminary molecular dynamics data of bound peptides, we expect that the kinetics is affected by peptide structural variations and differences in local binding contacts between the peptides and C3. We anticipate that compensatory effects, because of gain/loss of binding contacts, also contribute to the kinetic parameters.
Human C3b ELISA Studies
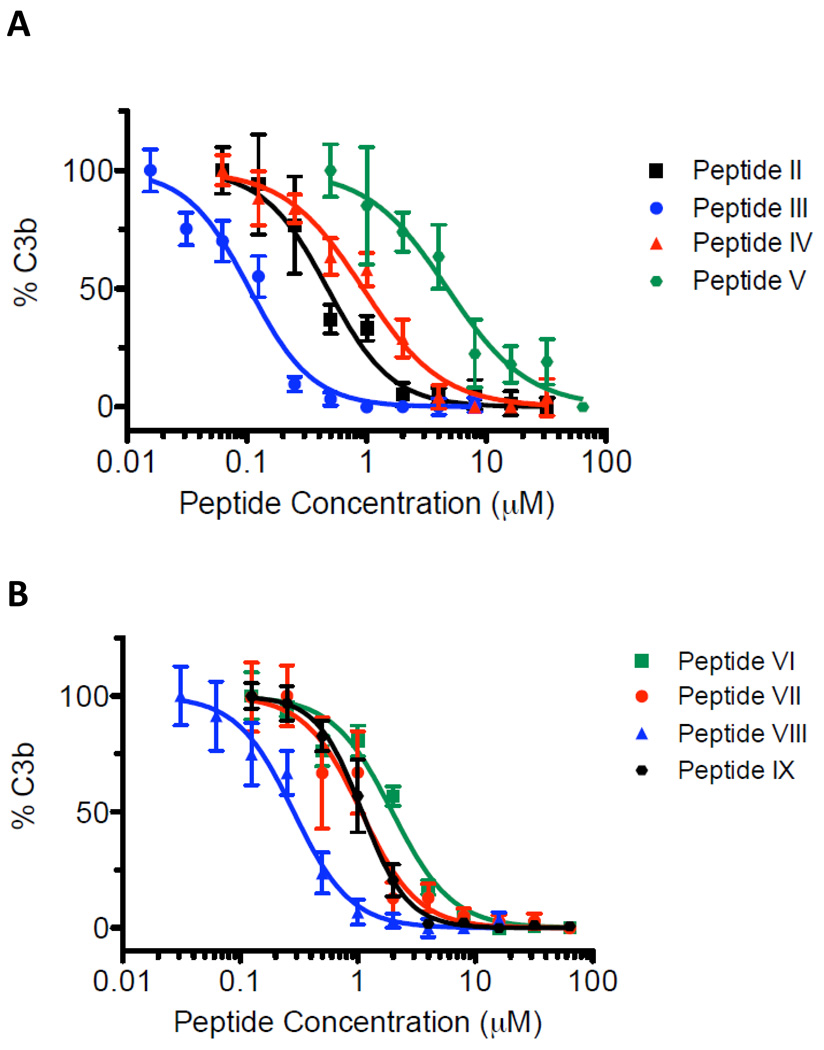
We have also performed C3b ELISA studies to determine IC50 values for inhibition of C3b formation at different peptide concentrations. Table 3 summarizes the determined IC50 values of Peptides II–IX. The inhibition order (from stronger to weaker) is Peptide III > Peptide VIII > Peptide II > Peptide IV > Peptide VII ~ Peptide IX > Peptide VI > Peptide V. We do not necessarily expect correlations between the SPR binding and ELISA inhibition results, because the SPR assays are binary with one component immobilized and the ELISA assays are multi-component detecting inhibition in NHS. However, there are notable differences in some of the peptides used for SPR and ELISA experiments. There are differences in the N-terminal acetylation patterns. In SPR studies, Peptides VII, VIII, and IX were acetylated, and the remaining peptides were not. In ELISA studies, all peptides were acetylated, with the exception of Peptides IV and VI which were not. Also, the studied peptides have different C-terminal attachments, given the pegylation and biotinylation additions in the SPR peptides (compare Tables 1 and 4). For the record, in the SPR experiments, acetylation was not used in the computationally designed peptides and controls for comparison with the computational predictions. The computational studies did not use acetylation because they are based on native amino acid libraries and parametrization. Subsequent to SPR studies, in the ELISA experiments we introduced acetylation, when was convenient for the peptide synthesis, cyclization, and purification processes. Finally, in the ELISA studies, peptides Peptides VII and IX were dissolved in DMSO because of poor solubility in aqueous solutions. Differences in solubilities of peptides that were used in ELISA studies compared to those used in SPR studies may reflect loss of the hydrophilic N-terminal backbone amine upon acetylation. Solubility was not an issue in the SPR studies, presumably due to the hydrophilic PEG groups in the long PEG-biotin chains at the C-termini. These data indicate that Trp at position 13 affects peptide solubility, perhaps due to increased hydrophobicity in the region. The relative IC50 values of the two positive controls, Peptides II and III, are in agreement with recently reported data, despite differences in their absolute values, owed possibly to differences in the assays and reagents used.30 Overall, the ELISA data are indicative of strong binding and activity for the newly designed peptides (Peptides IV–IX).
Table 3.
Compstatin peptide IC50 values from human C3b ELISA experiments.
| Peptide # |
Peptide Name | Amino Acid Sequencea | Mass Spectral Analysisb |
Concentration Range (µM) |
IC50 (µM) |
|---|---|---|---|---|---|
| II | Parent | Ac-I[CVVQDWGHHRC]T-NH2 | 1591.47 | 64-0.125 | 0.47 |
| III | W4/A9 | Ac-I[CVWQDWGAHRC]T-NH2 | 1613.20 | 8-0.016 | 0.11 |
| IV | SQ027 | W[CVWQDWGTNRC]W-NH2 | 1735.73 | 32-0.063 | 0.94 |
| V | SQ059 | Ac-D[CVWQDWGTNKC]W-NH2 | 1678.27 | 64-0.500 | 4.73 |
| VI | SQ086 | Q[CVWQDWGQNQC]W-NH2 | 1675.87 | 64-0.125 | 1.98 |
| VII | W4/A9/W13 | Ac-I[CVWQDWGAHRC]W-NH2 | 1697.87 | 64-0.125 | 1.08 |
| VIII | W1/W4/A9 | Ac-W[CVWQDWGAHRC]T-NH2 | 1685.60 | 16-0.031 | 0.29 |
| IX | W1/W4/A9/W13 | Ac-W[CVWQDWGAHRC]W-NH2 | 1771.20 | 64-0.125 | 1.09 |
Brackets denote cyclization. Sequence differences of various compstatin peptides from parent compstatin are shown in bold characters.
Molecular masses were provided by Abgent Inc.
Table 4.
Computational approximate binding affinity ranking for Peptides II–IX.a
| Peptide # |
Peptide Name | Approximate Binding Affinity Rankb |
K* |
|---|---|---|---|
| VIII | W1/W4/A9 | 1 | 3.89 × 10−8 |
| VII | W4/A9/W13 | 2 | 1.28 × 10−11 |
| II | Parent | 3 | 4.27 × 10−12 |
| IX | W1/W4/A9/W13 | 4 | 2.39 × 10−13 |
| I | Linear | 5 | 3.08 × 10−81 |
| III | W4/A9 | 1’ | 2.42 × 10−1 |
| VI | SQ086 | 2’ | 1.10 × 10−5 |
| V | SQ059 | 3’ | 6.56 × 10−6 |
| IV | SQ027 | 4’ | 1.50 × 10−7 |
The approximate binding affinity, K*, is a computational ranking parameter whose absolute value depends on statistical sampling and does not directly correspond to experimental binding affinities. For the new designs, Peptides VII, VIII, and IX, one of the positive controls, Peptide II, and the negative control Peptide I, 1000 peptide structures were predicted for each sequence, 1000 docked complexes were generated for each docking run, and the final peptide and complex ensembles numbered 22,000 each. A higher sampling rate was used for the previously computationally predicted Peptides IV, V, and VI and the other positive control, peptide III. 5000 peptide structures were predicted for each sequence, 5000 docked complexes were generated per docking run, and the final peptide and complex ensembles still numbered 22,000 each.
The two ranking orders (1’–5’ and 1–4) correspond to the different sampling rates used.
Computational Studies
The previously presented computational framework was capable of predicting new binding sequences.22 To evaluate the predicted binding ranking order compared to experimentally-derived binding and inhibition ranking orders, we present approximate binding affinities for Peptides II–IX (Table 4). The three computationally designed peptides (Peptides IV–VI) are predicted to bind better than the parent (Peptide II), but worse than the most potent inhibitor with natural amino acids (W4/A9, Peptide III). Peptides IV–VI were found experimentally to be strong binders to C3 and inhibitors of complement activation. It should be mentioned that the approximate binding affinity is used as a ranking measure only for the various compstatin peptides, and we are not attempting to directly compare computational approximate binding affinity values to experimental binding affinity values. The value of the approximate binding affinity depends on the statistical sampling used to evaluate Eqs. (1) and (2).
Significance of Results
Irrespective of ranking orders using binding or inhibition assays, our previous computational design22 and current rational design have produced six potent binders and inhibitors with novel features, such as the presence of Trp at positions 1 or 13, or both. From consensus rankings, Trp at position 1 (Peptide VIII) results in the most potent peptide derived from the studies presented here. Peptide VIII is free of solubility problems and has binding affinities and inhibitory activities close to that of Peptide III, whereas the soluble form of Peptide IX (for SPR studies, Table 1) has the closest binding affinity to that of Peptide III.
Discussion
Since the discovery of compstatin,7 optimization efforts using rational and combinatorial (both experimental and computational) and combination methods have persisted for many years.3,8–13 A major breakthrough in the process was the use of computational combinatorial methods16,31 which paved the way for large steps in inhibitory activity increases and established that Trp at position 4 was essential to produce the most active peptide comprised of natural amino acids (Peptide III).17 Subsequent optimization incorporated non-natural amino acids which further improved inhibitory activity.17,19 However, with the design of Peptide III, efforts to further optimize compstatin using natural amino acid replacements were stopped. The establishment of Peptide III as the most active peptide with natural amino acids reduced the original sequence template from X[CVXQDWGXXXC]X 3, 8– 9, 15, 16 to X[CVWQDWGXXXC]X, which we previously used for the design of Peptides IV–VI,22 and to X[CVWQDWGAHRC]X, which we used for the new design of Peptides VII–IX. This means that the six original optimizable positions, denoted with X, were subsequently reduced to five and then to two.
The new rationally designed peptides were made possible because of our earlier computational design, which consisted of two stages.22 The first stage used integer linear optimization to select novel sequences based upon a flexible backbone template and defined mutation set. The second stage ranked these sequences by calculating an approximate binding affinity. For the second stage, the structures of the sequences were predicted and clustered. Representative structures from the top clusters were docked against the target protein and the lowest energy structures were used to generate rotamerically-based conformation ensembles of the complex, free peptide, and free protein. These ensembles were used to calculate partition functions and then the final approximate binding affinity. The new sequence revealed by this computational study formed the basis for subsequent rational design, based on structural and physicochemical reasoning, of three more peptides.
In combination, our computational and rational designs used reduced optimizable sequence space of compstatin to design six strong binders and inhibitors. All of the new peptides incorporate a third Trp in the sequence of the previously most active compstatin variant, Peptide III, at position 13 (Peptides IV–VII, and IX) or at position 1 (Peptide IV, VIII, and IX). Two of these peptides incorporate Trp at positions 1 and 13 simultaneously, to a total of four Trp amino acids (Peptides IV and IX). Involvement of up to four Trp amino acids in the binding of compstatin to C3, paves the way for mechanistic binding studies which will exploit the importance of physicochemical properties of Trp, such as hydrophobicity (benzene ring), hydrogen bond donor capability (indole amide), and capability for pi-stacking or pi-cation interactions (pi electron system of the aromatic ring). Additionally, the data suggests that the retention of the polar character at residues 5–6/9–11 is necessary for both binding of compstatin to C3 and solubility of the peptide. Upon further optimization, the peptides presented here may become potential therapeutic candidates.
The identification of new strong binders renews interest for further optimization of compstatin by incorporating non-natural Trp-like amino acids, including methylated or halogenated derivatives, and cyclic-ring or fused-ring amino acids. Overall, Trp is shown to be a remarkable amino acid for C3 binding and complement system inhibition, when embedded in the sequence of compstatin. In this sense, Trp lives up to its reputation as the most druggable amino acid! Another avenue for further optimization is the use of molecular dynamics simulations. Such studies provide mechanistic arguments for binding, which can be used for peptide sequence alterations (including non-natural amino acids) to improve binding to C3. An example of such molecular dynamics study has compared the binding mechanism of W4/A9 (Peptide III) to human and rat C3c and has delineated the mechanism of species specificity of compstatin.32
Conclusion and Future Directions
Our study used two novel features to design three new and potent compstatin sequences. The novel features entail incorporation of Trp at positions 1 or/and 13. These features can be exploited further to improve potency and to fine-balance hydrophobicity and polarity. Improved hydrophobicity is much needed for increased binding to C3 and improved polarity, for storage and delivery of the compstatin peptides in the event they become therapeutic candidates. Ongoing molecular dynamics simulations will give insight into the mechanism of binding of these new sequences and C3, and facilitate the development of more potent compstatin peptides.
Figure 5.
Human C3b ELISA data used to extract the IC50 values of Table 3. Peptide concentration is plotted in the horizontal axis and percent C3b is plotted in the vertical axis. The plots depict the inhibition of cleavage of C3 to C3a and C3b by compstatin peptides, quantified as percent of generated C3b. Highest inhibition corresponds to 0% C3b generation.
Acknowledgments
This work was supported by grants from NSF (CAF and DM), NIH (R01 GM52032, CAF; R24 GM069736; CAF and D M; R01 AI063426, DDL), UCR Bioengineering Center seed fund (DM), and the US Environmental Protection Agency, EPA (R 832721-010, CAF). Although the research described in the article has been funded in part by the US EPA’s STAR program, it has not been subjected to any EPA review and therefore does not necessarily reflect the views of the Agency, and no official endorsement should be inferred. Portion of this research was made possible with Government support by DoD, Air Force Office of Scientific Research, National Defense Science and Engineering Graduate (NDSEG) Fellowship 32 CFR 168a (MLB).
Abbreviations
- C3
complement system protein C3
- C3b
the b-fragment of C3
- C3c
the c-fragment of C3
- FB
Factor B
- SPR
surface plasmon resonance
- PEG
polyethylene glycol
- PBS
phosphate buffer saline
- SDS
sodium dodecyl sulfate
- NHS
normal human serum
- ELISA
enzyme-linked immunosorbent assay
- LPS
lipopolysaccharides
- PBS-T
PBS with tween
- GVB
gelatin veronal buffer
- C3-HRP
horseradish peroxidase-conjugated antihuman C3 antibody
- ABTS-H2O2
2,2'-azino-bis(3-ethylbenzthiazoline-6-sulphonic acid) with peroxide
- DMSO
dimethyl sulfoxide HI-NHS, heat inactivated normal human serum
- PDB
Protein Data Bank
References
- 1.Walport MJ. Advances in immunology: complement (First of two parts) N Engl J Med. 2001;344:1058–1066. doi: 10.1056/NEJM200104053441406. [DOI] [PubMed] [Google Scholar]
- 2.Walport MJ. Advances in immunology: complement (Second of two parts) N Engl J Med. 2001;344:1140–1144. doi: 10.1056/NEJM200104123441506. [DOI] [PubMed] [Google Scholar]
- 3.Morikis D, Lambris JD. Structure, dynamics, activity, and function of compstatin and design of more potent analogs. In: Morikis D, Lambris JD, editors. Structural Biology of the Complement System. Boca Raton, FL: CRC Press/Taylor & Francis Group; 2005. pp. 317–340. [Google Scholar]
- 4.Mollnes TE, Kirschfink M. Strategies of therapeutic complement inhibition. Mol Immunol. 2006;43:107–121. doi: 10.1016/j.molimm.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 5.Morgan BP, Harris CL. Complement therapeutics; history and current progress. Mol Immunol. 2003;40:159–170. doi: 10.1016/s0161-5890(03)00111-1. [DOI] [PubMed] [Google Scholar]
- 6.Sahu A, Lambris JD. Complement inhibitors: a resurgent concept in anti-inflammatory therapeutics. Immunopharmacology. 2000;49:133–148. doi: 10.1016/s0162-3109(00)80299-4. [DOI] [PubMed] [Google Scholar]
- 7.Sahu A, Kay BK, Lambris JD. Inhibition of human complement by a C3-binding peptide isolated from a phage-displayed random peptide library. J Immunol. 1996;157:884–891. [PubMed] [Google Scholar]
- 8.Morikis D, Lambris JD. Structural aspects and design of low-molecular-mass complement inhibitors. Biochem Soc Trans. 2002;30:1026–1036. doi: 10.1042/bst0301026. [DOI] [PubMed] [Google Scholar]
- 9.Morikis D, Soulika AM, Mallik B, Klepeis JL, Floudas CA, Lambris JD. Improvement of the anti-C3 activity of compstatin using rational and combinatorial approaches. Biochem Soc Trans. 2004;32:28–32. doi: 10.1042/bst0320028. [DOI] [PubMed] [Google Scholar]
- 10.Holland MCH, Morikis D, Lambris JD. Synthetic small molecule complement inhibitors. Curr Opin Investig Drugs. 2004;5:1164–1173. [PubMed] [Google Scholar]
- 11.Floudas CA, Klepeis JL, Lambris JD, Morikis D. De novo protein design: an interplay of global optimization, mixed-integer optimization and experiments; FOCAPD 2004, Sixth International Conference on Foundations of Computer-Aided Process Design, Discovery through Product and Process Design; Floudas C. A., Agrawal R; 2004. pp. 133–146. [Google Scholar]
- 12.Morikis D, Floudas CA, Lambris JD. Structure-based integrative computational and experimental approach for the optimization of drug design. In: Sunderam VS, van Albada GD, Sloot PMA, Dongarra JJ, editors. ICCS 2005, Lecture Notes in Computer Science: Computational Science. Berlin-Heidelberg: Springer-Verlag; pp. 680–688. [Google Scholar]
- 13.Ricklin D, Lambris JD. Compstatin: a complement inhibitor on its way to clinical application. Current Topics in Complement II. 2008;632:273–292. doi: 10.1007/978-0-387-78952-1_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morikis D, Assa-Munt N, Sahu A, Lambris JD. Solution structure of compstatin, a potent complement inhibitor. Prot Sci. 1998;7:619–627. doi: 10.1002/pro.5560070311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morikis D, Roy M, Sahu A, Troganis A, Jennings PA, Tsokos GC, Lambris JD. The structural basis of compstatin activity examined by structure-function-based design of peptide analogs and NMR. J Biol Chem. 2002;277:14942–14953. doi: 10.1074/jbc.M200021200. [DOI] [PubMed] [Google Scholar]
- 16.Klepeis JL, Floudas CA, Morikis D, Tsokos CG, Argyropoulos E, Spruce L, Lambris JD. Integrated computational and experimental approach for lead optimization and design of compstatin variants with improved activity. J Am Chem Soc. 2003;125:8422–8423. doi: 10.1021/ja034846p. [DOI] [PubMed] [Google Scholar]
- 17.Mallik B, Katragadda M, Spruce LA, Carafides C, Tsokos CG, Morikis D, Lambris JD. Design and NMR characterization of active analogs of compstatin containing non-natural amino acids. J Med Chem. 2005;48:274–286. doi: 10.1021/jm0495531. [DOI] [PubMed] [Google Scholar]
- 18.Mallik B, Lambris JD, Morikis D. Conformational inter-conversion of compstatin probed with molecular dynamics simulations. Proteins. 2003;53:130–141. doi: 10.1002/prot.10491. [DOI] [PubMed] [Google Scholar]
- 19.Mallik B, Morikis D. Development of a quasi-dynamic pharmacophore model for anti-complement peptide analogs. J Am Chem Soc. 2005;127:10967–10976. doi: 10.1021/ja051004c. [DOI] [PubMed] [Google Scholar]
- 20.Katragadda M, Magotti P, Sfyroera G, Lambris JD. Hydrophobic effect and hydrogen bonds account for the improved activity of a complement inhibitor compstatin. J Med Chem. 2006;49:4616–4622. doi: 10.1021/jm0603419. [DOI] [PubMed] [Google Scholar]
- 21.Janssen BJC, Halff EF, Lambris JD, Gros P. Structure of compstatin in complex with complement component C3c reveals a new mechanism of complement inhibition. J Biol Chem. 2007;282:29241–29247. doi: 10.1074/jbc.M704587200. [DOI] [PubMed] [Google Scholar]
- 22.Bellows ML, Fung HK, Taylor MS, Floudas CA, López De Victoria A, Morikis D. New compstatin variants through two de novo protein design frameworks. Biophys J. 2010;98:2337–2346. doi: 10.1016/j.bpj.2010.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sahu A, Soulika A, Morikis D, Spruce L, Moore WT, Lambris JD. Binding kinetics, structure-activity relationship, and biotransformation of the complement inhibitor Compstatin. J Immunol. 2000;165:2491–2499. doi: 10.4049/jimmunol.165.5.2491. [DOI] [PubMed] [Google Scholar]
- 24.Sahu A, Morikis D, Lambris JD. Compstatin, a peptide inhibitor of complement, exhibits species-specific binding to complement component C3. Mol Immunol. 2003;39:557–566. doi: 10.1016/s0161-5890(02)00212-2. [DOI] [PubMed] [Google Scholar]
- 25.Soulika AM, Morikis D, Sarrias M-R, Roy M, Spruce LA, Sahu A, Lambris JD. Studies of structure-activity relations of complement inhibitor compstatin. J Immunol. 2003;170:1881–1890. doi: 10.4049/jimmunol.171.4.1881. Erratum: 2004, 172-5128. [DOI] [PubMed] [Google Scholar]
- 26.Ling J, Liao H, Clark R, Wong MS, Lo DD. Structural constraints for the binding of short peptides to claudin-4 revealed by surface plasmon resonance. J Biol Chem. 2008;283:30585–30595. doi: 10.1074/jbc.M803548200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karlsson R, Fält A. Experimental design for kinetic analysis of protein-protein interactions with surface plasmon resonance biosensors. J Immunol Meth. 1997;200:121–133. doi: 10.1016/s0022-1759(96)00195-0. [DOI] [PubMed] [Google Scholar]
- 28.Lilien RH, Stevens BW, Anderson AC, Donald BR. A novel ensemble-based scoring and search algorithm for protein redesign and its application to modify the substrate specificity of the Gramicidin Synthetase A Phenylalanine Adenylation enzyme. J Comp Biol. 2005;12:740–761. doi: 10.1089/cmb.2005.12.740. [DOI] [PubMed] [Google Scholar]
- 29.Katragadda M, Morikis D, Lambris JD. Thermodynamic studies on the interaction of the third complement component and its inhibitor compstatin. J Biol Chem. 2004;279:54987–54995. doi: 10.1074/jbc.M409963200. [DOI] [PubMed] [Google Scholar]
- 30.Magotti P, Ricklin D, Qu H, Wu Y-Q, Kaznessis YN, Lambris JD. Structure-kinetic relationship analysis of the therapeutic complement inhibitor compstatin. J Mol Recognit. 2009;22:495–505. doi: 10.1002/jmr.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klepeis JL, Floudas CA, Morikis D, Tsokos CG, Lambris JD. Design of peptide analogs with improved activity using a novel de novo protein design approach. Ind Eng Chem Res. 2004;43:3817–3826. [Google Scholar]
- 32.Tamamis P, Morikis D, Floudas CA, Archontis G. Species specificity of the complement inhibitor compstatin investigated by all-atom molecular dynamics simulations. Proteins. 2010;78:2655–2667. doi: 10.1002/prot.22780. [DOI] [PMC free article] [PubMed] [Google Scholar]