Abstract
Podosomes are ventral adhesion structures prominent in cells of the myeloid lineage. A common aspect of these cells is that they are highly motile and are required to traverse multiple tissue barriers in order to perform their functions. Recently podosomes have gathered attention from researchers as important cellular structures that can influence cell adhesion, motility and matrix remodeling. Adhesive and soluble ligands act via transmembrane receptors and propagate signals to the leukocyte cytoskeleton via small G proteins of the Rho family, tyrosine kinases and scaffold proteins and are able to induce podosome formation and rearrangements. Manipulation of the signals that regulate podosome formation and dynamics can therefore be a strategy to interfere with leukocyte functions in a multitude of pathological settings, such as infections, atherosclerosis and arthritis. Here, we review the major signaling molecules that act in the formation and regulation of podosomes.
Keywords: podosome, leukocyte, Rho GTPase, tyrosine kinase, transmembrane receptor
1. Introduction
Podosomes are adhesion structures prominent in cells of the myeloid lineage – monocytes, macrophages, osteoclasts and immature dendritic cells (DCs). Studies have also suggested the presence of podosomes or podosome-like structures in neutrophils [1] as well as in malignant lymphocytes of B cell chronic leukemia [2] and a range of leukocytes adhering to the luminal surface of endothelial cells during diapedesis [3]. They are distinguished from other adhesions structures, such as focal contacts, via their distinct localization and molecular arrangement (Figure 1). Unlike focal adhesions or the smaller focal complexes where actin bundles terminate in plaques of adhesion molecules, in podosomes a core of F-actin, presumably branched, is surrounded by a ring of adhesion-related proteins, such as talin, paxillin, vinculin, etc. Individual podosomes are, in turn, connected to each other via radial bundles of loose F-actin, termed the actin cloud. This supramolecular organization arranges podosomes into characteristic clusters. Additionally, unlike the more stable focal adhesions and focal complexes, podosomes are highly dynamic, with a lifetime of 2–10 minutes, with continuous actin turnover [4]. While the fine molecular organization of focal adhesions has recently been resolved using high-resolution light microscopy [5], revealing with impressive detail the arrangement of structural and signaling molecules at nm precision, such information is still lacking for podosomes. Arguably, however, podosomes are sites of signaling. Integrins, protein kinases, GTPases, the actin nucleation machinery, all converge to sense soluble and/or adhesive signals and influence cellular behavior.
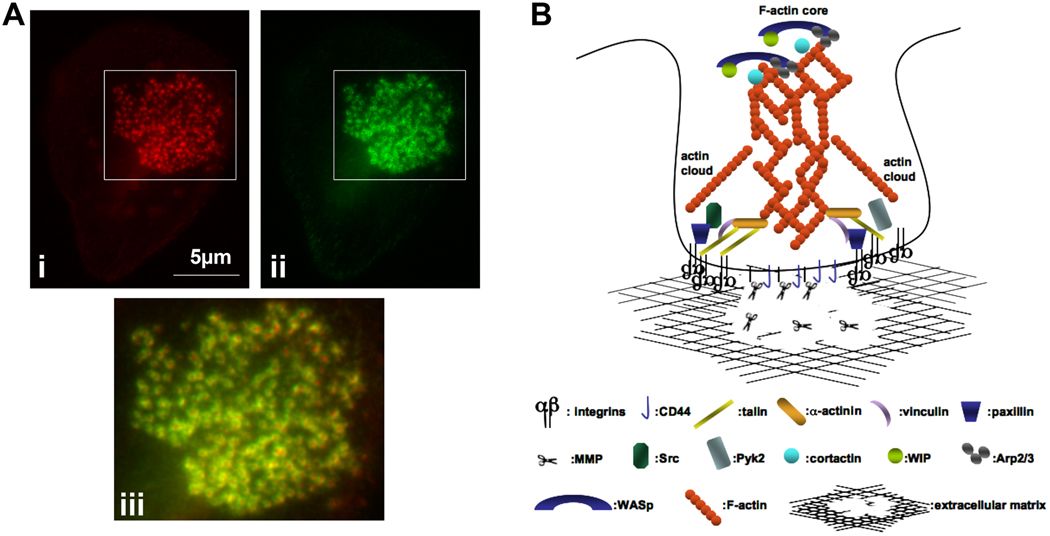
Figure 1. Structure of leukocyte podosomes.
(A). A human monocyte-derived macrophage stained for F-actin (i, red) and vinculin (ii, green). The boxed areas were merged and enlarged in (iii). Vinculin forms rings around the F-actin core of the podosome.
(B). Cartoon of a podosome. F-actin and actin regulatory molecules, such as WASP, cortactin, Arp2/3 are located in the podosome core, which is tethered to the extracellular matrix via CD44. The podosome ring contains integrins, which connect the extracellular matrix to the interior of the podosome via cytoskeletal linkers, such as talin and vinculin, and and regulate signaling cascades via tyrosine kinases, such as Src and Pyk2. Actin cables link the ring structure to the podosome core. Podosomes provide a focal point for MMP secretion and matrix degradation and remodeling.
1.1 Roles of podosomes
The most well-established function of leukocyte podosomes is matrix remodeling. Podosomes of macrophages and DCs define the sites of localized matrix degradation via the targeted delivery of MT1-MMP [6–8]. In addition, several signaling molecules that appear to be important for podosomes in vitro, such as the tyrosine kinase Hck [9] may also interfere with the ability of leukocytes to invade one or several tissue barriers in vivo, including endothelial monolayers and basement membranes. The recent characterization of macrophage podosomes in 3D environments [10] reinforces the idea that podosomes assist in cell invasion and motility, and we will address this by discussing the roles of key signaling components that regulate both podosome formation and cell invasion or motility in 3D.
Podosomes have also been implicated in cell migration on planar surfaces. Studies of macrophages obtained from Wiskott-Aldrich Syndrome (WAS) patients that lack distinctive podosomes due to the absence of the Wiskott-Aldrich Syndrome protein (WASP), revealed impaired chemotaxis to soluble cues including CSF-1, MCP-1 and fMLP, suggesting that they may be critical for directional migration [11, 12]. This may also be supported by the presence of podosomes into what is morphologically defined the leading edge of a migrating cell, however this link remains speculative with no direct evidence connecting podosomes to chemotaxis directly. Intriguingly, similar to studies in cells such as fibroblasts, adhesion is often correlated to decreased ability to migrate, and indeed signals that may result in podosome disassembly may promote increased migratory behavior of the cell. This has been observed in macrophages [13] but perhaps the most striking examples have been provided for immature DCs. Calle et al [14] have demonstrated that inhibition of the intracellular cysteine protease calpain stabilizes podosomes through accumulation of several podosome components such as WASP and talin, and prevents DC motility and transendothelial migration. Likewise, immature DCs contain podosomes and show enhanced β2 integrin-mediated adhesion to ICAM-1, compared with mature DCs, which were devoid of podosomes [15]. Lipopolysaccharide (LPS) or prostaglandin E2 (PGE2) induce the disassembly of podosomes [15–18] resulting in reduced adhesion strength to ICAM-1 [15] and increased DC migration [16]. Likewise, immature DC interactions with CD4+ T cells promoted dissolution of podosomes and stimulated migration towards several chemokines [19]. In vivo this correlates with ability of DCs to migrate to the lymph node following antigen binding. Even though the exact function of podosomes remains somewhat controversial, investigating the signaling mechanisms that regulate the dynamics of podosome assembly and disassembly are therefore essential in understanding the motility and migration mechanisms employed by macrophages and DCs.
Podosomes in osteoclasts are also very dynamic and represent more specialized entities with well-defined functions in bone metabolism. As osteoclasts mature in vitro, individual podosomes show remarkable rearrangements and transition from individual podosomes to podosome clusters to a podosome belt [20]. When in contact with apatite [20] or bone [21, 22], podosomes organize into a tightly packed array with a higher degree of interconnectivity between podosomes, forming the sealing zone, critical for the osteoclast’s bone resorptive properties. While osteoclast podosomes are capable of matrix degradation, their main role differs. Rather than the sealing zone itself mediating degradation of the mineralized matrix, it delimits the region of bone resorption within a tightly sealed compartment termed the resorption pit. This compartment is isolated from the extracellular environment and is highly acidified thus facilitating bone resorption via the activities of cathepsin K [23]. Several signaling molecules have been found to regulate podosome formation, dynamics and the transition to a sealing zone. The importance of podosomes in osteoclast functions in vivo is highlighted by the fact that animals deficient in key signaling molecules that regulate podosome dynamics tend to have abnormal bone mass and thus present with varying degrees of osteopetrosis. Among those that will be mentioned in this review, Src [24], Pyk2 [25], WASp [26], Syk [27], and Vav3 [28] are prominent examples.
Related structures to podosomes, such as invadopodia of carcinoma cells or rosettes of src-transformed cells have been described. These are distinct from leukocyte podosomes and several signaling molecules have been identified that may assist in the differentiation between these structures, apart from the more apparent morphological differences and relation to patho-physiological functions [29, 30]. Interestingly, several non-transformed cells such as endothelial cells or smooth muscle cells form podosomes that have the characteristic core-ring arrangement of leukocyte podosomes, or the rosette arrangement of src-transformed cells and may occur following several physiological stimuli. These structures will not be covered here, however, enzymes or scaffolding molecules from the same families are acting in all these structures (e.g. Src family kinases), highlighting common molecular themes in the formation and organization of ventral adhesion and invasive structures. For more details on invadopodia and podosome rosettes in non-myeloid cells the reader is referred to other reviews [6, 31].
While functional and molecular differences between podosomes, invadopodia and rosette-like structures of src-transformed cells or endothelial cells have been described and discussed elsewhere [29], here we provide a more detailed analysis of the signaling networks that govern the formation, dynamics and function of the leukocyte podosomes. These involve the co-ordination of many molecules including actin nucleating factors, small GTPases, kinases and transmembrane receptors. In particular, we will focus on the podosomes of myeloid cells – macrophages, DCs and osteoclasts – cells in which podosomes occur constitutively, at least in culture, and could be inherent to the functions of these cells, e.g. bone resorption and the ability to cross multiple heterogeneous barriers in order to encounter pathogens and present antigens.
2. Rho GTPases
Small GTPases of the Rho family have important and conserved roles in virtually every function of the cytoskeleton. Their activity is tightly regulated by three classes of molecules; guanine nucleotide exchange factors, GEFs, which exchange GDP for GTP therefore activating the GTPase [32]; GTPase activating proteins, GAPs, which assist in the hydrolysis of bound GTP to GDP thus contributing to inactivation [33]; and guanine dissociation inhibitors, GDIs, which sequester membrane-anchored GTPases into the cytosol and thus affect their distribution, localization and protein levels [34–36].
Initial experiments utilizing forced expression of constitutively active or dominant negative forms of the Rho GTPases, or treatment with bacterial toxins showed that RhoA, Cdc42 and Rac1 played important roles in podosome formation. In particular, expression of dominant negative Cdc42 induced podosome disassembly in DCs [37]) and macrophages [38], as did expression of constitutively active Cdc42 [38, 39]. This would suggest that precise control of the activity of Cdc42 is required for the correct regulation of podosomes; nevertheless, overexpression of such mutants may have several indirect effects. The importance of Cdc42 in macrophage podosome formation has also been confirmed by shRNA-mediated downregulation of endogenous Cdc42 [40]. Indeed, Cdc42 stands as a central player in the regulation of podosome dynamics as it orchestrates podosome actin polymerization via its canonical effector, WASP.
Rac1 and Rac2 may play distinct roles in macrophage podosome formation. Rac1 deficient macrophages show impaired podosome formation with fewer cells displaying podosomes that lacked the adhesion ring, while Rac2 deficiency results in complete loss of podosomes in macrophages [41]. Interestingly, Rac2-deficient cells were still able to migrate directionally, despite altered morphology, but were unable to invade through matrigel in vitro. This would suggest that podosomes are not required for efficient chemotaxis as previously suggested [11], but may still be required for invasion. DC podosomes were also found to depend on Rac1/2 activity, and PLCγ2 was important in regulating Rac1 activation [42]. Indeed, DCs from PLCγ2 deficient mice displayed abnormal podosome arrangement with loose clusters and lack of a discernible adhesion ring [42]. Rac1 and Rac2 are therefore essential in podosome formation and organization.
The role of RhoA has been somewhat controversial and its influence on podosomes may reflect cell type dependent differences or an effect of distinct RhoA effectors downstream of distinct signals. For example, inhibition with C3 transferase induced podosome disassembly in DCs [37], though this treatment may target other GTPases than just RhoA [43]. Acute podosome disassembly in immature DCs induced by PGE2 was dependent on RhoA via a cAMP-PKA pathway [17]. In osteoclasts, transduction of TAT-RhoA V14 induced podosome formation [44] while C3 transferase treatment on osteoclasts adhering on mineralized matrix prevented formation of the sealing zone [20]. This presumably works via a Rho kinase (ROCK)-dependent pathway, as the ROCK inhibitor Y-27632 inhibited bone resorption in vitro [45]. C3 treatment in osteoclasts plated on glass, however, stabilized the podosome belt following microtubule depolymerization [46] also implicating Rho in destabilization of the sealing zone. The same study reported disruption of the podosome belt when constitutively active RhoA was microinjected into osteoclasts. Indeed, RhoA inhibits microtubule acetylation and therefore stability of the podosome belt, an activity mediated by the effector mDia2 and the subsequent activation of the deacetylase HDAC6. RhoA is therefore involved in multiple aspects of podosome dynamics influencing both the formation and the stability of podosomes. This may be achieved by engaging with distinct RhoA effectors and their targets, or alternatively, by tuning of the amplitude of a signal such that above a threshold of activity RhoA can have either a positive or a negative effect on podosomes.
The above studies established a link between Rho GTPase activity status and the regulation of podosomes. However, the exact sequence that regulates the interplay between the Rho family GTPases is still missing.
2.1 Other GTPases
The roles of small GTPases other than the conventional RhoA, Rac1 and Cdc42 on leukocyte podosomes have not been explored in detail. However, Arf6 has been implicated in podosome formation in immature DCs as a constitutively active form (Q67L) or a GDP-locked mutant (T44N) decreased the number of cells displaying podosomes. Intriguingly, the nucleotide-free T27N mutant of Arf6, only induced a modest decrease in podosome numbers [47]. Similarly, in osteoclasts, Arf6 T27N did not affect sealing zone formation, unlike the constitutively active mutant Q67L [48], which was inhibitory. Depletion of the Arf-GAP GIT-2, but not GIT-1, similarly prevented sealing zone formation in osteoclasts. GIT-2 is a substrate for the Src tyrosine kinase, an enzyme with extensive functions in osteoclast podosome dynamics (discussed below).
The unconventional Rho GTPase Wrch1, also known as RhoU, localizes to the osteoclast podosome belt [49]. Overexpression of either wild-type, dominant negative or constitutively active Wrch1 altered the organization of the podosome belt but did not affect bone resorption in vitro [50]. The exact role of Wrch1 in osteoclast podosomes remains therefore uncharacterized [51]. Overall, these studies indicate that multiple small GTPases regulate podosome dynamics.
2.2 GTPase regulation
Despite the well-established role of Rho family GTPases in podosome formation and dynamics, GEFs that activate GTPases in the context of podosome regulation have not been extensively studied. αPIX, a Rac/Cdc42 GEF has been found to localize to podosome rings and regulates podosome size and number [52]. Interestingly, interfering with the GEF activity of αPIX did not affect podosome number significantly, implying that it may play another role in podosome dynamics independent of GEF activity. The localization of αPIX to macrophage podosome rings may be due to its association with the protein GIT-1, which is known to interact directly with paxillin, another podosome ring component [53]. GIT-1 is a GAP for the GTPase Arf6. It is possible that the interaction between GIT-1 and αPIX may link the Cdc42, Rac1 and Arf6 GTPases in podosome formation, or alternatively, that it restricts Arf6 activity from the podosome core thus preventing Arf6-induced podosome disassembly. The function of αPIX in podosomes remains controversial, especially due to the fact that siRNA against α or β PIX did not interfere with sealing zone formation of osteoclasts [48].
Recently a Rac1-specific GEF, FARP2, has been demonstrated as being important in podosome regulation of osteoclasts. Overexpression of a FARP2 construct lacking the GEF domain in osteoclasts abolished podosome formation while osteoclasts from FARP−/−mice failed to resorb bone efficiently in vitro [54]. Vav1, a GEF that regulates Rac1 also appears to be important in the regulation of leukocyte podosomes [55] although not required for their formation but instead for the actin dynamics inside the podosome. On the other hand, while Vav1 is not required for podosome formation and organization in osteoclasts, Vav3, which is expressed in abundance in osteoclasts and also regulates Rac1 is indispensable for podosome formation in osteoclasts [28].
Podosome regulation by Rho GAP activities is unknown. However, Cdc42GAP deficiency in neutrophils has been associated with a decrease in the presence of podosome-like structures in neutrophils [1], supporting earlier studies that suggested tight regulation of Cdc42 for podosome assembly [38].
The fact that interference with all three major Rho GTPases affects podosomes in macrophages and DCs suggests a co-operative or complementary function or, on occasion, antagonistic. For example, PGE2 stimulation, which induces podosome loss in DCs via RhoA-ROCK, increases RhoA-GTP levels while Rac1-GTP and Cdc42-GTP are decreased [17]. This can be achieved either through crosstalk between the three GTPases [56] (Figure 2), or alternatively, that each one acts independently of the other at distinct parts of podosomes or at separate times during the life-time of a podosome. The use of biosensors for reporting GTPase activity would be a useful tool to explore the time, location and interdependency of GTPase activities during conditions that promote podosome formation or disassembly [57, 58]. These, linked with identification of specific GEFs, GAPs and effector proteins will decipher the roles of all GTPases.
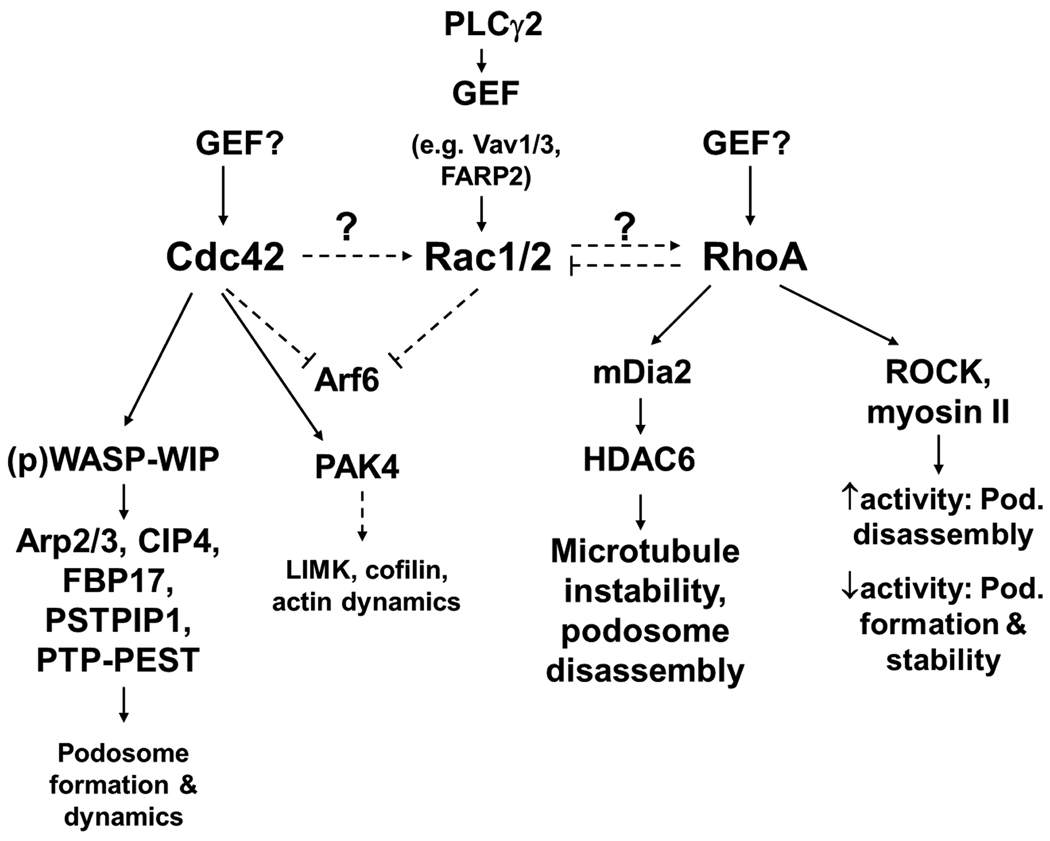
Figure 2. RhoGTPase signaling in podosome regulation.
The three major RhoGTPases, Cdc42, Rac and RhoA are essential for podosome formation and disassembly. All three GTPases potentially crosstalk (dashed arrows) and thus influence each other’s activity and ultimately, podosome dynamics. GEFs that activate Rac in podosome formation and regulation have been identified, but remain unknown for Cdc42 and RhoA. Cdc42 provides a major signal for podosome assembly via its effector WASP, which stimulates Arp2/3-dependent actin polymerization and acts as a scaffold to bring together signaling molecules. RhoA can play opposing roles in podosomes. Signaling via its effector mDia2 regulates microtubules and influences podosome stability. ROCK-dependent signaling may either contribute to podosome belt formation (in osteoclasts) or podosome disassembly and could be dependent on the strength of the RhoA-ROCK signal with low activity contributing to podosome assembly and high activity to disassembly.
Dashed lines represent speculative functions.
2.3 Effectors of Rho GTPases in podosome regulation
Despite numerous Rho effectors with catalytic or scaffolding activities, those with specific functions in podosome regulation are largely unexplored. Importantly, not all effectors that mediate cytoskeletal changes may be required for podosome formation. For example, the WAVE2 complex, which acts downstream of Rac, does not affect podosome formation in macrophages [59]. Of the known canonical effectors, the functions of ROCK, PAK4 and WASP are discussed.
2.3.1 ROCK
As mentioned above, RhoA mediates the dissolution of podosomes by PGE2. This is dependent on ROCK activity since pharmacological inhibition of ROCK prevented podosome disassembly by PGE2. This RhoA-ROCK effect was dependent on myosin IIA contractility as podosome disassembly is sensitive to blebbistatin [17]. The RhoA-ROCK signaling axis therefore promotes podosome dissolution through myosin-dependent contractility. Likewise, reduced ROCK activity is required for the stability of osteoclast podosomes. The two Rho associated kinases, ROCK I and ROCK II have distinct functions [60], and presently it is not known whether either or both affect podosomes in leukocytes. Interestingly in osteoclasts, ROCK II was found to phosphorylate CD44 [45], a transmembrane protein and component of the podosome core [61], and ROCK activity was required for efficient bone resorption [45]. These apparently conflicting functions of ROCK and myosin II-mediated contractility can be reconciled by assuming that a basal level of RhoA-ROCK-myosin II regulates podosome formation and function, but that increases in activity dissolve podosomes [62] (Figure 2).
2.3.2 PAK4
PAK4, a group II PAK family member, binds preferentially to Cdc42 and not to Rac1 [63]. It is localized to macrophage podosomes and forms part of a complex together with αPIX (see section 2.2, above). PAK4, unlike αPIX, localizes to the cores of macrophage podosomes. Its localization and activity in macrophage podosomes may therefore be independent of αPIX [52]. siRNA-mediated downregulation of PAK4 or expression of a truncation mutant lacking the GTPase binding domain severely impaired podosome formation. On the other hand formation of podosomes was independent of its kinase activity indicating a scaffolding role for PAK4 in podosome formation [52]. However, catalytic domain mutants of PAK4 affect actin in podosomes with a kinase-dead mutant resulting in smaller podosomes with less F-actin, opposite to a catalytically enhanced PAK4, that resulted in larger podosomes with more F-actin [52]. Substrates of PAK4 that may regulate actin content in podosomes have yet to be identified. However, a target for PAK4 LIMK phosphorylates and inactivates cofilin [64] and suggests that the effect of PAK4 on podosome F-actin could potentially involve the cofilin activity cycle [6, 65]. Active PAK4 may therefore inactivate cofilin via LIMK phosphorylation, resulting in decreased filament severing by cofilin and enhanced F-actin content. The above data point to two distinct functions for PAK4, one that is catalytically-independent and is involved in podosome formation, and another, catalytically-dependent and affects actin dynamics inside podosomes.
2.3.3 WASP-a critical intermediate in podosome formation
WASP is a scaffold protein and an actin nucleation promoting factor that acts via the Arp2/3 complex. It is an effector of Cdc42 and it plays a central role in nucleating nascent podosomes. WASP deficiency in humans with WAS or in mice with genetic deletion results in loss of podosomes in a variety of leukocytes, including macrophages [11], DCs [15] and osteoclasts [26] and disruption of WASP interaction with Cdc42 results in loss of podosomes in macrophages [40]. A multi-domain protein, WASP has several other critical binding partners necessary for its function in podosome formation (Figure 2). WASP interacting protein (WIP) is one such protein. This is highlighted by the fact that the majority of loci mutated in WAS or X-linked thrombocytopenia are found in the WH1 domain of WASP, which is responsible for its interaction with WIP [66]. Inhibition of the interaction of WASP with WIP results in a drop of the levels of WASP, suggesting that WIP acts as a chaperone for WASP [66]. Accordingly, loss of WIP in mice results in a dramatic drop of endogenous WASP levels, and defective podosome formation in DCs [67], and osteoclasts [61] (Sabadel 07). In addition to its interactions with WIP and Cdc42, further interactions of WASP are also important for its function in podosome formation. The Cdc42- and microtubule-interacting protein CIP4, member of the Pombe Cdc15 homology (PCH) family of proteins [68], is also required for WASP-dependent podosome formation though the CIP4-WASP interaction appears to be independent of Cdc42 [69]. Consistent with this theme, the related formin-binding protein FBP17 also recruits WASP/WIP to nascent podosomes and regulates podosome formation [70].
WASP is also a substrate for tyrosine kinases, including src-family kinases (SFKs; reviewed [71], and its tyrosine phosphorylation status affects podosome dynamics [40, 72]. In macrophages, WASP is phosphorylated by the SFK Hck (H. Park and D.C., personal communication), which also regulates podosomes and invasion [9, 39]. A WASP phosphomimetic (Y291E) expressed in macrophages increased the number of podosomes in macrophages, the rate of podosome nucleation and filament stability inside podosomes [40]. While biochemical studies have associated WASP tyrosine phosphorylation with enhanced activity [71, 73], a non-phosphorylatable WASP mutant (Y291F) was still active, but the active species was not localized correctly in macrophage podosomes as evidenced using FRET-based biosensors, and did not form podosomes efficiently when plated on fibronectin [40]. Also, Thrasher and coworkers have generated mice with a knock-in phosphomutant WASP and the phosphoabolishing Y291F mutation resulted in mice phenotypically similar to WASP null mice, with DCs not forming well-defined podosomes [72]. Interestingly, the phosphomimetic Y291E WASP also impaired podosome formation and dynamics. It was proposed that constitutive WASP phosphorylation resulted in proteasomal degradation of WASP, hence the result in decreased podosome formation.
Another protein that interacts with WASP [74] is Proline, serine, threonine phosphatase interacting protein (PSTPIP1) is a heavily phosphorylated adaptor molecule and member of the PCH family of proteins [68] together with CIP4, FBP17, and Toca-1, all of which interact with Cdc42 and/or WASp and have roles in podosome formation in macrophages, discussed above. While its homologue PSTPIP2 is involved in signaling from the CSF-1R [75], PSTPIP1 is not known to associate with CSF-1R and is likely phosphorylated by c-Abl [76] and/or SFKs [48]. Mutations in PSTPIP1 result in pyogenic arthritis, pyoderma gangrenosum and acne (PAPA) syndrome in humans. Recently it was demonstrated that macrophages from PAPA syndrome patients lack podosomes and cannot degrade matrix or invade matrigel [77], implicating PSTPIP1 in podosome function. Despite the interaction between the PSTPIP1 SH3 domain and the poly-proline region of WASP [74], it is not known whether the podosome phenotype in PAPA syndrome macrophages is due to an alteration in WASP association, despite a striking phenotypic similarity with WAS patient cells. The interaction between PSTPIP and WASP is increased in osteoclasts following osteopontin stimulation and results in the formation of a complex containing WASP, PSTPIP, the tyrosine kinases Src and Pyk2 and the tyrosine phosphatase PTP-PEST [78]. All the components of this signaling complex localize to sealing zones and could affect its dynamics. Importantly, several reports have suggested that the interaction of PSTPIP1 with PTP-PEST results in dephosphorylation of WASP and inhibition of WASP-dependent actin polymerization [79, 80]. Interestingly, while siRNA-mediated downregulation of PTP-PEST resulted in retention of a sealing zone, bone resorption stimulated by osteopontin was impaired. This would indicate that the complex between PTP-PEST and WASP, regulated by the scaffolding action of PSTPIP1, is required for dephosphorylation and thus inactivation of WASP and subsequent cycles of sealing zone disassembly, reassembly and resorption [78].
4. Tyrosine kinases
From the above example, it becomes clear that apart from GTPases, kinases are also active in podosome regulation. Historically, it was the transformation of fibroblasts with the constitutively active form of Src from Rous sarcoma virus, v-Src, which induced formation of ventral adhesion structures, variable referred to as invadopodia, podosomes or rosettes [81]. This turned the attention to tyrosine kinases, especially SFKs, in the regulation of podosomes in leukocytes. Pharmacological inhibition of SFKs using the inhibitor PP2 resulted in loss of podosomes in human macrophages [39, 82] suggesting they are important in the stability of native podosomes.
4.1 SFKs – Hck
Hck is a SFK expressed in leukocytes [83], where it plays important roles in motility and the regulation of podosomes. Hck is expressed as two isoforms, produced by alternative splicing of the same mRNA; p59Hck and p61Hck, and it is suggested that it is the later that localizes to macrophage podosomes [39] and osteoclast podosome belt [84]. Hck is activated by ligation of the urokinase plasminogen activator receptor and the integrin αMβ2 [85, 86] in macrophages, though it is unknown whether these modes of activation are relevant to podosome signaling. Hck is able to interact via its SH3 domain with the polyproline regions of WASP, WIP and ELMO1 [87] and phosphorylates both WASP [88] and ELMO1 [87] when these proteins are overexpressed in COS7 or CHO cells, respectively. Hck is also the major endogenous WASP kinase in macrophages, as shRNA against Hck results in dramatic decrease of WASP tyrosine phosphorylation in response to various stimulations (H. Park and D.C., unpublished data). ELMO1 interacts with Dock180, an unconventional GEF for Rac1 [89], suggesting that Hck can couple to GTPase signaling. This is further reinforced by the fact that another Hck substrate, the Rac GEF Vav1 [90], regulates podosome distribution and their F-actin content in DCs [55]. Importantly, while ectopic expression or constitutively active p61Hck induced rosette formation in fibroblasts, these were inhibited when cells were co-transfected with dominant negative Cdc42 or Rac1 – not RhoA – suggesting that p61Hck signals via these two GTPases [30]. It is assumed that nascent podosome biogenesis through p61Hck is achieved by actin polymerization that drives the fusion of lysosomes to podosomes [30] and this requires the catalytic activity of p61Hck and the concerted activity of WASP, Arp2/3 and Cdc42 in cell-free assays [91]. However, whether Hck phosphorylates WASP or other substrates in this setting is unknown.
4.2 SFKs – Src
Src has prominent roles to play in osteoclast podosome assembly and functions. Indeed, the knock-out mouse of the first ever proto-oncogene to be identified presented with severe osteopetrosis [24], highlighting the important role it played in bone metabolism. Osteoclasts from src−/− mice do not form a sealing zone [92, 93], while re-expression of Src in these null cells restored the phenotype [94]. Importantly, knock-out of other myeloid-expressed SFKs did not result in osteopetrosis [95], highlighting the specificity of Src in this process. Of note, while most double Src−/−Hck−/− mice die after birth, those that survived presented with severe osteopetrosis, suggesting that Hck may perform, at least in part, compensatory roles with Src in osteoclast functions [96]. This is further supported by the fact that Hck expression was increased in cells from src−/− mice [96]. Src in osteoclasts is activated following ligation of αvβ3 integrin via the tyrosine kinase Pyk2 (see 4.3, below) and the concerted activity of the cytosolic form of the tyrosine phosphatase PTPε [97]. Src deficient osteoclasts form individual podosomes, possibly through the action of Hck, however these podosomes lack the surrounding actin cloud, do not assemble a proper adhesion ring around the F-actin core, show altered actin dynamics inside the F-actin core, and do not form the podosome belt [98, 99].
Regulation of podosome dynamics by Src is achieved through multiple regulatory and signaling steps. In osteoclasts, Src can form a complex with the active αvβ3 integrin and the tyrosine kinase Syk [27, 100], where it phosphorylates and activates Syk. Syk in turn phosphorylates the Rac1 GEF Vav3 resulting in Rac1 activation and cytoskeletal reorganization [23] (Figure 3). Cortactin, a major Src kinase substrate is also required for osteoclast podosome dynamics and bone resorption [99, 101]. Indeed, Src-phosphorylated cortactin engages in vitro in a complex with WIP and N-WASP and results in enhanced Arp2/3-dependent actin polymerization compared to non-phosphorylated cortactin [102]. Importantly, all these proteins localize to osteoclast podosomes and the podosome belt [101, 102]. Depletion of endogenous cortactin resulted in loss of podosomes from differentiated osteoclasts and a mislocalization of WASP and N-WASP from podosome belts [101]. The requirement for Src-mediated cortactin phosphorylation in podosome dynamics is highlighted by studies where expression of cortactin with Y-to-F mutations of the three phosphorylation sites could not rescue for the absence of endogenous cortactin [101] while it also resulted in reduced life-span of the podosomes [99] in osteoclasts.
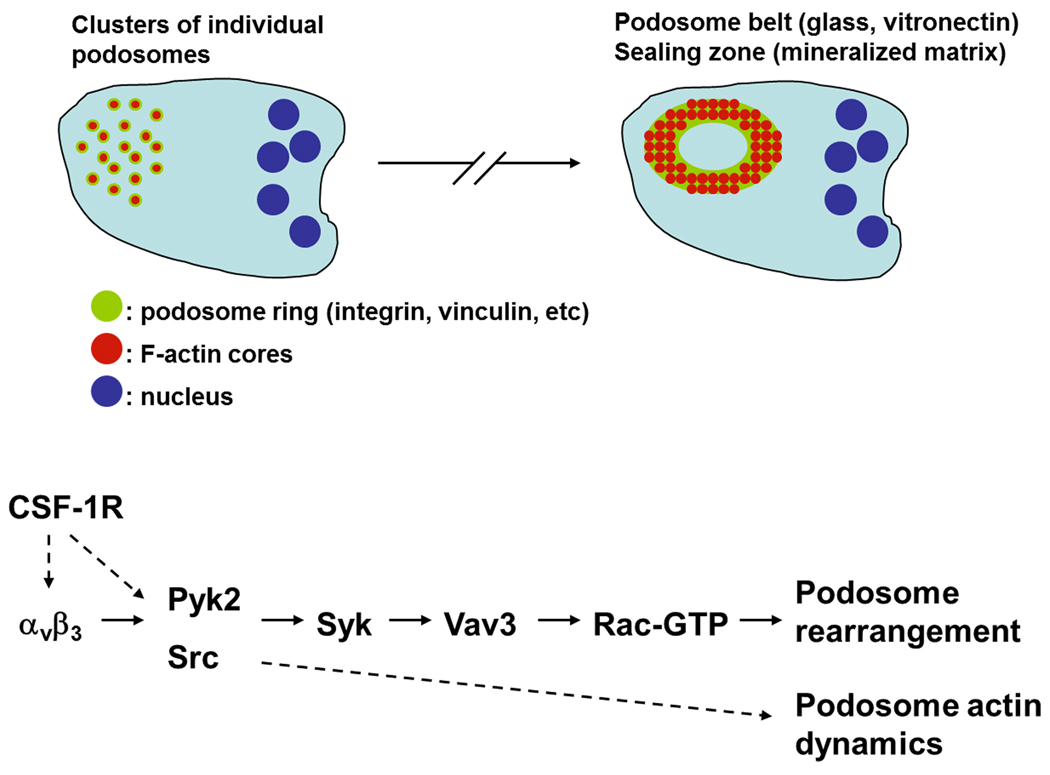
Figure 3. Signals regulating podosome cluster to podosome belt/sealing zone formation in osteoclasts.
Upon osteoclast maturation, podosomes transition from clusters to a podosome belt or a sealing zone, depending on the matrix onto which the cell adheres. This transition is dependent on signaling orchestrated by the αvβ3 integrin. Matrix adhesion and cross-talk with the CSF-1R promotes integrin-dependent activation of the kinases Pyk2 and Src. Src in turn phosphorylates and activates Syk, which phosphorylates and activates the Rac GEF Vav3 resulting in increased Rac-GTP levels and actin rearrangement.
Downstream of Src in osteoclast podosome signaling is also Cbl, a ubiquitin ligase that acts in down regulating several signaling pathways by targeting molecules for the ubiquitination machinery or via endocytosis [103, 104]. Cbl is phosphorylated by Src and other SFKs, including Hck, in osteoclasts [104] as well as in macrophages [105–107] and localizes to the podosome belt in osteoclasts [78, 103] (Tanaka 96; Sanjay 01). Osteoclasts express c-Cbl and Cbl-b and downregulation of c-Cbl in Cbl-b−/− osteoclasts disrupted formation of the podosome belt, and individual podosomes remained loosely connected at the cell periphery [108]. Cbl is therefore important for formation of the podosome belt in osteoclasts. Cbl participates in podosome belt regulation through association with dynamin [109, 110] and microtubules, where it displaces HDAC6 [108] thus resulting in microtubule stability, and potentially via PI3K (reviewed in [95]).
4.3 Pyk2
Pyk2 is a homolog of focal adhesion kinase (FAK), however unlike the ubiquitously expressed FAK, Pyk2 expression is restricted to neuronal and hematopoietic cells. Pyk2 localizes to the podosome ring in the mouse macrophage cell line IC-21 [111], suggesting it participates in adhesion-dependent signaling. Consistently, Pyk2 is phosphorylated downstream of β2 integrin ligation with clustering antibodies or following adhesion to various ECM proteins, such as fibronectin, vitronectin, fibrinogen, but not poly-L-lysine, collagen or laminin [111]. Pyk2 phosphorylation is dependent on the SFKs Hck and Fgr [112] and Pyk2 deficiency in macrophages results in defective activation of PI3K and RhoA pathways downstream of matrix adhesion [113].
In mature osteoclasts, Pyk2 is localized to the podosome belt and the sealing zone [93]. Ligation of the αvβ3 integrin results in elevation of intracellular Ca2+ and phosphorylation of Pyk2 on Y402 [104, 114]. Elevation of intracellular Ca2+ occurs potentially through the activity of PLCγ2 [115]. Autophosphorylation of Pyk2 on Y402 is a critical event as it provides a binding site for the SH2 domain of Src and activates Src via relief of its autoinhibitory constraints. The Y402F mutation of Pyk2 abolishes Src binding and impairs bone resorption [116]. Interestingly, a kinase-defective form of Pyk2, which otherwise acts as a dominant-negative, localizes normally to podosome belt and does not impair the ability of osteoclasts to resorb bone [25, 116]. This would suggest that another kinase trans-phosphorylates Pyk2 on Y402 following integrin ligation. The absence of Pyk2 in osteoclasts resulted in abnormal podosomes and absence of a podosome belt, while there were increased RhoA-GTP levels leading to destabilization of the microtubule network [25]. Importantly, expression of activated Src could not rescue for the absence of Pyk2 suggesting that Pyk2 signaling could involve two pathways, one affecting RhoA and the other proceeding via Src with both signals being required for bone degradation [25].
5. Transmembrane receptor signaling at podosomes
All cells communicate with their extracellular environment via transmembrane receptors that recognize soluble or immobilized ligands and transmit signals to the interior of the cell. Podosomes are the net result of such signals and both adhesion and chemokine and cytokine receptors participate in their formation or disassembly. Integrins present arguably the most important and well-studied class of adhesion receptor. The earliest signaling events initiated by integrins are activation of SFKs and Syk [117], and these are relevant in podosome formation and dynamics, as have been discussed above. In leukocyte podosomes the β2 and β3 integrins are found in the podosome ring and mediate adhesion signals [118]. The β1 integrin may be localized in the podosome core as in the case of B cells from B cell chronic lymphocytic leukemia [2] or the ring, as in the case of DCs [16]. In osteoclast podosomes β3 integrin localizes in rings of the podosome belt, while β1 integrin is altogether absent from the podosome belt [61]. Substrates onto which leukocytes are usually plated in order to study podosome formation include ICAM-1, which binds αLβ2, fibronectin, which binds a range of integrins, and vitronectin and bone sialic protein which bind αvβ3.
Integrins usually exist at a basal, low affinity or inactive state and activation occurs in a bi-directional manner [117]. Outside-in signaling following ligand occupancy of the extracellular part of the integrin induces clustering of the receptors and secondary changes further increasing the affinity of the integrin for its ligand. The second mode of activation, inside-out signaling, involves a signaling cascade initiated through a secondary receptor, usually a cytokine receptor, and the transmission of conformational changes from the intracellular part of the integrin to the extracellular. Thus the integrin is primed for ligand binding and the transmission of signals to the cell’s exterior, including those that regulate cytoskeletal organization. The exact conformation and activity state of integrins in podosomes has been largely unexplored. In osteoclasts, αvβ3 integrin inside the podosome belt appears to be in the basal state, as monoclonal antibodies against activation-induced epitopes could not react with the integrins [119]. By contrast, immunoreactivity against active αvβ3 was observed when osteoclasts were treated with CSF-1, and active integrins were localized to the leading edge and membrane ruffles of migrating osteoclasts [119].
Nevertheless, both modes of signaling appear to be required for integrin signaling in podosomes. Indeed, plating of DCs on poly-L-lysine did not induce formation of podosomes or other discernible adhesion structures, unless the media were supplemented with soluble factors such as SDF-1α [120]. Intriguingly, when cells were plated on the integrin ligands fibronectin or ICAM-1 alone, they formed large focal complexes but podosome formation could only be induced when soluble factors supplemented from serum were administered [120]. This would suggest that growth factors or cytokines induce inside-out signaling that activates integrins and promotes their specific inclusion in podosomes. This is important since given the actin core – adhesion ring architecture of podosomes, it is not yet established which is the initiating structure and what exactly is the inter-relationship, structural and biochemical, between those two components. Our data where RAW macrophage podosomes were disassembled by cytochalasin D and podosome reformation was induced by removal of the cytochalasin D suggested that formation of the F-actin core precedes formation of the vinculin ring ([40] and unpublished data). It is conceivable therefore that a core structure regulated by WASP may induce protrusion of the membrane enough to overcome barriers and position or cluster activated integrins thus enabling ligand binding and subsequent outside-in signaling to organize the adhesion ring.
Integrins do not always act alone and require the concerted activity of co-receptors on the cell surface. Integrin signaling modulation, at least in osteoclast podosomes, is also achieved via the urokinase plasminogen activator receptor (uPAR; [121], a membrane anchored glycoprotein acting as a vitronectin co-receptor with αvβ3 integrin. Specifically, osteoclasts from uPAR−/− mice fail to organize a correct podosome belt with podosomes clustering in rings and peripheral aggregates [121]. uPAR also physically interacts with integrin αMβ2 in monocytes and macrophages [85, 122] and regulates integrin-mediated adhesion and extravasation, a process recently described as requiring the extension of podosomes [3].
5.1 CSF-1R
The macrophage colony-stimulating factor (M-CSF or CSF-1) plays important roles in the development, homeostasis and motility of cells of the monocytic lineage [123]. CSF-1 signals to the cell via its transmembrane tyrosine kinase receptor, CSF-1R (also known as c-Fms). As such, it is able to affect cytoskeletal dynamics of macrophages and several signaling components have been identified to act downstream of the receptor. These include SFKs, adaptor proteins such as Grb2, PI3K etc. [124]. The precise role of CSF-1R signaling in podosome dynamics is not well known. For example, in the monocyte/macrophage cell line RAW 264.7, which grows independently of CSF-1, podosome assembly occurs spontaneously [40]. However, in CSF-1 – dependent bone marrow derived macrophages (BMM), withdrawal of CSF-1 resulted in reduction of podosomes [125]. When these CSF-1 deprived cells were stimulated with CSF-1, they gradually recovered their podosomes, indicating that a signaling pathway exists between CSF-1 and podosome formation. This was found to require at least PI3K, since pretreatment of BMMs with the inhibitor LY294002 prevented podosome formation by CSF-1 [125]. It is assumed that PI3K might act in the regulation of Cdc42, since several GEFs are regulated by PtdIns(3,4,5)P3, the product of PI3K [32]. Furthermore, WASP activation in macrophages following CSF-1 stimulation requires PI3K activity [126]. Activation and autophosphrylation of CSF-1R creates several binding sites for molecules such as Src and Cbl, important in the regulation of osteoclast podosomes as discussed earlier. CSF-1R also participates in inside-out signaling that activates the αvβ3 integrin enabling engagement of extracellular matrix ligands, and involves the MEK pathway (reviewed in [127]). Additionally, CSF-1R may further influence podosome/sealing zone organization since molecules such as Pyk2, Src, PLCγ and PI3K, all with established roles in osteoclast podosome organization, are either directly or indirectly activated by CSF-1R [124].
5.2 Syndecan-4
Syndecan-4 is a transmembrane heparan sulfate proteoglycan that has been known to act co-operatively with integrins in the regulation of focal adhesions [128]. Recently, it was also identified to influence DC podosomes [129]. While syndcan-4 was found to associate with α-actinin – positive dots in DCs stimulated for 24 hours with LPS, it is not clear whether these are bona fide podosomes [130]. Syndecan-4 is mulitimerized in the plasma membrane by PtdIns(4,5)P2 and can bind and activate PKCα [131]. This association is then able to influence RhoA [132] and Rac1 [133] signaling through RhoGDI phosphorylation [134]. Treatment of DCs with lysophosphatydilcholine (LysoPC) activates PKCδ, which phosphorylates syndecan-4 and influences its interaction with PKCα. This results in increased levels of phospho-syndecan-4 and decreased podosome numbers, adhesion and motility [129].
5.3 CD44
CD44 is a ubiquitously expressed transmembrane receptor for hyaluronic acid, collagen, osteopontin and laminin [135]. It is found in the core of osteoclast podosomes that form a podosome belt [61] (Figure 1). The presence of an extracellular matrix receptor at the actin core is important as it may potentially link a nascent protrusive podosome to the ECM and stabilize it before engagement of integrins and the formation of an adhesion plaque around the F-actin ring. This is, however, an unexplored possibility. Ligands for CD44 were able to stimulate podosome formation in WIP−/−osteoclasts, suggesting that CD44 is able to signal directly to WASP. The precise mechanism for this signaling pathway from CD44 to WASP is unknown, but it could potentially involve a direct interaction between the two molecules, since CD44 has been shown to interact directly with N-WASP [136]. This is further supported by the fact that CD44 activation in WIP−/− osteoclasts could stabilize the protein levels of WASP, which are otherwise decreased due to the absence of WIP [61]. CD44 is a signaling receptor in its own right and could potentially influence GTPase activation. For example, the cytoplasmic tail of CD44 binds ERM proteins, which sequester RhoGDI and release the bound Rho family GTPase [137]. Furthermore, CD44 interacts with the Cdc42 partner IQGAP1, which inhibits the GTPase activity of Cdc42 thus prolonging its activation [138]. A feedback loop between osteopontin and CD44 in osteoclasts has been described [45], which proceeds via ROCK II and results in CD44 surface expression, however, how this affects podosomes or sealing zone formation is unknown.
6. Conclusions and perspectives
Podosomes in leukocytes have long been recognized, yet signaling cascades that regulate their formation and dynamics are only recently becoming elucidated. The central roles played by integrins, Rho GTPases and Src family or other kinases are well established, and the fact that they are required for leukocyte motility (Baruzzi 08), establishes a firm connection between podosomes and motile or invasive behavior of leukocytes. Yet how these structures are required for motility is unclear. The exact interplay between the various adhesion and chemokine receptors is also another area that begs more attention. Podosomes are dynamic and could provide a platform for sensing local changes in the concentration, composition and mechanical properties of ligands, soluble or insoluble. The relative balance of extracellular signals, sensed via their respective receptors may then allow the cell to activate specific signaling pathways and alter its adhesive or motile behavior. For example, a chemokine gradient may signal the formation of podosomes towards that gradient and the establishment of polarity, while changes in matrix composition and/or rigidity may signal for pericellular proteolysis via the podosome.
Importantly, the evidence of podosome-like structures being formed in three-dimensional environments in vitro can stimulate investigation into their presence and functions in vivo. These “3D podosomes” may not be comparable in structure with their 2D counterparts, yet they show common molecular composition with podosomes formed on two-dimensional planar substrates, e.g. talin, cortactin, and are associated with pericellular proteolysis [10]. The relevance of podosomes in vivo though may not be easy to decipher. The fact that interstitial migration of leukocytes such as DCs can occur in the absence of proteolysis or integrin receptors [139] could imply that podosomes are redundant for motility in 3D or, alternatively, it may highlight the inherent plasticity of these cells in orchestrating migratory behaviors. Nevertheless, the activity of MT1-MMP appears to be a common essential element in tissue recruitment of macrophages [140] and other leukocytes [141] under inflammatory conditions. Experimentally, therefore, the presence of podosomes in vivo may have to be selected based on other considerations, for example, when a leukocyte encounters a distinct surface and displays a distinct polarity towards that surface such as when leukocytes encounter a luminal endothelial surface or a basement membrane. This will then allow for the proper definition of the context into which signaling components may be required for migration through distinct tissues.
Acknowledgements
Work in the laboratory of Dr Dianne Cox is supported by NIH Grant GM07828. We wish to thank Dr Hava Gil-Henn (Yale University) for critical reading of this manuscript and useful suggestions.
Abbreviations
- BMM
bone marrow-derived macrophage
- DC
dendritic cell
- CIP4
Cdc42 interacting protein 4
- CSF-1(R)
colony stimulating factor-1(receptor)
- GAP
GTPase activating protein
- GDI
guanine nucleotide dissociation inhibitor
- LPS
lipopolysaccharide
- mDia
mammalian Diaphanous
- (MT1)MMP
(membrane type1) matrix metalloprotease
- PAK
p21 associated kinase
- PGE2
prostaglandin E2
- PSTPIP
proline, serine, threonine phosphatase interacting protein
- PKC
protein kinase C
- PLC
phospholipase C
- ROCK
Rho associated kinase
- WASP
Wiskott-Aldrich Syndrome protein
- WIP
WASP interacting protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Szczur K, Xu H, Atkinson S, Zheng Y, Filippi M-D. Blood. 2006;108:4205–4213. doi: 10.1182/blood-2006-03-013789. [DOI] [PubMed] [Google Scholar]
- 2.Marchisio PC, Bergui L, Corbascio GC, Cremona O, D'Urso N, Schena M, Tesio L, Caligaris-Cappio F. Blood. 1988;72:830–833. [PubMed] [Google Scholar]
- 3.Carman CV, Sage PT, Sciuto TE, de la Fuente MA, Geha RS, Ochs HD, Dvorak AM, Springer TA. Immunity. 2007;26:784–797. doi: 10.1016/j.immuni.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Destaing O, Saltel F, Geminard J-C, Jurdic P, Bard F. Mol. Biol. Cell. 2003;14:407–416. doi: 10.1091/mbc.E02-07-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, Waterman CM. Nature. 2010;468:580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linder S. Trends Cell Biol. 2007;17:107–117. doi: 10.1016/j.tcb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Gawden-Bone C, Zhou Z, King E, Prescott A, Watts C, Lucocq J. J. Cell Sci. 2010;123:1427–1437. doi: 10.1242/jcs.056515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiesner C, Faix J, Himmel M, Bentzien F, Linder S. Blood. 2010;116:1559–1569. doi: 10.1182/blood-2009-12-257089. [DOI] [PubMed] [Google Scholar]
- 9.Cougoule C, Le Cabec V, Poincloux R, Al Saati T, Tabouret G, Lowell CA, Laviolette-Marilat N, Maridonneau-Parini I. Blood. 2010;115:1444–1452. doi: 10.1182/blood-2009-04-218735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Goethem E, Poincloux R, Gauffre F, Maridonneau-Parini I, Le Cabec V. J Immunol. 2010;184:1049–1061. doi: 10.4049/jimmunol.0902223. [DOI] [PubMed] [Google Scholar]
- 11.Zicha D, Allen WE, Brickell PM, Kinnon C, Dunn GA, Jones GE, Thrasher AJ. Br. J. Haematol. 1998;101:659–665. doi: 10.1046/j.1365-2141.1998.00767.x. [DOI] [PubMed] [Google Scholar]
- 12.Badolato R, Sozzani S, Malacarne F, Bresciani S, Fiorini M, Borsatti A, Albertini A, Mantovani A, Ugazio AG, Notarangelo LD. J Immunol. 1998;161:1026–1033. [PubMed] [Google Scholar]
- 13.Perri SR, Annabi B, Galipeau J. FASEB J. 2007;21:3928–3936. doi: 10.1096/fj.07-8158com. [DOI] [PubMed] [Google Scholar]
- 14.Calle Y, Carrager NO, Thrasher AJ, Jones GE. J. Cell Sci. 2006;119:2375–2385. doi: 10.1242/jcs.02939. [DOI] [PubMed] [Google Scholar]
- 15.Burns S, Hardy SJ, Buddle J, Yong KL, Jones GE, Thrasher AJ. Cell Motil Cytoskeleton. 2004;57:118–132. doi: 10.1002/cm.10163. [DOI] [PubMed] [Google Scholar]
- 16.van Helden SFG, Krooshoop DJEB, Broers KCM, Raymakers RAP, Figdor CG, van Leeuwen FN. J. Immunol. 2006;177:1567–1574. doi: 10.4049/jimmunol.177.3.1567. [DOI] [PubMed] [Google Scholar]
- 17.van Helden SF, Oud MM, Joosten B, Peterse N, Figdor CG, van Leeuwen FN. J Cell Sci. 2008;121:1096–1106. doi: 10.1242/jcs.020289. [DOI] [PubMed] [Google Scholar]
- 18.West MA, Prescott AR, Chan KM, Zhou Z, Rose-John S, Scheller J, Watts C. J Cell Biol. 2008;182:993–1005. doi: 10.1083/jcb.200801022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nobile C, Lind M, Miro F, Chemin K, Tourret M, Occhipinti G, Dogniaux S, Amigorena S, Hivroz C. Blood. 2008;111:3579–3590. doi: 10.1182/blood-2007-08-107755. [DOI] [PubMed] [Google Scholar]
- 20.Saltel F, Destaing O, Bard F, Eichert D, Jurdic P. Mol Biol Cell. 2004;15:5231–5241. doi: 10.1091/mbc.E04-06-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luxenburg C, Geblinger D, Klein E, Anderson K, Hanein D, Geiger B, Addadi L. PLoS One. 2007;2:e179. doi: 10.1371/journal.pone.0000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akisaka T, Yoshida H, Suzuki R, Takama K. Cell Tissue Res. 2008;331:625–641. doi: 10.1007/s00441-007-0552-x. [DOI] [PubMed] [Google Scholar]
- 23.Teitelbaum SL. Am J Pathol. 2007;170:427–435. doi: 10.2353/ajpath.2007.060834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soriano P, Montgomery C, Geske R, Bradley A. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- 25.Gil-Henn H, Destaing O, Sims NA, Aoki K, Alles N, Neff L, Sanjay A, Bruzzaniti A, De Camilli P, Baron R, Schlessinger J. J Cell Biol. 2007;178:1053–1064. doi: 10.1083/jcb.200701148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calle Y, Jones GE, Jagger C, Fuller K, Blundell MP, Chow J, Chambers T, Thrasher AJ. Blood. 2004;103:3552–3561. doi: 10.1182/blood-2003-04-1259. [DOI] [PubMed] [Google Scholar]
- 27.Zou W, Kitaura H, Reeve J, Long F, Tybulewicz VL, Shattil SJ, Ginsberg MH, Ross FP, Teitelbaum SL. J Cell Biol. 2007;176:877–888. doi: 10.1083/jcb.200611083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faccio R, Teitelbaum SL, Fujikawa K, Chappel J, Zallone A, Tybulewicz VL, Ross FP, Swat W. Nat Med. 2005;11:284–290. doi: 10.1038/nm1194. [DOI] [PubMed] [Google Scholar]
- 29.Oser M, Dovas A, Cox D, Condeelis J. Eur J Cell Biol. 2011;90:181–188. doi: 10.1016/j.ejcb.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Artym VV, Matsumoto K, Mueller SC, Yamada KM. Eur J Cell Biol. 2011;90:172–180. doi: 10.1016/j.ejcb.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Albiges-Rizo C, Destaing O, Fourcade B, Planus E, Block MR. J Cell Sci. 2009;122:3037–3049. doi: 10.1242/jcs.052704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossman KL, Der CJ, Sondek J. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 33.Moon SY, Zheng Y. Trends Cell Biol. 2003;13:13–22. doi: 10.1016/s0962-8924(02)00004-1. [DOI] [PubMed] [Google Scholar]
- 34.DerMardirossian C, Bokoch GM. Trends Cell Biol. 2005;15:356–363. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 35.Dovas A, Couchman JR. Biochem J. 2005;390:1–9. doi: 10.1042/BJ20050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boulter E, Garcia-Mata R, Guilluy C, Dubash A, Rossi G, Brennwald PJ, Burridge K. Nat Cell Biol. 2010;12:477–483. doi: 10.1038/ncb2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burns S, Thrasher AJ, Blundell MP, Machesky L, Jones GE. Blood. 2001;98:1142–1149. doi: 10.1182/blood.v98.4.1142. [DOI] [PubMed] [Google Scholar]
- 38.Linder S, Nelson D, Weiss M, Aepfelbacher M. Proc. Natl. Acad. Sci. USA. 1999;96:9648–9653. doi: 10.1073/pnas.96.17.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cougoule C, Carreno S, Castandet J, Labrousse A, Astarie-Dequeker C, Poincloux R, Le Cabec V, Maridonneau-Parini I. Traffic. 2005;6:682–694. doi: 10.1111/j.1600-0854.2005.00307.x. [DOI] [PubMed] [Google Scholar]
- 40.Dovas A, Gevrey J-C, Grossi A, Park H, Abou-Kheir W, Cox D. J. Cell Sci. 2009;122:3873–3882. doi: 10.1242/jcs.051755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wheeler AP, Wells CM, Smith SD, Vega FM, Henderson RB, Tybulewicz VL, Ridley AJ. J Cell Sci. 2006;119:2749–2757. doi: 10.1242/jcs.03024. [DOI] [PubMed] [Google Scholar]
- 42.Cremasco V, Benasciutti E, Cella M, Kisseleva M, Croke M, Faccio R. PLoS One. 2010;5:e8909. doi: 10.1371/journal.pone.0008909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aktories K, Just I. Curr Top Microbiol Immunol. 2005;291:113–145. doi: 10.1007/3-540-27511-8_7. [DOI] [PubMed] [Google Scholar]
- 44.Chellaiah MA, Soga N, Swanson S, McAllister S, Alvarez U, Wang D, Dowdy SF, Hruska KA. J Biol Chem. 2000;275:11993–12002. doi: 10.1074/jbc.275.16.11993. [DOI] [PubMed] [Google Scholar]
- 45.Chellaiah MA, Biswas RS, Rittling SR, Denhardt DT, Hruska KA. J Biol Chem. 2003;278:29086–29097. doi: 10.1074/jbc.M211074200. [DOI] [PubMed] [Google Scholar]
- 46.Destaing O, Saltel F, Gilquin B, Chabadel A, Khochbin S, Ory S, Jurdic P. J Cell Sci. 2005;118:2901–2911. doi: 10.1242/jcs.02425. [DOI] [PubMed] [Google Scholar]
- 47.Svensson HG, West MA, Mollahan P, Prescott AR, Zaru R, Watts C. Eur J Immunol. 2008;38:818–828. doi: 10.1002/eji.200737331. [DOI] [PubMed] [Google Scholar]
- 48.Heckel T, Czupalla C, Expirto Santo AI, Anitei M, Arantzazu Sanchez-Fernandez M, Mosch K, Krause E, Hoflack B. Proc Natl Acad Sci U S A. 2009;106:1451–1456. doi: 10.1073/pnas.0804464106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ory S, Brazier H, Blangy A. Biol Cell. 2007;99:701–716. doi: 10.1042/BC20070058. [DOI] [PubMed] [Google Scholar]
- 50.Brazier H, Pawlak G, Vives V, Blangy A. Int J Biochem Cell Biol. 2009;41:1391–1401. doi: 10.1016/j.biocel.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 51.Ory S, Brazier H, Pawlak G, Blangy A. Eur J Cell Biol. 2008;87:469–477. doi: 10.1016/j.ejcb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 52.Gringel A, Walz D, Rosenberger G, Minden A, Kutsche K, Kopp P, Linder S. J Cell Physiol. 2006;209:568–579. doi: 10.1002/jcp.20777. [DOI] [PubMed] [Google Scholar]
- 53.Hoefen RJ, Berk BC. J Cell Sci. 2006;119:1469–1475. doi: 10.1242/jcs.02925. [DOI] [PubMed] [Google Scholar]
- 54.Takegahara N, Kang S, Nojima S, Takamatsu H, Okuno T, Kikutani H, Toyofuku T, Kumanogoh A. FASEB J. 2010;24:4782–4792. doi: 10.1096/fj.10-158212. [DOI] [PubMed] [Google Scholar]
- 55.Spurrell DR, Luckashenak NA, Minney DC, Chaplin A, Penninger JM, Liwski RS, Clements JL, West KA. J Immunol. 2009;183:310–318. doi: 10.4049/jimmunol.0802096. [DOI] [PubMed] [Google Scholar]
- 56.Burridge K, Wennerberg K. Cell. 2004;116:167–179. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- 57.Bement WM, Miller AL, von Dassow G. Bioessays. 2006;28:983–993. doi: 10.1002/bies.20477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pertz O. J Cell Sci. 2010;123:1841–1850. doi: 10.1242/jcs.064345. [DOI] [PubMed] [Google Scholar]
- 59.Abou-Kheir W, Isaac B, Yamaguchi H, Cox D. J Cell Sci. 2008;121:379–390. doi: 10.1242/jcs.010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoneda A, Multhaupt HA, Couchman JR. J Cell Biol. 2005;170:443–453. doi: 10.1083/jcb.200412043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chabadel A, Banon-Rodriguez I, Cluet D, Rudkin BB, Wehrle-Haller B, Genot E, Jurdic P, Anton IM, Saltel F. Mol. Biol. Cell. 2007;18:4899–4910. doi: 10.1091/mbc.E07-04-0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Linder S. J Cell Sci. 2009;122:3009–3013. doi: 10.1242/jcs.032631. [DOI] [PubMed] [Google Scholar]
- 63.Wells CM, Jones GE. Biochem J. 425:465–473. doi: 10.1042/BJ20091173. [DOI] [PubMed] [Google Scholar]
- 64.Dan C, Kelly A, Bernard O, Minden A. J Biol Chem. 2001;276:32115–32121. doi: 10.1074/jbc.M100871200. [DOI] [PubMed] [Google Scholar]
- 65.van Rheenen J, Condeelis J, Glogauer M. J. Cell Sci. 2009;122:305–311. doi: 10.1242/jcs.031146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thrasher AJ, Burns SO. Nat Rev Immunol. 2010;10:182–192. doi: 10.1038/nri2724. [DOI] [PubMed] [Google Scholar]
- 67.Chou H-C, Antón IM, Holt MR, Curcio C, Lanzardo S, Worth A, Burns S, Thrasher AJ, Jones GE, Calle Y. Curr. Biol. 2006;16:2337–2344. doi: 10.1016/j.cub.2006.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chitu V, Stanley ER. Trends Cell Biol. 2007;17:145–156. doi: 10.1016/j.tcb.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 69.Linder S, Hüfner K, Wintergerst U, Aepfelbacher M. J. Cell Sci. 2000;113:4165–4176. doi: 10.1242/jcs.113.23.4165. [DOI] [PubMed] [Google Scholar]
- 70.Tsuboi S, Takada H, Hara T, Mochizuki N, Funyu T, Saitoh H, Terayama Y, Yamaya K, Ohyama C, Nonoyama S, Ochs HD. J Biol Chem. 2009;284:8548–8556. doi: 10.1074/jbc.M805638200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dovas A, Cox D. Commun. Integr. Biol. 2010:3. doi: 10.4161/cib.3.2.10759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blundell MP, Bouma G, Metelo J, Worth A, Calle Y, Cowell LA, Westerberg LS, Moulding DA, Mirando S, Kinnon C, Cory GO, Jones GE, Snapper SB, Burns SO, Thrasher AJ. Proc. Nat'l. Acad. Sci. 2009;106:15738–15743. doi: 10.1073/pnas.0904346106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Torres E, Rosen MK. Mol. Cell. 2003;11:1215–1227. doi: 10.1016/s1097-2765(03)00139-4. [DOI] [PubMed] [Google Scholar]
- 74.Wu Y, Spencer SD, Lasky LA. J Biol Chem. 1998;273:5765–5770. doi: 10.1074/jbc.273.10.5765. [DOI] [PubMed] [Google Scholar]
- 75.Yeung YG, Soldera S, Stanley ER. J Biol Chem. 1998;273:30638–30642. doi: 10.1074/jbc.273.46.30638. [DOI] [PubMed] [Google Scholar]
- 76.Cong F, Spencer S, Cote JF, Wu Y, Tremblay ML, Lasky LA, Goff SP. Mol Cell. 2000;6:1413–1423. doi: 10.1016/s1097-2765(00)00138-6. [DOI] [PubMed] [Google Scholar]
- 77.Cortesio CL, Wernimont SA, Kastner DL, Cooper KM, Huttenlocher A. Arthritis Rheum. 2010;62:2556–2558. doi: 10.1002/art.27521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chellaiah MA, Kuppuswamy D, Lasky L, Linder S. J. Biol. Chem. 2007;282:10104–10116. doi: 10.1074/jbc.M608957200. [DOI] [PubMed] [Google Scholar]
- 79.Cote JF, Chung PL, Theberge JF, Halle M, Spencer S, Lasky LA, Tremblay ML. J Biol Chem. 2002;277:2973–2986. doi: 10.1074/jbc.M106428200. [DOI] [PubMed] [Google Scholar]
- 80.Badour K, Zhang J, Shi F, Leng Y, Collins M, Siminovitch KA. J. Exp. Med. 2004;199:99–111. doi: 10.1084/jem.20030976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tarone G, Cirillo D, Giancotti FG, Comoglio PM, Marchisio PC. Exp Cell Res. 1985;159:141–157. doi: 10.1016/s0014-4827(85)80044-6. [DOI] [PubMed] [Google Scholar]
- 82.Linder S, Higgs H, Hüfner K, Schwarz K, Pannicke U, Aepfelbacher M. J. Immunol. 2000;165:221–225. doi: 10.4049/jimmunol.165.1.221. [DOI] [PubMed] [Google Scholar]
- 83.Guiet R, Poincloux R, Castandet J, Marois L, Labrousse A, Le Cabec V, Maridonneau-Parini I. Eur J Cell Biol. 2008;87:527–542. doi: 10.1016/j.ejcb.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 84.Poincloux R, Vincent C, Labrousse A, Castandet J, Rigo M, Cougoule C, Bordier C, Le Cabec V, Maridonneau-Parini I. Eur J Cell Biol. 2006;85:327–332. doi: 10.1016/j.ejcb.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 85.Bohuslav J, Horejsi V, Hansmann C, Stockl J, Weidle UH, Majdic O, Bartke I, Knapp W, Stockinger H. J Exp Med. 1995;181:1381–1390. doi: 10.1084/jem.181.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ren-Hong T, Law SKA, Suet-Mien T. FEBS letters. 2006;580:4435–4442. doi: 10.1016/j.febslet.2006.06.099. [DOI] [PubMed] [Google Scholar]
- 87.Scott MP, Zappacosta F, Kim EY, Annan RS, Miller WT. J Biol Chem. 2002;277:28238–28246. doi: 10.1074/jbc.M202783200. [DOI] [PubMed] [Google Scholar]
- 88.Cory GOC, Garg R, Cramer R, Ridley AJ. J. Biol. Chem. 2002;277:45115–45121. doi: 10.1074/jbc.M203346200. [DOI] [PubMed] [Google Scholar]
- 89.Meller N, Merlot S, Guda C. J Cell Sci. 2005;118:4937–4946. doi: 10.1242/jcs.02671. [DOI] [PubMed] [Google Scholar]
- 90.English BK, Orlicek SL, Mei Z, Meals EA. J Leukoc Biol. 1997;62:859–864. doi: 10.1002/jlb.62.6.859. [DOI] [PubMed] [Google Scholar]
- 91.Vincent C, Maridonneau-Parini I, Le Clainche C, Gounon P, Labrousse A. J Biol Chem. 2007;282:19565–19574. doi: 10.1074/jbc.M701501200. [DOI] [PubMed] [Google Scholar]
- 92.Boyce BF, Yoneda T, Lowe C, Soriano P, Mundy GR. J Clin Invest. 1992;90:1622–1627. doi: 10.1172/JCI116032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Duong LT, Lakkakorpi PT, Nakamura I, Machwate M, Nagy RM, Rodan GA. J Clin Invest. 1998;102:881–892. doi: 10.1172/JCI3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Miyazaki T, Sanjay A, Neff L, Tanaka S, Horne WC, Baron R. J Biol Chem. 2004;279:17660–17666. doi: 10.1074/jbc.M311032200. [DOI] [PubMed] [Google Scholar]
- 95.Horne WC, Sanjay A, Bruzzaniti A, Baron R. Immunol Rev. 2005;208:106–125. doi: 10.1111/j.0105-2896.2005.00335.x. [DOI] [PubMed] [Google Scholar]
- 96.Lowell CA, Niwa M, Soriano P, Varmus HE. Blood. 1996;87:1780–1792. [PubMed] [Google Scholar]
- 97.Granot-Attas S, Luxenburg C, Finkelshtein E, Elson A. Mol Biol Cell. 2009;20:4324–4334. doi: 10.1091/mbc.E08-11-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Destaing O, Sanjay A, Itzstein C, Horne WC, Toomre D, De Camilli P, Baron R. Mol. Biol. Cell. 2008;19:394–404. doi: 10.1091/mbc.E07-03-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Luxenburg C, Parsons JT, Addadi L, Geiger B. J. Cell Sci. 2006;119:4878–4888. doi: 10.1242/jcs.03271. [DOI] [PubMed] [Google Scholar]
- 100.Zou W, Reeve JL, Zhao H, Ross FP, Teitelbaum SL. J Biol Chem. 2009;284:18833–18839. doi: 10.1074/jbc.M109.012385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tehrani S, Faccio R, Chandrasekar I, Ross FP, Cooper JA. Mol. Biol. Cell. 2006;17:2882–2895. doi: 10.1091/mbc.E06-03-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tehrani S, Tomasevic N, Weed S, Sakowicz R, Cooper JA. Proc. Nat'l. Acad. Sci. 2007;104:11933–11938. doi: 10.1073/pnas.0701077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tanaka S, Amling M, Neff L, Peyman A, Uhlmann E, Levy JB, Baron R. Nature. 1996;383:528–531. doi: 10.1038/383528a0. [DOI] [PubMed] [Google Scholar]
- 104.Sanjay A, Houghton A, Neff L, DiDomenico E, Bardelay C, Antoine E, Levy J, Gailit J, Bowtell D, Horne WC, Baron R. J Cell Biol. 2001;152:181–195. doi: 10.1083/jcb.152.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Meng F, Lowell CA. EMBO J. 1998;17:4391–4403. doi: 10.1093/emboj/17.15.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Caveggion E, Continolo S, Pixley FJ, Stanley ER, Bowtell DD, Lowell CA, Berton G. J Cell Physiol. 2003;195:276–289. doi: 10.1002/jcp.10236. [DOI] [PubMed] [Google Scholar]
- 107.Dale BM, Traum D, Erdjument-Bromage H, Tempst P, Greenberg S. J Immunol. 2009;182:5654–5662. doi: 10.4049/jimmunol.0803942. [DOI] [PubMed] [Google Scholar]
- 108.Purev E, Neff L, Horne WC, Baron R. Mol Biol Cell. 2009;20:4021–4030. doi: 10.1091/mbc.E09-03-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bruzzaniti A, Neff L, Sanjay A, Horne WC, De Camilli P, Baron R. Mol Biol Cell. 2005;16:3301–3313. doi: 10.1091/mbc.E04-12-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bruzzaniti A, Neff L, Sandoval A, Du L, Horne WC, Baron R. Mol Cell Biol. 2009;29:3644–3656. doi: 10.1128/MCB.00851-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Duong LT, Rodan GA. Cell Motil Cytoskeleton. 2000;47:174–188. doi: 10.1002/1097-0169(200011)47:3<174::AID-CM2>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 112.Suen PW, Ilic D, Caveggion E, Berton G, Damsky CH, Lowell CA. J Cell Sci. 1999;112(Pt 22):4067–4078. doi: 10.1242/jcs.112.22.4067. [DOI] [PubMed] [Google Scholar]
- 113.Okigaki M, Davis C, Falasca M, Harroch S, Felsenfeld DP, Sheetz MP, Schlessinger J. Proc Natl Acad Sci U S A. 2003;100:10740–10745. doi: 10.1073/pnas.1834348100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Faccio R, Novack DV, Zallone A, Ross FP, Teitelbaum SL. J Cell Biol. 2003;162:499–509. doi: 10.1083/jcb.200212082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Epple H, Cremasco V, Zhang K, Mao D, Longmore GD, Faccio R. Mol Cell Biol. 2008;28:3610–3622. doi: 10.1128/MCB.00259-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lakkakorpi PT, Bett AJ, Lipfert L, Rodan GA, Duong le T. J Biol Chem. 2003;278:11502–11512. doi: 10.1074/jbc.M206579200. [DOI] [PubMed] [Google Scholar]
- 117.Abram CL, Lowell CA. Annu Rev Immunol. 2009;27:339–362. doi: 10.1146/annurev.immunol.021908.132554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Linder S, Aepfelbacher M. Trends Cell Biol. 2003;13:376–385. doi: 10.1016/s0962-8924(03)00128-4. [DOI] [PubMed] [Google Scholar]
- 119.Faccio R, Grano M, Colucci S, Villa A, Giannelli G, Quaranta V, Zallone A. J Cell Sci. 2002;115:2919–2929. doi: 10.1242/jcs.115.14.2919. [DOI] [PubMed] [Google Scholar]
- 120.Monypenny J, Chou HC, Banon-Rodriguez I, Thrasher AJ, Anton IM, Jones GE, Calle Y. Eur J Cell Biol. 2011;90:198–204. doi: 10.1016/j.ejcb.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Furlan F, Galbiati C, Jorgensen NR, Jensen JE, Mrak E, Rubinacci A, Talotta F, Verde P, Blasi F. J Bone Miner Res. 2007;22:1387–1396. doi: 10.1359/jbmr.070516. [DOI] [PubMed] [Google Scholar]
- 122.Sitrin RG, Todd RF, 3rd, Albrecht E, Gyetko MR. J Clin Invest. 1996;97:1942–1951. doi: 10.1172/JCI118626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pollard JW. Nat. Rev. Immunol. 2009;9:259–270. doi: 10.1038/nri2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pixley FJ, Stanley ER. Trends Cell Biol. 2004;14:628–638. doi: 10.1016/j.tcb.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 125.Wheeler AP, Smith SD, Ridley AJ. Cell Motil Cytoskeleton. 2006;63:132–140. doi: 10.1002/cm.20111. [DOI] [PubMed] [Google Scholar]
- 126.Cammer M, Gevrey J-C, Lorenz M, Dovas A, Condeelis J, Cox D. J. Biol. Chem. 2009;284:23302–23311. doi: 10.1074/jbc.M109.036384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ross FP, Teitelbaum SL. Immunol Rev. 2005;208:88–105. doi: 10.1111/j.0105-2896.2005.00331.x. [DOI] [PubMed] [Google Scholar]
- 128.Couchman JR. Annu Rev Cell Dev Biol. 2010;26:89–114. doi: 10.1146/annurev-cellbio-100109-104126. [DOI] [PubMed] [Google Scholar]
- 129.Buhligen J, Himmel M, Gebhardt C, Simon JC, Ziegler W, Averbeck M. J Cell Physiol. 2010;225:905–914. doi: 10.1002/jcp.22301. [DOI] [PubMed] [Google Scholar]
- 130.Averbeck M, Gebhardt C, Anderegg U, Termeer C, Sleeman JP, Simon JC. Exp Dermatol. 2007;16:580–589. doi: 10.1111/j.1600-0625.2007.00568.x. [DOI] [PubMed] [Google Scholar]
- 131.Oh ES, Woods A, Lim ST, Theibert AW, Couchman JR. J Biol Chem. 1998;273:10624–10629. doi: 10.1074/jbc.273.17.10624. [DOI] [PubMed] [Google Scholar]
- 132.Dovas A, Yoneda A, Couchman JR. J Cell Sci. 2006;119:2837–2846. doi: 10.1242/jcs.03020. [DOI] [PubMed] [Google Scholar]
- 133.Bass MD, Roach KA, Morgan MR, Mostafavi-Pour Z, Schoen T, Muramatsu T, Mayer U, Ballestrem C, Spatz JP, Humphries MJ. J Cell Biol. 2007;177:527–538. doi: 10.1083/jcb.200610076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dovas A, Choi Y, Yoneda A, Multhaupt HA, Kwon SH, Kang D, Oh ES, Couchman JR. J Biol Chem. 2010;285:23296–23308. doi: 10.1074/jbc.M109.098129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ponta H, Sherman L, Herrlich PA. Nat Rev Mol Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 136.Bourguignon LY, Peyrollier K, Gilad E, Brightman A. J Biol Chem. 2007;282:1265–1280. doi: 10.1074/jbc.M604672200. [DOI] [PubMed] [Google Scholar]
- 137.Hirao M, Sato N, Kondo T, Yonemura S, Monden M, Sasaki T, Takai Y, Tsukita S. J Cell Biol. 1996;135:37–51. doi: 10.1083/jcb.135.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bourguignon LY, Gilad E, Rothman K, Peyrollier K. J Biol Chem. 2005;280:11961–11972. doi: 10.1074/jbc.M411985200. [DOI] [PubMed] [Google Scholar]
- 139.Lämmermann T, Bader BL, Monkley SJ, Worbs T, Wedlich-Söldner R, Hirsch K, Keller M, Förster R, Critchley DR, Fässler R, Sixt M. Nature. 2008;453:51–55. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- 140.Sakamoto T, Seiki M. Genes Cells. 2009;14:617–627. doi: 10.1111/j.1365-2443.2009.01293.x. [DOI] [PubMed] [Google Scholar]
- 141.Rowe RG, Weiss SJ. Trends Cell Biol. 2008;18:560–574. doi: 10.1016/j.tcb.2008.08.007. [DOI] [PubMed] [Google Scholar]