Abstract
N-methyl-D-aspartate (NMDA) receptors are ligand-gated ion channels activated by the neurotransmitter glutamate. These channels are highly expressed by brain neurons and are critically involved in excitatory synaptic transmission. Results from previous studies show that both native and recombinant NMDA receptors are inhibited by ethanol at concentrations associated with signs of behavioral impairment and intoxication. Given the important role that NMDA receptors play in synaptic transmission and brain function, it is important to understand the factors that regulate the ethanol inhibition of these receptors. One dynamic mechanism for regulating ethanol action may be via phosphorylation of NMDA subunits by serine-threonine and tyrosine kinases. Both NR1 and NR2 subunits contain multiple sites of phosphorylation and in the NR1 subunit, most of these are contained within the C1 domain, a carboxy-terminal cassette that is subject to alternative splicing. While results from our previous studies suggest that single phosphorylation sites do not greatly affect ethanol sensitivity of NMDA receptors, it is likely that in vivo, these subunits are phosphorylated at multiple sites by different kinases. In the present study, we constructed a series of NMDA receptor mutants at serine (S) or threonine (T) residues proposed to be sites of phosphorylation by PKA and various isoforms of PKC. Ethanol (100 mM) inhibited currents from wild-type NR1/2A and NR1/2B receptors expressed in HEK293 cells by approximately 25% and 30% respectively. This inhibition was not different in single site mutants expressing alanine (A) or aspartate/glutamate (D/E) at positions T879, S896 or T900. The mutant NR1(S890D) showed greater ethanol inhibition than NR1(890A) containing receptors although this was only observed when it was combined with the NR2A subunit. Ethanol inhibition was not altered by aspartate substitution at four serines (positions 889, 890, 896, 897) or when T879D was added to the four serine-substituted mutant. Ethanol inhibition was increased when T900E was added to the five serine/threonine substituted mutant but again this was selective for NR2A containing receptors. Together with previously published data, these findings suggest that modification of putative phosphorylation sites could contribute to the overall acute ethanol sensitivity of recombinant NMDA receptors. Supported by R37 AA009986.
Keywords: PKA, PKC, phosphorylation, electrophysiology, alcohol
Introduction
N-methyl-D-aspartate receptors are glutamate-activated ion channels and are key regulators of excitability in the brain. These proteins are composed of multiple subunits including NR1 and NR2 that contain binding sites for glycine and glutamate, respectively (Dingledine et al., 1999). A third class of NMDA proteins are NR3 subunits that can subtly modulate receptor function and also form novel glycine-activated channels when combined with NR1 (Chatterton et al., 2002; Smothers and Woodward, 2007). NMDA receptors are highly calcium-permeable and are linked via cytoskeletal scaffolding proteins to intracellular signaling pathways that mediate various forms of synaptic plasticity (Malenka and Bear, 2004). Alterations in NMDA receptor function or expression as a result of disease or genetic mutation has been suggested to contribute to various neuropathologies including glutamate-induced neuron loss, schizophrenia and drug addiction (Tzschentke and Schmidt, 2003).
Numerous studies have demonstrated that NMDA receptors are inhibited by a variety of drugs including anesthetics, volatile solvents and ethanol (Cruz et al., 2000; Lovinger et al., 1989; Ogata et al., 2006; Woodward and Gonzales, 1990). The mechanism of action of these compounds has been most extensively studied for ethanol and data from these studies suggest that inhibition is not due to direct channel block or competition with glutamate or glycine binding sites (Masood et al., 1994; Mirshahi and Woodward, 1995; Peoples and Weight, 1992). Single channel studies show that ethanol influences receptor gating (Wright et al., 1996) and recent studies using mutagenesis to probe for physical sites of action suggest that ethanol may interact with key residues in transmembrane domains that contribute to channel function (Honse et al., 2004; Ren et al., 2003; Ronald et al., 2001; Smothers and Woodward, 2006). However, it is also clear that other factors can influence the receptor’s overall sensitivity to ethanol. These include differences in NR1 and NR2 subunit makeup, intracellular signaling molecules, and extracellular magnesium (Anders et al., 2000; Jin et al., 2008; Jin and Woodward, 2006; Masood et al., 1994; Mirshahi et al., 1998 ). Previous studies from this laboratory have also investigated whether phosphorylation can affect the ethanol sensitivity of NMDA receptors. The results from these studies demonstrate that no single kinase studied to date (Src, Fyn, PKA, CaMKII) imparts a robust or global alteration in the acute ethanol sensitivity of recombinant NMDA receptors (Anders et al., 1999a; Anders et al., 1999b; Xu et al., 2008; Xu and Woodward, 2006). In this study, we extend these studies to additional residues contained in the C1 cassette of the NR1 subunit that are predicted to be phosphorylated in vivo and explore whether multiple sites may combine to confer more significant effects on ethanol inhibition.
Materials and Methods
Molecular Biology, Cell Culture and Transfection
The NMDA receptor cDNAs used in these experiments were kindly provided by Drs. S. Nakanishi (Kyoto Univ, Kyoto, Japan) and P. Seeburg (Max-Planck Institute for Medical Research, Heidelberg, Germany). Predicted sites of phosphorylation on the rat NR1 subunit were identified using computer-generated screening programs including Scansite (http://www.scansite.mit.edu), NetPhos 2.0 (http://www.cbs.dtu.dk/services/NetPhos) and GPS2.1 (http://gps.biocuckoo.org/). Site-directed mutagenesis was performed using the Quik-Change mutagenesis kit (Invitrogen, Carlsbad, CA) and mutants were confirmed by DNA sequencing. Human embryonic kidney (HEK) 293 cells were obtained from ATCC (Manassas, VA). Cells were maintained in feeder flasks containing serum-supplemented DMEM in a humidified incubator supplied with 5% CO2 and were split weekly (Xu and Woodward, 2006). For recordings, cells were plated onto poly-ornithine coated 35 mm dishes and transfected with plasmids encoding various NMDA receptor subunits (typically 1 ug each) using Lipofectamine 2000 (Invitrogen, Inc., Carlsbad, CA) according to the manufacturer’s recommendation. In each set of transfections, at least one cDNA encoded the enhanced green fluorescent protein (eGFP) allowing for detection of transfected cells. Plasmids were used at a ratio of 1:1:1 unless otherwise indicated. Following transfection, the NMDA antagonist AP5 (200 μM) was added to the media to prevent glutamate-mediated excitotoxicity (Cik et al.). AP5 was removed by extensive washing prior to recording.
Electrophysiology
Dishes containing transfected cells were mounted on the stage of an Olympus IX50 inverted microscope and perfused with extracellular recording solution at 1–2 ml/min. The recording solution contained (in mM); NaCl (135), KCl (5.4), CaCl2 (1.8), HEPES (5), glucose (10), (pH adjusted to 7.4 and osmolarity adjusted to 310–325 mOsm with sucrose). Patch pipettes (2–5 mOhms) were pulled from borosilicate glass (1.5 × 0.86 mm) and filled with internal solution containing (in mM); KCl 140, MgCl2 6, CaCl2 1, EGTA 5, HEPES 10, tetraethylammonium chloride 2, and NaATP 4, (pH adjusted to 7.2 with KOH). Transfected cells were identified by eGFP fluorescence and whole-cell voltage clamp recordings were carried out at room temperature using an Axon 200B microamplifier (Molecular Devices, Union City, CA). Cells were held at −60 mV to monitor seal breakthrough and maintained at this potential unless otherwise noted. Whole-cell capacitance and series resistance were compensated for and access resistance was monitored over the course of the experiment. Cells with unstable holding currents or significant changes in series resistance were not used for analysis. NMDA receptor currents were evoked using a Warner FastStep multi-barrel perfusion system to switch between normal extracellular solution and those containing agonist (glutamate plus glycine; both at 10 μM) or agonist plus ethanol (10–500 mM). The order of solutions was interleaved to monitor current rundown. Data were filtered at 1–2 kHz and acquired at 5 kHz using an Instrutech ITC-16 digital interface (Instrutech Corp., Port Washington, NY) controlled by IgorPro software (Wavemetrics, Lake Oswego, OR) running the Pulse control acquisition module. Data were analyzed offline using Axograph software (Axograph, Sydney, New South Wales, Australia). Agonist-evoked currents were baseline subtracted and amplitudes were measured during the last 0.5 seconds of agonist application when currents had reached steady-state levels. Ethanol inhibition was calculated using the formula (1-(IGlutamate+Etoh/IControl)) x 100, where IGlutamate+Etoh represents the response to co-application of agonist + ethanol, and IControl represents the mean of two responses to agonist, one before and one after the co-application of ethanol. Ethanol was purchased from Aaper Alcohol and Chemical Company (Shelbyville, KY) while all other chemicals were purchased from Sigma Chemical Company (St. Louis, MO).
Data Analysis
Data are expressed as mean ± SEM and were analyzed by analysis of variance or t-test (as indicated) using Prism 4.0 software (Graphpad Software, San Diego, CA). Prism software was also used to generate concentration-response curves for ethanol inhibition using unweighted least-squares nonlinear regression of log concentration values versus percent inhibition.
Results
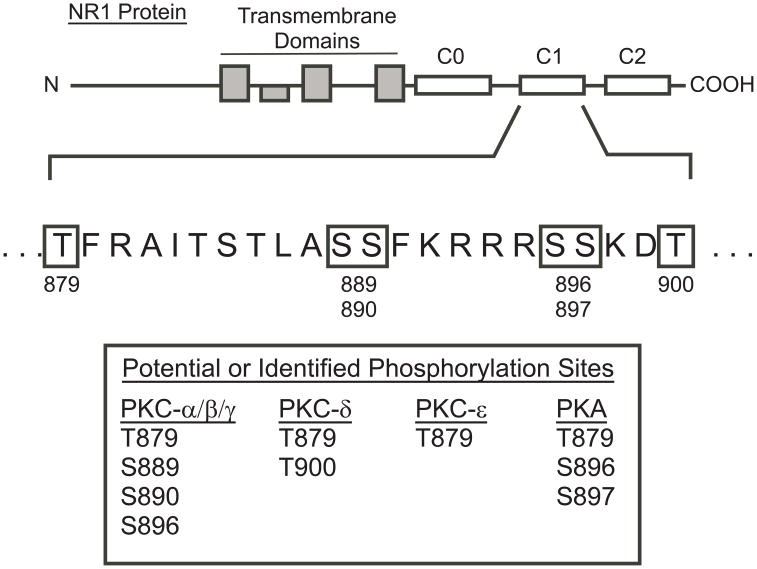
Figure 1 shows a schematic of the NR1 subunit and highlights the large N-terminal domain comprising the glycine binding site, four transmembrane domains and the intracellular C-terminal region of this protein. The NR1 C-terminus is divided into three domains with C1 and C2 cassettes being subject to alternative splicing. A previous study using both biochemical and immunological techniques (Tingley et al., 1997) identified several residues in the C1 domain that were phosphorylated by either purified cardiac PKC (T879, S889, S890, S896, S897) or brain PKA (S896, S897). Analysis of the C1 domain residues with sequence prediction software confirmed these sites and identified threonine 900 as an additional putative site of phosphorylation by PKC-δ (Figure 1, boxed residues). Results from our previous study (Xu et al., 2006) showed that treatments that enhanced or mimicked PKA phosphorylation of S897 slightly increased the degree of ethanol inhibition of recombinant NMDA receptors. To investigate whether other putative phosphorylation sites in the C1 domain also affect ethanol inhibition, a series of mutants were constructed to either mimic (aspartate/glutamate) or block (alanine) phosphorylation. We predicted that if these sites represent a major determinant of ethanol inhibition of NMDA receptors, there should be a significant difference in ethanol’s inhibition of currents generated by these phosphosite mutants.
Figure 1.
NR1 Subunit Phosphorylation Sites. The schematic diagram shows the organization of the NR1 subunit. Shaded boxes denote the transmembrane domains (note that the second transmembrane domain does not fully traverse the membrane) and open boxes show location of three discrete C-terminal domains. The amino acids shown below the diagram represent the sequence of a portion of the C1 domain with residues in boxes identified as potential phosphorylation sites for PKC or PKA. The table lists the various isoforms of PKC and PKA and their potential targets for phosphorylation within the C1 domain sequence.
NR1 mutant constructs were co-expressed with either NR2A or NR2B subunits in HEK293 cells and whole-cell patch-clamp electrophysiology was used to determine the degree of ethanol inhibition. Each pair of alanine/aspartate-glutamate mutants was tested on the same day using the same solutions along with control wild-type subunits. These experiments were then repeated 2–3 times and values from the control experiments were pooled to give an overall average. Table 1 shows the mean amplitude of peak and steady-state currents for each mutant tested and the steady-state to peak ratio that is a measure of current desensitization. All mutants showed fairly robust currents in response to 10 μM glutamate and 10 μM glycine although currents generated by NR1(S890D)/NR2B subunits were on average smaller than the others. It is not known whether this reflects a property of the NR1(890D)/NR2B combination or is just due to variability inherent in the HEK293 cell expression system since not all cells expressing this combination showed small currents. Steady-state to peak current ratios (SS/Pk) for all receptor combinations ranged between approximately 0.6 and 0.8. While there were no obvious effects of the mutations on this parameter for NR2B containing receptors, the SS/Pk ratios for the S890A and S890D mutants were significantly smaller than the value for wild-type NR1/NR2A receptors.
Table 1.
Functional Status of NMDA NR1 subunit phosphorylation site mutants.
| NR2A | NR2B | |||||
|---|---|---|---|---|---|---|
| Mutant Expressed | Peak Current, pA | Steady-state current, pA | SS:Pk ratio | Peak Current, pA | Steady-state current, pA | SS:Pk ratio |
| NR1-WT | 666.0±102.4 (31) | 492.1±70.5 (31) | 0.75±0.02 (31) | 591.7±165.5 (32) | 323.5±62.8 (32) | 0.66±0.03 (32) |
| T879A | 1574.1±453.99 (4) | 1000.6±209.5 (4) | 0.67±0.04 (4) | 398.3±138.4 (4) | 224.3±38.57 (4) | 0.65±0.09 (4) |
| T879D | 1379.5±443.6 (4) | 765.8±222.2 (4) | 0.63±0.11 (4) | 567.9±122.1 (4) | 413.8±91.3 (4) | 0.74±0.05 (4) |
| S890A | 1233.2±370.64 (7) | 677.8±227.4 (7) | 0.57±0.06** (7) | 593.4±130.2 (7) | 362.9±71.0 (7) | 0.63±0.03 (7) |
| S890D | 819.8±240.3 (7) | 425.0±86.3 (7) | 0.59±0.06* (7) | 143.9±63.45 (6) | 109.5±48.5 (6) | 0.75±0.03 (6) |
| S896A | 714.9±376.6 (7) | 387.2±205.7 (7) | 0.62±0.06 (7) | 352.6±163.5 (7) | 251.7±112.0 (7) | 0.71±0.04 (7) |
| S896D | 419.7±145.9 (6) | 209.8±68.8 (6) | 0.62±0.05 (6) | 376.8±133.7 (7) | 188.5±66.3 (7) | 0.58±0.06 (7) |
| T900A | 261.0±73.0 (8) | 211.7±61.9 (8) | 0.80±0.02 (8) | 732.0±168.2 (8) | 539.0±128.4 (8) | 0.71±0.04 (8) |
| T900E | 742.9±259.2 (12) | 589.2±212.3 (12) | 0.78±0.03 (12) | 756.5±277.4 (11) | 434.7±152.3 (11) | 0.65±0.04 (11) |
| SSDD | 455.5±99.7 (7) | 388.1±89.6 (7) | 0.83±0.03 (7) | 209.8±53.4 (7) | 135.9±16.6 (7) | 0.74±0.07 (7) |
| ATD | 1383.9±322.9 (4) | 915.0±123.9 (4) | 0.71±0.07 (4) | 384.3±185.1 (7) | 264.7±134.9 (7) | 0.70±0.04 (7) |
| ALLDE | 690.7±216.2 (7) | 487.1±152.5 (7) | 0.67±0.05 (7) | 853.0±205.9 (7) | 416.5±99.6 (7) | 0.59±0.06 (7) |
Values shown are mean peak and steady-state currents (±SEM, N) for each receptor combination.
Abbreviations: SSDD=S889D/S890D/S896D/S897D; ATD=T879D/S889D/S890D/S896D/S897D; ALLDE= T879D/S889D/S890D/S896D/S897D/T900E.
Symbol: value significantly different from control;
p<0.05,
p<0.01, Anova with Dunnett’s post-hoc comparison test.
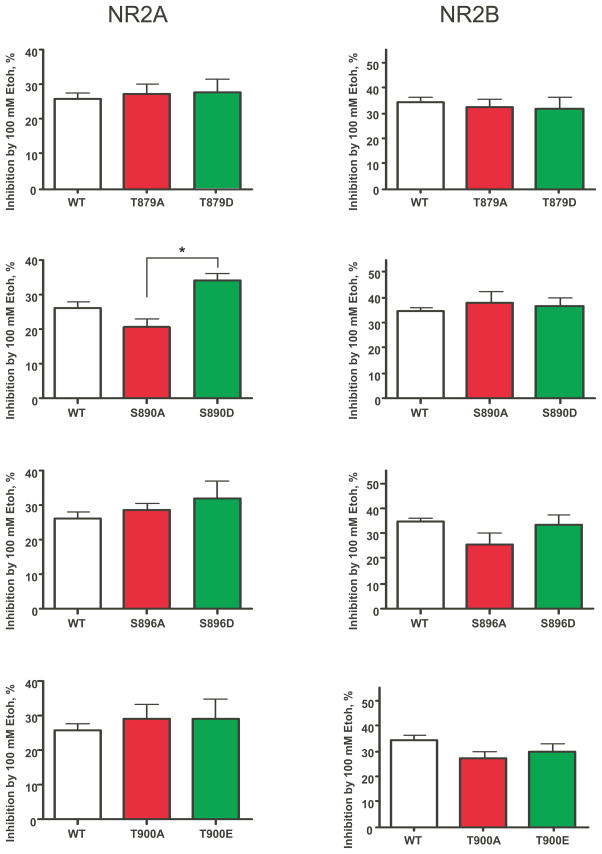
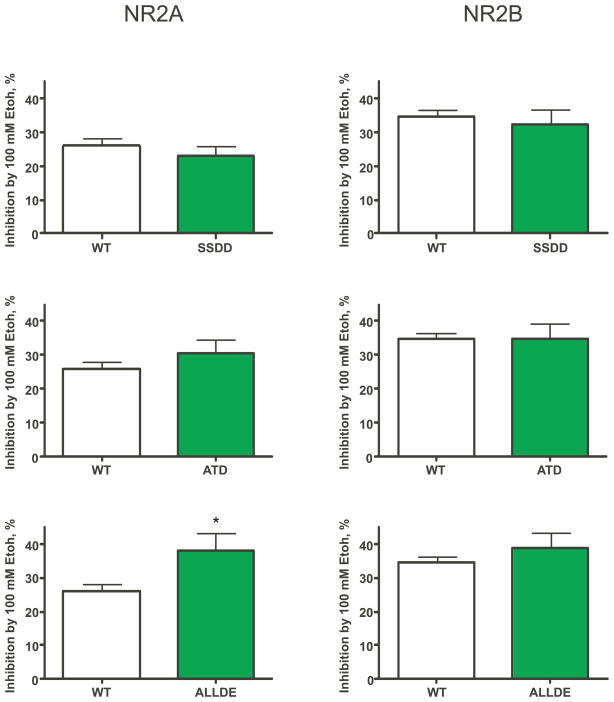
As shown in Figure 2, all single site mutants tested were signifcantly inhibited by 100 mM ethanol with NR2B containing receptors being overall slightly more affected than those with NR2A. There was a statistically significant difference in ethanol inhibition of the S890 mutants with the aspartate-substituted receptor (S890D) showing greater inhibition than alanine-substituted subunit (S890A). This difference was only observed when these NR1 mutants were co-expressed with the NR2A subunit. Results from previous studies suggest that under conditions that mimic complete dephosphorylation of the C1 cassette (eg. the NR1-2a splice variant that lacks C1 containing residues), ethanol inhibition of agonist-evoked currents is not significantly altered (Anders et al., 2000; Jin and Woodward, 2006). However, it is not known whether treatments that mimic phosphorylation of these residues alters ethanol’s effects on receptor function. To begin to address this question, we generated additional cDNA constructs carrying substitutions at multiple sites within the C1 domain of the NR1 subunit. Figure 3 shows that the ethanol inhibition of receptors with aspartate substituted at residues 889, 890, 896, and 897 (SSDD) of the NR1 subunit was not different from that of the wild-type control. Adding an additional mutation at threonine 879 (aspartate for threonine) to this construct (ATD) also did not affect ethanol inhibition. However, when the threonine 900 site was also modified to produce a mutant with six modified residues (ALLDE), 100 mM ethanol produced a small but statistically significant enhancement in ethanol inhibition in NR2A but not NR2B containing receptors.
Figure 2.
Effects of ethanol on wild-type and single-site mutant NMDA receptors. Graphs show the inhibition of glutamate-evoked currents (10 μM plus 10 μM glycine) by 100 mM ethanol for each of the mutant receptors tested. Results for alanine-substituted mutants are shown in red while those for aspartate/glutamate mutants are in green. Values represent the mean (± S.E.M.) percent inhibition by ethanol. Sample sizes for each mutant tested are shown in Table 1. Note that values for the controls for wild-type NR1/2A and NR1/2B receptors were pooled and are shown in each graph for comparison. Symbol(*): value significantly different from S890A; p<0.05, 1-way Anova with Bonferroni’s multiple comparison test.
Figure 3.
Effects of ethanol on wild-type and multiple-site mutant NMDA receptors. Graphs show the inhibition of glutamate-evoked currents (10 μM plus 10 μM glycine) by 100 mM ethanol for each of the mutant receptors tested. Values represent the mean (± S.E.M.) percent inhibition by ethanol. Sample sizes for each mutant are shown in Table 1. Note that values for the controls for NR1/2A and NR1/2B receptors were pooled and are shown in each graph for comparison. Symbol(*): value significantly different from control; p<0.05, unpaired t-test. Abbreviations: SSDD (S889D/S890D/S896D/S897D), ATD (T879D/S889D/S890D/S896D/S897D), ALLDE (T879D/S889D/S890D/S896D/S897D/T900E).
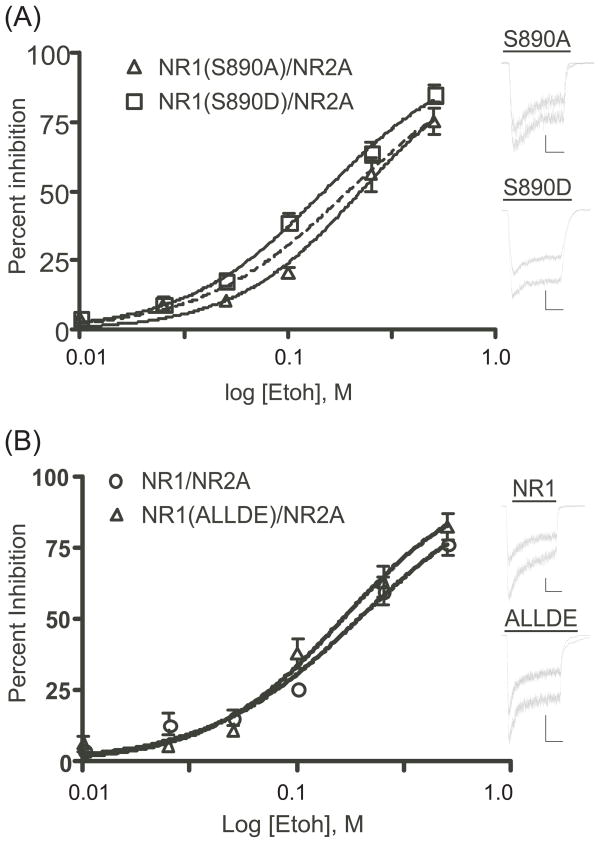
To further characterize the effects of C1 domain mutants on ethanol inhibition, the wild-type NR1 and S890A, S890D and ALLDE mutants were co-expressed with the NR2A subunit and tested with concentrations of ethanol ranging from 10–500 mM (Figure 4). Non-linear regression analysis of these data revealed an estimated IC50 value for ethanol inhibition of wild-type NR1/NR2A receptors of 193.5 mM (95% confidence intervals 153.2–244.4). The estimated IC50 values for the S890A (222.5 mM; 95% confidence intervals 183.9–269.1) and S890D (148.0 mM; 95% confidence intervals 132.2–165.7) mutants were statistically different from one another as judged by the non-overlapping confidence intervals. The ALLDE mutant showed a trend towards enhanced inhibition especially at higher ethanol concentrations but its estimated IC50 value (162.9 mM; 95% confidence interval 130.8–202.8) was not different from the control or those of the S890 mutants.
Figure 4.
Concentration-response relationship for ethanol inhibition of NR1 S890A, S890D and ALLDE mutant receptors. Graphs show the percent inhibition of glutamate-evoked currents (10 μM plus 10 μM glycine; N=6–10 cells for each concentration) by various concentrations of ethanol (10–500 mM). A) Ethanol inhibition of NR1(S890A) and NR1(S890D) mutants. The dashed line represents data for wild-type NR1/NR2A receptors (see next panel). B) Ethanol inhibition of NR1/NR2A and the NR1(ALLDE) mutant. Non-linear regression analysis resulted in calculated IC50 values for ethanol inhibition of 193.5 mM (NR1/NR2A; 95% confidence intervals 153.2–244.4), 225.5 mM (S890A; 95% confidence intervals 183.9–269.1), 148.0 mM (S890D; 95% confidence intervals 132.2–165.7), and 162.9 mM (ALLDE; 95% confidence intervals 130.8–202.8). Traces adjacent to each figure are representative examples of currents in the absence (bottom trace of each pair) and presence (top trace of each pair) of 100 mM ethanol. Currents after washout of the ethanol solution have been removed for clarity. Scale bars: X-axis (2 sec), Y-axis (S890A, 50 pA, S890D, 200 pA, NR1, 100 pA, ALLDE, 200 pA).
Discussion
In this study, we used site-directed mutagenesis to either block or mimic phosphorylation of residues within the C1 domain of the NR1 subunit that are thought to be targets for various serine/threonine kinases. Expression of these subunits in HEK293 cells in combination with either the NR2A or NR2B subunit generated glutamate-activated receptors that were all inhibited to some degree by ethanol. Of the mutants tested, both a single site mutant (S890A/D) and a multi-site mutant (ALLDE) showed changes in ethanol inhibition when expressed with the NR2A subunit. These results suggest that phosphorylation of specific residues contained within the C1 cassette of the NR1 subunit may contribute to the overall sensitivity of NMDA receptors to ethanol.
Previous studies have investigated the phosphorylation status of NR1 subunits and report that multiple residues within the C1 domain are targets of serine/threonine kinases including PKC and PKA. For example, phosphorylation of NR1 fusion proteins by purified PKC is dramatically reduced (but not eliminated) when serines at positions 889, 890, 896, and 897 are mutated to alanine (Tingley et al., 1997). Using a similar approach, these authors reported that PKA mediated phosphorylation is largely restricted to serines 896 and 897. Antibodies generated against specific phosphorylated residues of NR1 largely confirmed these findings with PKC activators increasing the intensity of the anti-phosphoserine 890 and 896 antibody signal while treatment with the PKA activator forskolin enhanced anti-phosphoserine 897 immunoreactivity (Tingley et al., 1997). Cellular imaging studies revealed that some of these sites also affect receptor clustering and surface expression. In particular, mutation of serine 890 to alanine was sufficient to prevent the re-distribution of NR1 clusters during treatment of transfected fibroblasts with PKC activators (Ehlers et al., 1995; Tingley et al., 1997). This observation is particularly interesting with respect to the present study as among the single phosphorylation sites tested, only the S890 pair of mutants showed a statistically significant difference in ethanol inhibition. It is not clear whether this difference reflects changes in receptor clustering or whether some other biophysical property that affects ethanol inhibition is involved. Clearly, this effect does not involve differences in macroscopic receptor kinetics as although both S890 mutants showed a similar reduction in SS/Pk ratios, the ethanol sensitivity of these two mutants was different. The effects of PKC activation on NMDA receptor currents are complex and appear to involve other kinases including Src family tyrosine kinases (Lu et al., 1999). In addition, PKC activators can potentiate NMDA receptor currents even in mutants lacking all known phosphorylation sites suggesting that other protein targets are involved (Liao et al., 2001; Zheng et al., 1999). Nonetheless, these results suggest that the phosphorylation status of S890 could influence the degree of ethanol inhibition of neuronal NMDA receptors.
Interestingly, the anti-phosphoserine 890 antibody has also been used to examine NMDA receptor expression in a subset of brain regions. This antibody gave strong signals on western blots from cultured cerebellar granule neurons (Sanchez-Perez and Felipo, 2005) and variable but discrete labeling of neurons within cortical and striatal regions of rat brain slices (Liu et al., 2004). In cerebellar granule neurons, the intensity of the anti-phosphoserine 890 antibody signal was reduced by inhibitors of PKC-γ or δ/θ while blockers of PKC-α/β isoforms had less effect (Sanchez-Perez and Felipo, 2005). In cortex and striata, the expression of phosphorylated S890 immunofluorescence was almost completely restricted to neurons that express the calcium binding protein parvalbumin (Liu et al., 2004). Parvalbumin positive neurons are typically GABAergic interneurons that show high-frequency patterns of firing during depolarization (Markram et al., 2004). Together with the results of the present study, these results suggest the possibility that modulation of ethanol inhibition of NMDA receptors by serine phosphorylation may be highly restricted to neurons that show strong expression of both PKC γ/δ/θ and phosphorylated NR1 serine 890.
The other receptor that displayed a change in ethanol inhibition, albeit modest, was the ALLDE mutant that contains aspartate or glutamate at 6 different positions including serines at positions 890 and 897. In a previous study, we showed that NR1 S897D mutants were also inhibited to a greater degree by ethanol than S897A mutants suggesting that in the ALLDE mutant, either S890D or S897D may confer additional ethanol inhibition. However, these aspartate substitutions were also contained in the SSDD and ATD mutants that in the present study showed no significant change in ethanol inhibition. These findings indicate that the ability of specific serine/threonine phosphorylation site mutants to enhance ethanol inhibition may be influenced by substitutions at neighboring sites.
An important caveat regarding the conclusions of the present study is that all experiments were conducted with mutants designed to block or mimic phosphorylation. While alanine substitution clearly precludes phosphorylation of that site by endogenous kinases, replacing serine/threonine residues with aspartate or glutamate can only approximate the additional negative charge associated with the phosphorylated residue. Despite this limitation, there is ample evidence from a variety of studies that phospho-site mutants produce effects that are consistent with those produced by a kinase. For example, introducing aspartate residues at S896 and S897 of the NR1 subunit enhances surface expression of this protein when it is expressed alone in HEK293 cells or neurons (Scott et al., 2003; Xia et al., 2001). This is presumably due to masking of an ER retention domain that serves as a quality control check for mis-folded or mismatched subunits. Adding the NR2 subunit also overcomes this retention signal even in non-phosphorylated (eg. alanine containing) NR1 mutants that would be retained intracellularly if expressed alone (see Table 1 in this study and Figure 1 in Xu et al, 2006). Similarly, in studies with the serotonin transporter (SERT), replacing threonine 276 with alanine had no effect of transporter function but substitution with aspartate to mimic phosphorylation enhanced 5-HT uptake similar to that observed after activation of PKG (Ramamoorthy et al., 2007). Finally, substitution of glutamate for threonine 107 in the BK potassium channel alpha subunit mimicked the effects of CaMKII phosphorylation on channel modulation and alcohol sensitivity (Liu et al., 2006). Together, the results from these studies suggest that amino acid substitution is a reasonable first step in testing the role that phosphorylation plays in regulating receptor function and ethanol sensitivity.
In summary, the findings of the present study complement those from previous reports and suggest that certain phosphorylation sites within the C1 domain of the NR1 subunit can affect the degree of ethanol inhibition of NR1/NR2 receptors. While these sites do not appear to define the major site of action for ethanol, the phosphorylation status of serines 890 and 897 in the NR1 subunit may be particularly important in determining the overall ethanol sensitivity of NMDA receptors.
Acknowledgments
This work was supported by NIH grant R37AA009986 to J.J.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- Anders DL, Blevins T, Smothers CT, Woodward JJ. Reduced ethanol inhibition of N-methyl-D-aspartate receptors by deletion of the NR1 C0 domain or overexpression of alpha-actinin-2 proteins. J Biol Chem. 2000;275:15019–15024. doi: 10.1074/jbc.275.20.15019. [DOI] [PubMed] [Google Scholar]
- Anders DL, Blevins T, Sutton G, Chandler LJ, Woodward JJ. Effects of c-Src tyrosine kinase on ethanol sensitivity of recombinant NMDA receptors expressed in HEK 293 cells. Alcohol Clin Exp Res. 1999a;23:357–362. [PubMed] [Google Scholar]
- Anders DL, Blevins T, Sutton G, Swope S, Chandler LJ, Woodward JJ. Fyn tyrosine kinase reduces the ethanol inhibition of recombinant NR1/NR2A but not NR1/NR2B NMDA receptors expressed in HEK 293 cells. J Neurochem. 1999b;72:1389–1393. doi: 10.1046/j.1471-4159.1999.721389.x. [DOI] [PubMed] [Google Scholar]
- Chatterton JE, Awobuluyi M, Premkumar LS, Takahashi H, Talantova M, Shin Y, Cui J, Tu S, Sevarino KA, Nakanishi N, et al. Excitatory glycine receptors containing the NR3 family of NMDA receptor subunits. Nature. 2002;415:793–798. doi: 10.1038/nature715. [DOI] [PubMed] [Google Scholar]
- Cik M, Chazot PL, Stephenson FA. Expression of NMDAR1–1a (N598Q)/NMDAR2A receptors results in decreased cell mortality. Eur J Pharmacol. 1994;266:R1–R3. doi: 10.1016/0922-4106(94)90146-5. [DOI] [PubMed] [Google Scholar]
- Cruz SL, Balster RL, Woodward JJ. Effects of volatile solvents on recombinant N-methyl-D-aspartate receptors expressed in Xenopus oocytes. Br J Pharmacol. 2000;131:1303–1308. doi: 10.1038/sj.bjp.0703666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- Ehlers MD, Tingley WG, Huganir RL. Regulated subcellular distribution of the NR1 subunit of the NMDA receptor. Science. 1995;269:1734–1737. doi: 10.1126/science.7569904. [DOI] [PubMed] [Google Scholar]
- Honse Y, Ren H, Lipsky RH, Peoples RW. Sites in the fourth membrane-associated domain regulate alcohol sensitivity of the NMDA receptor. Neuropharmacology. 2004;46:647–654. doi: 10.1016/j.neuropharm.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Jin C, Smothers CT, Woodward JJ. Enhanced ethanol inhibition of recombinant N-methyl-D-aspartate receptors by magnesium: role of NR3A subunits. Alcohol Clin Exp Res. 2008;32:1059–1066. doi: 10.1111/j.1530-0277.2008.00667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Woodward JJ. Effects of 8 different NR1 splice variants on the ethanol inhibition of recombinant NMDA receptors. Alcohol Clin Exp Res. 2006;30:673–679. doi: 10.1111/j.1530-0277.2006.00079.x. [DOI] [PubMed] [Google Scholar]
- Liao GY, Wagner DA, Hsu MH, Leonard JP. Evidence for direct protein kinase-C mediated modulation of N-methyl-D-aspartate receptor current. Mol Pharmacol. 2001;59:960–964. doi: 10.1124/mol.59.5.960. [DOI] [PubMed] [Google Scholar]
- Liu J, Asuncion-Chin M, Liu P, Dopico AM. CaM kinase II phosphorylation of slo Thr107 regulates activity and ethanol responses of BK channels. Nat Neurosci. 2006;9:41–49. doi: 10.1038/nn1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Mao L, Parelkar NK, Tang Q, Samdani S, Wang JQ. Distinct expression of phosphorylated N-methyl-D-aspartate receptor NR1 subunits by projection neurons and interneurons in the striatum of normal and amphetamine-treated rats. J Comp Neurol. 2004;474:393–406. doi: 10.1002/cne.20136. [DOI] [PubMed] [Google Scholar]
- Lovinger DM, White G, Weight FF. Ethanol inhibits NMDA-activated ion current in hippocampal neurons. Science. 1989;243:1721–1724. doi: 10.1126/science.2467382. [DOI] [PubMed] [Google Scholar]
- Lu WY, Xiong ZG, Lei S, Orser BA, Dudek E, Browning MD, MacDonald JF. G-protein-coupled receptors act via protein kinase C and Src to regulate NMDA receptors. Nat Neurosci. 1999;2:331–338. doi: 10.1038/7243. [DOI] [PubMed] [Google Scholar]
- Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- Masood K, Wu C, Brauneis U, Weight FF. Differential ethanol sensitivity of recombinant N-methyl-D-aspartate receptor subunits. Mol Pharmacol. 1994;45:324–329. [PubMed] [Google Scholar]
- Mirshahi T, Anders DL, Ronald KM, Woodward JJ. Intracellular calcium enhances the ethanol sensitivity of NMDA receptors through an interaction with the C0 domain of the NR1 subunit. J Neurochem. 1998;71:1095–1107. doi: 10.1046/j.1471-4159.1998.71031095.x. [DOI] [PubMed] [Google Scholar]
- Mirshahi T, Woodward JJ. Ethanol sensitivity of heteromeric NMDA receptors: Effects of subunit assembly, glycine and NMDAR1 Mg2+-insensitive mutants. Neuropharmacology. 1995;34:347–355. doi: 10.1016/0028-3908(94)00155-l. [DOI] [PubMed] [Google Scholar]
- Ogata J, Shiraishi M, Namba T, Smothers CT, Woodward JJ, Harris RA. Effects of anesthetics on mutant N-methyl-D-aspartate receptors expressed in Xenopus oocytes. J Pharmacol Exp Ther. 2006;318:434–443. doi: 10.1124/jpet.106.101691. [DOI] [PubMed] [Google Scholar]
- Peoples RW, Weight FF. Ethanol inhibition of N-methyl-D-aspartate-activated ion current in rat hippocampal neurons is not competitive with glycine. Brain Res. 1992;571:342–344. doi: 10.1016/0006-8993(92)90674-x. [DOI] [PubMed] [Google Scholar]
- Ramamoorthy S, Samuvel DJ, Buck ER, Rudnick G, Jayanthi LD. Phosphorylation of threonine residue 276 is required for acute regulation of serotonin transporter by cyclic GMP. J Biol Chem. 2007;282:11639–11647. doi: 10.1074/jbc.M611353200. [DOI] [PubMed] [Google Scholar]
- Ren H, Honse Y, Peoples RW. A site of alcohol action in the fourth membrane-associated domain of the N-methyl-D-aspartate receptor. J Biol Chem. 2003;278:48815–48820. doi: 10.1074/jbc.M302097200. [DOI] [PubMed] [Google Scholar]
- Ronald KM, Mirshahi T, Woodward JJ. Ethanol inhibition of N-methyl-D-aspartate receptors is reduced by site-directed mutagenesis of a transmembrane domain phenylalanine residue. J Biol Chem. 2001;276:44729–44735. doi: 10.1074/jbc.M102800200. [DOI] [PubMed] [Google Scholar]
- Sanchez-Perez AM, Felipo V. Serines 890 and 896 of the NMDA receptor subunit NR1 are differentially phosphorylated by protein kinase C isoforms. Neurochem Int. 2005;47:84–91. doi: 10.1016/j.neuint.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Scott DB, Blanpied TA, Ehlers MD. Coordinated PKA and PKC phosphorylation suppresses RXR-mediated ER retention and regulates the surface delivery of NMDA receptors. Neuropharmacology. 2003;45:755–767. doi: 10.1016/s0028-3908(03)00250-8. [DOI] [PubMed] [Google Scholar]
- Smothers CT, Woodward JJ. Effects of amino acid substitutions in transmembrane domains of the NR1 subunit on the ethanol inhibition of recombinant N-methyl-D-aspartate receptors. Alcohol Clin Exp Res. 2006;30:523–530. doi: 10.1111/j.1530-0277.2006.00058.x. [DOI] [PubMed] [Google Scholar]
- Smothers CT, Woodward JJ. Pharmacological characterization of glycine-activated currents in HEK 293 cells expressing N-methyl-D-aspartate NR1 and NR3 subunits. J Pharmacol Exp Ther. 2007;322:739–748. doi: 10.1124/jpet.107.123836. [DOI] [PubMed] [Google Scholar]
- Tingley WG, Ehlers MD, Kameyama K, Doherty C, Ptak JB, Riley CT, Huganir RL. Characterization of protein kinase A and protein kinase C phosphorylation of the N-methyl-D-aspartate receptor NR1 subunit using phosphorylation site-specific antibodies. J Biol Chem. 1997;272:5157–5166. doi: 10.1074/jbc.272.8.5157. [DOI] [PubMed] [Google Scholar]
- Tzschentke TM, Schmidt WJ. Glutamatergic mechanisms in addiction. Mol Psychiatry. 2003;8:373–382. doi: 10.1038/sj.mp.4001269. [DOI] [PubMed] [Google Scholar]
- Woodward JJ, Gonzales RA. Ethanol inhibition of N-methyl-D-aspartate-stimulated endogenous dopamine release from rat striatal slices: Reversal by glycine. J Neurochem. 1990;54:712–715. doi: 10.1111/j.1471-4159.1990.tb01931.x. [DOI] [PubMed] [Google Scholar]
- Wright JM, Peoples RW, Weight FF. Single-channel and whole-cell analysis of ethanol inhibition of NMDA-activated currents in cultured mouse cortical and hippocampal neurons. Brain Res. 1996;738:249–256. doi: 10.1016/s0006-8993(96)00780-9. [DOI] [PubMed] [Google Scholar]
- Xia H, Hornby ZD, Malenka RC. An ER retention signal explains differences in surface expression of NMDA and AMPA receptor subunits. Neuropharmacology. 2001;41:714–723. doi: 10.1016/s0028-3908(01)00103-4. [DOI] [PubMed] [Google Scholar]
- Xu M, Chandler LJ, Woodward JJ. Ethanol inhibition of recombinant NMDA receptors is not altered by coexpression of CaMKII-alpha or CaMKII-beta. Alcohol. 2008;42:425–432. doi: 10.1016/j.alcohol.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Woodward JJ. Ethanol inhibition of NMDA receptors under conditions of altered protein kinase A activity. J Neurochem. 2006;96:1760–1767. doi: 10.1111/j.1471-4159.2006.03703.x. [DOI] [PubMed] [Google Scholar]
- Zheng X, Zhang L, Wang AP, Bennett MV, Zukin RS. Protein kinase C potentiation of N-methyl-D-aspartate receptor activity is not mediated by phosphorylation of N-methyl-D-aspartate receptor subunits. Proc Natl Acad Sci U S A. 1999;96:15262–15267. doi: 10.1073/pnas.96.26.15262. [DOI] [PMC free article] [PubMed] [Google Scholar]