Abstract
In studies of somatic cell nuclear transfer (SCNT), the ability of factors within the oocyte to epigenetically reprogram transferred nuclei is essential for clone embryonic development to proceed. However, irregular patterns of X-chromosome inactivation, abnormal expression of imprinted genes and genomic DNA hypermethylation are frequently observed in reconstructed embryos suggesting abnormalities in this process. To better understand the epigenetic events underlying SCNT reprogramming, we sought to determine whether the abnormal DNA methylation levels observed in cloned embryos result from a failure of the oocyte to properly reprogram transcription versus differential biochemical regulation of the DNA methyltransferase family of enzymes (DNMTs) between embryonic and somatic nuclei. To address this question, we conducted real time quantitation of Dnmt transcripts in bovine preimplantation embryos generated though in vitro fertilization (IVF), parthenogentic activation and SCNT. By the 8-Cell stage, transcripts encoding Dnmt1 become significantly down-regulated in cloned embryos; likely in response to the state of genomic hypermethylation, while the de novo methyltranserases maintain an expression pattern indistinguishable from their IVF and parthenote counterparts. Depletion of embryonic / maternal Dnmt1 transcripts within IVF embryos using short-interfering RNAs, while able to lower genomic DNA methylation levels, resulted in developmental arrest at the 8/16-cell stage. In contrast, SCNT embryos derived from a stable, Dnmt1-depleted donor cell line develop to blastocyst stage but failed to carry to term. Our results indicate an essential role for Dnmt1 during bovine preimplantation development and suggest proper transcriptional reprogramming of this gene family in SCNT embryos.
Keywords: Epigenetics, DNA Methylation, Somatic Cell Nuclear Transfer, DNA Methyltransferase, Preimplantation Development
Introduction
Reprogramming of a somatic cell nucleus into a pluipotent or embryonic stem cell like state provides means to develop patient specific cells to be used in transplantation therapy (Jaenisch, 2002; Müller and Lengerke, 2009; Reik et al., 2001; Takahashi, 2010; Todd, 2009). Animals produced through somatic cell nuclear transfer (SCNT) represent the extreme of this reprogramming process wherein all tissues are derived from a single reprogrammed cell (Meissner and Jaenisch, 2006). Embryos produced through nuclear transfer display frequent developmental and metabolic abnormalities and have extremely low survival rates (Campbell et al., 2005). Studies of SCNT frequently cite a failure of the oocyte to properly epigenetically reprogram the donor nucleus as the sole cause of developmental failure (Bestor, 1998; Blelloch et al., 2006; Eilertsen et al., 2007; Reik et al., 2001). In support of this, cloned embryos exhibit a wide variety of epigenetic abnormalities including, altered patterns of X-chromosome inactivation, imprinted gene expression as well as unusually high levels of genomic DNA methylation, suggesting that the epigenome is not correctly established (Bourc'his et al., 2001; Dean et al., 2001; Kang et al., 2001; Kang et al., 2002; Santos et al., 2002; Santos et al., 2003; Xue et al., 2002). Understanding the consequences of nuclear reprogramming on the epigenome is key to proving the safety and stability of the cellular reprogramming process.
A family of structurally related proteins termed DNA (cytosine - 5) methyl-transferases (DNMTs) have been identified which catalyze the production and modulate the dynamics of mammalian patterns of global genomic DNA methylation (Bestor, 2000). DNMT1 is the most abundantly expressed methlytransferase and is thought to be largely responsible for maintaining methylation patterns through DNA replication; although it does exhibit de novo methylation when over-expressed (Leonhardt et al., 1992). DNMT3a and DNMT3b are both de novo methyltransferases, and act to transfer methyl groups to previously unmethylated CpG dinucleotides within the genome (Okano et al., 1999; Xie et al., 1999). Collectively these enzymes are the key modulators of DNA methylation and failed regulation could potentially lead to the observed hypermethylation and the aberrant patterns of X-chromosome inactivation frequently seen in animals produced using SCNT.
Immediately following fertilization, genomic DNA methylation patterns are partially erased (in a species dependent manner) and the epigenetic status of the zygotic genome reset to a totipotent reprogrammable state. As development proceeds the epigenome is programmed to direct development and differentiation of the embryo (Reik et al., 2001; Santos et al., 2002; Santos et al., 2003). In contrast, during preimplantation development of an SCNT embryo, genomic DNA methylation patterns are not fully erased and in fact increase as development proceeds (Kang et al., 2001; Kang et al., 2002). Accordingly, previous studies have demonstrated that preemptive reduction of methylation levels within the donor cell line increase the ability of reconstructed embryos to both develop to the blastocyst stage and produce competent embryonic stem cells (Eilertsen et al., 2007; Blelloch et al., 2006). Thus reducing SCNT embryo genomic DNA methylation levels may be a key step in improving the efficiency of SCNT. However, it is still not clear whether this hypermethylation phenomenon arises due differences between the abilities of the embryonic pronucleus and transferred somatic nuclei to regulate the biochemical activity of the DNMT family of enzymes, or is in fact a consequence of an inability of the ooplasm to direct their proper transcription.
Here, we utilized quantitative reverse transcription polymerase chain reaction (qRT-PCR) to measure Dnmt expression during IVF, parthenote and SCNT embryo development. Our results indicate that within cloned embryos, the Dnmt family is not over-expressed above and beyond the levels observed in IVF or parthenote controls, implicating inappropriate translational control, protein trafficking or enzymatic function as the basis for clone genomic hypermethylation. In attempting to develop protocols to reduce genomic methylation levels in early embryos using short interfering RNAs (siRNAs), we observed injection of Dnmt1 targeting siRNAs into IVF and parthenote embryos resulted in developmental arrest at the 8-16 cell stage. In contrast, SCNT embryos derived from a stable, Dnmt1-depleted donor cell line exhibited development rates indistinguishable from the control. Collectively our results indicate Dnmt transcription is properly programmed in preimplantation clone embryos and that epigenetic programming by maternal Dnmt1 is essential for bovine preimplantation development.
Results
Real Time Analysis of Dnmt expression in Nuclear Donor Cell Lines
Selection of cell type, age and even specific passage to be used as the nuclear donor can have a profound effect upon clone embryo development rates. Previous studies have postulated that DNA methylation may act as a mechanism in monitoring cellular aging wherein older cells accumulate higher levels of genomic methylation (Lopatina et al., 2002; Young and Smith, 2001). In support of this hypothesis, fetal cell lines generally display reduced genomic methylation levels as compared to adult lines and frequently yield greater development rates when used as nuclear donors (Monk et al., 1987; Hill et al., 2000). Likewise, bovine cloned embryo production rates drop as cell passage number increases (Liu et al., 2001). If increasing levels of DNA methylation correlate with a concomitant increase in enzyme expression, cellular age may have a significant impact upon the capacity of the ooplasm to reprogram Dnmt transcription. In order to investigate the possibility that Dnmt expression levels increase with cellular age, the wild type, bovine fibroblast cell line used as nuclear donors within the SCNT experiments reported in this study, were cultured as far as the cells would passage before senescence. At several key passages, RNA was isolated from a subset of cells and the remainder stained to examine morphology.
As cells passaged, their morphology shifted from the defined spindle shape characteristic of fibroblast cells to a more oblong spread out cell body. To measure Dnmt mRNA levels we employed qRT-PCR and normalized our results to the geometric mean of three independent housekeeping genes (Goossens et al., 2005). When Dnmt transcript levels were measured we were surprised to observe a steady state over the time course with the exception of Dnmt3b which showed a significant down-regulation (Fig. 1A). While some minor differences in Dnmt1 and Dnmt3a could be detected they were not as pronounced as the down-regulation of Dnmt3b. To determine if the observed down-regulation of Dnmt3b towards senescence was unique to the donor fibroblast cell line or is potentially a common phenomenon, we carried out similar analyses in cultured bovine cumulus cells which have also been used as nuclear donors in SCNT (Shin et al., 2002; Akshey et al., 2010). As with the donor fibroblasts the cumulus cells demonstrated a consistent level of Dnmt1 and Dnmt3a expression over the experimental time course whereas Dnmt3b again exhibited a marked reduction in transcript levels by passage ten (Fig. 1B). Whether Dnmt3b down-regulation occurs specifically as a mechanism inherent to the process of cellular senescence is unknown. However, our data do not indicate that Dnmt levels increase with cellular age.
Figure 1.
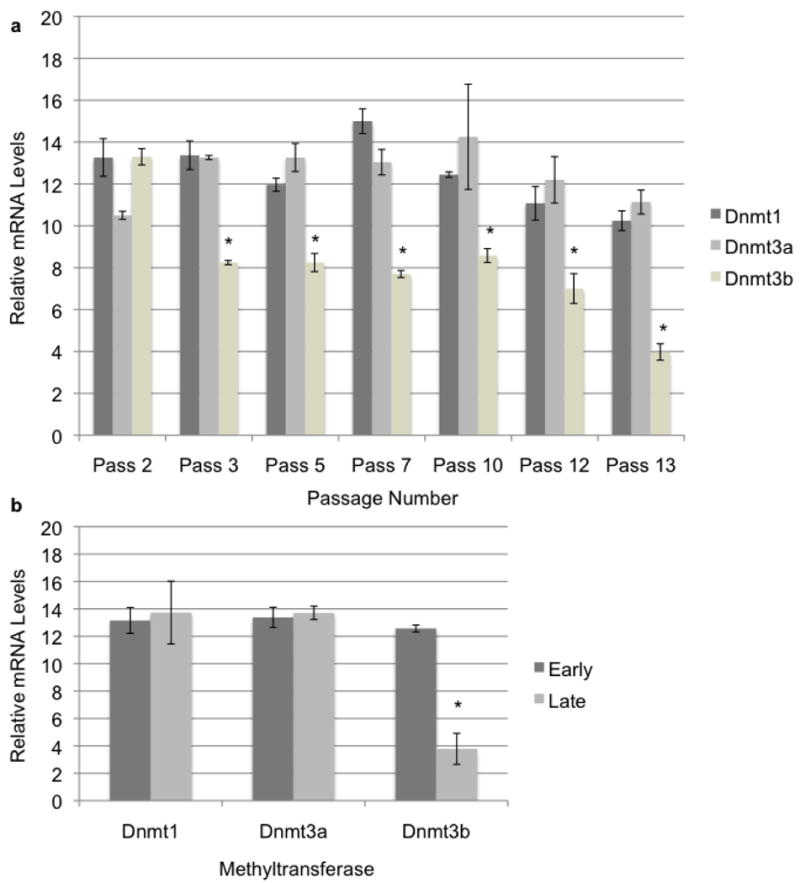
DNA Methyltransferase expression in aging donor cell lines. a) Dnmt expression in aging primary donor fibroblasts. Primary donor fibroblasts used as nuclear donors were cultured to senescence and RNA collected at each passage was analyzed for methyltransferase expression using quantitative RT-PCR. Error bars represent the SEM and * denote statistical significance P<0.01. b) Dnmt expression in aging cumulus cells. Primary cumulus cells were obtained from isolated cumulus-oocyte complexes and cultured to senescence. Measurements of transcripts encoding bovine Dnmts were measured using q-RT-PCR. Samples were normalized to the geometric mean of GAPDH, YWHAZ & SDHA. Error bars represent the SEM and * denote statistical significance P<0.01.
Quantitative Analysis of Bovine Dnmt Transcripts In Preimplantation Embryos Produced by IVF, Nuclear Transfer and Parthenogenetic Activation
In order to more accurately assess Dnmt trancription levels during preimplantation development of cloned embryos, qRT-PCR analysis was undertaken. In these experiments, comparisons were made between embryos produced via IVF, somatic cell nuclear transfer and parthenogenetic activation. Embryos activated parthenogenetically contain two female compliments of the genome, can develop as far as the second trimester of pregnancy but lack the ability to produce live offspring due to abnormal expression of imprinted genes (McGrath and Solter, 1984; McGrath and Solter, 1984). We hypothesized that examination of Dnmt expression profiles in embryos produced using these diverse methods may reveal specific differences imparted either by solely containing a female genome, or by containing a nucleus derived from a somatic cell. Embryos were produced using methods and a wild type bovine cell line described previously and development rates were in line with previously published studies from this laboratory (De Sousa et al., 1999; Golding and Westhusin, 2003; Hill et al., 2000; Winger et al., 2000).
A total of 160 embryos per experimental treatment were collected and RNA isolated from pools of 10 two-cell stage, 20 eight-cell stage and 10 blastocyst stage embryos. Approximately 50 ng of total RNA for each pool was seeded into two independent reactions measuring the panel of reference genes as well as an individual methyltransferase (Goossens et al., 2005). We postulated examination of these smaller pools individually may more accurately reveal minor differences between the experimental groups. In total, 4 independent measurements from each experimental group were taken at each of the 2-cell, 8-cell and blastocyst stages. Given the large disparity between different Dnmt transcript levels when graphed on a exponential scale, we have represented the data from this analysis as normalized delta C(T) values instead of relative expression levels (Fig. 2).
Figure 2.
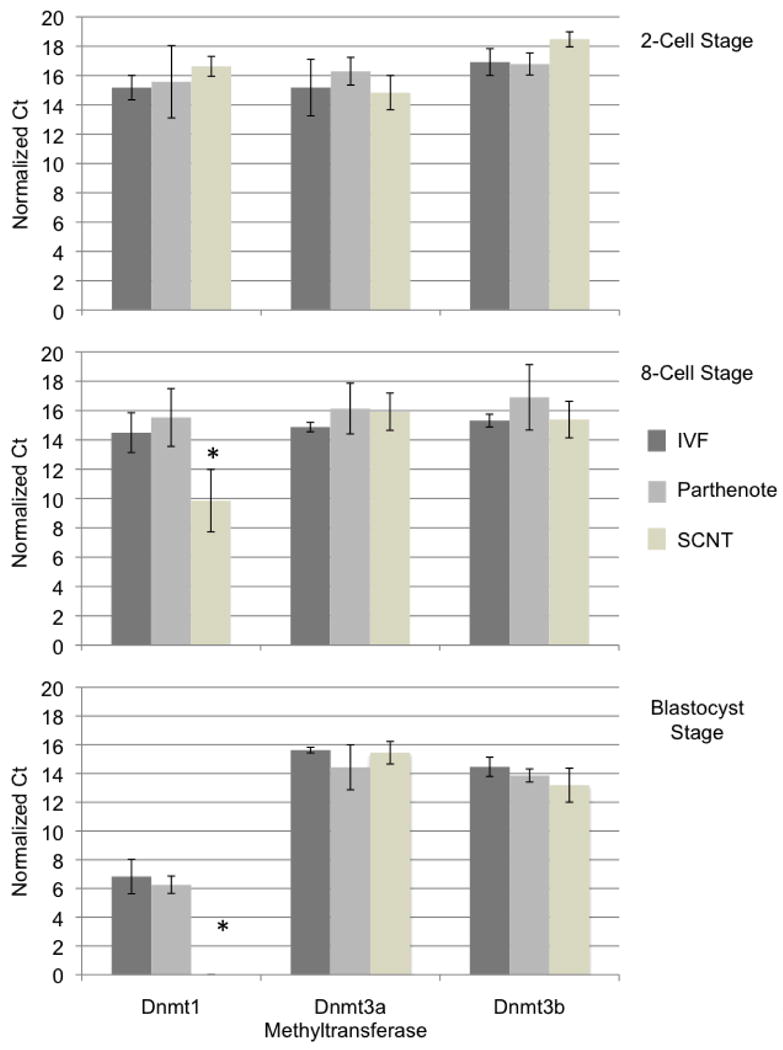
DNA Methyltransferase expression in two-cell, eight-cell and Blastocyst stage embryos produced through in vitro fertilization (IVF), parthenogenetic activation and somatic cell nuclear transfer (SCNT). Embryos produced using the described techniques were cultured to the indicated developmental stage, collected in pools of 10 two-cell stage, 20 eight-cell stage and 10 blastocyst stage embryos. RNA was isolated, and seeded into qRT-PCR reactions measuring levels of each of the Dnmts. Data was normalized to the geometric mean of GAPDH, YWHAZ & SDHA and values graphed as a normalized ΔCT. Error bars represent the SEM and * denote statistical significance of at least P<0.01.
During the two-cell stage, no differences in expression of any of the Dnmt transcripts can be discerned (Fig. 2A; P>0.05). Embryonic genome activation occurs at the eight cell stage during bovine embryonic development and thus consistent Dnmt measurements at the two-cell stage are likely reflective of maternal transcripts. However, during the eight-cell stage, a significant reduction in the expression of Dnmt1 in NT as compared to IVF embryos was clearly observed (Fig. 2B; P<0.01). A similar reduction in the expression of Dnmt1 between parthenotes and NT embryos also exists (P<0.0001) whereas the expression of neither Dnmt3a nor Dnmt3b was statistically different during the 8-cell stage between all experimental groups examined.
In embryos reconstructed using somatic cell nuclear transfer, Dnmt1 exhibits a dramatically different expression pattern during the blastocyst stage of development (Fig. 2C). Transcript levels of Dnmt1 in NT embryos as compared to both IVF and parthenote development were lower (P<0.001). Again, expression of Dnmt3a and Dnmt3b measured at the blastocyst stage was not different in any of the experimental groups. Taken together these results suggest that subsequent to embryonic genome activation the transcriptional activity of Dnmt3a and 3b is properly reprogrammed where as Dnmt1 transcription is suppressed, likely as a response to the levels of hypermethylation previously reported in NT embryos and their potential impact upon methylation responsive control elements within the Dnmt1 promoter (Kang et al., 2001; Kang et al., 2002; Dean et al., 2001; Bigey et al., 2000; Slack et al., 1999; Slack et al., 2001).
RNAi Depletion of Dnmt1
Regardless of the biochemical origin, clone embryo genomic hypermethylation represents a substantive block to SCNT reprogramming and thus clone embryonic development. Previous studies have tested a variety of genetic, biochemical and pharmacological strategies to induce genomic demethylation both in donor cell lines and reconstructed embryos (Simonsson and Gurdon, 2004; Eilertsen et al., 2007; Blelloch et al., 2006; Enright et al., 2003). However, no study has yet attempted to mimic the process of passive DNA demethylation during preimplantation development by specifically blocking Dnmt1 expression. Therefore, we opted to deplete Dnmt1 transcripts within preimplantation stage embryos using RNA interference (RNAi). We hypothesized that reduced maternal Dnmt1 transcript levels would result in passive demethylation of the transferred genome and improve SCNT development rates. To develop protocols in order to test this hypothesis, we began by injecting siRNAs targeting Dnmt1 into either bovine IVF zygotes at the one-cell stage or ova which were subsequently parthenogentically activated and cultured in vitro. The siRNAs utilized targeted both both of the reported isoforms for bovine Dnmt1 (Dnmt1a and Dnmt1b){Russell 2008}. As a control, Dnmt1-siRNA injected embryos were compared to non-injected controls and oocytes injected with a fluorescently labelled scrambled siRNA. To validate siRNA mediated depletion of Dnmt1 transcripts, 8-cell stage embryos were collected, RNA isolated and analyzed using qRT-PCR. Injection of Dnmt1 targeting siRNAs produced a significant reduction in target transcripts (<0.0001) as compared to uninjected and scrambled siRNA injected controls (Fig. 3A). Unexpectedly, the injection process produced a significant (P<0.05) increase in Dnmt1 transcript levels in the scrambled siRNA injected group as compared to uninjected controls, likely as a stress response. Collectively, these results indicate our siRNA treatment is capable of over a 95% reduction in bovine Dnmt1 transcripts. To examine potential compensatory effects of Dnmt1 depletion by Dnmt3a and Dnmt3b, 8-cell stage embryos were again collected and analyzed by qRT-PCR. siRNA mediated depletion of Dnmt1 had no observable effect on Dnmt3a/3b transcript levels, although again the injection process produced a measurable increase in Dnmt3b levels in both siRNA injected groups as compared to uninjected controls (Fig. 3B).
Figure 3.
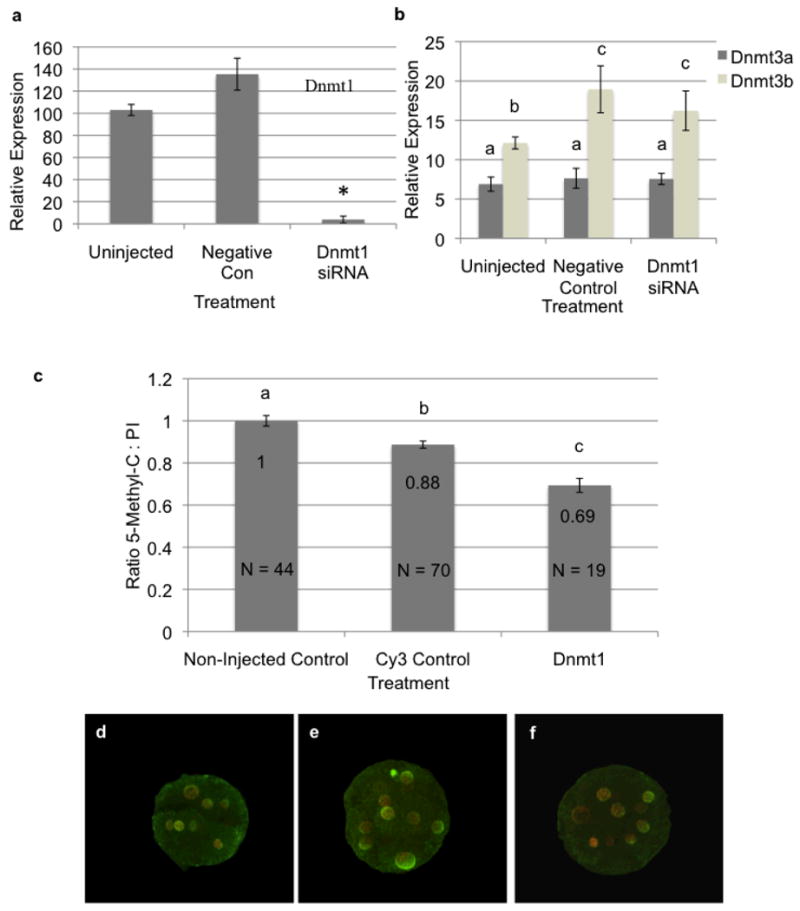
Depletion of maternal transcripts encoding Dnmt1. a) Depletion of Dnmt1 in control and siRNA injected eight-cell stage embryos was measured using qRT-PCR. Error bars represent the SEM and * denote statistical significance P<0.0001. b) Measurements of transcripts encoding Dnmt3a and Dnmt3b in non-injected and siRNA injected eight cell stage embryos using qRT-PCR. Error bars represent the SEM and letters denote statistical significance P<0.01. All q_RT-PCR measurements were normalized to the geometric mean of GAPDH, YWHAZ & SDHA. c) Reduction of genomic methylation in Dnmt1-siRNA injected embryos. Non-injected, control and Dnmt1-siRNA injected embryos were fixed and stained using an antibody recognizing methylated cytosine. 5MC staining was normalized using propidium iodine and graphed relative to non-injected controls. Error bars represent the SEM and letters denote statistical significance P<0.05. Raw numbers used in this analysis are available in supplementary figure 1a. d-f) Confocal images of eight-cell stage non-injected (d), control (e) and Dnmt1-siRNA (f) injected embryos double-stained with the 5MC antibody (green signal) and propidium iodine (red signal).
To evaluate the impact of siRNA mediated depletion of Dnmt1 on embryonic DNA methylation levels, 8-cell stage parthenote embryos were isolated, fixed and assayed using immunocytochemistry as described previously (Dean et al., 2001). Dnmt1-siRNA injected parthenotes displayed a 40% reduction in methylation levels as compared to non-injected controls (Fig. 3C & D). This difference was significant at P<0.05. Interestingly, control siRNA injected embryos displayed a 10% reduction in 5M-C staining suggesting that the micromanipulation / injection procedure itself can influence DNA methylation levels (P<0.05). However, the reduction observed in controls was minimal as compared to the 40% reduction seen with the Dnmt1-siRNA. Previous studies in mice have demonstrated that Dnmt1 is not essential for development to the blastocyst stage (Li et al., 1992). Surprisingly, while non-injected and control siRNA parthenote and IVF bovine embryos consistently yielded 37% and 26-30% blastocyst rates, embryos injected with the Dnmt1-siRNA displayed developmental arrest at the 8-16 cell stage (Table 1). In experiments utilizing parthenote embryos, a small number of very poor quality blastocysts developed while no Dnmt1-siRNA injected IVF blastocysts were produced. This unexpected reduction in embryo survival was consistently reproducible and significant (Chi Square Analysis P<0.05). Recently, another group reported developmental arrest of ovine embryos at the morula stage when Dnmt1 transcripts were targeted using siRNAs (Taylor et al., 2009).
TABLE 1.
Depletion of maternal Dnmt1 results in developmental arrest.
| Parthenote | |||||
|---|---|---|---|---|---|
| Treatment | n | Cleaved | % Cleaved | Blastocysts | % Blastocysts |
| no siRNA | 182 | 152 | 74%a | 40 | 37.7%a |
| non-target siRNA | 375 | 183 | 57.9%b | 38 | 29.2%b |
| DNMT1 siRNAs | 525 | 182 | 46.3%c | 8 | 5.8%c |
| IVF | |||||
|---|---|---|---|---|---|
| Treatment | n | Cleaved | % Cleaved | Blastocysts | % Blastocysts |
| no siRNA | 250 | 185 | 74%a | 69 | 37.3%a |
| non-target siRNA | 190 | 110 | 57.9%b | 29 | 26.4%a |
| DNMT1 siRNAs | 188 | 87 | 46.3%c | 0 | 0%b |
In order to further examine this block to bovine preimplantation development, we sought to generate a donor cell line containing a stably expressed short hairpin RNA targeting bovine Dnmt1 for use as a nuclear donor. Utilization of Polymerase II driven constructs would induce demethylation of the donor genome in cell culture but transgene transcription within reconstructed embryos would not be induced until zygotic genome activation at the 8-cell stage and thus should not impact stores of maternal mRNAs. To achieve stable depletion of Dnmt1 transcripts over a prolonged culture period, lentiviral constructs containing microRNA based short hairpin RNAs (shRNAMirs) targeting bovine Dnmt1 were cloned. Again, we designed these shRNAs to target both reported isolforms of the bovine Dnmt1 transcript{Russell 2008}. We utilized the previously described PEG lentiviral vector expressing the shRNAMir from within the 3′ UTR of the green fluorescent protein (GFP) transgene (Fig. 4A) (Golding et al., 2010). To achieve stable integration and expression of the Dnmt1 targeting shRNAMirs, these constructs were used to generate infectious, replication deficient lentiviral particles and delivered into the same primary bovine fibroblasts utilized as nuclear donors above, using methods previously described (Golding et al., 2006; Golding et al., 2010). Stable integration and expression of these constructs was verified by observation of GFP fluorescence within transgenic cells. Dnmt1 shRNAMir expressing cell lines, along with a non-targeting control, were selected to homogeneity using puromycin and Dnmt1 depletion verified using a combination of qRT-PCR and western blot analysis. Four independent shRNAMirs were tested and the results of the two best candidates can be seen in Figures 4B (mRNA) and 4C (protein). Bovine shRNAMirs Dnmt1-3 and Dnmt1-4 consistently produced a significant reduction in DNMT1 expression, with Dnmt1-4 yielding greater than a 95% reduction in levels of both Dnmt1 message and protein.
Figure 4.
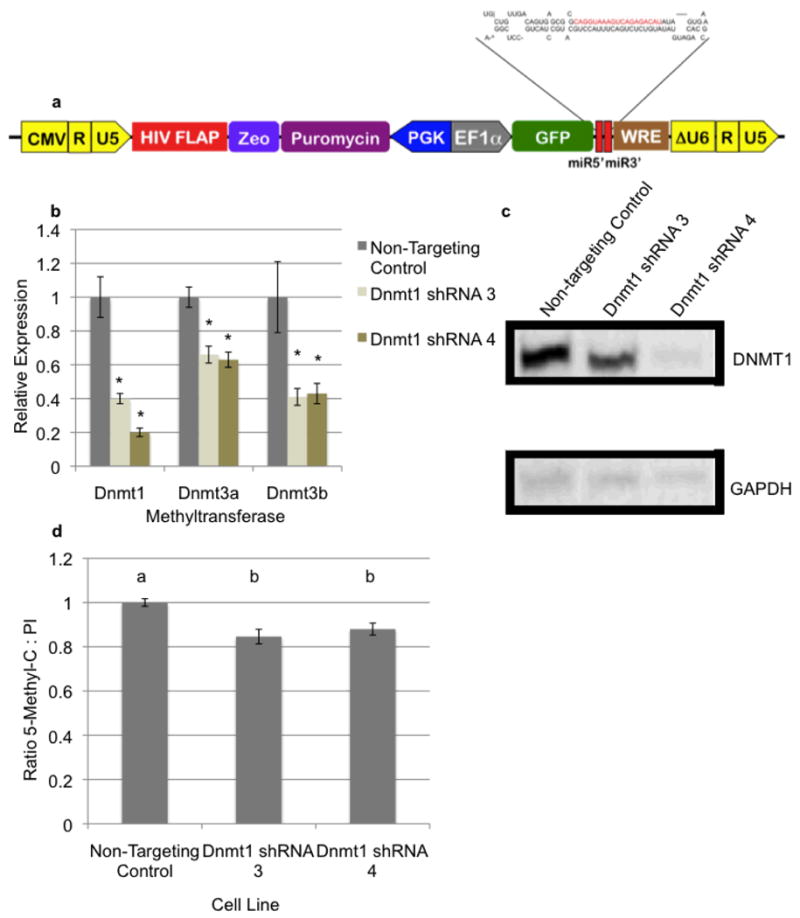
Stable depletion of Dnmt1 in transgenic donor fibroblasts. a) Cartoon map of the PGK Puromycin EF1a GFP containing (PEG) lentiviral shRNA expression constructs used to deliver shRNAs targeting Dnmt1. b) Reduced expression of the bovine Dnmts in transgenic cell lines. Primary bovine fibroblasts were transduced with either non-targeting control or Dnmt1 targeting shRNAs. RNA from transgenic cells was isolated and seeded into qRT-PCR reactions measuring each of the Dnmts. Data was normalized to the geometric mean of GAPDH, YWHAZ & SDHA. Error bars represent the SEM and * denote statistical significance P<0.001. c) Verification of DNMT1 depletion using Western analysis. Protein extracts from shRNA containing cell lines were isolated and probed using antibodies recognizing DNMT1. To normalize samples, blots were stripped and probed for GAPDH. d) Reduced levels of genomic methylation in transgenic cell lines. Fibroblasts containing non-targeting control or Dnmt1 targeting shRNAs were fixed and stained using an antibody recognizing methylated cytosine. 5MC staining was normalized using propidium iodine. Measurements of the non-targeting control cell line were set to 1 and relative values graphed. Error bars represent the SEM and letters denote statistical significance P<0.05. Raw numbers used in this analysis are available in supplementary figure 1b.
To examine if depletion of Dnmt1 influences expression of either Dnmt3a or Dnmt3b, RNA preparations were analyzed using qRT-PCR. Unexpectedly, as can be seen in Fig. 4B, depletion of Dnmt1 is associated with a significant drop in levels of both Dnmt3a and Dnmt3b message (P<0.01). Next, cell lines containing Dnmt1 or control shRNAMirs were analyzed for levels of genomic DNA methylation. After 7 passages in culture, cell lines were analyzed using immunocytochemistry as previously described (Dean et al., 2001). Depletion of Dnmt1 and corresponding drop in Dnmt3a/3b levels was associated with a statistically significant (P<0.05) 15% drop in genomic methylation relative to transgenic control cells (Fig. 4D). To determine the effect stable Dnmt1 depletion has on SCNT development rates, Dnmt1-shRNA4 and control cell lines were used as nuclear donors. In contrast to experiments reported by Eilertsen et al., (2007) which saw increased development rates using transient siRNA mediated depletion of Dnmt1 in the donor cell lines, no significant differences in blastocyst development rates were observed using stably Dnmt1 depleted cell lines as compared to controls (Table 2). Importantly however, embryonic development was not arrested at the 8-16 cell stage and produced blastocyst development rates indistinguishable from the controls. These results suggest maternal transcripts encoding Dnmt1 can carry development to at least the blastocyst stage and that in contrast to the shRNA expressing cell line, protocols using siRNAs deplete these maternal transcripts and arrest development.
TABLE 2.
Embryo development rates for SCNT embryos generated using transgenic cells expressing either the non-targeting control shRNA or Dnmt1-shRNA 4.
| Cell Line | Oocyte | Enucleated | Fused (%) | Blastocysts (%) | Pregnancy Rate Day 40 |
|---|---|---|---|---|---|
| Non-Targeting Control | 200 | 186 | 160 (86%) | 54 (34%) | 33% |
| Dnmt1 shRNA 4 | 200 | 138 | 116 (84%) | 37 (32%) | 0% |
To further examine the impact of stable shRNA mediated Dnmt1 depletion of bovine embryonic development, we opted to transfer 20 Dnmt1 shRNAMir blastocyst stage embryos into 5 recipients. As a control, blastocysts containing non-targeting shRNAMir s were also transferred. As can be seen in Table 2, control embryos resulted in a 33% pregnancy rate as measured on day 40 as compared to 0% for the Dnmt1-depleted embryos. These results are consistent with previous studies in mice which demonstrate DNMT1 activity is dispensable for in vivo preimplantation development, yet essential for postimplantation development (Li et al., 1992). These above results collectively suggest that Dnmt1 is essential for preimplantation development in ruminants and siRNA depletion of maternal Dnmt1 transcripts or the persistence of Dnmt1-siRNA treatment past the 8-cell stage interferes with the epigenetic programming function of Dnmt1 and results in developmental arrest. As our siRNA treatment arrested our control embryos, no attempts were made to test Dnmt1-siRNA injection in embryos produced through SCNT.
Discussion
Abnormal patterns of gene expression are a hallmark of cloned embryos and studies in cloned cattle and mice demonstrating genomic hypermethylation have begun to provide some explanation to these transcriptional abnormalities (Bourc'his et al., 2001; Dean et al., 2001; Kang et al., 2001; Xue et al., 2002). The key modulators of DNA methylation are the DNMTs and over-expression of this enzyme family has been hypothesized to lead to the observed hypermethylation and the aberrant patterns of X-chromosome inactivation and imprinted gene expression frequently seen in animals produced using nuclear transfer (Bestor, 1998; Golding and Westhusin, 2003). To distinguish between DNMT over-expression versus differing capacities of somatic and embryonic nuclei to biochemically regulate this enzyme family, we conducted real time quantitation of bovine Dnmt transcripts during IVF, NT and parthenote development. Results from this study demonstrate that within cloned embryos, members of the Dnmt family are not over-expressed above and beyond levels seen in IVF or parthenote embryos but rather demonstrate drastically reduced transcript levels encoding the most abundant methyltransferase, Dnmt1. The promoter of human dnmt1 contains several methylation responsive elements that have been hypothesized to modulate transcription in accordance with local levels of genomic methylation (Bigey et al., 2000; Slack et al., 1999; Slack et al., 2001). Given the hypermethylated status of the genome in cloned embryos it is possible that transcription of Dnmt1 in clones becomes attenuated as a result of the high levels of CpG methylation within the donor genome.
During early development of cloned mice removal of the female pronucleus from the oocyte precludes any ability of the zygote to demethylate the donor genome and removes key elements controlling the trafficking of the oocyte specific isoform of Dnmt1 (Dnmt1o) (Howell et al., 2001; Chung et al., 2003; Kang et al., 2001; Oswald et al., 2000). Similarly, studies of nuclear breakdown and the dynamics of murine Dnmt1o in reconstructed embryos support the assertion that DNMT trafficking is disrupted in NT embryos. (Gonda et al., 2003; Chung et al., 2003). Given our observed down-regulation of bovine Dnmt1 transcription along with previous observations of early murine NT development, we postulate that the increasing levels of genomic methylation observed in cloned cattle may result from either abnormal trafficking or biochemical regulation of this gene family. Thus within the unique environment of an oocyte, somatic nuclei lack key elements controlling enzyme access or activity resulting in genomic hypermethylation.
In an effort to reduce genomic methylation levels we sought to suppress Dnmt1 activity in donor cell lines and in early embryos using RNAi. Utilizing siRNAs to block DNMT1 expression during preimplantation development we sought to develop protocols aimed at lowering levels of genomic methylation by inducing passive demethylation. However, similar to previously described studies using Dnmt1 targeting siRNAs in sheep, depletion of the most abundant methyltransferase within IVF and parthenote embryos resulted in a developmental arrest before the blastocyst stage (Taylor et al., 2009). In contrast using a stably expressed shRNA to interfere with Dnmt1 expression in cloned embryos did not induce developmental arrest and produced development rates identical to the controls. We hypothesize that these observations can be explained by the differences in RNAi inducing molecule dosage and timing. SCNT embryos containing a shRNA expression cassette will not be transcribed until embryonic genome activation at the 8-cell stage where as siRNAs will immediately prime the RNAi machinery upon injection and suppress homologous transcripts. Moreover siRNAs were injected in picomolar amounts where as shRNAs will exhibit similar kinetics and abundance to other Polymerase II transcripts (Silva et al., 2005; Stegmeier et al., 2005). Thus in SCNT embryos generated using a Dnmt1-shRNA expressing cell line, maternal transcripts were likely sufficient to carry development beyond the blastocyst stage into post-implantation development at which point the depletion of DNMT1 was lethal. While ineffective at improving SCNT embryo development rates, our results clearly show that the techniques described above are effective as a tool to study functional genomics in a large animal model.
We were surprised that the reduction in global DNA methylation within the donor cell line was only on the order of 15% given the potent depletion of Dnmt1 transcripts to levels less than 5% of controls. Given the number of passages the transgenic fibroblasts were grown before analysis we fully expected to see a reduction in methylation levels by at least half. It is possible that DNMT3b, which does have some reported maintenance activity, could be compensating and maintaining methylation levels (Eilertsen et al., 2007; Chen et al., 2003). However our observations that levels of both Dnmt3a and Dnmt3b transcripts were reduced in Dnmt1-shRNAMir cell lines do not wholly support this assertion. Moreover we are at a loss as to explain the down-regulation of both Dnmt3a and 3b in the absence of Dnmt1.
As the mammalian embryo develops, progressive changes in epigenetic modifications to DNA and histones gradually program cell specific patterns of gene expression and restrict both lineage potential and cell identity (Goldberg et al., 2007). However, in contrast to cells within the inner cell mass of in vivo preimplatation embryos, both ES and IPS cells in culture are relatively hypermethylated; yet clearly retain the capacity to generate all the somatic lineages. Moreover, while increased levels of genomic methylation are an impediment of SCNT reprogramming they are clearly not insurmountable. Therefore methods aimed at partially reducing these high levels should improve the “programability” of the donor nucleus and thus embryo survival. Conversely, a recent report suggests that bovine cloning efficiency is not correlated with altered levels of key epigenetic modifiers, including the Dnmts 1, 3a and 3b in the donor cell line (Zhou et al., 2009). Our studies indicate that targeting maternal Dnmt1 transcripts within the preimplantation embryo using siRNAs does not likely represent a viable strategy for improving SCNT development rates as presumably the affect of RNAi persists past the 8-cell stage and block Dnmt1's essential role in developmental programming (Howell et al., 2001). In contrast, transient siRNA mediated depletion of Dnmt1 within donor cell lines produces a ∼30% increase in SCNT development rates to the blastocyst stage (Eilertsen et al., 2007). It will be interesting to see whether this increase in development rates correlates with an increased ability to produce competent ES cells or live offspring.
The birth of Dolly and more recently the discovery of methods for the generation of induced pluripotent stem cells have raised fundamental questions concerning the irreversibility of the differentiated state (Campbell et al., 1996; Takahashi and Yamanaka, 2006). Since its discovery, induction of pluripotency within somatic cells in vitro have shifted studies of the cellular reprogramming process from those restricted by the scale of embryo culture to large-scale studies in cell culture and greatly accelerated the pace of research within this field. It is now becoming clear that remodeling the epigenetic landscape of a cell represents the key to success for this reprogramming process and accordingly much effort has gone into identifying crucial factors involved in this process. Critical roles for genomic demethylating enzymes, polycomb proteins and chromatin remodeling factors in this process speak to the critical nature of epigenetics in IPS cell generation and highlight how essential a firm understanding of these processes are to IPS stability and therapeutic potential (Kim et al., 2010; Singhal et al., 2010; Pereira et al., 2010; Bhutani et al., 2009). Thus, it is very likely that methods aimed at priming somatic cells for IPS programing will also serve to improve cell suitability for the SCNT process. The recent success in demethylating cellular genomes using proteins involved in elements of base excision repair is one example of potential methods that my be used to improve SCNT efficiency (Kim et al., 2010).
Materials & Methods
Embryo Production and siRNA Injection
Mature bovine oocytes were obtained (Ovitra Biotechnology, Inc. Hereford, TX) and utilized to produce IVF, parthenote and SCNT embryos using methods previously described (De Sousa et al., 1999; Golding and Westhusin, 2003; Hill et al., 2000; Winger et al., 2000). siRNAs were diluted prior to injection to 50 mM in TE buffer. Both injection groups contained Cy3 labelled siRNAs for a visual conformation of injection into the cytoplasm. The zygotes or activated ova were subsequently placed in G1™ -Version 3 media (Vitrolife 10091) for 3 days then G2™ -Version 3 media (Vitrolife 10092) for culture from the 8-cell to the blastocyst stage. siRNA sequences were: Dnmt1 852 GCACAGAAGUCAACCCAAAtt Dnmt1 1296 CCUCUUUUCUGGUUCAGCAtt and were injected as a 50/50 mixture. The sequences shown correspond to the sense strand and the numbers indicate the position of the Dnmt1 (Accession NM_182651) transcript targeted.
qRT-PCR & Western Analysis
RNA isolation and RT-PCR were performed as previously described (Golding and Westhusin, 2003). Relative gene expression levels from each sample were calculated in triplicate using the SYBR Green comparative Ct method (Applied Biosystems, USA), adjusted according to individual PCR efficiencies for each primer pair and normalized to the geometric mean Ct of 3 endogenous controls (Gapdh, Ywhaz & Sdha) as described previously (Goossens et al., 2005). Primer sequences used were: Gapdh (Fwd 5′-CTGCCCGTTCGACAGATAG-3′, Rev 5′-CTCCGACCTTCACCATCTTG-3′) Ywhaz (Fwd 5′-CTGAACTCCCCTGAGAAAGC-3′ Rev 5′-CCTTCTCCTGCTTCAGCTTC-3′) Sdha (Fwd 5′-ACCTGATGCTTTGTGCTCTG-3′ Rev 5′-TCGTACTCGTCAACCCTCTC-3′) Dnmt1Fwd (5′-TGACTCCACCTACGAAGACC-3′ Rev 5′-TCTCTACTTGCTCCACCACG-3′) Dnmt3a (Fwd 5′-CAACGGAGAAGCCTAAGGTCAA-3′ Rev 5′-TTGAGGCTCCCACAAGAGATG-3′) Dnmt3b (Fwd 5′-AGTATCAGGATGGGAAGGAGTTTG-3′ Rev 5′-CCAGGAGAAACCCTTGATCTTTC-3′). For Western analysis DNMT1 goat polyclonal antibody (Santa Cruz Biotech sc-10221) was diluted 1 μg/ml in 2.0% skim milk/TBST.
Generation of Transgenic Primary Fibroblasts
Primary bovine fibroblasts were isolated using previously described procedures (Hill et al., 2000) and seeded into 24-well plates to give a density of 50% confluence after 12-18 hours of growth. The next day, recombinant retroviral particles were passed through a 0.45um filter and added to the bovine fibroblast culture media along with a final concentration of 8 μg/ml polybrene. Cells were sub-passed via standard protocol and selected using 2 μg/ml Puromycin (Sigma Aldrich). Methods detailing the production of recombinant lenti class retroviral particles have been described previously (Lois et al., 2002; Golding et al., 2006; Golding et al., 2010). Bovine Dnmt1 shRNAs were designed to target bases 1082-1111 and 1789-1818 (Dnmt1-shRNA3 and shRNA4 respectively) of the full length Dnmt1 transcript (Accession NM_182651).
Immunocytochemistry
Parthenote and IVF embryos at the 8-16 cell stage (84 hours post activation) and blastocyst stage (168 hours post activation) were removed from culture, washed twice with 1 × PBS and fixed with -20°C methanol for 5 minutes. Embryos were washed in PBS and permeabilized for 30 minutes at room temperature with 0.2% Triton X-100 in PBS and treated wtih 3 M HCL diluted in ddH2O with 0.1% Triton X-100 to denature the DNA and allow binding of the primary antibody to methylated DNA. The HCL was removed after 13 minutes and a 100 mM Trizma Hydrochloride buffer, pH 8.5, was added for 30 minutes to neutralize the HCL. Embryos were incubated in blocking buffer with 3.0% BSA (Sigma 9022) and 0.1% Triton X-100 in PBS for four hours at room temperature to block non-specific binding of antibodies. Primary antibody to 5-methylcytidine (Eurogentec San Diago CA) was diluted 1:200 in blocking buffer and embryos were incubated in 150 μl drops for one hour at 37°C. Embryos were washed by washing embryos through blocking buffer six times for five minutes each. An Alexa 488 (Invitrogen) secondary antibody was diluted 1:200 in PBS with 0.1% Tween-20 and embryos were incubated for one hour at 37°C. Secondary antibody was washed three times with PBS with 0.1% Tween-20 for five minutes each time. RNase A was diluted to 25 μg/ml in PBS and incubated for 20 minutes at 37°C. Propidium Iodide (PI) was diluted 25 μg/ml in PBS and incubated for 30 minutes at 37°C. Cover slips were mounted with a 50/50 solution of anti-fade and glycerol on glass slides and sealed with clear fingernail polish. Cell lines grown on glass cover slips were subjected to similar procedures.
Images were taken using a BioRad Radiance 2003 multiphoton/confocal microscope and a 60X water immersion objective using Laser Sharp 2000 imaging software. A z-series was taken from individual embryos with 5 microns between each section. There were approximately 20-35 images per embryo depending on their shape and size. Each section contained an image of the 5-methylcytosine label and one for the PI nuclear stain. Mean intensity measurements of the embryo images were taken using NIS Elements 3.0 software. 5-methylcytosine intensity measurements were divided by the intensity of the PI stain for each individual blastomere nuclei to give us the ratio of methylated DNA to total DNA for each cell in an embryo.
Statistical Analysis
For qRT-PCR analysis comparisons between groups were made using the students t-test. For analysis of differences in embryo development rates, Chi-Square analysis was performed to determine significant differences (p < 0.05) between three independent groups of embryos. Expected values for each group were estimated according to the chi-square test, compared with the observed values, and used to calculate p-values for each pairwise comparison. Blastocyst rates were significantly different for all comparisons with the exception of non-injected controls to non-targeting siRNA injected controls.
Supplementary Material
Ratios of 5-methylcytosine staining normalized to propidium iodine for Dnmt1-siRNA injected embryos (a) and Dnmt1-shRNA transgenic cells. Letters denote statistical significance P<0.05.
Acknowledgments
The authors would like to thank Kim Tessanne, Kim Green, Suzanne Menges, Alison Wilkerson and Jane Pryor for their technical support during this project. This project was supported by NIH NICHD grants 1R21HD055631-01A2 and 5R01HD058969-02.
Abbreviations
- DNMT
DNA Methyltransferase
- SCNT
Somatic Cell Nuclear Transfer
- RNAi
RNA Interference
- siRNA
short interfering RNA
- shRNA
short hairpin RNA
- IVF
in vitro fertilization
- NT
Nuclear Transfer
- qRT-PCR
Quantitative Reverse Transcriptase Polymerase Chain Reaction
- ICC
Immnuocytochemistry
References
- Akshey YS, Malakar D, De AK, Jena MK, Garg S, Dutta R, Pawar SK, Mukesh M. Hand-made cloned goat (Capra hircus) embryos—a comparison of different donor cells and culture systems. Cell Reprogram. 2010;12:581–588. doi: 10.1089/cell.2009.0120. [DOI] [PubMed] [Google Scholar]
- Bestor TH. Cytosine methylation and the unequal developmental potentials of the oocyte and sperm genomes. Am J Hum Genet. 1998;62:1269–1273. doi: 10.1086/301891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2009 doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigey P, Ramchandani S, Theberge J, Araujo FD, Szyf M. Transcriptional regulation of the human DNA Methyltransferase (dnmt1) gene. Gene. 2000;242:407–418. doi: 10.1016/s0378-1119(99)00501-6. [DOI] [PubMed] [Google Scholar]
- Blelloch R, Wang Z, Meissner A, Pollard S, Smith A, Jaenisch R. Reprogramming efficiency following somatic cell nuclear transfer is influenced by the differentiation and methylation state of the donor nucleus. Stem Cells. 2006;24:2007–2013. doi: 10.1634/stemcells.2006-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc'his D, Le Bourhis D, Patin D, Niveleau A, Comizzoli P, Renard JP, Viegas-Péquignot E. Delayed and incomplete reprogramming of chromosome methylation patterns in bovine cloned embryos. Curr Biol. 2001;11:1542–1546. doi: 10.1016/s0960-9822(01)00480-8. [DOI] [PubMed] [Google Scholar]
- Campbell KH, Alberio R, Choi I, Fisher P, Kelly RD, Lee JH, Maalouf W. Cloning: eight years after Dolly. Reprod Domest Anim. 2005;40:256–268. doi: 10.1111/j.1439-0531.2005.00591.x. [DOI] [PubMed] [Google Scholar]
- Campbell KH, McWhir J, Ritchie WA, Wilmut I. Sheep cloned by nuclear transfer from a cultured cell line. Nature. 1996;380:64–66. doi: 10.1038/380064a0. [DOI] [PubMed] [Google Scholar]
- Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung YG, Ratnam S, Chaillet JR, Latham KE. Abnormal regulation of DNA methyltransferase expression in cloned mouse embryos. Biol Reprod. 2003;69:146–153. doi: 10.1095/biolreprod.102.014076. [DOI] [PubMed] [Google Scholar]
- Dean W, Santos F, Stojkovic M, Zakhartchenko V, Walter J, Wolf E, Reik W. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc Natl Acad Sci U S A. 2001;98:13734–13738. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sousa PA, Winger Q, Hill JR, Jones K, Watson AJ, Westhusin ME. Reprogramming of fibroblast nuclei after transfer into bovine oocytes. Cloning. 1999;1:63–69. doi: 10.1089/15204559950020102. [DOI] [PubMed] [Google Scholar]
- Eilertsen KJ, Power RA, Harkins LL, Misica P. Targeting cellular memory to reprogram the epigenome, restore potential, and improve somatic cell nuclear transfer. Anim Reprod Sci. 2007;98:129–146. doi: 10.1016/j.anireprosci.2006.10.019. [DOI] [PubMed] [Google Scholar]
- Enright BP, Kubota C, Yang X, Tian XC. Epigenetic characteristics and development of embryos cloned from donor cells treated by trichostatin A or 5-aza-2′-deoxycytidine. Biol Reprod. 2003;69:896–901. doi: 10.1095/biolreprod.103.017954. [DOI] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Golding MC, Long CR, Carmell MA, Hannon GJ, Westhusin ME. Suppression of prion protein in livestock by RNA interference. Proc Natl Acad Sci U S A. 2006;103:5285–5290. doi: 10.1073/pnas.0600813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding MC, Westhusin ME. Analysis of DNA (cytosine 5) methyltransferase mRNA sequence and expression in bovine preimplantation embryos, fetal and adult tissues. Gene Expr Patterns. 2003;3:551–558. doi: 10.1016/s1567-133x(03)00121-2. [DOI] [PubMed] [Google Scholar]
- Golding MC, Zhang L, Mann MR. Multiple epigenetic modifiers induce aggressive viral extinction in extraembryonic endoderm stem cells. Cell Stem Cell. 2010;6:457–467. doi: 10.1016/j.stem.2010.03.014. [DOI] [PubMed] [Google Scholar]
- Gonda K, Fowler J, Katoku-Kikyo N, Haroldson J, Wudel J, Kikyo N. Reversible disassembly of somatic nucleoli by the germ cell proteins FRGY2a and FRGY2b. Nat Cell Biol. 2003;5:205–210. doi: 10.1038/ncb939. [DOI] [PubMed] [Google Scholar]
- Goossens K, Van Poucke M, Van Soom A, Vandesompele J, Van Zeveren A, Peelman LJ. Selection of reference genes for quantitative real-time PCR in bovine preimplantation embryos. BMC Dev Biol. 2005;5:27. doi: 10.1186/1471-213X-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JR, Winger QA, Long CR, Looney CR, Thompson JA, Westhusin ME. Development rates of male bovine nuclear transfer embryos derived from adult and fetal cells. Biol Reprod. 2000;62:1135–1140. doi: 10.1095/biolreprod62.5.1135. [DOI] [PubMed] [Google Scholar]
- Howell CY, Bestor TH, Ding F, Latham KE, Mertineit C, Trasler JM, Chaillet JR. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001;104:829–838. doi: 10.1016/s0092-8674(01)00280-x. [DOI] [PubMed] [Google Scholar]
- Jaenisch R. Nuclear cloning, embryonic stem cells, and transplantation therapy. Harvey Lect. 2002;98:145–171. [PubMed] [Google Scholar]
- Kang YK, Koo DB, Park JS, Choi YH, Chung AS, Lee KK, Han YM. Aberrant methylation of donor genome in cloned bovine embryos. Nat Genet. 2001;28:173–177. doi: 10.1038/88903. [DOI] [PubMed] [Google Scholar]
- Kang YK, Koo DB, Park JS, Choi YH, Kim HN, Chang WK, Lee KK, Han YM. Typical demethylation events in cloned pig embryos. Clues on species-specific differences in epigenetic reprogramming of a cloned donor genome. J Biol Chem. 2001;276:39980–39984. doi: 10.1074/jbc.M106516200. [DOI] [PubMed] [Google Scholar]
- Kang YK, Park JS, Koo DB, Choi YH, Kim SU, Lee KK, Han YM. Limited demethylation leaves mosaic-type methylation states in cloned bovine pre-implantation embryos. EMBO J. 2002;21:1092–1100. doi: 10.1093/emboj/21.5.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Liu L, Shin T, Pryor JH, Kraemer D, Westhusin M. Regenerated bovine fetal fibroblasts support high blastocyst development following nuclear transfer. Cloning. 2001;3:51–58. doi: 10.1089/15204550152475554. [DOI] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Lopatina N, Haskell JF, Andrews LG, Poole JC, Saldanha S, Tollefsbol T. Differential maintenance and de novo methylating activity by three DNA methyltransferases in aging and immortalized fibroblasts. J Cell Biochem. 2002;84:324–334. doi: 10.1002/jcb.10015. [DOI] [PubMed] [Google Scholar]
- McGrath J, Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37:179–183. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- McGrath J, Solter D. Inability of mouse blastomere nuclei transferred to enucleated zygotes to support development in vitro. Science. 1984;226:1317–1319. doi: 10.1126/science.6542249. [DOI] [PubMed] [Google Scholar]
- Meissner A, Jaenisch R. Mammalian nuclear transfer. Dev Dyn. 2006;235:2460–2469. doi: 10.1002/dvdy.20915. [DOI] [PubMed] [Google Scholar]
- Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- Müller R, Lengerke C. Patient-specific pluripotent stem cells: promises and challenges. Nat Rev Endocrinol. 2009;5:195–203. doi: 10.1038/nrendo.2009.18. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- Pereira CF, Piccolo FM, Tsubouchi T, Sauer S, Ryan NK, Bruno L, Landeira D, Santos J, Banito A, Gil J, Koseki H, Merkenschlager M, Fisher AG. ESCs require PRC2 to direct the successful reprogramming of differentiated cells toward pluripotency. Cell Stem Cell. 2010;6:547–556. doi: 10.1016/j.stem.2010.04.013. [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- Santos F, Zakhartchenko V, Stojkovic M, Peters A, Jenuwein T, Wolf E, Reik W, Dean W. Epigenetic marking correlates with developmental potential in cloned bovine preimplantation embryos. Curr Biol. 2003;13:1116–1121. doi: 10.1016/s0960-9822(03)00419-6. [DOI] [PubMed] [Google Scholar]
- Shin T, Kraemer D, Pryor J, Liu L, Rugila J, Howe L, Buck S, Murphy K, Lyons L, Westhusin M. A cat cloned by nuclear transplantation. Nature. 2002;415:859. doi: 10.1038/nature723. [DOI] [PubMed] [Google Scholar]
- Simonsson S, Gurdon J. DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei. Nat Cell Biol. 2004;6:984–990. doi: 10.1038/ncb1176. [DOI] [PubMed] [Google Scholar]
- Singhal N, Graumann J, Wu G, Araúzo-Bravo MJ, Han DW, Greber B, Gentile L, Mann M, Schöler HR. Chromatin-Remodeling Components of the BAF Complex Facilitate Reprogramming. Cell. 2010;141:943–955. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- Slack A, Cervoni N, Pinard M, Szyf M. Feedback regulation of DNA methyltransferase gene expression by methylation. Eur J Biochem. 1999;264:191–199. doi: 10.1046/j.1432-1327.1999.00603.x. [DOI] [PubMed] [Google Scholar]
- Slack A, Pinard M, Araujo FD, Szyf M. A novel regulatory element in the dnmt1 gene that responds to co-activation by Rb and c-Jun. Gene. 2001;268:87–96. doi: 10.1016/s0378-1119(01)00427-9. [DOI] [PubMed] [Google Scholar]
- Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, Sheth N, Bradshaw J, Burchard J, Kulkarni A, Cavet G, Sachidanandam R, McCombie WR, Cleary MA, Elledge SJ, Hannon GJ. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc Natl Acad Sci U S A. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K. Direct reprogramming 101. Dev Growth Differ. 2010;52:319–333. doi: 10.1111/j.1440-169X.2010.01169.x. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Taylor J, Moore H, Beaujean N, Gardner J, Wilmut I, Meehan R, Young L. Cloning and expression of sheep DNA methyltransferase 1 and its development-specific isoform. Mol Reprod Dev. 2009;76:501–513. doi: 10.1002/mrd.20968. [DOI] [PubMed] [Google Scholar]
- Todd JA. Stem cells and a cure for type 1 diabetes? Proc Natl Acad Sci U S A. 2009;106:15523–15524. doi: 10.1073/pnas.0908373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winger QA, Hill JR, Shin T, Watson AJ, Kraemer DC, Westhusin ME. Genetic reprogramming of lactate dehydrogenase, citrate synthase, and phosphofructokinase mRNA in bovine nuclear transfer embryos produced using bovine fibroblast cell nuclei. Mol Reprod Dev. 2000;56:458–464. doi: 10.1002/1098-2795(200008)56:4<458::AID-MRD3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Xie S, Wang Z, Okano M, Nogami M, Li Y, He WW, Okumura K, Li E. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene. 1999;236:87–95. doi: 10.1016/s0378-1119(99)00252-8. [DOI] [PubMed] [Google Scholar]
- Xue F, Tian XC, Du F, Kubota C, Taneja M, Dinnyes A, Dai Y, Levine H, Pereira LV, Yang X. Aberrant patterns of X chromosome inactivation in bovine clones. Nat Genet. 2002;31:216–220. doi: 10.1038/ng900. [DOI] [PubMed] [Google Scholar]
- Yoon TJ, Irizarry RA, Jung N, Seita J, Hanna J, Murakami P, Jaenisch R, Weissleder R, Orkin SH, Weissman IL, Feinberg AP, Daley GQ. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JI, Smith JR. DNA methyltransferase inhibition in normal human fibroblasts induces a p21-dependent cell cycle withdrawal. J Biol Chem. 2001;276:19610–19616. doi: 10.1074/jbc.M009470200. [DOI] [PubMed] [Google Scholar]
- Zhou W, Sadeghieh S, Abruzzese R, Uppada S, Meredith J, Ohlrichs C, Broek D, Polejaeva I. Transcript levels of several epigenome regulatory genes in bovine somatic donor cells are not correlated with their cloning efficiency. Cloning Stem Cells. 2009;11:397–405. doi: 10.1089/clo.2009.0016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ratios of 5-methylcytosine staining normalized to propidium iodine for Dnmt1-siRNA injected embryos (a) and Dnmt1-shRNA transgenic cells. Letters denote statistical significance P<0.05.


