Abstract
Membrane protrusion or ruffling is the formation of actin rich membrane protrusions, essential for cell motility. The exact mechanism of ruffling is not fully known. Using YFP and CFP fluorescent chimeras, we show for the first time a co-localization of Phospholipase D2 (PLD2) and Growth factor Receptor Bound protein-2 (Grb2) with actin-rich membrane protrusions of macrophages. Grb2 cooperates with PLD2 in enhancing membrane ruffling, whether in resting cells or in cells stimulated with the growth factor M-CSF, although in the latter an increase in dorsal ruffles was observed, consistent with receptor-ligand internalization. Cells transfected with PLD2 mutated in the PH domain (Y169F) or with Grb2 mutated in the SH2 site (R86K) negate this effect, indicating an association PLD2(Y169)-SH2-Grb2 that was confirmed by immunoprecipitation and Western blotting. The association results in enhanced PLD activity, and the lipase activity that can only partially explain the formation of membrane ruffles in vivo. A third component involves the Rho-GTPase Rac2 and it is only when Rac2 is overexpressed that a full biological effect, including actin polymerization in vivo, is obtained. The mechanism involved is, then, as follows: PLD enzymatic action, after been increased due to the binding to Grb2-SH2 via Y169, cooperates with Rac2, and the three molecules stimulate actin polymerization and consequently, membrane ruffle formation. Since membrane ruffling precedes cell migration, the results herein provide a novel mechanism for control of membrane dynamics, crucial for the physiology of leukocytes.
1. INTRODUCTION
Membrane ruffling (also known as cell ruffling) is the formation of actin rich membrane protrusions. This occurs in cellular zones undergoing rapid reorganization of the plasma membrane and often precedes the formation of a lamellipodium. Lamellipodia, filopodia and membrane ruffles are essential among other events for cell motility. There is a regulated recruitment of molecular scaffolds to their tips (where actin polymerization localizes) coupled to the coordinated organization of actin filaments into lamella networks and bundled arrays [1]. Stimulation by EGF results in the recruitment of Grb2 and Shc to cellular compartments that contain the receptor for EGF (EGFR) followed by the formation of endosomes [2].
Fast plasma membrane movements (FPMM) are involved in ruffling, blebbing, fast shape change and fast translocation. A certain level of FPMM is a prerequisite for invasive capacity. Invasive epithelial cells show faster plasma membrane movements than related or parental non-invasive cells [3]. Abl kinases are activated downstream of growth factor receptors, Src family kinases and phospholipase Cγ1 in fibroblasts and influence growth factor-mediated proliferation, membrane ruffling, migration and cell invasion of cancer cells [4]. The exact mechanisms by which ruffling is regulated are the subject of much study but both Rac and WAVE have been implicated. Also, amphiphysin 1, an endocytic adaptor concentrated at synapses interacts with N-WASP and stimulates N-WASP and Arp2/3-dependent actin polymerization [5–7]. Growth factor-stimulated cells show ruffles that are Rac- and WAVE2-dependent [8]. Other pathways are surely involved in membrane ruffling.
Our laboratory has demonstrated previously that a mutation of phospholipase D2 (PLD2) in residue Y179 leads to an upregulation of DNA synthesis mediated by PI3K and Akt [9]. Y179 is located upstream of Y169, another tyrosine that, like Y179, exists within the context of a site recognizable by the docking protein Growth factor-Receptor Bound protein-2 (Grb2). As this second site remains under-studied, we wondered if it would mediate a biological effect different from DNA synthesis and cell growth. We discovered a role in cell protrusion in leukocytes. Moreover, the present study uncovered for the first time the participation of two signaling molecules, PLD2 and Grb2 during in vivo formation of membrane ruffles and shows the requirement of a third one, Rho-GTPase Rac2, to fully explain actin polymerization and the full biological effect of membrane protrusions or cell ruffling in macrophages.
2. MATERIALS AND METHODS
2.1. Materials
Reduced sodium bicarbonate DMEM and COS-7 cells were obtained from American Type Culture Collection (ATCC) (Rockville, MD). RAW 264.7/LR-5 cells ("RAW/LR5") were developed at the laboratory of one of the authors (DC). Histopaque-1077 was obtained from Sigma Aldrich (St. Louis, MO). RPMI 1640 1× was obtained from Mediatech (Manassas, VA). Superfect transfection reagent was obtained from Qiagen (Valencia, CA). M-CSF and EGF were from PeproTech Inc. (Rocky Hill, NJ). Antibodies were from Millipore (Temecula, CA). Triton X- 100, phalloidin-FITC conjugate (from Amanita phalloides) were purchased from Sigma (St. Louis, MO). pRK5-myc-Rac2 and pcDNA3-HA-Rac1 plasmids were generous gifts from and Dr. Mary Dinauer and Dr. Floria Peng.
2.2. RAW 264.7/LR5 and COS-7 Cell Culture and Transfection
LR5 cells derived from the murine monocyte/macrophage RAW 264.7 cell line [10] (henceforth called "RAW/LR5") were resuspended in fresh complete growth medium (CGM) consisting of reduced sodium bicarbonate DMEM, 20% FCS (fetal calf serum) and 1% gentamycin and transferred to tissue culture coated flasks and maintained at 37 °C in 5% CO2. Approximately 3×106 cells per transfection were used. A mixture of superfect reagent, plasmid DNA (1–5 µg) and CGM (no serum) was incubated for 10 minutes at room temperature. Cells were incubated with transfection reagent and plasmid for ~3 hours after which time cells were washed 3 times with CGM and incubated an additional 28 hours in the presence of CGM prior to experimental use. COS-7 cells were transfected with lipofectamine in Opti-MEM (serum free media).
2.3. Actin Polymerization
Cells used in these experiments were between passages ten and fifteen. Adherent cells were detached using 0.25% Trypsin/EDTA. Cells were incubated with FITC-Phalloidin for 1 hour and flow cytometry conducted as indicated in Ref. [11]. FITC-phalloidin reports the presence of polymerized actin versus a background of monomeric actin.
2.4. Cell ruffling assay
This was performed as indicated in [12]. Approximately 1×105 RAW/LR5 cells were transfected with fluorescent chimeras Cerulean-N1-Grb2-WT, Citrine-N1-PLD2-WT or both. After 48 hours, cells were plated on round glass coverslips in a 24-well plate and were cultured overnight at 37°C with 5% CO2 in RPMI media with 10% fetal bovine serum. Prior to stimulation with M-CSF, the cells were serum-starved with BWD buffer (20 mM HEPES, 125 mM NaCl, 5 mM KCl, 5 mM dextrose, 10 mM NaHCO3, 1 mM KH2PO4, 1 mM CaCl2, 1mM MgCl2; pH 7.4) for 2 h. Cells were then stimulated with 1 nM M-CSF for 5 min. Stimulation was promptly stopped by fixing the cells with 3.7% formaldehyde, permeabilization (0.2% Triton X-100) and then stained to determine F-actin using phalloidin conjugated with Alexa Fluor-568. The fluorescent signals from YFP-PLD2 and from CFP-Grb2 were detected with YFP and CFP filters, respectively. All images were taken using the 60× oil/1.40 phase3 objective of an Olympus IX71 microscope coupled to a Sensicam cooled charge-coupled device camera. Cell ruffles were quantified as described previously [8]. Briery, the cell ruffles were scored using a scale from 0 to 3, with 0 indicating no protrusion, 1 indicating protrusions in one area of the cell, 2 indicating protrusions in two distinct areas of the cell and 3 indicating protrusions in more than two distinct areas of the cell. The ruffling index was calculated by determining the average of the ruffling scores of at least 50 cells.
2.5. Preparation of CFP-N1-Grb2 and YFP-N1-PLD2 fluorescent chimeras
For pCerulean(CFP)-N1-Grb2, construction of the chimeric plasmid began with Homo sapiens growth factor receptor-bound protein 2 (Grb2) cDNA. The backbone CFP-N1 plasmid was a gift from Dr. Joel Swanson, University of Michigan. Grb2 was cloned into pcDNA 3.1 to yield pcDNA-Grb2 WT. The Grb2-WT fragments were amplified using Pfu polymerase (forward: GAGTCGACATGGAAGCCATC; reverse: ATCCGCGGTTAGACGTTC). The PCR product was gel purified with QIAquick Gel Extraction Kit, then ligated to pCR-Blunt II-TOPO with Zero Blunt TOPO PCR cloning kit and used to transform DH5α competent cells. Colonies positive for the plasmids were screened on LB/K+ agar plates. Plasmid verification was performed using PCR and restriction enzyme digestion with SalI and SacII, sequentially. The CFP and Grb2 fragments were ligated overnight at 4 °C with T4 DNA ligase. Size and restriction map of each of the plasmids was verified by RED (~ 5300 bp). Molecular identity of pCerulean-N1-Grb2 was confirmed by direct sequence analysis (Agencourt Bioscience Corporation, Beverly MA). Similar procedures were followed for the preparation of pCitrine(YFP)-N1-PLD2.
2.6. PLD activity
The measurement of PLD activity in vitro was conducted in liposomes with short-chain phosphatidylcholine (PC8) and [3H]-Butanol as indicated in (11).
2.7. Statistical Analyses
Data are presented as the mean ± SEM. The difference between means was assessed by the Single Factor Analysis of Variance (ANOVA) test or by t-test, as indicated in the legends of the figures. In either case, probability of p<0.05 was considered to indicate a significant difference.
3. RESULTS
3.1. PLD2 and by Grb2 co-localize in membrane ruffles
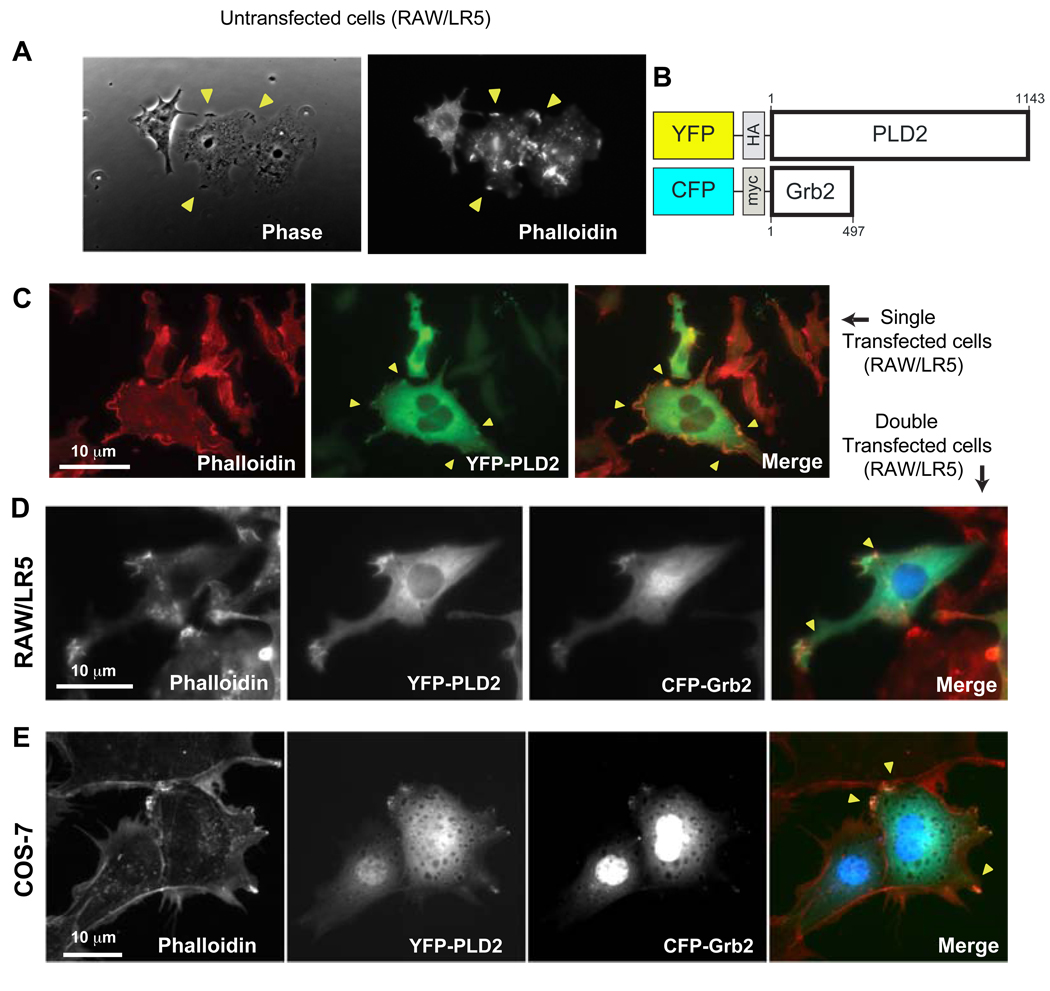
As cell ruffling (F-actin enriched membrane protrusions) is a prelude to chemotaxis [10], we investigated the molecular basis for the former function. Membrane ruffles are curled structures located on the edge of cell surfaces in lamellipodia and participate in motility and are shown in Fig. 1A in untransfected cells (yellow arrows that coincide with actin-rich zones as seen with phalloidin labeling). Since our laboratory and others has reported a role for PLD2 in cell migration [13–17] and also Grb2 is known to modulate PLD2 [18], we created plasmids encoding for fluorescent chimeras: CFP-Grb2 and YFP-PLD2 (Fig. 1B), intended to follow the intracellular localization of these two molecules along with the process of cell ruffling. The cell ruffling structures are present in PLD2-transfected cells (Fig. 1C) indicating that the transfection procedure was not harmful for the formation of these structures (also, viability of cells remained >95% after transfection). Ruffling activity (yellow arrows) was observed with double transfection of cells with YFP-PLD2 and CFP-Grb2. PLD2 localizes in cytosol and plasma membrane, whereas Rac2 localizes also in the cytosol and in the nucleus. The co-localization of YFP-PLD2 and CFP-Grb2 to membrane ruffles (actin-rich zones stained with phalloidin) was evident in either RAW/LR5 (Fig. 1D) or COS-7 (Fig. 1E).
Figure 1.
(A) Yellow arrows point to cell ruffles in untransfected RAW/LR5 cells, as seen by phase contrast microscopy and after Alexa Fluor 568-phalloidin staining. (B) Scheme of YFP-PLD2 and CFP-Grb2 chimeras. (C) YFP-PLD2-transfected RAW/LR5 cells. Cells observed under an Alexa Fluor filter to detect actin or under a YFP filter to detect PLD2 expression and the merged images. The yellow arrows indicate F-actin-enriched membrane protrusions (ruffles). RAW/LR5 cells (D) or COS-7 cells (E) were co-transfected with YFP-PLD2 and CFP-Grb2. Two days post-transfection, F-actin-rich protrusions were visualized by Alexa-568-phalloidin staining. YFP-PLD2 and CFP-Grb2 were visualized in the YFP and CFP channels, respectively. Scale bar=10 µm. The images are representative of at least 5 independent experimental repeats.
3.2. Cell membrane ruffling is activated by PLD2 and by Grb2 in resting cells
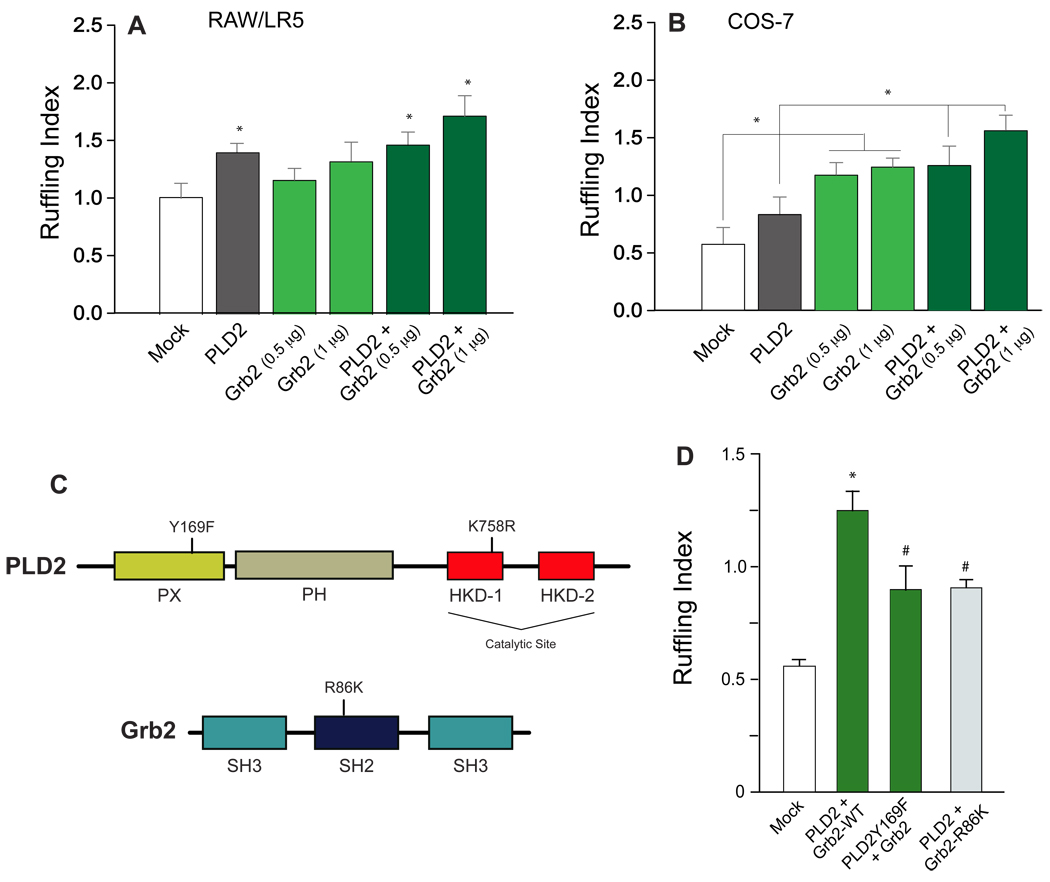
Membrane ruffles were visual indicators of LR5 and COS-7 migration even in basal conditions (i.e., no chemoattractant added to the cells). F-actin-rich membrane protrusions in cells overexpresing either Grb2- or PLD2-transfected samples or a combination of the two plasmids allowed cell quantitation (Fig. 2). A ruffling index was calculated by determining the average of the ruffling scores and indicated that overexpression of PLD2 or Grb2 enhanced a cell’s ability to undergo F-actin-enriched membrane protrusions in RAW/LR5 cells (Fig. 2A) and more so in COS-7 cells (Fig. 2B). The optimal DNA amount needed to transfect cells was found to be 1 µg for Grb2 and 2 µg for PLD2. In COS-7, the combination of Grb2 and PLD2 elicited a higher number of membrane ruffles than either one (Grb2 or PLD2) alone (Fig. 2B). Thus, overexpression of PLD2 or Grb2 enhanced a cell’s ability to undergo F-actin-enriched membrane protrusions.
Figure 2. Quantification of cell ruffling and PLD2/Grb2 mutants.
F-actin-rich membrane protrusions in RAW/LR5 (A) or COS-7 cells (B) overexpressing YFP-PLD2, CFP-Grb2 or both proteins. PLD2 and Grb2 were visualized in the YFP and CFP channels, respectively. (C) Schemes representing the domains of both PLD2 and Rac2 and the location of the point mutants used in this study. (D) Effect of several mutants of PLD2 and Grb2 on cell ruffling quantitation in COS-7 cells. In A, B and D, visualization of cell ruffling was aided by Alexa-568-phalloidin staining. The number of protrusions was quantified using a scoring scale from 0 to 3 with 0 indicating no protrusion, 1 indicating protrusions in one area of the cell, 2 indicating protrusions in two distinct areas of the cell and 3 indicating protrusions in more than two distinct areas of the cell. The ruffling index was calculated by determining the average of the ruffling scores of at least 50 cells. (*) denotes statistical significance (p<0.05 by ANOVA) between Mock controls and means of indicated data points. (#) in panel (D) indicates an inhibition of the mutants over the PLD2-wild type transfections.
3.3. PLD2 and Grb2 site of interaction and functional result
The results presented in Fig. 2A,B where Grb2 and PLD2 additively induce cell ruffling, led us to investigate if these two proteins were physically associated. To determine the site of binding between PLD2 and Grb2, we utilized the following mutants (Fig. 2C): for PLD2, a Y169F mutant (Y169 is within the PH domain of PLD2) known to be unable to bind to Grb2 in vitro [18]; and for Grb2, an SH2-deficient Grb2, “Grb2-noSH2” with the R86K mutation that makes it unable to bind to tyrosine-phosphorylated targets. The results of Fig. 2D show that the full extent of membrane ruffling is only attained with WT-PLD2 and WT-Grb2. Mutants lacking the indicated activities resulted in a diminished extent of membrane ruffling. This indicates that the full biological effect is mediated by a PLD2-Grb2 interaction at the level of Y169 (for PLD2) and the SH2 domain (for Grb2).
3.4. Stimulated macrophages: PLD2 and Grb2 also enhance membrane ruffling
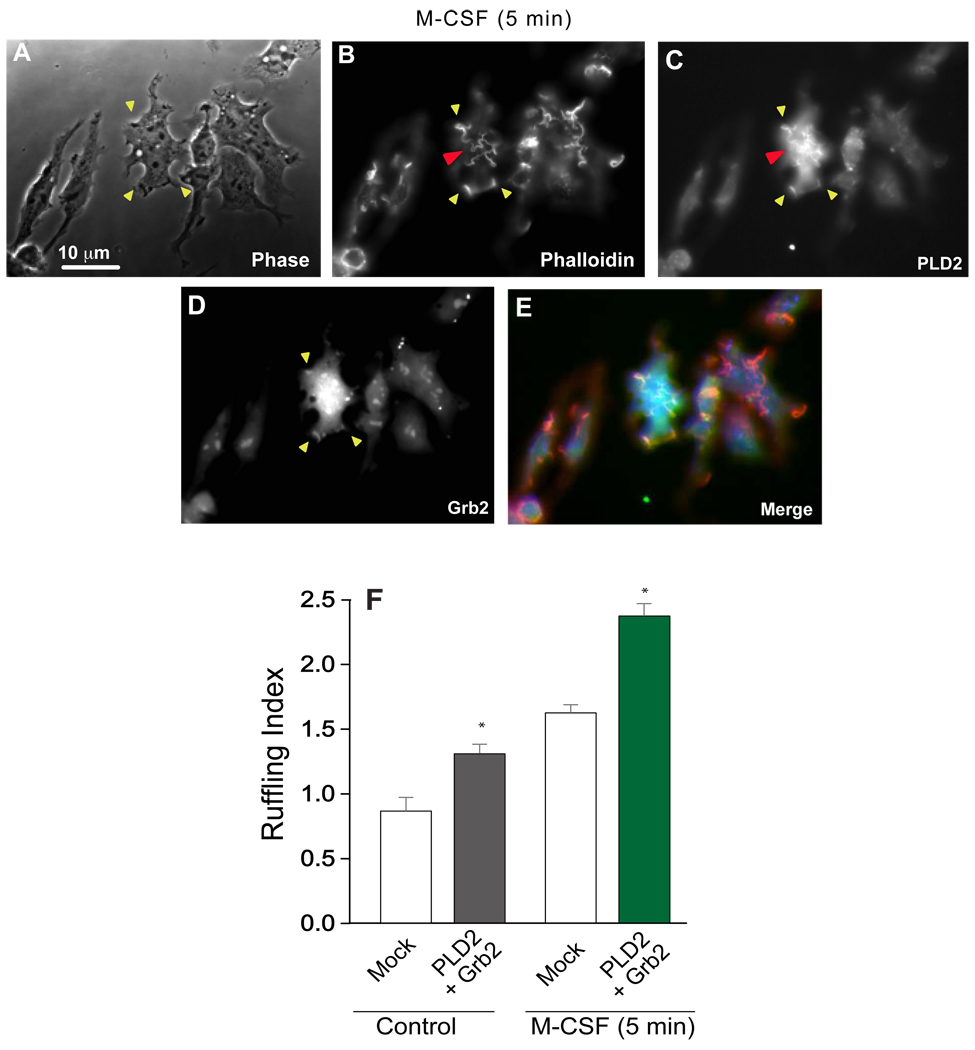
A new set of experiments was designed to study membrane ruffling in CSF-stimulated cells. For this, RAW/LR5 transfected cells were incubated in the presence of M-CSF (Fig. 3). Co-expression of Grb2 and PLD2 followed by stimulation with M-CSF for 5 min led to the formation of membrane ruffles (Fig. 3A–E) and, again, PLD2 and Grb2 colocalized in the ruffles. We noted that in these CSF-stimulated cells the presence of dorsal ruffles was increased (red arrows in Fig 3B,C). These ruffles are associated with membrane endocytosis and indicated that the receptor-ligand (M-CSFR/M-CSF) was internalized. Quantitation indicated that Grb2 + PLD2 increased ruffle formation almost 2-fold over controls in CSF-stimulated cells (Fig. 3F), which is slightly more than what we observed in basal cells. This led us to postulate the existence of a stimuli-dependent regulatory mechanism for binding activity in vivo.
Figure 3. Effect of PLD2 and Grb2 on cell ruffling during M-CSF stimulation.
(A-E) Effect of the growth factor (M-CSF, 3 nM) stimulation in PLD2- or Grb2-double transfected RAW/LR5 cells. Yellow arrows point to the actin-rich cell membrane ruffles. The red arrow points to the presence of dorsal ruffles indicative of receptor-ligand internalization. (F) Membrane ruffle quantitation.
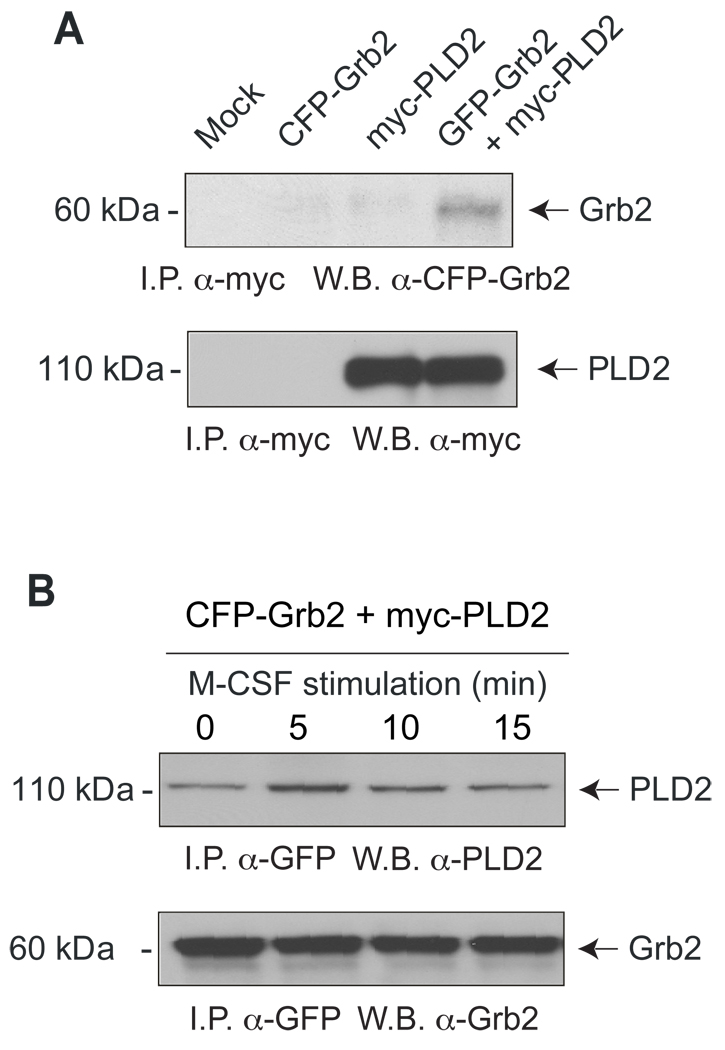
The results of previous figures point at a PLD2-Grb2 interaction but to confirm this, we carried over immunoprecipitation/Western blotting analyses. In resting macrophages, it was possible to observe an association Grb2-PLD2 as Grb2 was pulled down with antibodies directed to the PLD2 tag (Fig. 4A). We also investigated if the stimulation of cells with M-CSF, which according to Figure 3 led to an increased level of membrane ruffling, had any effect on the physical association of PLD2 and Grb2. We were also able to pull down PLD2 using antibodies against the fluorescent tag of Grb2 (Fig. 4B) also in resting cells (time zero), indicating again a PLD2-Grb2 association as was seen with the reverse antibodies in Fig. 4A. Interestingly, this band was more intense in the case of cells that had been stimulated with M-CSF prior to the preparation of cell lysates for 5–15 minutes with a maximal time of 5 minutes, which coincides with full activation of the receptor. Thus, full-length PLD2 co-immunoprecipitates with Grb2 maximally when cells are stimulated with growth factors, confirming the presence of a stimuli-dependent regulatory mechanism.
Figure 4. Molecular association between PLD2 and Grb2.
Co-immunoprecipitation of Grb2 and PLD2 from samples of cells that were either resting (A) or stimulated with the growth factor M-CSF for the indicated lengths of time (B). Cells were initially transfected with GFP-Grb2 and myc-PLD2. Cell lysates were immunoprecipitated with the indicated antibodies, and Western blots were obtained. Experiment typical among three total experiments.
3.5. Analysis of the enzymatic activity of PLD2 and on membrane ruffling
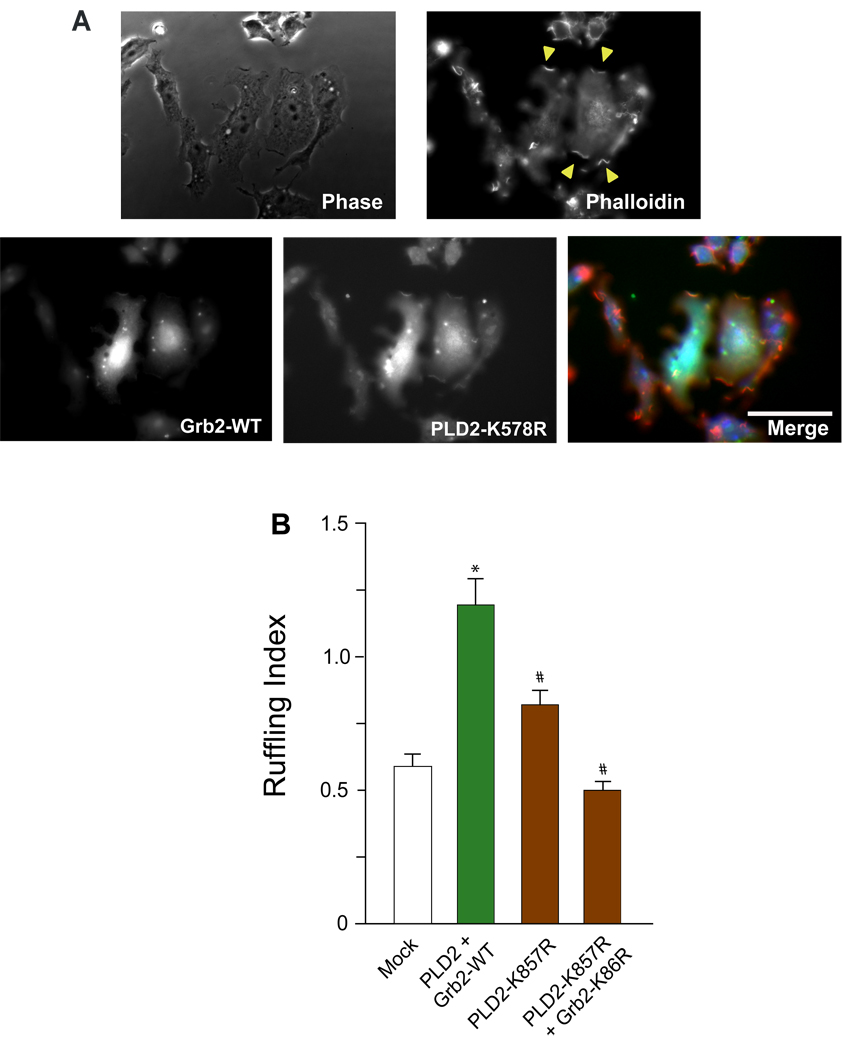
We next investigated if the enzymatic activity of PLD2 (i.e., the production of phosphatidic acid, PA) was required for the biological effect of membrane ruffles formation. For this, we used a lipase-inactive mutant of PLD2 (PLD2-K758R). Figure 5A indicates that transfection of macrophages with this mutant did not alter the subcellular localization of PLD2 and membrane ruffles could be readily observed and quantified. The ruffling index presented in Fig. 5B indicates nevertheless a decrease in the level of membrane ruffling with the PLD2-K758R mutant indicating that the enzymatic activity of PLD does play a role in the biological effect. However, the inhibition was only about 50%. A decrease it to basal levels only occurred only when the PLD-activity lacking mutant was combined with the SH2-impaired form of Grb2 (right most bar in Figure 5B). This indicates that PA production, as measured by PLD activity, is necessary but not sufficient to elicit membrane ruffling.
Figure 5. Effect of the lipase-inactive mutant PLD2 and Grb2 on cell ruffling.
(A) Show co-localization of PLD2-K758r mutant and Grb2 in membrane ruffles in PLD2- and Grb2-double transfected RAW/LR5 cells. Yellow arrows point to the actin-rich cell membrane ruffles. (B) Membrane ruffle quantitation.
3.6. The third factor for full membrane ruffling induction: Rac2
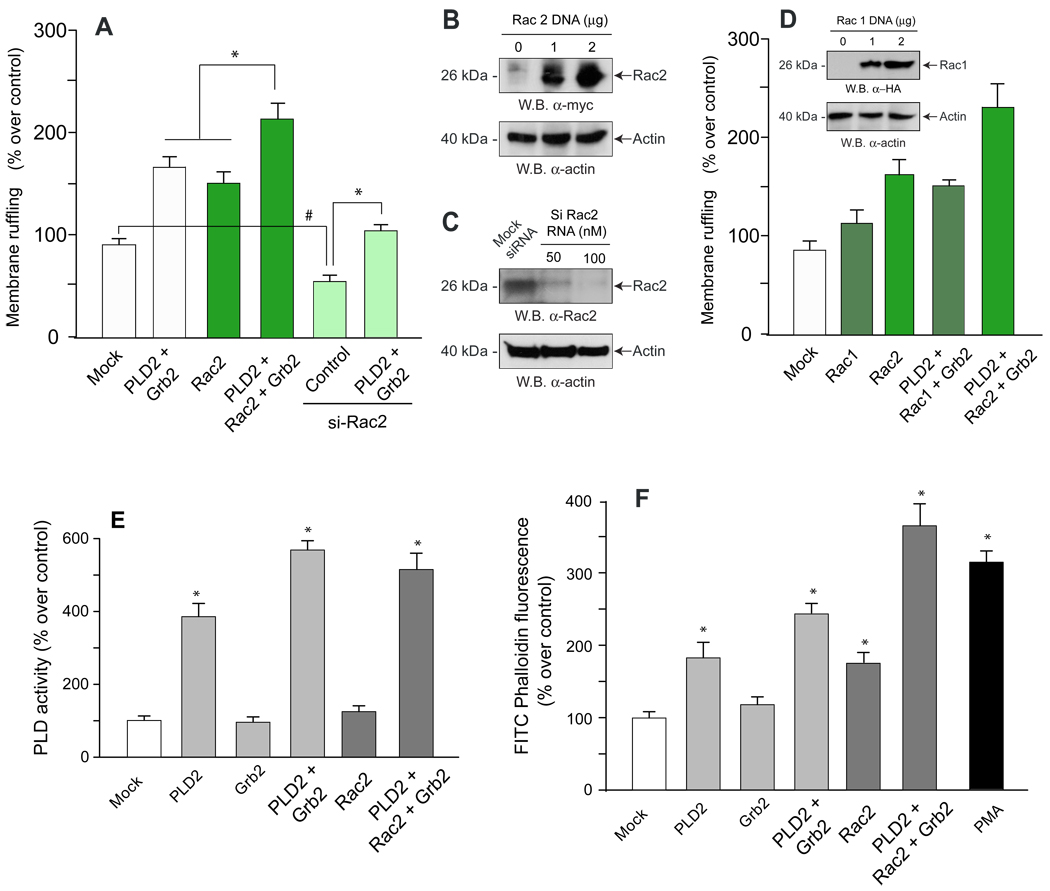
We decided to investigate what other factor(s) might be needed for a full physiological stimulation and found that it was the Rho-family GTPase Rac2. The rationale for this is that Rac2 is an activator of Arp2/3, the direct modulator of actin polymerization [19] that is necessary for cell membrane ruffling and formation of cell protrusions. As indicated in Fig. 6A, co-transfection of PLD2 and Grb2 benefited from the co-expression of Rac2. Additionally, Rac2 silencing of macrophages reduced by about 50% membrane ruffling, which could be somewhat rescued by Grb2 and PLD2 transfection, but not fully. Fig. 6B indicates that the expression of protein encoded by the pRK5-myc-Rac2 plasmid is very robust, and Fig. 6C shows that silencing of endogenous Rac2 was fully accomplished with siRNA. A test of specificity was devised since the other isoform of Rac, Rac1 is also known to activate actin polymerization. Rac1 overexpression also increase membrane ruffling (Fig. 6D and inset, for levels of protein expression of HA-Rac1), especially when in combination with Grb2 and PLD2 expression but the level of membrane ruffling was at least 50% lower than that accomplished with Rac2 and therefore we concentrated in Rac2 for subsequent studies. We next measured PLD’s enzymatic activity aimed at investigating the role that each transfection (individual or in combination) might have in modulating PLD2. Fig. 6E indicates that neither Grb2 nor Rac2 alone had an effect on PLD activity. Grb2, but not Rac2, in combination with PLD2 resulted in an enhanced activity, indicated that in the mechanism underlying membrane ruffling Grb2 acts at the level of PLD activity but Rac2 has to work at a different level, presumably after it. The fact that PA and related lipids bind to Rac2 [20] lends credibility to this scenario.
Figure 6. The presence of a third element, Rac2, confers full membrane ruffling.
(A) After transfection of LR5/RAW cells with either over-expressing plasmids or silencing RNA (si), cells were quantified for membrane ruffling. In the last experimental condition (bar at the outer right) both overexpresion and silencing were conducted. (B,C) Cell lysates were prepared to determine by immunoblot the level of expression of the pRK5-myc-Rac2 plasmid and also to detect the level of endogenous Rac2 protein after silencing with siRNA (D). A test of specificity was conducted with Rac1 (pcDNA3-HA-Rac1 plasmid) and both cell membrane ruffling and immunobloting (inset) were conducted). (E) In parallel to experiments of panels A,B and C, cell lysates were also prepared from which PLD enzyme activity was measured. (F) Flow cytometry. Cell suspensions were labeled with phalloidin-FITC and analyzed by flow cytometry. Fluorescence on gated macrophage populations were measured and plotted in the Y-axis as relative fluorescence with respect to the control. Results are the mean ± SEM from 3 independent experiments conducted in duplicate. (*) Denotes difference between means that were statistically significant (p<0.05) by ANOVA. (#) in panel A denotes inhibition that is significant (p<0.05) with respect to mock controls. pRK5-myc-Rac2 and pcDNA3-HA-Rac1
A further importance of Rac2 is that it brings closure to the PLD2/Grb2 mechanism of membrane ruffling by drawing in the ability of Rac to affect actin polymerization. Therefore, we analyzed the effect of PLD2/Grb2/Rac2 in actin polymerization measured by flow cytometry of phalloidin-loaded cells. As indicated in Fig. 6F, these three molecules had a robust enhancing potential of actin polymerization, close in quantity to that seen with the positive control PMA (bar to the furthest right). Thus, the cooperation between PLD2 and Grb2 and the presence of Rac2 lead to full actin polymerization, the molecular basis for membrane ruffling.
4. DISCUSSION
Our results show for the fist time a co-localization of the enzyme phospholipase D2 and the docking protein Grb2 in actin-rich membrane ruffles that has the physiological consequence of enhanced membrane ruffle formation in macrophages. PLD2 has the property to modulate the functionality of these cells even in the absence of any chemoatractant ("basal" conditions) and Grb2 further enhances that. Moreover, the intensity of membrane ruffle formation can be further increased in M-CSF-stimulated cells. This effect was demonstrated by biochemical analysis in parallel with physical observation of membrane ruffle formation with two fluorescent chimeras we prepared in the laboratory: CFP-Rac2 and YFP-PLD2.
As seen in Figure 5, a lipase-inactive mutant of PLD2 was able to reduce by about 50% membrane ruffle formation. But it was not until both the lipase-inactive mutant and a Grb2 mutant that has the SH2 domain compromised that the levels of membrane ruffling were equivalent to those of mock-transfected cells. This clearly indicated that another mechanism in addition to PLD activity (formation of phoshatidic acid, PA) was necessary to confer a full biological effect. This other pathway was found to be one utilizing Rac2, as confirmed in Figure 6 using both overexpression and silencing with RNA intereference (siRNA).
The cooperation between PA (due to an increased Grb2/PLD2 association) and Rac2 seems like a mechanism that the cell can use to its own advantage. Although there are no reports on this particular cooperation, there are some precedents for PLD and small GTPases (such as Rac and others) that function cooperatively. As known, Rac2 is involved in membrane ruffling [21]. On the other hand, GTPase ARF6 activates PI5-Kinase in a process dependent on PA, while both PLD2 and ARF6 translocate to membrane ruffles [21]. Further, continual production of PLD-derived PA is essential for antigen-stimulated membrane ruffling in mast cells [23]. Our study is in agreement with those two previous studies regarding the activity of PLD, but provides two new mechanistic elements, with uncovering of the particular residues involved: (a) the presence of a Grb2 anchor that serves to attract other signaling molecules through its SH2 domain to the actin-rich environment (specifically PLD2 via the residue Y169), which was observed at the cellular and at the molecular levels, and (b) the presence of the Rho-family small GTPase Rac2 that links the PLD2-Grb2 dimer to actin polymerization. All these three molecules, when combined, are capable of enhancing actin polymerization in a very powerful way, which is a prerequisite for membrane protrusion/cell ruffling. As far as the activation of Rac, there have been described guanine nucleotide exchange factors (GEFs) such as SWAP-70 that can activate Rac, independently of Ras but dependently on phosphatidylinositol(3,4,5) trisphosphate and move together to membrane ruffles [24].
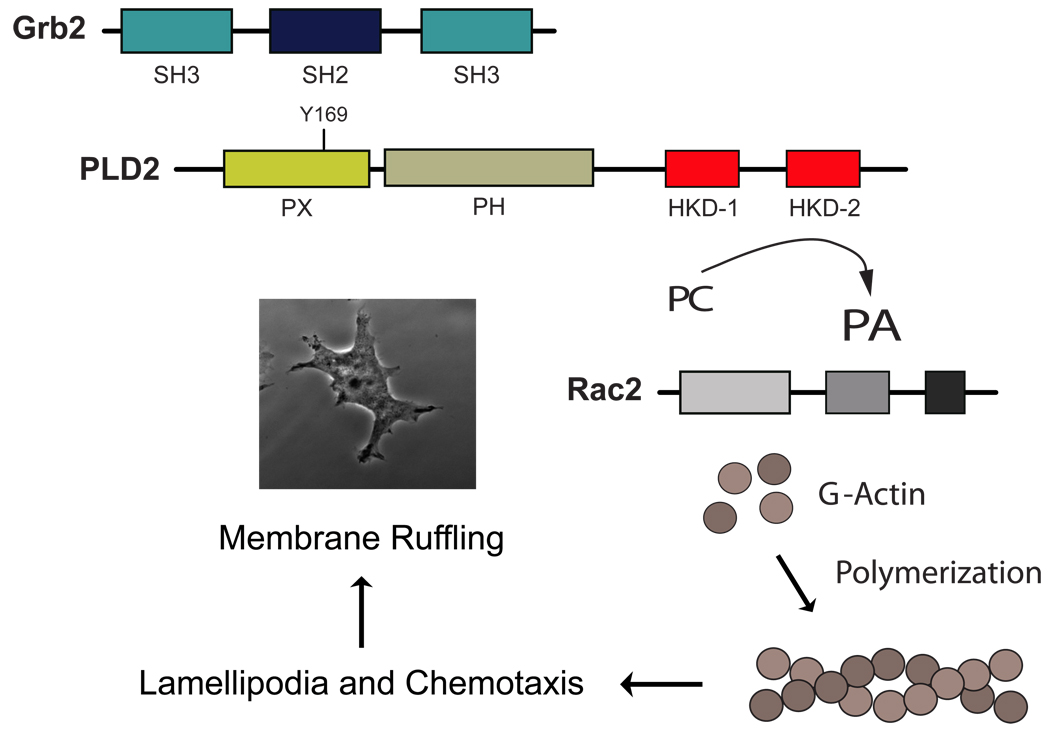
As presented in the model of Fig. 7, PLD after binding to Grb2-SH2 to the Y169 residue within the PH domain of PLD2, enhanced the activity of the lipase by increasing the production of phosphatidic acid (PA). Although this increase in PA is necessary, it is not sufficient for full biological action. A further cooperation with Rac2 is needed to fully stimulate actin polymerization, which is key to the formation of membrane ruffling and related migratory events. As membrane ruffling precedes lamellipodia formation and the initiation of cell migration, a PLD2-PA/Grb2/Rac2 interaction opens new mechanistic pathways for membrane remodeling in leukocytes and other important migratory cells.
Figure 7. Model summarizing the new findings of this study.
PLD enzymatic action, after been increased due to the binding to Grb2-SH2 via Y169, cooperates with Rac2, and the three molecules stimulate actin polymerization and consequently, membrane ruffle formation. Since membrane ruffling precedes cell migration, the results herein provide a novel mechanism for control of membrane dynamics, crucial for the physiology of leukocytes.
ACKNOWLEDGMENTS
This work has been supported by the National Institutes of Health grants HL056653 (JGC) and GM071828 (DC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Small JV, Stradal T, Vigna E, Rottner K. The lamellipodium: where motility begins. Trends Cell Biol. 2002;12:112–120. doi: 10.1016/s0962-8924(01)02237-1. [DOI] [PubMed] [Google Scholar]
- 2.Sorkin A. Internalization of the epidermal growth factor receptor: role in signaling. Biochem Soc Trans. 2001;29:480–484. doi: 10.1042/bst0290480. [DOI] [PubMed] [Google Scholar]
- 3.van Larebeke NA, Bracke ME, Mareel MM. Invasive epithelial cells show more fast plasma membrane movements than related or parental non-invasive cells. Cytometry. 1992;13:9–14. doi: 10.1002/cyto.990130104. [DOI] [PubMed] [Google Scholar]
- 4.Srinivasan D, Plattner R. Activation of Abl tyrosine kinases promotes invasion of aggressive breast cancer cells. Cancer Res. 2006;66:5648–5655. doi: 10.1158/0008-5472.CAN-06-0734. [DOI] [PubMed] [Google Scholar]
- 5.Yamada H, Padilla-Parra S, Park SJ, Itoh T, Chaineau M, Monaldi I, Cremona O, Benfenati F, DeCamilli P, Coppey-Moisan M, Tramier M, Galli T, Takei K. Dynamic interaction of amphiphysin with N-RAC2 regulates actin assembly. J Biol Chem. 2009;284:34244–34256. doi: 10.1074/jbc.M109.064204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 7.Miki H, Suetsugu S, Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 1998;17:6932–6941. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kheir WA, Gevrey JC, Yamaguchi H, Isaac B, Cox D. A WAVE2-Abi1 complex mediates CSF-1-induced F-actin-rich membrane protrusions and migration in macrophages. J Cell Sci. 2005;118:5369–5379. doi: 10.1242/jcs.02638. [DOI] [PubMed] [Google Scholar]
- 9.Di Fulvio M, Frondorf K, Gomez-Cambronero J. Mutation of Y179 on phospholipase D2 (PLD2) affects DNA synthesis in a PI3K-dependent manner. Cell Signaling. 2008;20:176–185. doi: 10.1016/j.cellsig.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox D, Chang P, Zhang Q, Reddy PG, Bokoch GM, Greenberg S. Requirements for both Rac1 and Cdc42 in membrane ruffling and phagocytosis in leukocytes. J. Exp. Med. 1997;186:1487–1494. doi: 10.1084/jem.186.9.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frondorf K, Henkels KM, Frohman MA, Gomez-Cambronero J. Phosphatidic acid is a leukocyte chemoattractant that acts through S6 kinase signaling. J Biol Chem. 2010;285:15837–15847. doi: 10.1074/jbc.M109.070524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo Y, Isaac BM, Casadevall A, Cox D. Phagocytosis inhibits F-actin-enriched membrane protrusions stimulated by fractalkine (CX3CL1) and colony-stimulating factor 1. Infect Immun. 2009;77:4487–4495. doi: 10.1128/IAI.00530-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lehman N, Di Fulvio M, McCray N, Campos I, Tabatabaian F, Gomez-Cambronero J. Phagocyte cell migration is mediated by phospholipases PLD1 and PLD2. Blood. 2006;108:3564–3572. doi: 10.1182/blood-2006-02-005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powner DJ, Pettitt TR, Anderson R, Nash GB, Wakelam MJ. Stable adhesion and migration of human neutrophils requires phospholipase D-mediated activation of the integrin CD11b/CD18. Mol Immunol. 2007;44:3211–3221. doi: 10.1016/j.molimm.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 15.Gorshkova I, He D, Berdyshev E, Usatuyk P, Burns M, Kalari S, Zhao Y, Pendyala S, Garcia JG, Pyne NJ, Brindley DN, Natarajan V. Protein kinase C-epsilon regulates sphingosine 1-phosphate-mediated migration of human lung endothelial cells through activation of phospholipase D2, protein kinase C-zeta, and Rac1. J Biol Chem. 2008;283:11794–11806. doi: 10.1074/jbc.M800250200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knoepp SM, Chahal MS, Xie Y, Zhang Z, Brauner DJ, Hallman MA, Robinson SA, Han S, Imai M, Tomlinson S, Meier KE. Effects of active and inactive phospholipase D2 on signal transduction, adhesion, migration, invasion, and metastasis in EL4 lymphoma cells. Mol Pharmacol. 2008;74:574–584. doi: 10.1124/mol.107.040105. [DOI] [PubMed] [Google Scholar]
- 17.Itoh T, Hasegawa J, Tsujita K, Kanaho Y, Takenawa T. The tyrosine kinase Fer is a downstream target of the PLD-PA pathway that regulates cell migration. Sci Signal. 2009;2:52. doi: 10.1126/scisignal.2000393. [DOI] [PubMed] [Google Scholar]
- 18.Di Fulvio M, Lehman N, Lin X, Lopez I, Gomez-Cambronero J. The elucidation of novel SH2 binding sites on PLD2. Oncogene. 2006;25:3032–3040. doi: 10.1038/sj.onc.1209340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim DJ, Kim SH, Lim CS, Choi KY, Park CS, Sung BH, Yeo MG, Chang S, Kim JK, Song WK. Interaction of SPIN90 with the Arp2/3 complex mediates lamellipodia and actin comet tail formation. J Biol Chem. 2006;281:617–625. doi: 10.1074/jbc.M504450200. [DOI] [PubMed] [Google Scholar]
- 20.Bourdon DM, Wing MR, Edwards EB, Sondek J, Harden TK. Quantification of isozyme-specific activation of phospholipase C-beta2 by Rac GTPases and phospholipase C-epsilon by Rho GTPases in an intact cell assay system. Methods Enzymol. 2006;406:489–499. doi: 10.1016/S0076-6879(06)06037-X. [DOI] [PubMed] [Google Scholar]
- 21.Miki H, Takenawa T. Regulation of actin dynamics by RAC2 family proteins. J Biochem. 2003;134:309–313. doi: 10.1093/jb/mvg146. [DOI] [PubMed] [Google Scholar]
- 22.Honda A, Nogami M, Yokozeki T, Yamazaki M, Nakamura H, Watanabe H, Kawamoto K, Nakayama K, Morris AJ, Frohman MA, Kanaho Y. Phosphatidylinositol 4- phosphate 5-kinase alpha is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell. 1999;99:521–532. doi: 10.1016/s0092-8674(00)81540-8. [DOI] [PubMed] [Google Scholar]
- 23.O'Luanaigh N, Pardo R, Fensome A, Allen-Baume V, Jones D, Holt MR, Cockcroft S. Continual production of phosphatidic acid by phospholipase D is essential for antigen-stimulated membrane ruffling in cultured mast cells. Mol Biol Cell. 2002;13:3730–3746. doi: 10.1091/mbc.E02-04-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shinohara M, Terada Y, Iwamatsu A, Shinohara A, Mochizuki N, Higuchi M, Gotoh Y, Ihara S, Nagata S, Itoh H, Fukui Y, Jessberger R. SWAP-70 is a guanine-nucleotide-exchange factor that mediates signaling of membrane ruffling. Nature. 2002;416:759–763. doi: 10.1038/416759a. [DOI] [PubMed] [Google Scholar]