Abstract
Cationic polymers are potential intracellular carriers for small interfering RNA (siRNA). The short and rigid nature of an siRNA chain often results in larger and more loosely packed particles compared to plasmid DNA (pDNA) after complexing with carrier polycations, and in turn, poor silencing effects are seen against the target mRNAs. A helper polyanion, pDNA, was incorporated along with siRNA to form compact nanosized polyplexes. At C/A (cation/anion) ratios of 2 and 5, poly(L-lysine) (PLL)/siRNA-pGFP and PLL/siRNA-pGFP-OSDZ (oligomeric sulfadiazine (OSDZ) for endosomolysis) complexes produced particles 90–150 nm in size with a 15–45 mV surface charge, while PLL/siRNA complexes yielded particles 1–2 μm in size at the same C/A ratios. The PLL/siRNA-pGFP (C/A 2) complexes showed significantly higher specific gene silencing (50–90% vs. 10–25%) than the complexes formed at C/A 5. PLL/siRNA-pGFP-OSDZ (C/A 2) complexes improved the specific gene silencing (90%) more dramatically than PLL/siRNA-pGFP (C/A 2) complexes (50%), demonstrating a potential role for OSDZ. PLL/siRNA-pGFP-OSDZ (C/A 2) complexes sustained higher specific gene silencing compared with PLL/siRNA-pGFP (C/A 2) complexes. Other oligomeric sulfonamides (OSA) with varying pKa used in PLL/siRNA-pGFP-OSA complexes also caused effective gene silencing. The pGFP in the PLL/siRNA-pGFP complexes successfully expressed GFP protein without interfering with the siRNA. In conclusion, this study demonstrates that long pDNA helps effectively form nanosized siRNA particles and that OSA enhances specific gene silencing. In a single nucleic acid carrier formulation, co-delivery of siRNA and pDNA is feasible to maximize therapeutic effects or to include therapeutic or diagnostic functionalities.
1. Introduction
RNA interference (RNAi) has been recognized as a promising therapy to induce sequence-specific and post-transcriptional silencing via the cleavage of homologous mRNA [1, 2] to treat cancers, infectious diseases, and CNS diseases [1]. RNAi is most often mediated by short hairpin RNAs (shRNA) or small interfering RNAs (siRNA) [2]. The shRNA is expressed after nuclear delivery of shRNA-expressing plasmid DNA (pDNA) and the duration of shRNA expression depends on the use of viral or non-viral vectors [2, 3]. The transient expression of shRNA carried by non-viral vectors is strongly influenced by dilution of the vector following cell proliferation and by the stability of siRNA in the cytosol [2, 3]. The use of siRNA has several advantages over shRNA. siRNA delivery avoids the barrier of the nuclear membrane as it acts in the cytosol, unlike shRNA-expressing pDNA [2, 3]. More importantly, siRNA duplexes are readily available from various sources via custom or pre-designed siRNAs targeting a particular mRNA of interest.
As unaided delivery of naked siRNA is very limited due to its anionic nature and resulting low permeability to plasma membranes [2, 4], polymer-based (polyplex and polymersome) or lipid-based (lipoplex and liposome) non-viral vectors are often used to aid in siRNA delivery [2, 5–8]. While lipoplexes often aggregate or are unstable over time [9], polymeric siRNA vectors are more attractive due to their chemical diversity and ease of modifications [4, 10] for cell targeting, endosomal escape [11], and cytosolic release [8, 12, 13] of siRNA.
However when cationic polymers are used, siRNA frequently results in more loosely condensed and larger complexed particles than are seen under similar complexing conditions when used for pDNA polyplexes of 100–200 nm in size. Along with size concerns for use in in vivo tests, these less stable polymeric siRNA polyplexes were prone to premature dissociation of siRNA from the counterpart polycation [14]. This is the result of the siRNA chain being short and rigid [14], unlike long and flexible pDNA. In general, the electrostatic strength between polycations and polyanions (here, nucleic acids) increases with higher molecular weights (MW) [14, 15]. These short siRNAs (usually duplexes of 21–23 nucleotides) may result in one hundredth of the electrostatic strength of pDNA (more than 2000 base pairs) in an association with a given polycation [16]. Thus, to form stable and nanoscale siRNA polyplexes, one should adopt higher molecular weight (MW) polymers or high polycation/siRNA ratios [10]. However, excess polycation induces polycation-mediated cytotoxicity [17]. Alternatively, siRNA can be elongated by concatemerization with sticky overhangs [16] or multimerization via chemical linkages [18]. To avoid off-target silencing effects of lengthy siRNAs, these multimeric siRNAs can be designed to be cleaved into monomeric siRNAs in the cytoplasm.
Instead of modifying siRNAs, some groups have incorporated polyanions as an additive [19, 20]. The Huang group added calf thymus DNA or hyaluronic acid (HA) when formulating siRNA polyplexes with protamine and then the siRNA/protamine/DNA (or HA) nanoparticles were encapsulated in liposomes [19, 20]. This strategy resulted in a particle size reduction by 10–30% and improved silencing efficiency 20–80% when compared with siRNA formulations without polyanion additives [5, 19–21].
In this study, polymer-based siRNA carriers were used to exclude the instability of liposome-based formulations. Poly(L-lysine) (PLL) was selected as a representative polycation. Oligomeric sulfonamides (OSA) with known endosomolytic activity that have been developed by our group [22] were also introduced into the PLL-based siRNA complexes. In addition, to enhance the stability of polymeric siRNA complexes as well as to examine the potential of including multiple therapeutics in a single drug carrier, a reporter pDNA was employed as a lengthy polyanion instead of calf thymus DNA or HA. This study aimed to elucidate how the addition of pDNA in PLL/siRNA-pDNA and PLL/siRNA-pDNA-OSA complexes modulates their physicochemical, biological, and therapeutic characteristics.
2. Materials and methods
2.1. Materials and cell culture
Poly(L-lysine) hydrogen bromide (PLL·HBr; MW 24 kDa), branched polyethyleneimine (bPEI; Mw 25 kDa, Mn 10 kDa), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), Dulbecco’s Modified Eagle’s Medium (DMEM), Ca2+-free and Mg2+-free Dulbecco’s phosphate buffered saline (DPBS), dimethyl sulfoxide (DMSO), 4-(2-hydroxy-ethyl)-1-piperazine (HEPES), D-glucose, sodium bicarbonate, ethidium bromide (EtBr), aphidicolin, heparin sodium salts (149 USP units/mg), 4′,6-diamidino-2-phenylindole (DAPI), paraformaldehyde (PFA), fetal bovine serum (FBS), penicillin-streptomycin antibiotics, and trypsin-EDTA solution were purchased from Sigma-Aldrich (St. Louis, MO). YOYO-1 was purchased from Invitrogen, Inc. (Carlsbad, CA). Plasmid DNA (pDNA) encoding firefly luciferase (gWiz-Luc or pLuc) or green fluorescent protein (gWiz-GFP or pGFP) was purchased from Aldevron, Inc. (Fargo, ND). Fluorescein-labeled pDNA (F-pDNA) (i.e., pGeneGrip Fluorescein/β-galactosidase (F-pGal)) was purchased from Genlantis, Inc. (San Diego, CA). Small interfering RNA (siRNA) such as Silencer® firefly luciferase (GL2+GL3) siRNA (siLuc), Silencer® negative control siRNA #1 (siCtrl), and Silencer® Cy™3-labeled negative control siRNA #1 (Cy3-siRNA) were bought from Applied Biosystems/Ambion, Inc. (Austin, TX). Luciferase assay kit and BCA™ protein assay kit were purchased from Promega Corporation (Madison, WI) and Pierce Biotechnology, Inc. (Rockford, IL), respectively. Oligosulfonamides (OSA) such as oligomeric sulfamerazine (OSMZ), oligomeric sulfadiazine (OSDZ), oligomeric sulfadimethoxine (OSDM), and oligomeric sulfamethizole (OSMT) were synthesized as previously reported and their MWs were 1.8–2.5 kDa [22].
HEK293 cells (human embryonic kidney cell line) were cultured in DMEM supplemented with D-glucose (4.5 g/L) and 10% FBS in humidified air containing 5% CO2 at 37°C. The proliferating cells are called P-HEK293. P-HEK293 cells treated with aphidicolin (1 μg/mL) to arrest their growth are called GA-HEK293. GA-HEK293 cells were prepared 1 day after being treated with aphidicolin and their growth-arrested status was maintained with continuous treatment of aphidicolin (1 μg/mL).
2.2. Physicochemical characterizations of PLL/siRNA, PLL/siRNA-pDNA, and PLL/siRNA-pDNA-OSA complexes
Varying amounts of PLL (in 10 μL) were mixed with siRNA (50 pmol in 10 μL) in a HEPES solution (pH 7.4, 20 mM). The mixture (20 μL per 50 pmol of siRNA) was incubated for 30 min at room temperature (RT), resulting in PLL/siRNA complexes. For PLL/siRNA-pDNA complexes, 0.5 μg of pDNA was added to the siRNA solution (50 pmol). The resulting nucleic acid solution (10 μL) was mixed with the PLL solution (10 μL) to form PLL/siRNA-pDNA complexes. In the cases of PLL/siRNA-pDNA-OSA complexes, 0.5–7.5 nmol (based on monomeric sulfonamides) of anionic OSA was additionally added to the nucleic acid solutions (50 pmol of siRNA or a mixture of siRNA (50 pmol) and pDNA (0.5 μg)). The C/A ratio (cations (C) from polycation per anion (A) from siRNA, pDNA and OSA) was used as the complexation ratio in this study.
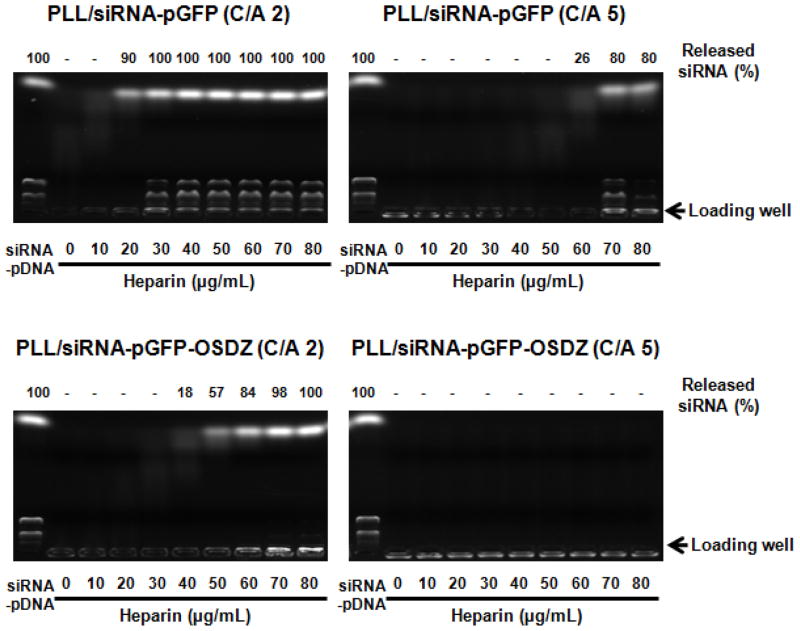
The condensation of nucleic acids with PLL was monitored using a gel electrophoresis assay. The PLL/siRNA, PLL/siRNA-pDNA and PLL/siRNA-pDNA-OSA complexes (25 μM of siRNA) formed at differing complexation ratios were loaded into the wells of a 0.8% wt/vol agarose gel (100 mL) supplemented with EtBr (20 μL of 0.625 mg/mL). A constant voltage (50 V) was applied to the polyplex-loaded gel in 0.5X TBE buffer for 100 min. Uncomplexed or exposed siRNA and pDNA in the polyplexes were detected with a UV illuminator. In addition, to evaluate the exposure and release of siRNA and pDNA from the polymeric nucleic acid complexes following polyanion challenge, the polyplexes were exposed to a model polyanion (i.e., heparin) in 150 mM NaCl for 1 hr, and then the mixed solution (12.5 μM of siRNA) was subjected to electrophoresis.
The surface charge and particle size of the polyplexes were measured with a Zetasizer 3000HS (Malvern Instrument, Inc, Worcestershire, UK) at a wavelength of 677 nm and a constant angle of 90° at RT. The concentration of siRNA in the polyplex solution was set to 125 nM.
2.3. Biological characterizations of PLL and PLL/siRNA, PLL/siRNA-pDNA, and PLL/siRNA-pDNA-OSA complexes
To evaluate the efficiency of gene silencing, we used sequential co-transfection to first express luciferase in HEK293 cells using a luciferase plasmid (pLuc) followed by silencing the mRNA of luciferase using a luciferase siRNA (siLuc). The siCtrl was applied to pLuc-transfected HEK293 cells to characterize any non-specific silencing (off-target) effect. In detail, HEK293 cells were seeded at a density of 5×105 cells/well (in 6-well plates) and the cells were cultured for 24 hr. One hour before transfection, the complete culture medium was replaced with serum-free medium. The bPEI/pLuc (C/A 5) complexes (1 μg of pLuc per well) were then added to HEK293 cells. After a 10 min or 24 hr, time interval of co-transfection (TICO), the cells were treated with PLL or PLL/siRNA, PLL/siRNA-pDNA, or PLL/siRNA-pDNA-OSA complexes, followed by incubation for 4 hr in serum-free medium. After the culture medium was replaced with serum-containing medium, the cells were further incubated for 20–68 hr. For TICO 24 hr, the transfected cells were cultured in complete culture medium for the time between bPEI/pLuc (C/A 5) transfection and siRNA polyplex addition. When the silencing experiments were complete, the cells were rinsed with DPBS and lysed in a lysis buffer. The relative luminescence units (RLU) of the expressed luciferase and the whole protein content of the transfected cells were measured according to Promega’s luciferase assay protocol and Pierce’s BCA™ protein assay protocol.
Following treatment with PLL and siRNA polyplexes, an MTT-based cytotoxicity assay was carried out on the bPEI/pLuc-transfected HEK293 cells similarly to the in vitro silencing studies. However, HEK293 cells were seeded in 12-well plates at a density of 2.5×105 cells/well and then the polyplexes were applied to the cells incubated in 1 mL of culture medium. After completion of the silencing studies, 0.1 mL of the MTT solution (5 mg/mL) was added into the cells. After a 4 hr incubation, the cytotoxicity of the polyplexes was measured as previously described [23].
The cellular uptake of bPEI/pDNA, PLL/siRNA-pDNA, or PLL/siRNA-pDNA-OSA complexes was evaluated using YOYO-1-intercalated nucleic acids as previously described [23, 24]. These experiments were performed similarly to in vitro silencing studies. After a 4 hr incubation with polyplexes, including YOYO-1-intercalated nucleic acids, cells were detached from plates and fixed with a 4% PFA solution. The cells containing fluorescent polyplexes were monitored using a FACSCANTO flow cytometry system (Becton Dickinson, Franklin Lakes, NJ) equipped with a primary argon laser (488 nm) and a fluorescence detector (530±15 nm) for detecting YOYO-1. The uptake of the polyplexes into the cells was analyzed using a gated population of at least 5,000 cells. In particular, to determine whether siRNA-pGFP(-OSA) polyplexes can interfere with the cellular uptake of bPEI/pLuc complexes, YOYO-1 was intercalated into pLuc (abbreviated as pYOYO). In addition, to evaluate cellular uptake of siRNA-pGFP(-OSA) polyplexes in bPEI/pLuc-transfected HEK293 cells, the dye was intercalated into the siRNA (abbreviated as siYOYO).
To identify the subcellular localization of siRNA and pDNA delivered by PLL/siRNA-pDNA or PLL/siRNA-pDNA-OSA complexes (50 pmol siRNA and 0.5 μg pDNA in 20 μL), HEK293 cells were seeded on coverslips as previously described in the silencing experiments [23, 24]. The PLL/siRNA-pDNA and PLL/siRNA-pDNA/OSA complexes were prepared using Cy3-siRNA and F-pDNA and then the complexes were added to the cells. After a 4 hr incubation, the cells were rinsed with DPBS and fixed with 4% PFA. Thirty min prior to completing the 4 hr incubation, the nuclei were stained with DAPI. The cells were evaluated with a laser scanning confocal microscope equipped with excitation lasers (408 nm for the diode, 488 nm for Ar, and 543 nm for HeNe) and variable band-pass emission filters. Confocal images were collected in 500-nm sections and used to construct images of the entire cell.
To evaluate the potential for dual nucleic acid delivery, pGFP expression was also induced. The preparation was similar to the previously described in vitro transfection and intracellular trafficking studies [23, 24] except for the cell number (1×105 cells). Forty-eight hr after loading polyplexes, GFP expression was evaluated using laser scanning confocal microscopy at a 488 nm excitation wavelength.
The statistical significance of the data was evaluated by conducting an unpaired Student t-test at a confidence level of p<0.05.
3. Results and discussion
3.1. Physicochemical properties and transfection of PLL/siRNA complexes
Prior to the investigation of PLL/siRNA-pDNA complexes, the physicochemical and transfection properties of the PLL/siRNA complexes were studied as a reference. PLL was used to condense siRNA over a complexation range of 2 ≤ C/A (cation/anion) ≤ 20 (Fig. 1(A)). The PLL/siRNA complexes were retained well when run on an agarose gel. However, a weak EtBr intensity (approximately 10–20% of the EtBr intensity when EtBr is intercalated with all of the siRNA) from the polyplexes was still detected. This is likely caused by either siRNA exposed on the surface of the polyplexes or by loosely condensed polyplexes.
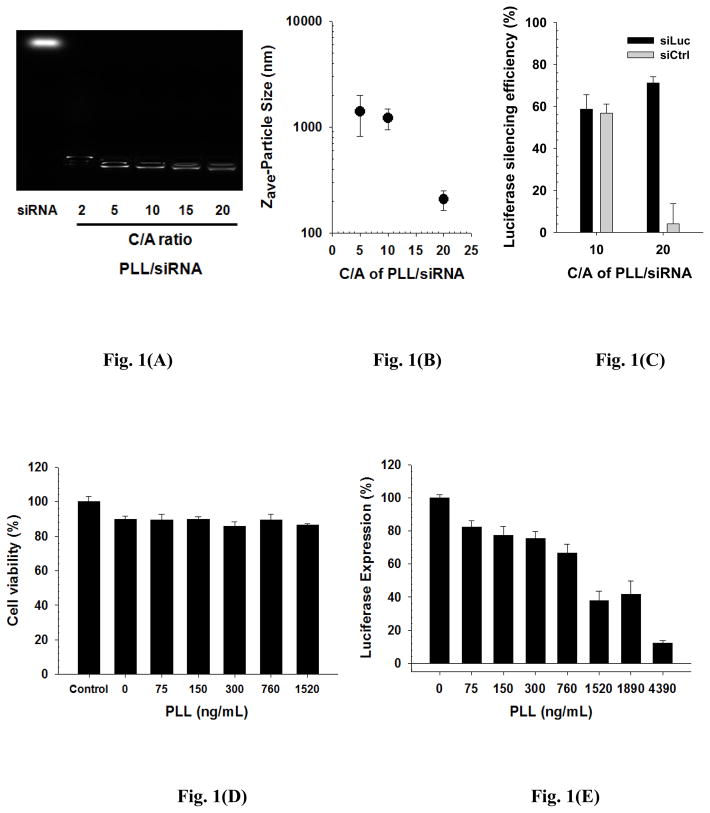
Fig. 1.
Physicochemical and transfection characteristics of PLL/siRNA complexes ((A) gene condensation assay in an agarose gel, (B) particle sizes (n=3; Mean±SD), and (C) luciferase silencing efficiency (25 nM of siRNA; n=3; Mean±SEM)) and transfection characteristics of uncomplexed PLL ((D) cytotoxicity (n=4; Mean±SEM)) and (E) luciferase expression (n=4; Mean±SEM)). PLL/siRNA complexes were prepared in a HEPES solution (pH 7.4; 20 mM). At 1 day post-silencing, transfection characteristics (C, D, and E) were evaluated using bPEI/pLuc-transfected P-HEK293 cells (TICO 10 min).
At lower C/A (i.e., 5 and 10) ratios the particles were 1–2 μm in diameter, whereas PLL/siRNA complexes at C/A 20 reached approximately 200 nm (Fig. 1(B)). In general when preparing PLL/pDNA complexes, a low C/A (for example C/A 3) will result in 100–200 nm polyplexes [25]. Thus, the much larger size of PLL/siRNA complexes arises from the lower binding strength of PLL to nucleic acids (short siRNA vs. long pDNA) as previously discussed [16].
To understand the effects of the size of PLL/siRNA complexes on silencing, two PLL/siRNA complexes formed at either C/A 10 or C/A 20 (25 nM of siRNA) were delivered to bPEI/pLuc-transfected P-HEK293 cells (TICO 10 min). When a specific siRNA (siLuc) targeting the mRNA of luciferase was used, the luciferase silencing efficiencies were approximately 59% and 71%, respectively, and the difference between their silencing efficiencies was not statistically significant (p=0.11) (Fig. 1(C)). Interestingly, however, PLL/siRNA complexes with control siRNA (siCtrl) showed approximately 57% and 4% luciferase silencing efficiencies at C/A 10 and C/A 20, respectively (p<0.001). By subtraction, this means that the specific luciferase silencing efficiencies of the PLL/siLuc complexes were approximately 2% and 67% at C/A 10 and C/A 20. This indicates that the nanosized siRNA polyplexes (C/A 20) are significantly more effective for specific silencing than the microparticle siRNA polyplexes (C/A 10), whereas the micron-sized siRNA polyplexes mostly caused non-specific silencing.
In general, looser polyplexes (lower C/A) can be more easily dissociated than tighter polyplexes (higher C/A). Dissociation consequently increases the concentration of free polycations, which are more toxic than polycations complexed with nucleic acids [26]. Thus, non-specific gene silencing may be induced by uncomplexed polycations (i.e., polymer-mediated off-target effects). To investigate this hypothesis, bPEI/pLuc-transfected P-HEK293 cells (TICO 10 min) were exposed to varying amounts of PLL as a model of uncomplexed polycations. That is, when bPEI/pLuc (C/A 5) complexes and uncomplexed PLL were co-delivered, the effects on cytotoxicity and luciferase expression were monitored. As shown in Fig. 1(D), PLL-mediated cytotoxicity at concentrations up to 1520 ng/mL of PLL (86%-90% cell viability) was not different (no statistical significance (p=0.1–0.9)) from the cytotoxicity of bPEI/pLuc-transfected P-HEK293 cells (90% cell viability). However, free PLL reduced the expression level of luciferase in a dose-dependent manner (e.g., 82% at 75 ng/mL (p=0.001) and 10% at 4390 ng/mL (p<0.001) compared to 100% (no reduction) at 0 ng/mL) as shown in Fig. 1(E). These results suggest that free or dissociated polycations can cause non-specific gene silencing in the presence of pDNA polyplexes.
To understand clearly whether the off-target effects or non-specific silencing of uncomplexed polycations are influenced by the existence of polyplexes, different amounts of PLL were applied to bPEI/pLuc-transfected P-HEK293 cells (TICO 24 hr) because this experimental condition allows us to understand the non-specific silencing effects of PLL in the absence of polyplexes, unlike the TICO 10 min condition. As shown in Fig. S1(A), free PLL (up to 8 μg/mL) was not cytotoxic to bPEI/pLuc-transfected P-HEK293 cells. However, free PLL still caused approximately 20% non-specific silencing (Fig. S1(B)).
These TICO-dependent non-specific silencing results indicate that PLL in addition to polyplexes can cause more non-specific silencing than PLL alone. It is unclear why PLL induces non-specific gene silencing. More detailed investigation will be required to understand this finding. However, based on the observed PLL/siRNA-mediated silencing effects, we suggest that polymeric siRNA complexes should be compact and that the amount of polycation used for complexation and the free polycations after complexation should be minimized to avoid off-target effects.
3.2. Physicochemical characteristics of PLL/siRNA-pDNA complexes
High MW polyanions have previously been introduced into siRNA polyplexes to obtain stable and compact siRNA polyplexes. In this study, pDNA was selected as the polyanion due to its potential for use in dual therapeutic delivery and imaging in disease models in small animals. The complexes were further supplemented by adding OSA, developed by Bae’s group [22], to compensate for the lack of endosomolytic activity of PLL. Using siRNA and the pDNA pGFP as a reporter gene, PLL/siRNA-pGFP and PLL/siRNA-pGFP-OSDZ complexes were prepared and characterized in terms of condensation, particle size, and surface charge as shown in Fig. 2.
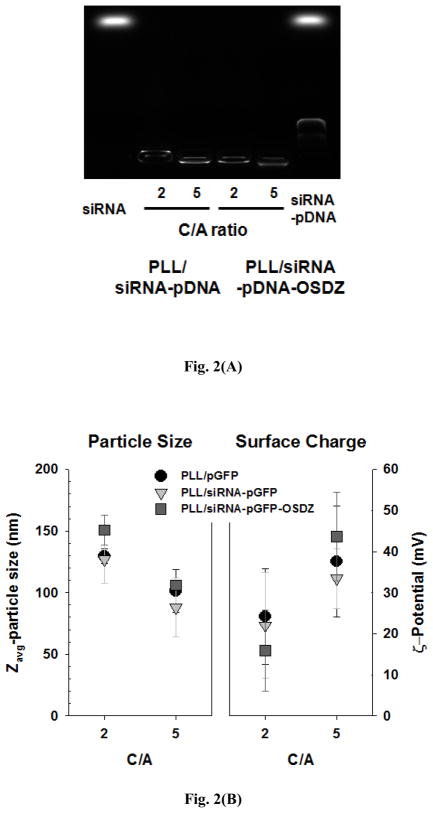
Fig. 2.
Physicochemical characteristics of PLL/siRNA-pGFP and PLL/siRNA-pGFP-OSDZ complexes: (A) gene condensation assay in an agarose gel and (B) particle sizes (n=3; Mean±SD) and surface charges (n=3; Mean±SD). The polyplexes were prepared in a HEPES solution (pH 7.4; 20 mM).
The siRNA-pGFP(-OSDZ) polyplexes prepared at C/A 2 and C/A 5 were retained in the wells of the agarose gel. Approximately 10%-20% of total nucleic acid (i.e., siRNA and pDNA) in the complexes was detected (Fig. 2(A)). The size and surface charge of the PLL/siRNA-pGFP and PLL/siRNA-pGFP-OSDZ complexes were 90–150 nm and 15–45 mV (Fig. 2(B)). The C/A 5 polyplexes were smaller by 30–40 nm and had a surface charge 10–30 mV higher than the C/A 2 polyplexes. At a constant C/A, the surface charges and particle sizes of PLL/siRNA-pGFP complexes were similar to those of the PLL/pGFP complexes. However, the PLL/siRNA-pGFP-OSDZ complexes were 10–20 nm larger than the PLL/siRNA-pGFP complexes. The PLL/siRNA-pGFP-OSDZ complexes had approximately a 10 mV lower positive surface charge at C/A 2 but approximately a 10 mV higher positive surface charge at C/A 5 compared to the surface charges of the PLL/pGFP complexes. It seems that the PLL/siRNA-pGFP-OSDZ complexes may have little or no uncomplexed PLL at C/A 2, unlike at C/A 5. Most importantly, even when low C/A conditions were applied, adding pDNA to siRNA polyplexes dramatically decreased the particle sizes to 1/20 to 1/10 of the diameter of pDNA-free PLL/siRNA complexes (1–2 μm).
3.3. Gene silencing of PLL/siRNA-pDNA complexes
The siCtrl was employed as a control for the evaluation of the non-specific silencing effect. The PLL/siRNA-pGFP(-OSDZ) complexes were formed at C/A 2 and C/A 5. As shown in Fig. 3, the PLL/siLuc-pGFP and PLL/siLuc-pGFP-OSDZ complexes at C/A 2 induced more luciferase silencing (83% and 88%, respectively) than the C/A 5 polyplexes (35% and 65%). This result was contradictory to the general observation that higher complexation ratios (e.g., weight ratio, +/− ratio) induce better gene silencing [27, 28].
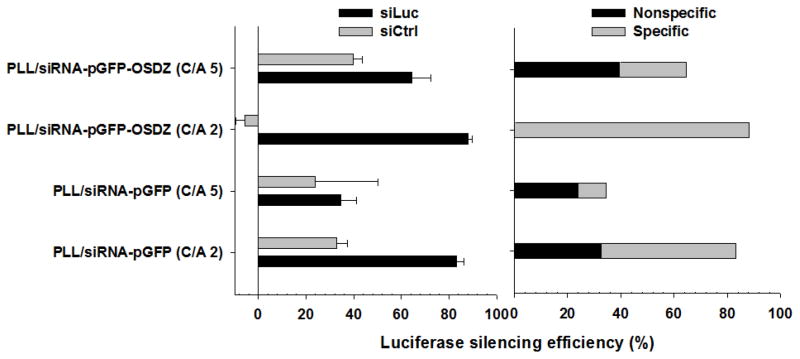
Fig. 3.
siLuc/siCtrl-mediated and specific or non-specific luciferase silencing efficiency of PLL/siRNA-pGFP and PLL/siRNA-pGFP-OSDZ complexes at 1 day post-silencing in bPEI/pLuc-transfected P-HEK293 cells (TICO 10 min). The siRNA concentration was 25 nM. (n≥4; Mean±SEM)
When an siRNA that was mismatched to the luciferase mRNA was used, the PLL/siCtrl-pGFP-OSDZ (C/A 2) complexes did not induce luciferase silencing (Fig. 3). However, three other siCtrl-pGFP(-OSDZ) polyplexes silenced luciferase activity 25%–40%. If the siCtrl was able to induce non-specific gene silencing, all siCtrl-pGFP(-OSDZ) polyplexes should silence gene expression, regardless of the polyplex composition. Thus, it is feasible that the luciferase silencing of the siCtrl-pGFP(-OSDZ) polyplexes may not be the result of the siCtrl, but the result of the polycations uncomplexed with or dissociated from the siRNA-pGFP(-OSDZ) polyplexes as shown in Fig. 1(E).
The overall gene silencing was divided into specific and non-luciferase specific effects based on the results from the control counterparts of the PLL/siCtrl-pGFP(-OSDZ) complexes. The PLL/siLuc-pGFP-OSDZ (C/A 2) complexes showed the most effective silencing (88% total and 100% specific silencing); PLL/siLuc-pGFP-OSDZ (C/A 5) (25% total and 39% specific); PLL/siLuc-pGFP (C/A 2) (50% total and 60% specific); PLL/siLuc-pGFP (C/A 5) (11% total and 31% specific). These results showed that the C/A 2-polyplexes induced a more specific effect than the C/A 5-polyplexes and that the addition of OSDZ improved specific silencing. One possible explanation for this observation is that relatively less dense polyplexes (e.g., PLL/siRNA-pGFP complexes prepared at C/A 2) containing less free PLL could silence target genes with fewer off-target effects. One the other hand, tight polyplex formulations (e.g., PLL/siRNA-pGFP (C/A 5) and PLL/siRNA-pGFP-OSDZ (C/A 5)) including a significant fraction of uncomplexed PLL could ineffectively silence the specific target gene due to reduced bioavailability (i.e., slow cytosolic release) of the specific siRNA. However, the free PLL in the polyplex solution could potentially induce significant non-specific gene silencing. The presence of OSDZ in PLL/siRNA-pGFP-OSDZ (C/A 2) complexes further minimized the amount of free PLL in the polyplex formulations and promoted specific effects due to endosomolytic activity.
3.4. Cellular uptake, decomplexation, and intracellular location of PLL/siRNA-pDNA(-OSDZ) complexes
With regard to the silencing induced by PLL/siRNA-pGFP(-OSDZ) complexes, it is possible that conflicting data obtained using bPEI/pLuc (C/A 5) and PLL/siRNA-pGFP(-OSDZ) complexes may be due to sequential co-transfection, which is the currently recommended test protocol for various commercial siRNA transfection reagents [29–33] and was confirmed in vivo [34]. One concern is that PLL/siRNA-pGFP(-OSDZ) complexes could interfere with the cellular uptake of the bPEI/pLuc complexes, further influencing their luciferase expression. Thus, the cellular uptake of bPEI/pYOYO complexes (C/A 5) prepared with YOYO-1-intercalated pLuc was monitored with and without sequential co-transfection with PLL/siRNA-pGFP (C/A 2) or PLL/siRNA-pGFP-OSDZ (C/A 2) complexes by flow cytometry. As shown in Fig. 4(A), the PLL/siRNA-pGFP(-OSDZ) complexes did not significantly affect the cellular uptake of the bPEI/pYOYO complexes. In addition, the effects of sequential co-transfection on the luciferase expression of the bPEI/pLuc complexes is further supported by the silencing results (almost no luciferase silencing efficiency) of the PLL/siCtrl (C/A 20) complexes (Fig. 1(C)) and the PLL/siCtrl-pGFP-OSDZ (C/A 2) complexes (Fig. 3). These results support the conclusion that the PLL/siRNA-pGFP(-OSDZ) complexes (or PLL/siRNA complexes) did not interfere with the transgene expression of bPEI/pLuc complexes because these polyplexes did not cause polymer-induced luciferase silencing effects.
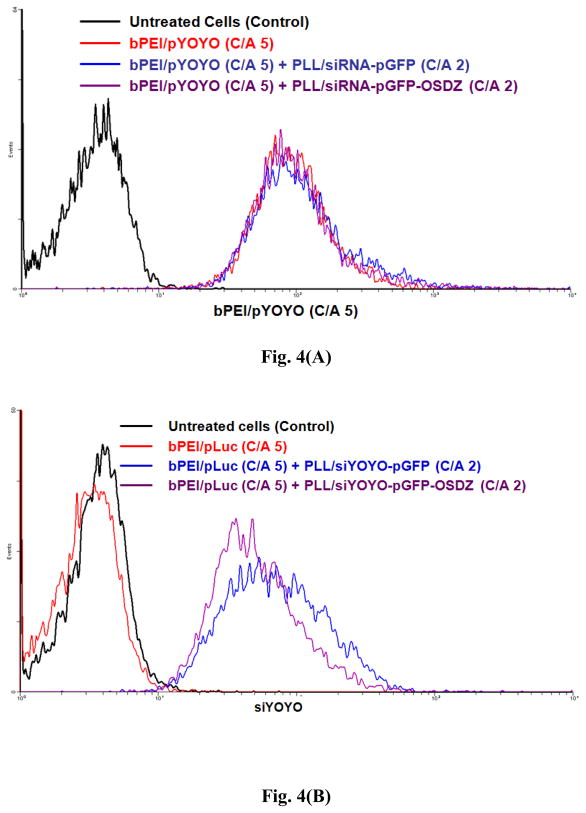
Fig. 4.
(A) Cellular uptake of bPEI/pYOYO complexes (C/A 5) during sequential co-transfection (TICO 10 min) at 4 hr post-transfection in P-HEK293 cells and (B) cellular uptake of siYOYO-pGFP polyplexes (C/A 2) at 4 hr post-transfection in bPEI/pLuc-transfected P-HEK293 cells (TICO 10 min). The siRNA concentration was 25 nM.
Another concern is the different cellular uptake of siRNA when PLL/siRNA-pGFP and PLL/siRNA-pGFP-OSDZ complexes are used. To investigate this issue, siRNA intercalated with YOYO-1 (siYOYO) was loaded into the PLL/siRNA-pGFP (C/A 2) and PLL/siRNA-pGFP-OSDZ (C/A 2) complexes. The cellular uptake of the PLL/siYOYO-pGFP(-OSDZ) complexes was monitored as shown in Fig. 4(B). The PLL/siYOYO-pGFP complexes showed more cellular uptake than the PLL/siYOYO-pGFP-OSDZ complexes in bPEI/pLuc-transfected P-HEK293 cells (TICO 10 min). This might be the result of different surface charge characteristics of the two polyplexes because the former complex had more positive surface charges than the latter (Fig. 2(B)). However, although a lower amount of the PLL/siLuc-pGFP-OSDZ complexes was taken up by the cells, these polyplexes showed a higher luciferase silencing efficiency than the PLL/siLuc-pGFP complexes. These silencing results may result from OSDZ-induced endosomal release of the PLL/siLuc-pGFP-OSDZ complexes.
After cellular internalization of the PLL/siRNA-pGFP(-OSDZ) complexes, the polyplexes should release the loaded siRNA into the intracellular environment, particularly the cytoplasm, to silence the target mRNA. Thus, polyplex stability was monitored by measuring polyanion (i.e., heparin)-induced decomplexation in 150 mM NaCl. As shown in Fig. 5, 20 μg/mL of heparin caused the release of approximately 90% of the loaded siRNA from the PLL/siRNA-pGFP (C/A 2) complexes. Most of the siRNA and pGFP was unloaded from the complexes following the addition of 30 μg/mL of heparin. In the case of the PLL/siRNA-pGFP-OSDZ (C/A 2) complexes, approximately 18% of the loaded siRNA was released at 40 μg/mL of heparin and the amount of released siRNA increased with increasing heparin concentrations. Almost all of the siRNA was decomplexed from the complexes at 70–80 μg/mL of heparin. Interestingly however, most of the pGFP was exposed on the surface of the polyplexes without being released. In the C/A 2-polyplexes, it is possible that the PLL/siRNA-pGFP complexes were more easily dissociated to release the siRNA from the PLL than were PLL/siRNA-pGFP-OSDZ complexes. In the polyplexes at C/A 5, more heparin was required to release the siRNA from the polyplexes. Comparing the two groups (C/A 2 and 5) at 60 μg/mL of heparin, the PLL/siRNA-pGFP-OSDZ (C/A 2) complexes released more siRNA (84% vs. 26%) than the PLL/siRNA-pGFP (C/A 5) complexes. These decomplexation results may indicate why the C/A 2-polyplexes were able to show better gene silencing than the C/A 5-polyplexes.
Fig. 5.
Heparin-induced decomplexation of PLL/siRNA-pGFP, and PLL/siRNA-pGFP-OSDZ complexes (C/A 2 and C/A 5). The polyplexes prepared in a HEPES solution (pH 7.4; 20 mM) were incubated in heparin-containing 150 mM NaCl solution. The siRNA concentration was 12.5 nM.
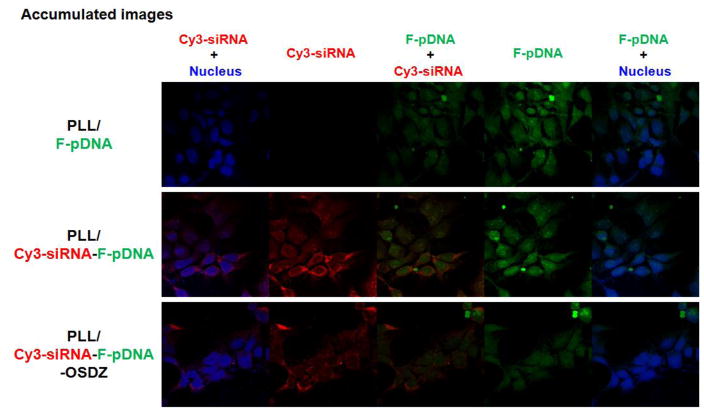
The intracellular localization of the nucleic acids delivered by polymers is critical to the delivery efficacy. Thus, the nucleic acids were labeled with fluorescent dyes (Cy3-siRNA and F-pGal) to monitor the intracellular locations of the nucleic acids. As opposed to pGFP, F-pGal (i.e., F-pDNA) was used to avoid confusion between GFP expression and the fluorescein dye. In addition, the PLL/Cy3-siRNA (C/A 2) was not tested because the polyplex particle was too large to enter the cells (Fig. 1(B)). As shown in Fig. 6, more PLL/pDNA (C/A 2) and PLL/siRNA-pDNA (C/A 2) complexes were taken up by bPEI/pLuc-transfected P-HEK293 cells compared to PLL/siRNA-pDNA-OSDZ (C/A 2) complexes, based on the fluorescence intensities of the delivered nucleic acids. These results are consistent with the flow cytometry-based polyplex uptake results (Fig. 4(B)).
Fig. 6.
Intracellular localization of siRNA and pDNA delivered with PLL-based polyplexes at 4 hr post-transfection in bPEI/pLuc-transfected P-HEK293 cells.
The PLL/F-pDNA complexes evenly delivered F-pDNA to both the nucleus and the cytoplasm. Similarly, the F-pDNA and Cy3-siRNA delivered with PLL/Cy3-siRNA-F-pDNA complexes were distributed in both intracellular compartments. However, a fraction of the delivered Cy3-siRNA was strongly localized to the perinuclear region, and the siRNA seemed to be sequestrated in the endolysosomes. When the PLL/Cy3-siRNA-F-pDNA-OSDZ complexes were delivered, the F-pDNA was mostly localized in the nucleus. However, relatively more of the siRNA accumulated in the cytoplasm than in the nucleus, despite the relatively small amount of siRNA that was sequestrated in likely endolysosomal compartments when compared to the PLL/Cy3-siRNA-F-pDNA complexes. The reduced sequestration of siRNA in the endosomes and lysosomes may be attributed to the presence of OSDZ.
3.5. Various effectors for gene silencing of PLL/siRNA-pDNA(-OSA) complexes
The gene silencing effects of the PLL/siRNA-pGFP(-OSA) complexes can be influenced by various factors, including siRNA dose, post-silencing time, TICO, cell proliferation rates, and OSAs. Thus, the PLL/siRNA-pGFP (C/A 2) and the PLL/siRNA-pGFP-OSDZ (C/A 2) complexes, which showed the most specific silencing, were delivered into bPEI/pLuc-transfected HEK293 cells to monitor the effects of these factors on the luciferase silencing efficiency of siRNA.
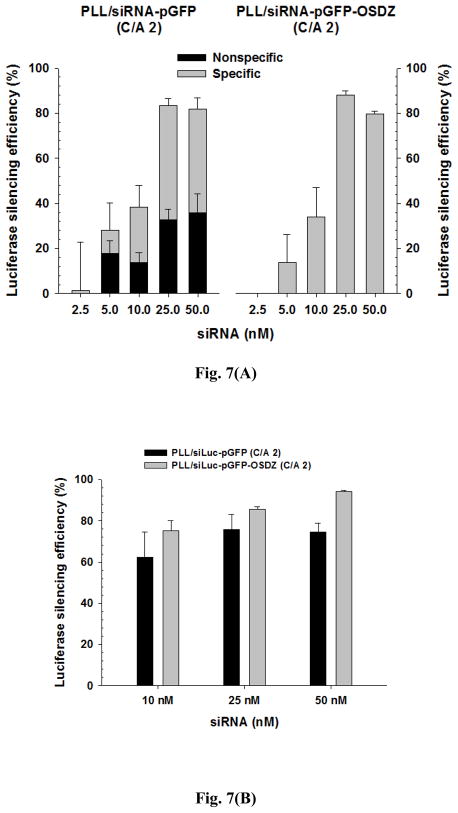
The first factor is the siRNA dose. As shown in Fig. 7(A), both polyplexes showed similar siLuc concentration-dependent luciferase silencing efficiencies with negligible cellular toxicity (Fig. S2). Their maximum luciferase silencing efficiencies were approximately 80–90% at 25–50 nM siLuc. When considering the specific and non-specific luciferase silencing of the PLL/siLuc-pGFP complexes based on the silencing result of the PLL/siCtrl-pGFP complexes, the PLL/siLuc-pGFP complexes showed 10%-50% specific silencing (i.e., 36%-63% of total luciferase silencing) and their non-specific silencing might be from polycation-related off-target effects. However, the OSDZ-containing polyplexes (i.e., PLL/siRNA-pDNA-OSDZ) specifically silenced up to 88% of the luciferase expression without any non-specific luciferase silencing.
Fig. 7.
siRNA concentration-dependent luciferase silencing efficiency of PLL/siRNA-pGFP and PLL/siRNA-pGFP-OSDZ complexes (C/A 2) at 1 day post-silencing in bPEI/pLuc-transfected (A) P-HEK293 cells and (B) GA-HEK293 cells (TICO 10 min). (n≥4; Mean±SEM)
Next, GA-HEK293 cells were used instead of P-HEK293 cells to understand the different silencing effects depending on different cell proliferation rates. As shown in Fig. 7(B), the PLL/siLuc-pGFP (C/A 2) and the PLL/siLuc-pGFP-OSDZ (C/A 2) complexes with 10 nM of siLuc dramatically increased the luciferase silencing in GA-HEK293 cells (approximately 60% and 75%, respectively) compared with the silencing results (approximately 40% for both) with P-HEK293 cells. The PLL/siLuc-pGFP (C/A 2) complexes induced a saturated silencing efficiency of approximately 75% at 25 nM and 50 nM of siLuc, whereas the PLL/siLuc-pGFP-OSDZ (C/A 2) complexes induced a linear increase in luciferase silencing up to 95% at 50 nM of siLuc. These findings may result from both lower cell numbers when the polyplexes are added and the lack of an increase in cell numbers during polymeric silencing [35].
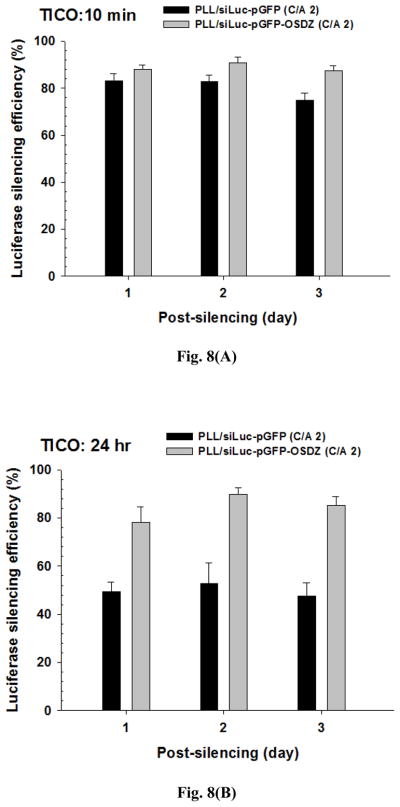
The silencing effects of siRNAs are often delayed until target proteins are turned over. In proliferating cells, the gene silencing effects of siRNA are continuously diluted due to cellular division [35]. Thus, as with post-transfection time, it is important to evaluate the maintenance of the gene silencing effects is to evaluate the efficiency of polymeric siRNA carriers. The silencing effects of the PLL/siLuc-pGFP(-OSDZ) complexes at two different time interval of co-transfection (TICO) were monitored in the same manner as post-silencing time. In addition, the post-silencing period was limited to 4 days after bPEI/pLuc transfection because the bPEI/pLuc-transfected P-HEK293 cells stably expressed luciferase proteins for only this time period (Fig. S3). As shown in Fig. 8(A), PLL/siLuc-pGFP-OSDZ (C/A 2) complexes with TICO 10 min induced better luciferase silencing than the PLL/siLuc-pGFP (C/A 2) complexes. Importantly, the luciferase silencing efficiencies of PLL/siLuc-pGFP-OSDZ (C/A 2) complexes remained approximately 90% for 3 days. However, the PLL/siLuc-pGFP (C/A 2) complexes showed 83% luciferase silencing for 2 days and then their silencing efficiencies dropped to 75% at 3 days.
Fig. 8.
TICO- and post-silencing time-dependent luciferase silencing efficiency of PLL/siLuc-pGFP and PLL/siLuc-pGFP-OSDZ complexes (C/A 2) in bPEI/pLuc-transfected P-HEK293 cells: (A) TICO 10 min (B) TICO 24 hr. (n≥4; Mean±SEM)
However, the silencing conditions of TICO 10 min complexes does not reflect the actual silencing conditions as no expression of target proteins is observed when bPEI/pLuc and PLL/siRNA-pGFP(-OSDZ) complexes are co-transfected into P-HEK293 cells. Thus, when testing TICO at 24 hours, the luciferase gene silencing of PLL/siRNA-pGFP(-OSDZ) complexes was evaluated as shown in Fig. 8(B). The PLL/siLuc-pGFP-OSDZ (C/A 2) complexes showed much better silencing efficiencies than the PLL/siLuc-pGFP (C/A 2) complexes over 3 days. The PLL/siLuc-pGFP-OSDZ (C/A 2) complexes induced no noticeable differences in silencing efficiency from those at TICO 10 min, although their silencing effect was slightly delayed at day 1. However, the PLL/siLuc-pGFP (C/A 2) complexes induced significantly less silencing efficiency than their TICO 10 min counterparts. This may have resulted from reduced polymer-mediated non-specific silencing as explained in Fig. 1(E) and Fig. S1(B). The TICO 24 hr results emphasize that the PLL/siLuc-pGFP-OSDZ (C/A 2) complexes are a much more effective silencing system than the PLL/siLuc-pGFP (C/A 2) complexes because OSDZ can assist in the endosomal release of PLL/siLuc-pGFP-OSDZ complexes.
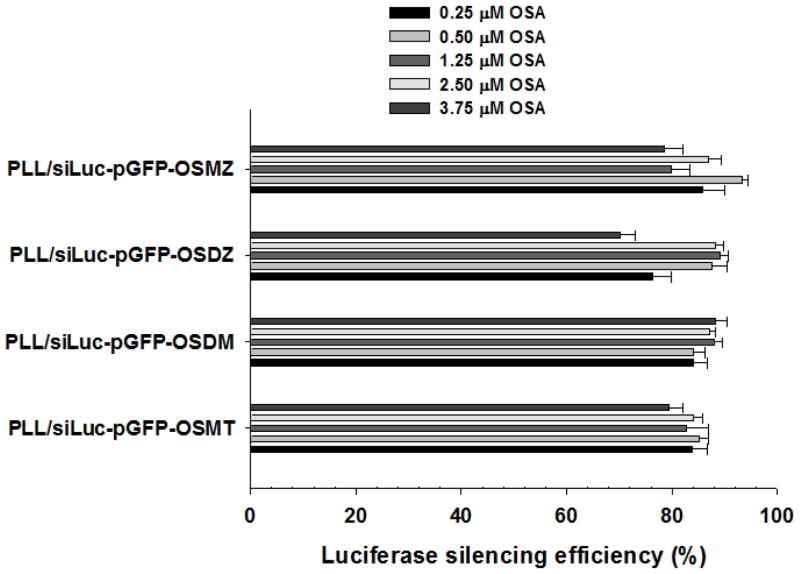
In this study, OSDZ (2.5 μM), with an apparent pKa of 6.5, assisted in the effective silencing of the PLL/siRNA-pGFP-OSDZ complexes because OSDZ could compensate for the weak endosomolytic activity of PLL. Like OSDZ, other OSAs such as OSMZ (pKa 7.3), OSDM (pKa 6.5) and OSMT (pKa 5.7) [22] are available to induce the endolysosomal release of the PLL/siRNA-pGFP-OSA complexes. However, depending on the pKas and concentrations in the complexes, the cytosolic release locations and release times of these PLL/siRNA-pGFP-OSA complexes from the endolysosomal compartments could be modulated. The higher pKas and higher concentrations of the OSAs could cause earlier endosomal escape of the polyplexes. Thus, PLL/siLuc-pGFP-OSA complexes having different concentrations of OSAs (e.g., OSMZ, OSDZ, OSDM, and OSMT) were introduced into bPEI/pLuc-transfected P-HEK293 cells (TICO 10 min). All of the PLL/siRNA-pGFP-OSA complexes induced 80–90% luciferase silencing (Fig. 9). The luciferase silencing results seem not to depend on concentrations or type of OSA in the polyplexes. This indicates that either the cytosolic release location or time point of the polyplex release does not affect the luciferase silencing or that the OSA in the polyplexes may disrupt the endosomal membrane to silence the target mRNA very quickly.
Fig. 9.
OSA effects on luciferase silencing efficiency of PLL/siLuc-pGFP-OSA complexes (C/A 2) at 1 day post-silencing in bPEI/pLuc-transfected P-HEK293 cells (TICO 10 min). (n≥4; Mean±SEM)
3.6. GFP expression of PLL/siRNA-pGFP(-OSDZ) complexes
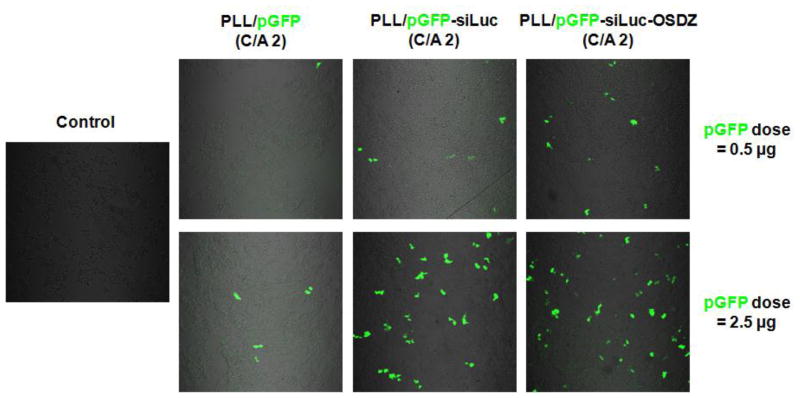
This study introduced long polyanions into polyplexes to generate nanosized and stable siRNA complexes with polycations. The functional pDNA pGFP was selected as a polyanion due to increasing interest in the delivery of multiple therapeutics or diagnostics in a single carrier. Thus, the GFP expression of the PLL/siRNA-pGFP(-OSDZ) complexes was evaluated as shown in Fig. 10. When dosing 0.5 μg of pDNA by polyplexes to the cells, the PLL/pGFP (C/A 2) complexes did not induce GFP expression because the complexation condition was lower not an optimal condition (e.g., C/A 5). Although the PLL/siRNA-pGFP (C/A 2) and PLL/siRNA-pGFP-OSDZ (C/A 2) complexes induced weak GFP expression, the OSDZ-containing polyplexes resulted in more GFP-expressing cells than did OSDZ-free polyplexes. This result might be supported by differential nuclear localization of the pDNA delivered by the polyplexes. An additional reason for these results may be that OSDZ helps the polyplexes to escape from the endolysosomal compartments.
Fig. 10.
GFP expression of PLL/pGFP, PLL/siLuc-pGFP, and PLL/siLuc-pGFP-OSDZ complexes (C/A 2) at 2 days post-transfection in bPEI/pLuc-transfected P-HEK293 cells (TICO 10 min).
Compared to 0.5 μg of pDNA, a higher pDNA dose (2.5 μg) induced better GFP expression levels when transfected in PLL/siRNA-pGFP and PLL/siRNA-pGFP-OSDZ complexes. However, unlike the previous observation that OSDZ-containing polyplexes were better transfection systems than OSDZ-free polyplexes, the GFP expression levels of these two types of complexes were similar as shown in Fig. 10. This result might be caused by pDNA doses higher than the optimum dose. The large amount of polycations needed to deliver such high pDNA doses could cause cytotoxicity, leading to the inhibition of transcription and reduced transgene expression. However, pGFP expression was still not high, due to the moderate transfection efficiency of PLL. Thus, the current model polycation, PLL, should be replaced with new polycations that have good biocompatibility and good transfection activity.
In this study, a helper polyanion, pDNA, was used to assist in the delivery of nanosized polyplexes with siRNA and lead to effective gene silencing. Simultaneous pDNA delivery in a single carrier is an attractive addition to specific gene silencing using siRNA to increase the potential of these delivery systems. First, the silencing of the target mRNA can be extended. For example, if the pDNA is an shRNA-expressing pDNA for the target mRNA, the siRNA-pDNA polyplexes can elongate the silencing effects, with siRNA inducing early silencing and the expressed shRNA inducing intermediate and late silencing. In addition, if pDNA the results in the expression fluorescent proteins, the location of the polyplexes can be monitored. The combinational delivery of nucleic acids may allow for the maximum therapeutic effects and dual effects of therapeutics and diagnostics.
4. Conclusion
This study was designed to produce stable and dense nanocomplexes by complexing PLL with siRNA and pGFP at low C/A ratios. The resulting PLL/siRNA-pGFP complexes were compact and induced effective gene silencing. OSA-containing PLL/siRNA-pGFP complexes were better at inducing silencing than their OSA-free counterparts. In addition to silencing a target mRNA, the polyplexes were able to express GFP. Thus, the polyplexes investigated in this study may have the potential to silence an mRNA of interest as well as to express therapeutic, diagnostic, or imaging proteins.
Supplementary Material
Acknowledgments
This work was partially supported by National Institutes of Health, USA (NIH GM82866).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kurreck J. RNA interference: from basic research to therapeutic applications. Angew Chem Int Ed Engl. 2009;48:1378–1398. doi: 10.1002/anie.200802092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takahashi Y, Nishikawa M, Takakura Y. Nonviral vector-mediated RNA interference: its gene silencing characteristics and important factors to achieve RNAi-based gene therapy. Adv Drug Deliv Rev. 2009;61:760–766. doi: 10.1016/j.addr.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Stokman G, Qin Y, Racz Z, Hamar P, Price LS. Application of siRNA in targeting protein expression in kidney disease. Adv Drug Deliv Rev. 2010;62:1378–1389. doi: 10.1016/j.addr.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Kim WJ, Kim SW. Efficient siRNA delivery with non-viral polymeric vehicles. Pharm Res. 2009;26:657–666. doi: 10.1007/s11095-008-9774-1. [DOI] [PubMed] [Google Scholar]
- 5.Gao K, Huang L. Nonviral methods for siRNA delivery. Mol Pharmaceutics. 2009;6:651–658. doi: 10.1021/mp800134q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nam HY, McGinn A, Kim PH, Kim SW, Bull DA. Primary cardiomyocyte-targeted bioreducible polymer for efficient gene delivery to the myocardium. Biomaterials. 2010;31:8081–8087. doi: 10.1016/j.biomaterials.2010.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim SH, Jeong JH, Ou M, Yockman JW, Kim SW, Bull DA. Cardiomyocyte-targeted siRNA delivery by prostaglandin E(2)-Fas siRNA polyplexes formulated with reducible poly(amido amine) for preventing cardiomyocyte apoptosis. Biomaterials. 2008;29:4439–4446. doi: 10.1016/j.biomaterials.2008.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeong JH, Christensen LV, Yockman JW, Zhong Z, Engbersen JF, Kim WJ, et al. Reducible poly(amido ethylenimine) directed to enhance RNA interference. Biomaterials. 2007;28:1912–1917. doi: 10.1016/j.biomaterials.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 9.de Fougerolles AR. Delivery vehicles for small interfering RNA in vivo. Hum Gene Ther. 2008;19:125–132. doi: 10.1089/hum.2008.928. [DOI] [PubMed] [Google Scholar]
- 10.Howard KA. Delivery of RNA interference therapeutics using polycation-based nanoparticles. Adv Drug Deliv Rev. 2009;61:710–720. doi: 10.1016/j.addr.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Dehousse V, Garbacki N, Colige A, Evrard B. Development of pH-responsive nanocarriers using trimethylchitosans and methacrylic acid copolymer for siRNA delivery. Biomaterials. 2010;31:1839–1849. doi: 10.1016/j.biomaterials.2009.11.028. [DOI] [PubMed] [Google Scholar]
- 12.Breunig M, Hozsa C, Lungwitz U, Watanabe K, Umeda I, Kato H, et al. Mechanistic investigation of poly(ethylene imine)-based siRNA delivery: disulfide bonds boost intracellular release of the cargo. J Control Release. 2008;130:57–63. doi: 10.1016/j.jconrel.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 13.Creusat G, Rinaldi AS, Weiss E, Elbaghdadi R, Remy JS, Mulherkar R, et al. Proton sponge trick for pH-sensitive disassembly of polyethylenimine-based siRNA delivery systems. Bioconjug Chem. 2010;21:994–1002. doi: 10.1021/bc100010k. [DOI] [PubMed] [Google Scholar]
- 14.Gary DJ, Puri N, Won YY. Polymer-based siRNA delivery: perspectives on the fundamental and phenomenological distinctions from polymer-based DNA delivery. J Control Release. 2007;121:64–73. doi: 10.1016/j.jconrel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 15.Danielsen S, Maurstad G, Stokke BT. DNA-polycation complexation and polyplex stability in the presence of competing polyanions. Biopolymers. 2005;77:86–97. doi: 10.1002/bip.20170. [DOI] [PubMed] [Google Scholar]
- 16.Bolcato-Bellemin AL, Bonnet ME, Creusat G, Erbacher P, Behr JP. Sticky overhangs enhance siRNA-mediated gene silencing. Proc Natl Acad Sci USA. 2007;104:16050–16055. doi: 10.1073/pnas.0707831104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang HC, Lee M, Bae YH. Polymeric gene carriers. Crit Rev Eukaryot Gene Expr. 2005;15:317–342. doi: 10.1615/critreveukargeneexpr.v15.i4.30. [DOI] [PubMed] [Google Scholar]
- 18.Mok H, Lee SH, Park JW, Park TG. Multimeric small interfering ribonucleic acid for highly efficient sequence-specific gene silencing. Nat Mater. 2010;9:272–278. doi: 10.1038/nmat2626. [DOI] [PubMed] [Google Scholar]
- 19.Li SD, Huang L. Targeted delivery of antisense oligodeoxynucleotide and small interference RNA into lung cancer cells. Mol Pharmaceutics. 2006;3:579–588. doi: 10.1021/mp060039w. [DOI] [PubMed] [Google Scholar]
- 20.Chono S, Li SD, Conwell CC, Huang L. An efficient and low immunostimulatory nanoparticle formulation for systemic siRNA delivery to the tumor. J Control Release. 2008;131:64–69. doi: 10.1016/j.jconrel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li SD, Chen YC, Hackett MJ, Huang L. Tumor-targeted delivery of siRNA by self-assembled nanoparticles. Mol Ther. 2008;16:163–169. doi: 10.1038/sj.mt.6300323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang HC, Bae YH. pH-Tunable endosomolytic oligomers for enhanced nucleic acid delivery. Adv Funct Mater. 2007;17:1263–1272. [Google Scholar]
- 23.Kang HC, Kang HJ, Bae YH. A reducible polycationic gene vector derived from thiolated low molecular weight branched polyethyleneimine linked by 2-iminothiolane. Biomaterials. 2011;32:1193–1203. doi: 10.1016/j.biomaterials.2010.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang HC, Samsonova O, Bae YH. Trafficking microenvironmental pH of gene vector polycation in drug-sensitive and multidrug-resistant MCF7 breast cancer cell. Biomaterials. 2010;31:3071–3078. doi: 10.1016/j.biomaterials.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang HC, Kim S, Lee M, Bae YH. Polymeric gene carrier for insulin secreting cells: Poly(l-lysine)-g-sulfonylurea for receptor mediated transfection. J Control Release. 2005;105:164–176. doi: 10.1016/j.jconrel.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 26.Fahrmeir J, Gunther M, Tietze N, Wagner E, Ogris M. Electrophoretic purification of tumor-targeted polyethylenimine-based polyplexes reduces toxic side effects in vivo. J Control Release. 2007;122:236–245. doi: 10.1016/j.jconrel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 27.Tarcha PJ, Pelisek J, Merdan T, Waters J, Cheung K, von Gersdorff K, et al. Synthesis and characterization of chemically condensed oligoethylenimine containing beta-aminopropionamide linkages for siRNA delivery. Biomaterials. 2007;28:3731–3740. doi: 10.1016/j.biomaterials.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 28.Philipp A, Zhao X, Tarcha P, Wagner E, Zintchenko A. Hydrophobically modified oligoethylenimines as highly efficient transfection agents for siRNA delivery. Bioconjug Chem. 2009;20:2055–2061. doi: 10.1021/bc9001536. [DOI] [PubMed] [Google Scholar]
- 29.Calvin S, Pitz S, Jacobsen L. High-performance shRNA-based gene knockdown using FuGENE HD transfection reagent. Biochemica. 2008:15–18. [Google Scholar]
- 30.Andreou I, Weber M, Valer M, Buhlmann C. Confirming gene silencing mechanism by pGFP/GFP22-siRNA co-transfection. Agilent Technologies; 2003. [Google Scholar]
- 31.TransIT-TKO® Transfection Reagent™. Mirus Bio Protocol [Google Scholar]
- 32.Guidelines for co-transfection of adherent cells with siRNA and plasmid DNA using TransMessenger Transfection Reagent. QIAGEN Supplementary Protocol [Google Scholar]
- 33.Polyplus transfection reagent for transfection. Polyplus-transfection protocol [Google Scholar]
- 34.Mukai H, Kawakami S, Hashida M. Renal press-mediated transfection method for plasmid DNA and siRNA to the kidney. Biochem Biophys Res Commun. 2008;372:383–387. doi: 10.1016/j.bbrc.2008.04.097. [DOI] [PubMed] [Google Scholar]
- 35.De Paula D, Bentley MV, Mahato RI. Hydrophobization and bioconjugation for enhanced siRNA delivery and targeting. RNA. 2007;13:431–456. doi: 10.1261/rna.459807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.