Abstract
The E1A gene of species C human adenovirus is an intensely investigated model viral oncogene that immortalizes primary cells and mediates oncogenic cell transformation in cooperation with other viral or cellular oncogenes. Investigations using E1A proteins have illuminated important paradigms in cell proliferation and the functions of cellular proteins such as the retinoblastoma protein. Studies with E1A have led to the surprising discovery that E1A also suppresses cell transformation and oncogenesis. Here, I review our current understanding of the transforming and tumor suppressive functions of E1A, and how E1A studies led to the discovery of a related tumor suppressive function in benign human papillomaviruses. The potential role of these opposing functions in viral replication in epithelial cells is also discussed.
Transforming proteins of DNA tumor viruses
During the past three decades, research with the transforming proteins of small DNA tumor viruses such as human adenoviruses (HAdvs), SV40 and human papillomaviruses (HPVs) has illuminated critical cellular pathways that control proliferation and oncogenic transformation of mammalian cells. Due to their small genome size, these viruses are heavily dependent on the host cell machineries to express their genes and replicate their DNA. Since these viruses generally replicate in terminally differentiated quiescent target cells, the viral genes expressed during the early phase of their life cycle subvert the cell cycle to induce transient proliferation of the infected cells to generate a permissive S-phase state to facilitate viral replication. In non-permissive cells, these viruses express only their early genes that induce cell proliferation resulting in abortive infection. A fraction of the infected cells recover and assume oncogenic properties as a result of continued expression of a subset of viral early genes referred to as transforming genes. Transduction of subgenomic DNA fragments containing the transforming genes also achieves transformation of the target cells in significant numbers. The use of defined viral mutants and transduction of isolated genes have been widely used for molecular genetic analysis of viral transforming genes.
Researchers have used biochemical approaches to identify cellular proteins associated with viral proteins in order to elucidate the mechanisms through which the transforming proteins of small DNA tumor viruses subvert the cell cycle and elicit oncogenic cell transformation. This approach revealed the interaction of viral transforming proteins with the tumor suppressor proteins such as p53 and the retinoblastoma (pRb) protein (reviewed in [1]). The transforming proteins of HAdvs, SV40 and HPVs share common mechanisms of cell transformation as they target the same cell cycle regulatory proteins such as p53 and the pRb family proteins. The genomes of HAdvs contain three oncogene-coding regions, E1A, E1B and E4 (Figure 1). The E1A gene has been intensely studied as a model dominant oncogene and has been instrumental in unraveling the functions of pRb and elucidating the E2F pathway of cell proliferation (reviewed in [2]). In this article, I review our current understanding of the transforming activity of HAdv E1A and how studies on E1A led to the identification of an unexpected tumor suppressive activity of E1A and the E6 protein of benign HPVs.
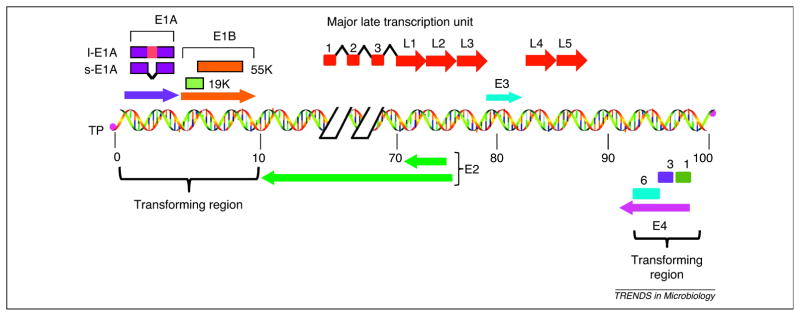
Figure 1.
Schematic illustration of HAdv5 genome. The HAdv5 genome consists of a 35 kb linear double stranded DNA that is covalently linked to viral terminal proteins (TP) at the 3′ ends. The viral genome contains five early transcription units (E1A, E1B, E3, E4 and E2) and one major late transcription unit. The major late transcript is differentially processed into five groups of late mRNAs (L1–L5) to which three leader sequences are attached by RNA splicing. The late mRNAs code for various viral structural proteins. The left most 14% of the genome contains the E1A and E1B genes that constitute the transforming genes. l-E1A indicates the large E1A protein and s-E1A indicates the small E1A protein. The early region E4 of different HAdv species encodes multiple proteins and at least three of them, Orf 1, Orf 3 and Orf 6 (indicated by 1, 3 and 6) exhibit transforming activities.
Transforming activity of E1A
The E1A region codes for two major protein isoforms that are expressed from two differentially spliced messenger RNAs (mRNAs, 13S and 12S) (Figure 2). The 13S mRNA encodes an E1A protein of 289 amino acids (289R, l-E1A) while the 12S mRNA encodes a smaller E1A protein of 243 amino acids (243R, s-E1A). These proteins differ by the inclusion of a unique 46 amino acid region in the larger E1A that plays an essential role in viral replication through transcriptional activation of other essential viral early genes. Additionally, both E1A proteins can also deregulate the cell cycle by transcriptional reprogramming of the cell to generate a cellular S-phase environment conducive for viral replication. The smaller E1A protein (243R) is sufficient to induce a transient proliferative state of the host cell and induce oncogenic cell transformation. Thus, the 243R E1A protein (hereafter referred to as E1A) of species C HAdv (types 2 and 5) has been extensively investigated as a model oncoprotein. Although several earlier studies employed rodent embryonic fibroblasts to study the transforming activities of HAdv genes, most of our current knowledge on cell transformation by E1A is based on results obtained with primary neonatal rat kidney (BRK) cells (see Glossary) [3]. Investigators preferred the BRK cell model since the results could be extrapolated to human epithelial cancers and the assays using these cells result in high frequency transformation by the transforming genes with only very low levels of background transformation. In BRK cells, expression of E1A induces efficient (transient) cell proliferation and low frequency immortalization. Transduction of a second oncogene, such as the activated Ras oncogene [4] or E1B [3], leads to high frequency transformation resulting in cells that are capable of forming tumors in immunodeficient mice. The E1A-Ras cooperative transformation assay was used by various investigators to map the functional domains of E1A required to elicit transformation and to elucidate the mechanisms of cell transformation. Detailed mutagenesis studies by several groups localized the transforming activity of E1A within the N-terminal half (coded by exon 1 of 12S mRNA, Figure 1). Specifically, these studies identified two regions, the N-terminal 80 amino acid region that encompasses a conserved domain designated as conserved region 1(CR1) and the amino acid region between 120 to 140 that contains CR2 (reviewed in [5]).
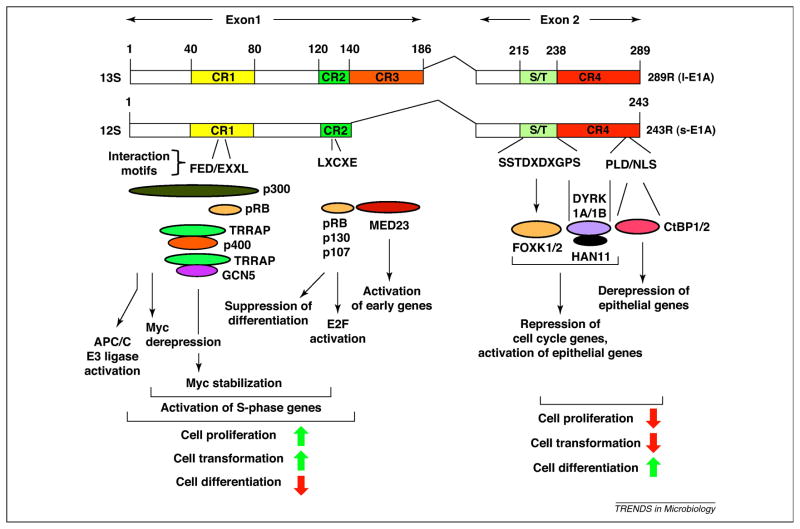
Figure 2. Schematic illustration of interaction of cellular proteins with HAdv5 E1A proteins.
The functional consequences of such protein interactions are also indicated. The N-terminal region of E1A exhibits a dominant oncogenic activity while the C-terminal region contributes suppression of oncogenesis. The green upward arrows indicate activation of various cellular processes and the red downward arrows indicate suppression of such processes.
Interaction of cellular proteins and transformation
To determine the mechanism by which E1A mediates cell transformation, investigators analyzed cellular protein complexes associated with E1A domains required to mediate E1A-Ras cooperative transformation. These studies revealed that E1A is a mosaic of several protein interaction sequence motifs located within four conserved regions (reviewed in [5]). Although the N-terminal 80 amino acid region has been reported to interact with a large number of cellular proteins [6], mutational analyses have linked E1A-Ras cooperative transformation with the interaction of E1A with two major cellular protein complexes. These protein complexes are the histone acetyltransferases, p300/CBP [7–9], and the TRRAP/p400 multi-protein chromatin remodeling complex [10–11]. In addition, the TRRAP-interacting module of E1A has also been reported to form a complex with an acetyltransferase GCN5 in mammalian cells [12] or the GCN5-containing protein complex known as the SAGA complex in yeast [13]. The CR2 region is indispensable for the transforming activity of E1A and interacts with pRb and pRb family members p107 and p130 (reviewed in [2]). The cellular protein complexes targeted by E1A to facilitate Ras transformation of primary mammalian cells are remarkably similar to those protein complexes that regulate Ras signaling in Caenorhabditis elegans. The vulval development pathway in C. elegans is modulated by Ras and perturbation of the pathway results in a multivulval phenotype called synMuv. Genetic studies have identified three classes of synMuv genes (A, B and C)[14]. The group B genes encode proteins that are homologs of the mammalian pRb transcriptional repression pathway [15]. The class C genes include homologs of the constituents present in the mammalian TRRAP/p400 complex [16]. Thus, E1A appears to subvert certain conserved cellular signaling pathways to mediate oncogenic transformation.
Subversion of p300/CBP functions
p300/CBP are considered to be global transcriptional activators [17]. However, they have also been implicated in context-dependent transcriptional repression. Although it is generally believed that interaction of p300 and CBP by E1A contributes to cell proliferation and the oncogenic activity of E1A, the precise mechanisms have come into focus only recently. Berk and colleagues showed that the interaction of E1A with p300/CBP was quantitative and resulted in global reduction of p300/CBP-mediated acetylation of histone H3 lysine 18 (H3K18Ac) [18]. E1A mutants that were defective in transformation and in interaction with p300/CBP did not cause global H3K18 deacetylation, suggesting that H3K18 deacetylation is linked to the transforming activity of E1A. A genome wide ChIP-chip analysis has also revealed that p300/CBP are retargeted to promoters of various cellular genes that drive cell proliferation by E1A to activate transcription (H3K8 hyperacetylation) [19]. Thimmapaya and colleagues have shown that p300 represses the transcription of the c-Myc oncogene in association with the repressor YY1 and histone deacetylase (HDAC) 3 [20]. Expression of E1A relieved c-Myc repression through interaction with p300, resulting in cell proliferation [21]. SV40 T antigen (T Ag) also activated c-Myc expression through interaction with p300 [22], suggesting that this may be one of the general mechanisms (i.e. activation of c-Myc transcription) by which DNA tumor viruses might induce cell proliferation.
In addition to subversion of the transcriptional activities of p300/CBP, it appears that E1A interaction with p300/CBP might promote cell cycle progression via a non-transcriptional mechanism [23]. Turnell and coinvestigators discovered that two constituents (APC 5 and APC7) of the anaphase-promoting complex/cyclosome (APC/C) E3 ubiquitin ligase interact with CBP through a sequence motif similar to the E1A motif (FXD/EXXXL). E1A was shown to target a complex between CBP and APC/C resulting in the activation of APC/C E3 ligase activity. As the APC/C activity is involved in maintaining the fidelity of sister-chromatid separation during mitosis, the sequestration of p300/CBP might liberate APC/C to promote aberrant mitotic exit. The deregulated E3 ligase activity of APC/C might then promote selective ubiquitination of cell cycle regulatory molecules such as cyclin B1 that function in G2 and the inhibitor of sister chromatid separation (Securin) leading to genome instability and aneuploidy.
Subversion of TRRAP and p400 complexes
The E1A N-terminal region spanning residues 26 to 35 interacts with the scaffolding protein TRRAP [11, 24] as well as the chromatin remodeling ATPase p400 [10]. Both TRRAP and p400 are constituents of the Tip60 acetyltransferase complex [25]. TRRAP is also a constituent of a second acetyltransferase complex known as the SAGA complex that contains the acetyltransferase GCN5 (reviewed in [26]). The E1A mutant (Δ26–35) defective in interaction with p400 is also defective in interaction with GCN5 in mammalian cells [12]. E1A also interacts with the SAGA complex in yeast [13]. The presence of Tip60 with E1A-associated TRRAP/p400 complex has not been detected thus far. Both TRRAP/GCN5 [27–28] and TRRAP/Tip60 [29–30] complexes also interact with c-Myc to activate Myc-target genes. The interaction of c-Myc with the acetyltransferases GCN5 and Tip60 was reported to result in substantial increase in the stability of c-Myc [31].
During HAdv5 infection, E1A has also been shown to stabilize c-Myc [32]. Specifically, it appears that E1A interaction with p400 is required for the stabilization of c-Myc as the E1A mutant Δ26–35 was defective in this activity [33]. Expression of E1A enhanced stable association of p400 with c-Myc resulting in the attenuation of Myc ubiquitination. The expression of E1A also enhanced the transcriptional activity of c-Myc. Although the molecular basis of E1A-mediated enhancement of p400 interaction with c-Myc is unknown, c-Myc appears to be a downstream target for E1A. This was also substantiated by the demonstration of transformation of rodent fibroblasts by E1A-Myc chimeric gene constructs in which the E1A region that interacts with TRRAP/p400/GCN5 was fused to the DNA-binding domain of c-Myc [11, 24]. Thus, the N-terminal 80 amino acid region of E1A could mediate cell proliferation and transformation activities through transcriptional activation (via E1A-mediated relief of repression by p300/CBP) and protein stabilization (via interaction of E1A with p400) of c-Myc resulting in increased transcriptional activity of c-Myc.
Subversion of pRb family proteins
The CR2 region is indispensable for the transforming activity of E1A. One of the most important discoveries in tumor virology is that the transforming proteins of small DNA tumor virus proteins such as SV40 T Ag, the E7 proteins of high-risk HPVs such as HPV16 and 18 and HAdv E1A interact with pRb family members (pRb, p107 and p130) through a conserved sequence motif Leu-X-Cys-X-Glu (LXCXE) to deregulate the cell cycle (reviewed in [2, 34]). While E1A and T Ag form stable complexes with pRb family members, the E7 proteins target pRb family proteins for proteolysis. Although E1A interacts with all three pRb family proteins through the high affinity motif LXCXE, it interacts with pRb through a second low affinity motif in the CR1 region [35–36]. One of the well established consequences of E1A interaction with pRb family proteins is relief of pRb-mediated transcriptional repression and the activation of the E2F pathway to activate S-phase genes. E1A interaction with pRb family members strips them from E2F family transcriptional activators to stimulate transcription of S phase genes [37]. Since E1A is unable to strip pRb from E2F1 [38–39], it appears that E1A activates other trans-activating members of the E2F family (such as E2F2-4). Genome wide ChIP-chip analysis [19] revealed that the E1A-p300/CBP complex transiently binds to the promoters of different growth and cell cycle genes that are repressed by pRb family proteins to activate gene expression by removing the Rb family members from the target promoters.
In addition to the activation of cell proliferation genes, a major effect of the E1A N-terminal region is suppression of cell differentiation. E1A is a potent suppressor of myoblast differentiation [40] and this activity was mapped to the N-terminal region of E1A [41]. ChIP-chip analysis also revealed that E1A and the Rb family member p107 were found at the repressed promoters for various differentiation and development genes [19]. Thus, the N-terminal half of E1A mediates oncogenic transformation by activating the transcriptional pathways mediated by c-Myc and Rb-E2F to activate cell proliferation and by suppressing genes that mediate cell differentiation.
Tumor suppression by E1A C-terminal region
In contrast to the intense interest that E1A N-terminal region (exon 1) has received, the functions of E1A C-terminal region (exon 2) was obscure until two large deletion mutants of E1A were used to broadly probe the functions of the C-terminal region in E1A-Ras cooperative transformation [42]. These studies revealed a surprising hyper-transforming phenotype associated with these mutants. The E1A C-terminal mutants cooperated with Ras to transform cells at enhanced frequencies compared with wild-type E1A. The transformed cells exhibited aggressive growth properties and induced relatively large tumors in immunodeficient mice. The tumors induced by the transformed cells expressing mutant E1A were highly metastatic while transformed cells expressing wild-type E1A formed smaller non-metastatic tumors. In immune-competent rats, the mutant-transformed cells readily formed tumors leading to the demise of animals within one to two weeks while the wild-type-transformed cells were non-tumorigenic. From these experiments, it was concluded that E1A possesses a tumor suppressive activity in addition to the tumor promoting activity [43]. Subsequently, Margaret Quinlan and colleagues described amino acid substitution mutants within the E1A C-terminal region that also exhibited a hyper-transforming phenotype in Ras-cooperative transformation assays [44–45]. In independent studies, Steve Frisch and colleagues showed that transduction of E1A into several human cancer cell lines reversed their oncogenic properties [46–47]. These experiments by three different groups have expanded the function of E1A to include a tumor suppressive activity, in addition to its widely known dominant oncogenic function.
Regulation of epithelial to mesenchymal transition by E1A
Rodent kidney cells transformed with wild-type E1A and Ras are highly adherent and retain predominantly an epithelial phenotype while cells transformed with E1A C-terminal mutants and Ras are less adherent and exhibit a fibroblast-like (mesenchymal) phenotype [42, 48]. These observations suggested that the E1A C-terminal region might exert its inhibitory effect on cell transformation by promoting cell differentiation. Frisch and colleagues reported that E1A was able to suppress the tumorigenic activity of human cancer cell lines by activating several epithelial genes [46]. They demonstrated that expression of E1A in human cancer cells converted them into epithelial-like cells. Fischer and Quinlan reported that rodent epithelial (BRK) cells immortalized with wild-type E1A retained the morphology and growth characteristics of epithelial cells while cells expressing C-terminal mutants were less epithelial and were defective in tight and adheren junction complexes and expressed stress fibers and filopodia similar to fibroblasts [48]. Thus, the E1A N-terminus appears to promote oncogenesis through epithelial to mesenchymal transitions while the E1A C-terminus appears to restrain oncogenesis by inducing mesenchymal to epithelial transitions. In transformed cells that express the entire E1A, there could be a balance between these two activities resulting in moderation of the extent of transformation.
Interaction of cellular proteins with E1A C-terminus
In order to elucidate the mechanisms by which the E1A C-terminus negatively modulates oncogenic transformation, cellular proteins that interact with E1A C-terminal region were searched for. One search identified and cloned the C-terminal binding protein (CtBP) that interacts with a conserved sequence motif PLDLS within E1A CR4 [49–50]. A more recent proteomics approach identified the interaction of two other protein complexes - the forkhead family transcription factors, FOXK1 and FOXK2, and the dual-specificity Ser/Thr kinases DYRK1A/1B and their substrate HAN11 [51]. The E1A C-terminal region was also reported to interact with the yeast homolog of DYRK1A/1B, Yak1p [52]. It appears that a defect in interaction with all three cellular protein complexes might additively contribute to the strong hyper-transformation and tumorigenic phenotypes observed in E1A-Ras cooperative transformation assays that employed deletion mutants encompassing these protein binding regions [42].
Subversion of CtBP
Among the interactions of cellular proteins with the E1A C-terminal region, the functional consequence of interaction of E1A with CtBP is the best understood. There are two major members of the CtBP family, CtBP1 and CtBP2 (collectively referred here as CtBP) [53]. Both proteins function predominantly as transcriptional corepressors [54] and E1A interacts with both CtBP1 and CtBP2 in HAdv5-infected cells [51]. Grooteclaes and Frisch reported that wild-type E1A activated the expression of epithelial cell adhesion molecules such as E-cadherin, desmoglein-2 and plakoglobin while an E1A mutant defective in interaction with CtBP did not when transduced into a human melanoma cell line, implicating CtBP in the repression of the epithelial genes [55]. This study further identified the CtBP-interacting E-box repressor Zeb-1 as the negative regulator of E-cadherin, suggesting that E1A might activate E-cadherin expression by relieving transcriptional repression by disrupting CtBP interaction with Zeb-1 [55]. CtBP1 has also been identified as a specific inhibitor of differentiation of colon epithelium in the zebrafish model and in humans [56].
The interaction of E1A with CtBP was also implicated in conferring anoikis sensitivity [55]. Consistent with these results, a gene expression profiling study using fibroblasts derived from CtBP knockout mouse embryo fibroblasts (lacking both CtBP1 and CtBP2) and RNA interference (RNAi)-mediated depletion of CtBP2 in tumor cells identified activation of several pro-apoptotic genes such as Bax, Noxa, Perp and p21 [57] and Bik [58]. Additionally, CtBP was also identified as a negative regulator of the tumor suppressor PTEN, which plays important roles in limiting cell proliferation and promoting apoptosis by inhibiting the activation of the cell survival promoting protein kinase Akt [57]. CtBP might contribute to suppression of apoptosis by preventing phosphorylation of pro-apoptotic Akt targets, such as BAD and caspase-9 [59]. Thus, CtBP functions as a specific antagonist of epithelial phenotypes and anoikis and E1A could interact with CtBP to confer epithelial characteristics to mesenchymal cells and enhance sensitivity to anoikis.
Subversion of FOXK1/K2
Recent proteomic analysis of E1A-associated cellular proteins identified the interaction of two forkhead family transcription factors, FOXK1 and FOXK2 with E1A [51]. FOXK1/K2 (referred here as FOXK) are unique among the forkhead family transcription factors as they contain the forkhead-associated (FHA) domain (reviewed in [60]). FHA domains are protein interaction modules that recognize phosphorylated residues of different target proteins (reviewed in [61]). E1A interacts with FOXK1 through a Ser/Thr containing sequence motif conserved in species C (non-oncogenic) HAdvs. An E1A mutant deficient in interaction with FOXK induced increased proliferation of quiescent epithelial (BRK) cells and enhanced frequency of Ras-cooperative transformation. The tumorigenic activity (in immune deficient mice) of the transformed cells expressing the mutant E1A and Ras was also enhanced, suggesting that the E1A domain involved in interaction with FOXK contributes to suppression of cell proliferation and oncogenic transformation [51].
As with CtBP, the FOXK proteins might also regulate cell proliferation, differentiation and apoptosis pathways. In mammalian cells, FOXK1 has been reported to function as a stem cell proliferation and maintenance factor (reviewed in [62]). In Foxk1 knockout mice, proliferation of myogenic progenitor cells were severely impaired resulting in a defect in skeletal muscle regeneration [63] which was rescued by the combined deficiency of both Foxk1 and p21(CIP) loci. In a muscle stem cell population derived from Foxk1 null mice, differentiation related genes were also upregulated [64]. FOXK2 is believed to be functionally similar to FOXK1 and has been implicated in the regulation of p21 expression during the cell cycle [65]. However, yeast proteins Fkh1 and Fkh2 appear to be homologs of the mammalian FOXK proteins since they also possess the FHA domain. The yeast Fkh1/2 factors have been implicated in the transcriptional regulation of genes involved in G2/M transition and yeast Fkh1/2 mutants display aberrant mitotic exit and constitutive pseudohyphal growth [66]. Although transcriptional repression and activation functions of mammalian FOXK1/K2 are possible, only the transcriptional repression activity of FOXK1 has been studied in some detail [67–68]. Thus, the interaction of E1A with FOXK factors could deregulate their transcriptional activities causing cell cycle withdrawal and cell differentiation contributing to suppression of oncogenic transformation.
Subversion of DYRK1A/1B/HAN11 complex
Proteomics analysis also identified the interaction of the dual specificity kinases DYRK1A and DYRK1B and their target molecule HAN11 with the CR4 region of E1A C-terminus [51]. Unlike the interaction of CtBP and FOXK proteins, the interaction of DYRK1A/1B appears to depend on more extended sequences within the CR4 region [51–52]. Like the CtBP-binding sequence, the CR4 region involved in interaction with DYRK1A/1B is also highly conserved among the E1A proteins of primate adenoviruses. E1A mutants deficient in interaction with these proteins also induced enhanced cell proliferation and caused hyper-transformation [51]. The transformed cells were more tumorigenic than transformed cells expressing wild-type E1A. In general, these mutants exhibited more potent hyper-transforming and tumorigenic activities than the E1A mutant defective in interaction with FOXK proteins. The possibility that the hyper-transforming E1A mutants previously identified by the Quinlan laboratory [44–45] could map within the CR4 region broadly involved in DYRK(1A/1B)/HAN11 interaction remains to be investigated.
Similar to the other two protein complexes that interact with the E1A C-terminal region, the DYRK(1A/1B)/HAN11 complex might also regulate cell proliferation and differentiation. In mammalian cells, DYRK1A has been implicated in the activation of transcription factors such as NFAT, Gli and FoxO1 and the Notch signaling pathway that are associated with cell fate and differentiation [69–72]. DYRK1B has been shown to regulate differentiation and apoptosis (reviewed in [73]). As pointed out above, Yak1p is linked to pseudohyphal growth in yeast [52]. In zebrafish, the DYRK1A cofactor HAN11 is linked to the endothelin-1 signaling pathway that controls the craniofacial developmental program [74]. Therefore, the E1A C-terminal region could exert its tumor restraining activity by disrupting the normal differentiation activities of the DYRK1A/1B/HAN11 complex. Genetic studies in yeast also suggest there is significant crosstalk between the Yak1p and Fkh1/2 pathways [75]. Therefore, there might be crosstalk among all three pathways deregulated by the E1A C-terminal region to exert a coordinated effect on cell cycle exit and promotion of epithelial differentiation to inhibit cell transformation and oncogenesis.
Functional similarity between E1A and the E6 proteins of beta-HPVs
As small DNA tumor viruses promote cell proliferation and oncogenic transformation via common pathways, it was investigated whether proteins of other epithelial tropic viruses could also inhibit oncogenic transformation similar to E1A. A search for protein-interaction sequence motifs similar to those present in the E1A C-terminal region revealed the presence of a Ser/Thr-rich motif (required for the interaction of FOXK1/K2) in the N-terminus of the E6 proteins of two benign cutaneous beta-HPVs, HPV14 and HPV21 [51] (Figure 3) (see Box 1 for HPV classification and biology). The E6 protein of another beta-HPV, HPV20, which is found in some skin cancers (along with HPV5 and HPV8) contains the unique N-terminal domain similar to the E6 protein of HPV14 and HPV21; but the N-terminal domain of HPV20 E6 contains a single amino acid mutation in a conserved Thr residue (Thr→Glu) (Figure 3). In coimmunoprecipitation studies, the E6 proteins of HPV14and HPV21 interacted efficiently with FOXK1/K2 while the Thr→Glu mutation (present in HPV20 E6) prevented such interaction [51]. Thus, the E6 proteins of more benign beta-HPVs interact with of FOXK1/K2 while the E6 proteins of other beta-HPVs such as HPV5 and HPV8 associated with skin cancer do not contain the unique N-terminal domain present in HPV14/and HPV21. In the E1A-Ras cooperative transformation assay, the E6 protein of HPV21 reduced the transforming activity of an E1A hyper-transforming mutant that is defective in interaction FOXK1/K2 while a mutant E6 defective in interaction with FOXK1/K2 did not inhibit E1A-Ras transformation [51]. The E6 domain also efficiently substituted for the corresponding domain in E1A in interaction with FOXK and in inhibition of transformation. These results suggest that HPV14 and HPV21 E6 proteins could possess a transformation suppression function by virtue of sequestering FOXK1/K2 transcription factors. The potential effect of E6 interaction with FOXK1/K2 transcription factors on epithelial differentiation during virus replication remains to be determined. Such studies with beta-HPVs could now be a reality as efficient in vitro raft tissue culture systems that faithfully replicate HPV genomes in primary keratinocytes have been developed.
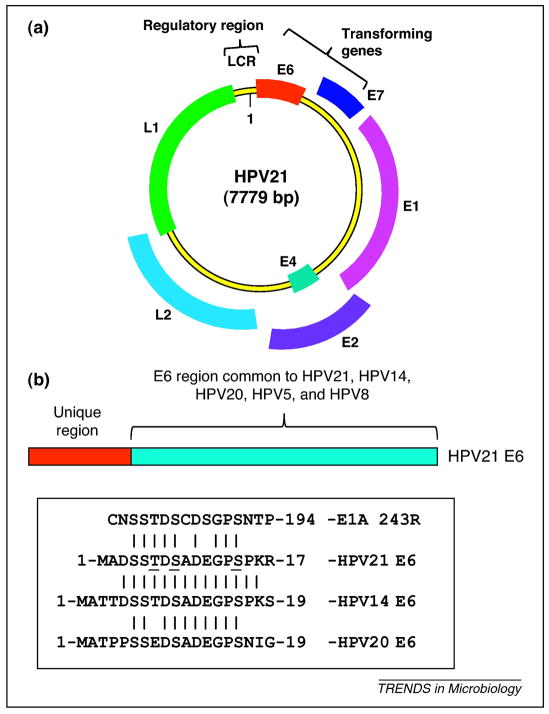
Figure 3.
Genome and proteins of HPV21. [GT1] (a) Schematic illustration of HPV21 genome. The genome structures of beta-HPVs conform to the overall genome structures of alpha-HPVs except that beta-HPVs do not encode an early protein, E5. The genome of HPV21 consists of 7.9 kb circular double stranded DNA. The viral genome encode five early proteins (designated as E1, E2, E4, E6 and E7) and two late proteins (designated as L1 and L2) that are viral capsid proteins. The viral genome contains a non-coding region (NCR) that contains the major promoter for the transcription of early genes. The early genes E6 and E7 constitute the transforming genes. (b) Schematic illustration of the E6 proteins of EV-associated beta-HPVs. The unique N-terminal domains of the E6 proteins of HPV14, HPV21 and HPV20 are compared with the FOXK1/K2-interacting motif of HAdv5 E1A in the boxed area.[GT2]
Box 1. Benign HPVs: natural-born tumor suppressive agents?
Papillomaviruses are small DNA viruses that exhibit stringent epithelial tropism. There are over 200 HPVs and a vast majority of HPVs are ubiquitous pathogens that mostly cause benign skin lesions such as warts. A subset of them, alpha-HPVs, are designated as high-risk and infect the anogential tract and head and neck mucosa where they contribute to benign warts and low-grade epithelial lesions that can progress into high-grade carcinomas (reviewed in [82]). Beta-HPVs are associated with prevalent skin infections that are generally asymptomatic in the general population. Immunosuppressed patients and those suffering from an inherited disorder known as epidermodyplasia verruciformis (EV) are prone to infection by beta-HPVs. In EV patents, a subspecies of related beta-HPVs such as HPV5, HPV8, HPV20 and HPV47 are generally more prevalent and are linked to the development of premalignant skin lesions that progress to squamous cell carcinomas in sun-exposed areas [83]. The EV-associated beta-HPVs have been divided into two clusters – those that are more frequently associated with squamous cell carcinomas (e.g. HPV5, HPV8, HPV20 and HPV47) and those less frequently associated with such neoplasm (e.g. HPV14 and HPV21). While the E6 proteins of beta-HPVs share significant amino acid homology, the E6 proteins of HPV14 and HPV21 contain an additional N-terminal domain that mediates interaction with FOXK proteins [51].
HPVs infect the basal layer of the epithelium through surface wounds. The viral DNA replication is initiated in virus-harboring basal cells and the viral genome is amplified only in post-mitotic replicating and differentiating squamous epithelial cells. The E7 gene of high-risk HPVs (which is generally considered as a functional equivalent of E1A) promotes S phase entry of keratinocytes by targeting pRb and pRb family members p107 and p130 for degradation to deregulate the E2F transcriptional pathway that activates the expression of S phase genes (reviewed in [34]). The E6 protein of high-risk HPVs targets the tumor suppressor p53 for degradation to facilitate cell cycle entry (reviewed in [84]). The E7 and E6 proteins of high-risk HPVs contribute to the oncogenic activity of these viruses. Although the E7 gene of high-risk HPV has been linked to epithelial to mesenchymal transition [85], there is no significant information linking any of the viral genes to the promotion of epithelial differentiation.
In contrast to extensive studies on the E7 and E6 proteins of high-risk HPVs, the E7 and E6 proteins of beta-HPVs have not been intensely investigated. However, it appears that the E7 proteins of beta-HPVs could facilitate S phase entry of differentiated keratinocytes as effectively as the E7 proteins of alpha-HPVs. Several beta-HPV E7 proteins have been reported to bind pRb and also target pRb for degradation. The E6 genes of beta-HPVs that are associated with the development of skin cancer (HPV5, HPV8 and HPV47) have been shown to possess a more potent in vitro transforming activity in an established rodent fibroblast cell line than the more benign HPVs such as HPV14, HPV21 and HPV25 [86]. Transgenic mice expressing the E6 protein of HPV8 have also been shown to develop spontaneous skin cancer [87]. Although the E6 proteins of beta-HPVs do not affect p53 protein levels, it appears that at least some of the oncogenic activities the E6 proteins of beta-HPVs might be linked to their ability to protect human keratinocytes against UV-induced apoptosis by targeting the pro-apoptotic molecule BAK for degradation [88]. As HPV replication is stringently dependent on epithelial differentiation and is restricted in squamous cell carcinomas [82], the natural history of HPVs suggests that they might be ‘natural-born’ tumor (epithelial) suppressive agents driving epithelial cells to terminal differentiation. The oncogenic events induced by these viruses might be rare consequences of viral infection that is also influenced by stochastic environmental and behavioral conditions.
Concluding remarks and perspectives
Adenovirus E1A oncoprotein continues to serve as a cornucopia of paradigms on cell proliferation and oncogenesis. Now E1A is on the cusp of illuminating novel pathways of restraining oncogenesis that could be exploited to restrict and reverse epithelial cancers. Recent studies have revealed that the ‘oncoproteins’ of certain benign HPVs are also endowed with such a tumor suppressive function. Considering that the vast majority of papillomaviruses are benign and replicate in differentiating epithelial cells (Box 1), it is possible that some of the proteins encoded by these viruses could possess novel tumor suppressive functions by promoting epithelial differentiation. Several new polyomaviruses such as Merkel cell polyomavirus (MCV)[76], WU polyomavirus [77] and KI polyomavirus [78] and human polyomaviruses 6–7 [79] have been isolated from epithelial tissues. Among them, MCV has been associated with the epithelial cancer Merkel cell carcinoma (MCC) [76] which contains clonally integrated MCV sequences that only encode the N-terminal region of the T Ag [80]. It would be interesting to determine whether the transforming proteins of these epithelial tropic viruses encode any tumor suppressive and cell differentiation functions. The possibility that viruses which replicate in other differentiated tissues might also possess the capacity to suppress or reverse oncogenesis remains to be explored (Box 2). Why have viral proteins evolved this function? As discussed earlier, the epithelial tropic viruses such as HAdv and HPVs induce transient cell proliferation to facilitate their DNA replication (Figure 4). The early viral proteins such as E1A (N-terminal region) and E7 that promote cell proliferation also inhibit differentiation. The tumor suppressive activity of the viral transforming proteins might have evolved to counteract the differentiation inhibitory activity to enhance viral multiplication resulting in a homeostasis between two opposing activities.
Box 2. Outstanding questions.
Does interaction of the E1A C-terminal region with CtBP, FOXK1/K2 and DYRK1A/1B/HAN11 sequester these factors and/or retarget them to new target sites on the cell genome?
Do other viral genes target CtBP and DYRK1A/1B/HAN11 complexes to suppress oncogenesis?
Are there cellular targets other than CtBP, FOXK1/K2 and DYRK1A/1B/HAN11 for epithelial tropic viral genes to suppress oncogenesis?
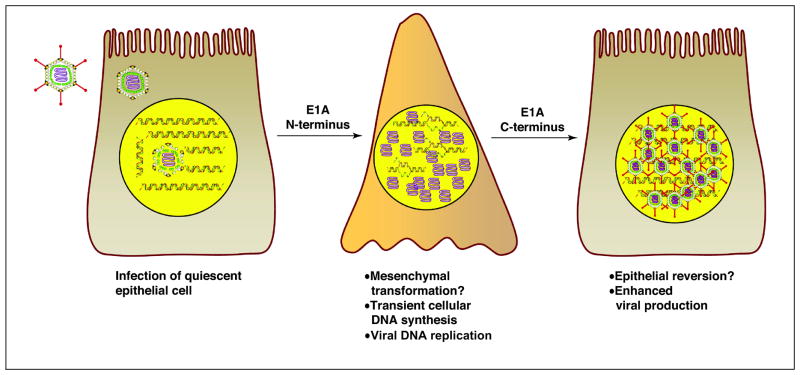
Figure 4.
Potential consequences of epithelial differentiation on virus multiplication. The activities of the E1A N-terminal region are depicted promoting cell proliferation, viral DNA replication and possibly [GT3] epithelial to mesenchymal cell transformation. The activities of the E1A C-terminal region are depicted as possibly [GT4] promoting mesenchymal to epithelial transformation thereby providing a cellular environment that facilitates further steps in virus multiplication.
Studies on E1A have revealed that subverting the pathways controlled by three different cellular protein complexes - CtBP, DYRK1A/1B/HAN11 and FOXK1/K2 - leads to tumor suppression. Detailed investigations of these pathways could reveal crucial regulatory check points to inhibit cancer. Recently, a proof-of-principle report has shown that inhibition of the activities of CtBP with a small molecule leads to suppression of tumorigenesis by human colon cancer cells [81]. These results suggest that CtBP could be a potential anti-cancer drug target. Similarly, the possibility of inhibiting human epithelial cancers by targeting FOXK1/K2 transcription factors and DYRK1A/1B kinases remain attractive goals. It is hoped that investigations with other viral transforming genes could identify other potential targets by which they promote epithelial differentiation and suppress oncogenesis.
Acknowledgments
The author thanks Bill Wold, Ling Zhao and Denise Galloway for their comments on the manuscript and S. Vijayalingam for the figures. The author received support from research grants by the National Cancer Institute (CA-084941 and CA-033616).
Glossary
- Anoikis
a form of apoptosis induced by detachment of adherent mammalian cells
- Baby rat kidney (BRK) cells
are prepared from 2–4 day-old neonatal rats. These are used as model epithelial cells for in vitro transformation by viral oncogenes
- ChIP-chip
an experimental approach used to identify the DNA binding sites for transcription factors on a genome-wide basis. It involves chromatin immunoprecipitation (ChIP) and microarray (chip) analysis
- CR1-4
conserved region 1–4, the domains conserved among the E1A proteins of all primate adenoviruses
- Transcription/transformation domain-associated protein (TRRAP)
a scaffolding protein that is part of two different histone acetyl transferase (HAT) protein complexes – Tip (Tat-interactive protein) 60 complex and SAGA (Spt-Ada-Gcn5) complex
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Javier RT, Butel JS. The history of tumor virology. Cancer Res. 2008;68:7693–7706. doi: 10.1158/0008-5472.CAN-08-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeCaprio JA. How the Rb tumor suppressor structure and function was revealed by the study of Adenovirus and SV40. Virology. 2009;384:274–284. doi: 10.1016/j.virol.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 3.Graham FL, et al. Size and location of the transforming region in human adenovirus type 5 DNA. Nature. 1974;251:687–691. doi: 10.1038/251687a0. [DOI] [PubMed] [Google Scholar]
- 4.Ruley HE. Adenovirus early region 1A enables viral and cellular transforming genes to transform primary cells in culture. Nature. 1983;304:602–606. doi: 10.1038/304602a0. [DOI] [PubMed] [Google Scholar]
- 5.Pelka P, et al. Intrinsic structural disorder in adenovirus E1A: a viral molecular hub linking multiple diverse processes. J Virol. 2008;82:7252–7263. doi: 10.1128/JVI.00104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallimore PH, Turnell AS. Adenovirus E1A: remodelling the host cell, a life or death experience. Oncogene. 2001;20:7824–7835. doi: 10.1038/sj.onc.1204913. [DOI] [PubMed] [Google Scholar]
- 7.Wang HG, et al. Identification of specific adenovirus E1A N-terminal residues critical to the binding of cellular proteins and to the control of cell growth. J Virol. 1993;67:476–488. doi: 10.1128/jvi.67.1.476-488.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckner R, et al. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 9.Arany Z, et al. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 10.Fuchs M, et al. The p400 complex is an essential E1A transformation target. Cell. 2001;106:297–307. doi: 10.1016/s0092-8674(01)00450-0. [DOI] [PubMed] [Google Scholar]
- 11.Deleu L, et al. Recruitment of TRRAP required for oncogenic transformation by E1A. Oncogene. 2001;20:8270–8275. doi: 10.1038/sj.onc.1205159. [DOI] [PubMed] [Google Scholar]
- 12.Lang SE, Hearing P. The adenovirus E1A oncoprotein recruits the cellular TRRAP/GCN5 histone acetyltransferase complex. Oncogene. 2003;22:2836–2841. doi: 10.1038/sj.onc.1206376. [DOI] [PubMed] [Google Scholar]
- 13.Kulesza CA, et al. Adenovirus E1A requires the yeast SAGA histone acetyltransferase complex and associates with SAGA components Gcn5 and Tra1. Oncogene. 2002;21:1411–1422. doi: 10.1038/sj.onc.1205201. [DOI] [PubMed] [Google Scholar]
- 14.Ferguson EL, Horvitz HR. The multivulva phenotype of certain Caenorhabditis elegans mutants results from defects in two functionally redundant pathways. Genetics. 1989;123:109–121. doi: 10.1093/genetics/123.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ceol CJ, Horvitz HR. dpl-1 DP and efl-1 E2F act with lin-35 Rb to antagonize Ras signaling in C elegans vulval development. Mol Cell. 2001;7:461–473. doi: 10.1016/s1097-2765(01)00194-0. [DOI] [PubMed] [Google Scholar]
- 16.Ceol CJ, Horvitz HR. A new class of C elegans synMuv genes implicates a Tip60/NuA4-like HAT complex as a negative regulator of Ras signaling. Dev Cell. 2004;6:563–576. doi: 10.1016/s1534-5807(04)00065-6. [DOI] [PubMed] [Google Scholar]
- 17.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 18.Horwitz GA, et al. Adenovirus small e1a alters global patterns of histone modification. Science. 2008;321:1084–1085. doi: 10.1126/science.1155544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferrari R, et al. Epigenetic reprogramming by adenovirus e1a. Science. 2008;321:1086–1088. doi: 10.1126/science.1155546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sankar N, et al. p300 provides a corepressor function by cooperating with YY1 and HDAC3 to repress c-Myc. Oncogene. 2008;27:5717–5728. doi: 10.1038/onc.2008.181. [DOI] [PubMed] [Google Scholar]
- 21.Kadeppagari RK, et al. Adenovirus transforming protein E1A induces c-Myc in quiescent cells by a novel mechanism. J Virol. 2009;83:4810–4822. doi: 10.1128/JVI.02145-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singhal G, et al. Simian virus 40 large T overcomes p300 repression of c-Myc. Virology. 2008;377:227–232. doi: 10.1016/j.virol.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turnell AS, et al. The APC/C and CBP/p300 cooperate to regulate transcription and cell-cycle progression. Nature. 2005;438:690–695. doi: 10.1038/nature04151. [DOI] [PubMed] [Google Scholar]
- 24.Nikiforov MA, et al. TRRAP-dependent and TRRAP-independent transcriptional activation by Myc family oncoproteins. Mol Cell Biol. 2002;22:5054–5063. doi: 10.1128/MCB.22.14.5054-5063.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ikura T, et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 26.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMahon SB, et al. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 28.McMahon SB, et al. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frank SR, et al. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frank SR, et al. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 2003;4:575–580. doi: 10.1038/sj.embor.embor861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patel JH, et al. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol Cell Biol. 2004;24:10826–10834. doi: 10.1128/MCB.24.24.10826-10834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lohr K, et al. Mutual interference of adenovirus infection and myc expression. J Virol. 2003;77:7936–7944. doi: 10.1128/JVI.77.14.7936-7944.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tworkowski KA, et al. Adenovirus E1A targets p400 to induce the cellular oncoprotein Myc. Proc Natl Acad Sci U S A. 2008;105:6103–6108. doi: 10.1073/pnas.0802095105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McLaughlin-Drubin ME, Munger K. The human papillomavirus E7 oncoprotein. Virology. 2009;384:335–344. doi: 10.1016/j.virol.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whyte P, et al. Cellular targets for transformation by the adenovirus E1A proteins. Cell. 1989;56:67–75. doi: 10.1016/0092-8674(89)90984-7. [DOI] [PubMed] [Google Scholar]
- 36.Liu X, Marmorstein R. Structure of the retinoblastoma protein bound to adenovirus E1A reveals the molecular basis for viral oncoprotein inactivation of a tumor suppressor. Genes Dev. 2007;21:2711–2716. doi: 10.1101/gad.1590607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nevins JR. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 38.Seifried LA, et al. pRB-E2F1 complexes are resistant to adenovirus E1A-mediated disruption. J Virol. 2008;82:4511–4520. doi: 10.1128/JVI.02713-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nemajerova A, et al. Rb function is required for E1A-induced S-phase checkpoint activation. Cell Death Differ. 2008;15:1440–1449. doi: 10.1038/cdd.2008.66. [DOI] [PubMed] [Google Scholar]
- 40.Webster KA, et al. Adenovirus E1A products suppress myogenic differentiation and inhibit transcription from muscle-specific promoters. Nature. 1988;332:553–557. doi: 10.1038/332553a0. [DOI] [PubMed] [Google Scholar]
- 41.Mymryk JS, et al. Ability of adenovirus 5 E1A proteins to suppress differentiation of BC3H1 myoblasts correlates with their binding to a 300 kDa cellular protein. Mol Biol Cell. 1992;3:1107–1115. doi: 10.1091/mbc.3.10.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subramanian T, et al. Enhanced ras oncogene mediated cell transformation and tumorigenesis by adenovirus 2 mutants lacking the C-terminal region of E1a protein. Oncogene. 1989;4:415–420. [PubMed] [Google Scholar]
- 43.Chinnadurai G. Adenovirus E1a as a tumor-suppressor gene. Oncogene. 1992;7:1255–1258. [PubMed] [Google Scholar]
- 44.Douglas JL, et al. Modulation of transformation of primary epithelial cells by the second exon of the Ad5 E1A12S gene. Oncogene. 1991;6:2093–2103. [PubMed] [Google Scholar]
- 45.Quinlan MP, Douglas JL. Immortalization of primary epithelial cells requires first- and second-exon functions of adenovirus type 5 12S. J Virol. 1992;66:2020–2030. doi: 10.1128/jvi.66.4.2020-2030.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frisch SM. Antioncogenic effect of adenovirus E1A in human tumor cells. Proc Natl Acad Sci U S A. 1991;88:9077–9081. doi: 10.1073/pnas.88.20.9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frisch SM. E1a induces the expression of epithelial characteristics. J Cell Biol. 1994;127:1085–1096. doi: 10.1083/jcb.127.4.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fischer RS, Quinlan MP. The C terminus of E1A regulates tumor progression and epithelial cell differentiation. Virology. 1998;249:427–439. doi: 10.1006/viro.1998.9337. [DOI] [PubMed] [Google Scholar]
- 49.Boyd JM, et al. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 1993;12:469–478. doi: 10.1002/j.1460-2075.1993.tb05679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schaeper U, et al. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc Natl Acad Sci U S A. 1995;92:10467–10471. doi: 10.1073/pnas.92.23.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Komorek J, et al. Adenovirus type 5 E1A and E6 proteins of low-risk cutaneous beta-human papillomaviruses suppress cell transformation through interaction with FOXK1/K2 transcription factors. J Virol. 84:2719–2731. doi: 10.1128/JVI.02119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Z, et al. Interaction of the E1A oncoprotein with Yak1p, a novel regulator of yeast pseudohyphal differentiation, and related mammalian kinases. Mol Biol Cell. 2001;12:699–710. doi: 10.1091/mbc.12.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chinnadurai G. CtBP, an Unconventional Transcriptional Corepressor in Development and Oncogenesis. Mol Cell. 2002;9:213–224. doi: 10.1016/s1097-2765(02)00443-4. [DOI] [PubMed] [Google Scholar]
- 54.Chinnadurai G. Transcriptional regulation by C-terminal binding proteins. Int J Biochem Cell Biol. 2007;39:1593–1607. doi: 10.1016/j.biocel.2007.01.025. [DOI] [PubMed] [Google Scholar]
- 55.Grooteclaes ML, Frisch SM. Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene. 2000;19:3823–3828. doi: 10.1038/sj.onc.1203721. [DOI] [PubMed] [Google Scholar]
- 56.Phelps RA, et al. A two-step model for colon adenoma initiation and progression caused by APC loss. Cell. 2009;137:623–634. doi: 10.1016/j.cell.2009.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grooteclaes M, et al. C-terminal-binding protein corepresses epithelial and proapoptotic gene expression programs. Proc Natl Acad Sci U S A. 2003;100:4568–4573. doi: 10.1073/pnas.0830998100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kovi RC, et al. An ARF/CtBP2 complex regulates BH3-only gene expression and p53-independent apoptosis. Cell Death Differ. 17:513–521. doi: 10.1038/cdd.2009.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paez J, Sellers WR. PI3K/PTEN/AKT pathway A critical mediator of oncogenic signaling. Cancer Treat Res. 2003;115:145–167. [PubMed] [Google Scholar]
- 60.Durocher D, Jackson SP. The FHA domain. FEBS Lett. 2002;513:58–66. doi: 10.1016/s0014-5793(01)03294-x. [DOI] [PubMed] [Google Scholar]
- 61.Mahajan A, et al. Structure and function of the phosphothreonine-specific FHA domain. Sci Signal. 2008;1:re12. doi: 10.1126/scisignal.151re12. [DOI] [PubMed] [Google Scholar]
- 62.Martin CM, et al. Molecular signatures define myogenic stem cell populations. Stem Cell Rev. 2006;2:37–42. doi: 10.1007/s12015-006-0007-x. [DOI] [PubMed] [Google Scholar]
- 63.Hawke TJ, et al. Absence of p21CIP rescues myogenic progenitor cell proliferative and regenerative capacity in Foxk1 null mice. J Biol Chem. 2003;278:4015–4020. doi: 10.1074/jbc.M209200200. [DOI] [PubMed] [Google Scholar]
- 64.Meeson AP, et al. Cellular and molecular regulation of skeletal muscle side population cells. Stem Cells. 2004;22:1305–1320. doi: 10.1634/stemcells.2004-0077. [DOI] [PubMed] [Google Scholar]
- 65.Marais A, et al. Cell cycle-dependent regulation of the forkhead transcription factor FOXK2 by CDK.cyclin complexes. J Biol Chem. 2010;285:35728–35739. doi: 10.1074/jbc.M110.154005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu G, et al. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature. 2000;406:90–94. doi: 10.1038/35017581. [DOI] [PubMed] [Google Scholar]
- 67.Yang Q, et al. The winged-helix/forkhead protein myocyte nuclear factor beta (MNF-beta) forms a co-repressor complex with mammalian sin3B. Biochem J. 2000;345(Pt 2):335–343. [PMC free article] [PubMed] [Google Scholar]
- 68.Freddie CT, et al. Functional interactions between the Forkhead transcription factor FOXK1 and the MADS-box protein SRF. Nucleic Acids Res. 2007;35:5203–5212. doi: 10.1093/nar/gkm528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fernandez-Martinez J, et al. Attenuation of Notch signalling by the Down-syndrome-associated kinase DYRK1A. J Cell Sci. 2009;122:1574–1583. doi: 10.1242/jcs.044354. [DOI] [PubMed] [Google Scholar]
- 70.Arron JR, et al. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature. 2006;441:595–600. doi: 10.1038/nature04678. [DOI] [PubMed] [Google Scholar]
- 71.Gwack Y, et al. A genome-wide Drosophila RNAi screen identifies DYRK-family kinases as regulators of NFAT. Nature. 2006;441:646–650. doi: 10.1038/nature04631. [DOI] [PubMed] [Google Scholar]
- 72.Mao J, et al. Regulation of Gli1 transcriptional activity in the nucleus by Dyrk1. J Biol Chem. 2002;277:35156–35161. doi: 10.1074/jbc.M206743200. [DOI] [PubMed] [Google Scholar]
- 73.Friedman E. Mirk/Dyrk1B in cancer. J Cell Biochem. 2007;102:274–279. doi: 10.1002/jcb.21451. [DOI] [PubMed] [Google Scholar]
- 74.Nissen RM, et al. A zebrafish screen for craniofacial mutants identifies wdr68 as a highly conserved gene required for endothelin-1 expression. BMC Dev Biol. 2006;6:28. doi: 10.1186/1471-213X-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jaspersen SL, et al. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:2803–2817. doi: 10.1091/mbc.9.10.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Feng H, et al. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Le BM, et al. Clinical and epidemiologic characterization of WU polyomavirus infection, St Louis, Missouri. Emerg Infect Dis. 2007;13:1936–1938. doi: 10.3201/eid1312.070977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Allander T, et al. Identification of a third human polyomavirus. J Virol. 2007;81:4130–4136. doi: 10.1128/JVI.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schowalter RM, et al. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe. 2010;7:509–515. doi: 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shuda M, et al. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc Natl Acad Sci U S A. 2008;105:16272–16277. doi: 10.1073/pnas.0806526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Straza MW, et al. Therapeutic targeting of C-terminal binding protein in human cancer. Cell Cycle. 2010;9:3740–3750. doi: 10.4161/cc.9.18.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chow LT, et al. The natural history of human papillomavirus infections of the mucosal epithelia. APMIS. 118:422–449. doi: 10.1111/j.1600-0463.2010.02625.x. [DOI] [PubMed] [Google Scholar]
- 83.Orth G, et al. Characteristics of the lesions and risk of malignant conversion associated with the type of human papillomavirus involved in epidermodysplasia verruciformis. Cancer Res. 1979;39:1074–1082. [PubMed] [Google Scholar]
- 84.Howie HL, et al. Papillomavirus E6 proteins. Virology. 2009;384:324–334. doi: 10.1016/j.virol.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hellner K, et al. HPV16 E7 oncogene expression in normal human epithelial cells causes molecular changes indicative of an epithelial to mesenchymal transition. Virology. 2009;391:57–63. doi: 10.1016/j.virol.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kiyono T, et al. Differences in transforming activity and coded amino acid sequence among E6 genes of several papillomaviruses associated with epidermodysplasia verruciformis. Virology. 1992;186:628–639. doi: 10.1016/0042-6822(92)90029-o. [DOI] [PubMed] [Google Scholar]
- 87.Marcuzzi GP, et al. Spontaneous tumour development in human papillomavirus type 8 E6 transgenic mice and rapid induction by UV-light exposure and wounding. J Gen Virol. 2009;90:2855–2864. doi: 10.1099/vir.0.012872-0. [DOI] [PubMed] [Google Scholar]
- 88.Jackson S, et al. Role of Bak in UV-induced apoptosis in skin cancer and abrogation by HPV E6 proteins. Genes Dev. 2000;14:3065–3073. doi: 10.1101/gad.182100. [DOI] [PMC free article] [PubMed] [Google Scholar]