Abstract
The sulphilimine cross-link of the Goodpasture (GP) autoantigen is a novel molecular mechanism (structural constraint) for conferring immune privilege to a site which otherwise is susceptible to structural changes that induce an immunogenic and pathogenic conformation. Perturbation of the assembly or cleavage of the sulphilimine cross-links could be a key factor in the aetiology of Goodpasture's disease in susceptible individuals.
Keywords: Goodpasture disease, autoantigen, Sulphilimine cross-links
Introduction
Goodpasture's (GP) disease is an autoimmune disease mediated by autoantibodies that attack the glomerular and alveolar basement membranes. GP autoantibodies target the non-collagenous domain (NC1) of the α3 and α5 chain of collagen IV, both of which are part of the α345 NC1 hexamer, the GP autoantigen [1]. A three-dimensional model of the GP autoantigen has been built by homology modelling using the crystal structure of the α121 NC1 hexamer (Fig. 1) [2,3]. The hexamer is formed by the interaction of the trimeric NC1 domains of two adjoining triple-helical collagen IV molecules, and is stabilized by novel sulphilimine bonds that cross-link the interface of the trimeric NC1 domains [3,4]. Understanding the structure of the autoantigen and its epitopes is of pivotal importance in deciphering clues about aetiology of GP disease.
Fig. 1.
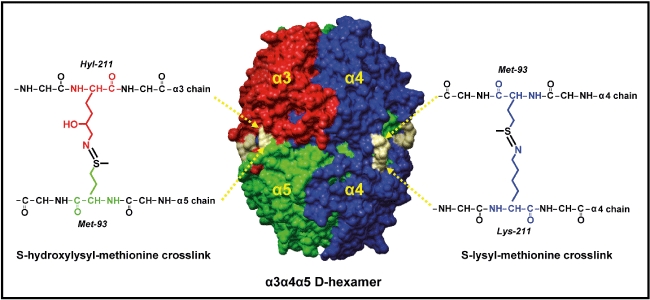
Sulphilimine bonds reinforce the structure of the α345 non-collagenous domain (NC1) hexamer, the Goodpasture (GP) autoantigen. The illustration shows a space-filling representation of the three-dimensional structure of the α345 NC1 hexamer, the GP autoantigen. It is composed of two trimeric caps, each composed of α3 (red), α4 (blue) and α5 (green) NC1 domains. Residues Met93 and Lys211 (or its hydroxylated form Hyl211) are connected through a sulphilimine bond, shown in grey, forming two different types of cross-links: s-hydroxyl-methionine (sHM, left) and s-lysyl-methionine (sLM, right).
Sulphilimine cross-links and Goodpasture's (GP) disease epitopes
The α345 NC1 hexamer requires dissociation into subunits for epitope formation [1,5,6]. Cross-linked hexamer is inert to binding of GP antibody, but upon dissociation into dimer and monomer subunits by low pH or denaturants, the subunits bind antibodies. In contrast, un-cross-linked hexamer can also be dissociated by GP antibody into monomer subunits that bind the antibody, revealing that sulphilimine cross-links provide a constraint against dissociation (Fig. 2). Hexamer dissociation involves conformational changes that unlock domain-swapping interactions between subunits and exposes residues sequestered by neighbouring subunits [1,7,8]. Concomitantly, hexamer dissociation transforms the non-pathogenic Ea and Eb regions of the α3 and α5 NC1 subunits into pathogenic conformations (epitopes) that confer antibody binding within the dissociated subunits. The sulphilimine cross-links, together with other quaternary interactions, represent a novel molecular mechanism (structural constraints) for conferring immune privilege to sites which otherwise form an immunogenic and pathogenic conformation.
Fig. 2.
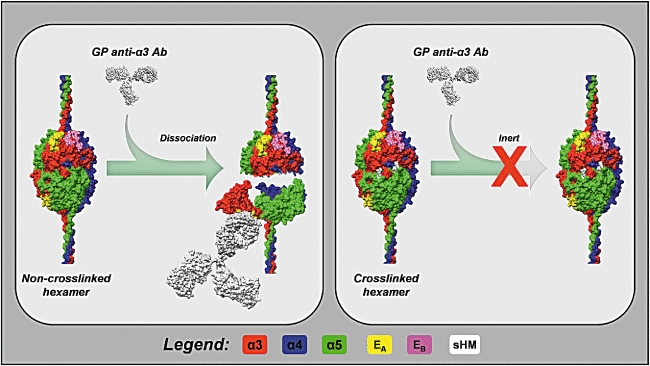
Sulphilimine cross-links provide a structural constraint against Goodpasture (GP) antibody binding. The α345 non-collagenous domain (NC1) hexamer is shown in the un-cross-linked (left panel) and cross-linked (right panel) forms. Ea and Eb regions are shown in yellow and magenta, respectively, whereas the sulphilimine cross-links are shown in white. The left panel shows an immunoglobulin G (IgG) molecule (white) binding the α3NC1 domain inducing hexamer dissociation and transitioning the Ea region into an epitope. Experimental evidence suggests that hexamer dissociation involves a conformational change within the NC1 domains, which unlocks domain-swapping interactions between subunits. The sulphilimine cross-link provides a constraint against dissociation, rendering the hexamer inert to antibody dissociation and binding.
Sulphilimine cross-links and B and T cell epitopes
GP disease is presumed to result from activation of ‘ignorant’ lymphocytes by an unknown stimulant coupled with epitope presentation and kidney and/or lung susceptibility. B cells are activated upon binding of the B cell receptors (BCR) to the soluble antigen, with assistance from T cells. BCRs presumably do not bind to the native form of the GP autoantigen, the cross-linked hexamer, but bind subunits upon hexamer dissociation into subunits. The un-cross-linked hexamer is more susceptible to dissociation; therefore, the loss of sulphilimine cross-links may be a predisposing factor for binding of BCR to hexamer subunits.
The sulphilimine cross-links may also influence the generation of T cell epitopes. Unlike B cells, T helper cells (CD4+) recognize GP antigens in the form of linear peptides bound to human leucocyte antigen (HLA) class II molecules on the surface of antigen-presenting cells (APC). GP disease has strong positive and negative major histocompatibility complex (MHC)-II associations [9]. Previous studies on the processing of GP antigen by feeding human B cells homozygous for HLA-DR15 with recombinant α3NC1 monomer have determined that enzymatic processing, rather than affinity of peptides for HLA molecules, drive GP antigen presentation [10]. The α3NC1 peptides were presented as two core sequences with medium affinity for HLA molecules and their presentation was enhanced by inhibition of the lysosomal protease asparagine endopeptidase.
As a natural mechanism to prevent the activation of T cells, sulphilimine bonds may interfere with the generation of α3NC1 sequences at different stages of proteolytic processing, peptide binding to MHC class II molecules and/or the presentation of peptides–MHC complexes for recognition by CD4+ T cells. However, it is plausible that under pathophysiological conditions that favour the absence of sulphilimine cross-links, antigen processing and presentation may be enhanced, and therefore contribute to the aetiology of GP disease.
Acknowledgments
B. G. H. is supported by a grant DK 18381 from the National Institute of Diabetes and Digestive and Kidney Diseases.
Disclosure
None.
References
- 1.Pedchenko V, Bondar O, Fogo AB, et al. Molecular architecture of the Goodpasture autoantigen in anti-GBM nephritis. N Engl J Med. 2010;363:343–54. doi: 10.1056/NEJMoa0910500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sundaramoorthy M, Meiyappan M, Todd P, Hudson BG. Crystal structure of NC1 domains. Structural basis for type IV collagen assembly in basement membranes. J Biol Chem. 2002;277:31142–53. doi: 10.1074/jbc.M201740200. [DOI] [PubMed] [Google Scholar]
- 3.Vanacore RM, Ham AJ, Cartailler JP, et al. A role for collagen IV cross-links in conferring immune privilege to the Goodpasture autoantigen: structural basis for the crypticity of B cell epitopes. J Biol Chem. 2008;283:22737–48. doi: 10.1074/jbc.M803451200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vanacore R, Ham AJ, Voehler M, et al. A sulfilimine bond identified in collagen IV. Science. 2009;325:1230–4. doi: 10.1126/science.1176811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.David M, Borza DB, Leinonen A, Belmont JM, Hudson BG. Hydrophobic amino acid residues are critical for the immunodominant epitope of the Goodpasture autoantigen. A molecular basis for the cryptic nature of the epitope. J Biol Chem. 2001;276:6370–7. doi: 10.1074/jbc.M008956200. [DOI] [PubMed] [Google Scholar]
- 6.Borza D-B, Bondar O, Todd P, et al. Quaternary organization of the Goodpasture autoantigen, the alpha 3(IV) collagen chain. Sequestration of two cryptic autoepitopes by intraprotomer interactions with the α4 and α5 NC1 domains. J Biol Chem. 2002;277:40075–83. doi: 10.1074/jbc.M207769200. [DOI] [PubMed] [Google Scholar]
- 7.Wieslander J, Langeveld J, Butkowski R, Jodlowski M, Noelken M, Hudson BG. Physical and immunochemical studies of the globular domain of type IV collagen. Cryptic properties of the Goodpasture antigen. J Biol Chem. 1985;260:8564–70. [PubMed] [Google Scholar]
- 8.Borza DB, Bondar O, Colon S, et al. Goodpasture autoantibodies unmask cryptic epitopes by selectively dissociating autoantigen complexes lacking structural reinforcement: novel mechanism for immune privilege and autoimmune pathogenesis. J Biol Chem. 2005;280:27147–54. doi: 10.1074/jbc.M504050200. [DOI] [PubMed] [Google Scholar]
- 9.Phelps RG, Rees AJ. The HLA complex in Goodpasture's disease: a model for analyzing susceptibility to autoimmunity. Kidney Int. 1999;56:1638–53. doi: 10.1046/j.1523-1755.1999.00720.x. [DOI] [PubMed] [Google Scholar]
- 10.Zou J, Henderson L, Thomas V, Swan P, Turner AN, Phelps RG. Presentation of the Goodpasture autoantigen requires proteolytic unlocking steps that destroy prominent T cell epitopes. J Am Soc Nephrol. 2007;18:771–9. doi: 10.1681/ASN.2006091056. [DOI] [PubMed] [Google Scholar]


