Abstract
Alveolar epithelial cells are directly exposed to acute and chronic fluctuations in alveolar oxygen tension. Previously, we found that the oxygen-binding protein hemoglobin is expressed in alveolar Type II cells (ATII). Here, we report that ATII cells also express a number of highly specific transcription factors and other genes normally associated with hemoglobin biosynthesis in erythroid precursors. Because hypoxia-inducible factors (HIFs) were shown to play a role in hypoxia-induced changes in ATII homeostasis, we hypothesized that the hypoxia-induced increase in intracellular HIF exerts a concomitant effect on ATII hemoglobin expression. Treatment of cells from the ATII-like immortalized mouse lung epithelial cell line-15 (MLE-15) with hypoxia for 20 hours resulted in dramatic increases in cellular levels of HIF-2α protein and parallel significant increases in hemoglobin messenger RNA (mRNA) and protein expression, as compared with that of control cells cultured in normoxia. Significant increases in the mRNA of globin-associated transcription factors were also observed, and RNA interference (RNAi) experiments demonstrated that the expression of hemoglobin is at least partially dependent on the cellular levels of globin-associated transcription factor isoform 1 (GATA-1). Conversely, levels of prosurfactant proteins B and C significantly decreased in the same cells after exposure to hypoxia. The treatment of MLE-15 cells cultured in normoxia with prolyl 4-hydroxylase inhibitors, which mimic the effects of hypoxia, resulted in increases of hemoglobin and decreases of surfactant proteins. Taken together, these results suggest a relationship between hypoxia, HIFs, and the expression of hemoglobin, and imply that hemoglobin may be involved in the oxygen-sensing pathway in alveolar epithelial cells.
Keywords: hemoglobin, alveolar epithelial cells, hypoxia, hypoxia-inducible factor
CLINICAL RELEVANCE.
Alveolar hypoxia is a common characteristic of a number of lung diseases, including sleep apnea, chronic obstructive pulmonary disease, and pulmonary edema resulting from lung injury. Our study identifies globins as oxygen-sensitive genes in alveolar epithelial cells, providing additional insights into the hypoxic response of these cells as well as a possible role for hemoglobin during alveolar development.
Pulmonary hypoxia occurs under both physiologic and pathologic conditions. In fact, a hypoxic environment is necessary for proper embryonic lung development, by promoting the formation of microvasculature and epithelial branching morphogenesis (the average fetal blood O2 fraction is ∼2–5%) (1–3). However, postnatal decreases in alveolar oxygen tension as a result of pulmonary disease disrupt alveolar homeostasis. High-altitude ascent, pathologic conditions resulting in inadequate respiration, pulmonary edema after acute lung injury, or congestive heart failure may all result in decreased oxygen tension. Alveolar Type II (ATII) cells represent approximately two thirds of epithelial cell numbers, and are of special clinical interest because of their role in the production, secretion, and recycling of pulmonary surfactant (4). In addition, ATII cells differentiate into Type I (ATI) cells upon epithelial injury, and also act to clear fluid from the alveolar space. Although numerous studies evaluated the effects of hypoxia on the pulmonary endothelium, few sought to identify hypoxia-regulated genes in alveolar epithelial cells.
Our previous studies conclusively demonstrated the expression of messenger RNA (mRNA) transcripts and polypeptides for the α-globin (HBA) and β-globin (HBB) subunits of adult hemoglobin in ATII-like cell lines and rodent ATII primary cells, as determined by a combination of RT-PCR, immunodetection, and protein sequencing (5). This discovery spurred numerous questions concerning the role of hemoglobin expression in the alveolar epithelium, particularly regarding possible effects on alveolar function and hypoxic response, as well as in protecting alveolar cells against oxidative and nitrosative stress.
Heme-binding proteins are ubiquitously found in both prokaryotes and eukaryotes, and are traditionally believed to play a role in oxygen storage or transport, as well as in binding and transporting nitric oxide (NO) and products of NO metabolism (6). Adult vertebrate hemoglobin is a tetrameric hemeprotein [α(2):β(2)], with each of the four peptide subunits binding an iron-containing heme prosthetic group (6). In our previous study, hemoglobin RNA and protein were shown to be expressed in a number of alveolar cell lines, including A549 cells (human ATII-like adenocarcinoma) and immortalized mouse lung epithelial (MLE-15) cells (transformed murine ATII), as well as in primary ATII cells purified from murine and rat lungs. Similar results were obtained by another group, demonstrating that the expression of hemoglobin appears to be absent from fully differentiated ATI cells (7). The expression of nonerythroid hemoglobins was also reported in activated macrophages from adult mice, cells of the ocular lens, and putatively in embryonic neurons (8–11). Here, we further characterize the expression of hemoglobin by ATII cells, and identify the mRNAs of a number of transcription factors and other genes that are traditionally associated with hemoglobin biosynthesis in erythroid precursors.
The signaling pathways and molecular mechanisms involved in ATII oxygen homeostasis remain largely undefined. However, a number of reports implicate that hypoxia-inducible transcription factors (HIFs) have a critical role in the hypoxic response of the lung and alveolar epithelial cells (12–15). HIF, a vital component of the mammalian oxygen-sensing pathway that mediates adaptive responses to low oxygen availability through transcriptional activation, is a heterodimer comprised of a tightly regulated α subunit and a constitutively expressed β subunit (also called ARNT) (16–18). During periods of normoxia, cellular HIF-α protein subunits are controlled by ubiquitin-targeted degradation, mediated through the hydroxylation of key proline residues. Hypoxia inhibits these hydroxylation events, allowing HIF-α to enter the nucleus, bind to ARNT, and form a functional transcriptional complex (16, 19, 20). Among the three types of α subunits, HIF-1α and HIF-2α possess defined roles in the molecular and physiologic responses to hypoxia. Conflicting reports have been published regarding the presence and hypoxic induction of HIF-1α expression in the alveolar epithelium (15, 21–24). However, the hypoxic induction of HIF-2α in ATII was confirmed in vitro and in vivo, and was implicated in a defined role in lung development during the last phase of fetal lung maturation (22, 23, 25–27). The present study suggests a role for HIFs and known globin-associated transcription factors, particularly globin-associated transcription factor isoform 1 (GATA1), in the hypoxic regulation of ATII hemoglobin.
MATERIALS AND METHODS
Cell Culture
The murine ATII cell line MLE-15 (28) was kindly provided by Dr. Jeffrey Whitsett (Children's Hospital Medical Center, Cincinnati, OH) and cultured in HITES (RPMI-1640 with 5 μg/ml insulin, 10 μg/ml transferrin, 30 nM sodium selenite, 10 nM hydrocortisone, 10 nM β-estradiol, and 10 mM Hepes) with 2% FBS (Hyclone, Provo, UT). The murine erythroleukemia (BB88) (29) and alveolar macrophage (MH-S) (30) cell lines (ATCC, Rockville, MD) were cultured in RPMI-1640 (with 10 mM Hepes, 50 μM 2-mercaptoethanol, and 10% FBS). BB88 cells were induced to differentiate terminally by adding 1.5% dimethyl sulfoxide for 72 hours. Purified murine ATII cells were kindly provided by Dr. Steven Glasser (Children's Hospital Medical Center, Cincinnati, OH). Unless otherwise noted, all culture reagents were from Invitrogen (Carlsbad, CA). Cells were cultured at 37°C in a humidified atmosphere of air (normoxia) with 5% CO2. For hypoxia experiments, culture flasks were transferred to a Bactron I Anaerobic Environmental Chamber (Sheldon, Cornelius, OR) and exposed to a humidified atmosphere of 1.0% O2/5.0% CO2 in N2 for specified time periods. When noted, prolyl-4-hydroxylase inhibitors (PHIs) were added to a medium consisting of 800 μM L-mimosine (L-mim), 500 μM dimethyloxaloylglycine (DMOG), 250 μM ethyl-3,4-dihydroxybenzoate (EDHB), or 200 μM deferoxamine (31, 32). As noted, cells were treated with 1 μg/ml cycloheximide (CHX; Sigma, St. Louis, MO) added to the medium 20 hours before RNA extraction (CHX was added 1 hour before PHI in cotreatments). GATA1 knockdown experiments were performed according to the manufacturer's instructions for transient transfection (using their GATA1 and their control short, interfering RNAs [siRNAs]; Santa Cruz Biotechnology, Santa Cruz, CA) or stable transfection with the pSingle-tTS-shRNA vector and accessory reagents (Clontech, Mountain View, CA), selection in 400 μg/ml G418 antibiotic (Clonetech), and treatment with 1 μg/ml doxycycline to induce GATA1 short hairpin RNA (shRNA).
Immunohistochemistry
Thin sections of alveolar tissue were prepared from normal mouse lungs perfused with saline to clear blood before removal from the animal. Frozen sections of normal human distal lung were obtained via biopsy, treated with red blood cell lysis solution, and washed with saline before fixation. Thin sections were paraformaldehyde-fixed and costained with anti-hemoglobin antibodies (either anti-pan hemoglobin antibody from Bethyl Laboratories, Montgomery, TX, or anti-HBA antibody from Santa Cruz Biotechnology) and anti–proSP-C antibody (Chemicon/Millipore, Billerica, MA), followed by Alexa Fluor 488–conjugated and Fluor 555–conjugated secondary antibodies (Invitrogen), respectively. According to Chemicon/Millipore, the proSP-C antibody reacts specifically with the preprocessed form of surfactant protein C (SP-C). Immunofluorescent studies defined proSP-C staining in Golgi/endosomal compartments as a standard marker of ATII cells (33–35). Cell nuclei were stained with TO-PRO-3 (Invitrogen). Antibodies specific for CD144/VE-cadherin (vascular endothelial; B.D. Pharmingen, Franklin Lakes, NJ), aquaporin-5, or CC10 (both from Santa Cruz Biotechnology) were used to identify other cell types in thin sections.
Quantitative PCR and Real-Time PCR
RNA was purified from cells using an RNeasy Mini Kit (Qiagen, Valencia, CA), complementary DNA (cDNA) was generated from 2 μg total RNA using a First-Strand Synthesis kit (GE Healthcare/Amersham, Piscataway, NJ), and quantitative PCR (qPCR) reactions and data analyses were performed using iQ SYBR Green Supermix and a MyiQ thermal cycler (Bio-Rad, Hercules, CA). All oligonucleotide primers (Table 1) were synthesized by Integrated DNA Technologies (Coralville, IA).
TABLE 1.
OLIGONUCLEOTIDE PRIMERS USED FOR PCR IN THESE STUDIES
| Primer Pair | Oligonucleotide Sequence | GenBank Identification |
|---|---|---|
| HBA | F: 5′-ATGTTTGCTAGCTTCCCCACCACCAAG-3′ | BC043020 |
| R: 5′-GCTTAACGGTATTTGGAGGTCAGCACG-3′ | ||
| HBB | F: 5′-TGATGCTGAGAAGGCTGCTGTCTCTG-3′ | NM_008220 |
| R: 5′-GTGCCCTTGAGGCTGTCCAAGTGA-3′ | ||
| SP-B | F: 5′-TGCCAAGAGTGTGAGGATATTGTCCAC-3′ | NM_147779 |
| R: 5′-CCAGCTTGTCCAGCAGAGGGTTTG-3′ | ||
| SP-C | F: 5′-CCGGATTACTCGGCAGGTCCCAG-3′ | NM_011359 |
| R: 5′-ATGCCAGTGGAGCCGATGGAAAAGG-3′ | ||
| HIF1α | F: 5′-CTTCTGGATGCCGGTGGTCTAGAC-3′ | NM_010431 |
| R: 5′-CTCACTGGGCCATTTCTGTGTGTAAG-3′ | ||
| HIF2α | F: 5′-AGCTCCTGTCCTCAGTCTGCTCTG-3′ | NM_010137 |
| R: 5′-CCTGTTAGTTCTACCTGAGTAAGTCCC-3′ | ||
| ACT | F: 5′-TCCTTCTTGGGTATGGAATCCTGTGG-3′ | NM_007393 |
| R: 5′-CGCTCAGGAGGAGCAATGATCTTG-3′ | ||
| GATA1 | F: 5′-GTCCTCACCATCAGATTCCACAGG-3′ | NM_008089 |
| R: 5′-ATTCCCTCCATACTGTTGAGCAGTGG-3′ | ||
| GATA2 | F: 5′-GTTCCCAAGACACAGTAGTGGACC-3′ | NM_008090 |
| R: 5′-CTGCGAGTCGAGATGGTTGAAGAAG-3′ | ||
| NFE2 | F: 5′-GCAGGAATCCTTTGTGCTTGTGGAG-3′ | NM_008685 |
| R: 5′-TAGACCCTGCAGCTCAGTAATGGAC-3′ | ||
| KLF | F: 5′-TCTTGAATTGTCTGGGACCTGGGAC-3′ | NM_010635 |
| R: 5′-AGGTGCGAGCTCTTGGAGTAGCTC-3′ | ||
| CP2 | F: 5′-GGAACCTGGCACATCTGAATGTTGG-3′ | NM_033476 |
| R: 5′-CATTACCACTCTGCCCTCTACAAACC-3′ | ||
| ALAS2 | F: 5′-ACTCACCGTCTTTGGTTCGTCCTC-3′ | NM_009653 |
| R: 5′-TGAGAACAGGTTGGTCCTTGAGTGG-3′ | ||
| TTF-1/Nkx2-1 | F: 5′-GCGCCATGTCTTGTTCTACCTTGC-3′ | NM_009385 |
| R: 5′-GTCGTCCAGCAGTTTGGTCTTTGTG-3′ | ||
| SLC4A1/band3 | F: 5′-TGGCTGCTGTCATCTTCATCTAC-3′ | NM_011403 |
| R: 5′-TTTGGGCTTCATCACAACAGG-3′ |
Definition of abbreviations: F, forward (sense); R, reverse (antisense); HBA, hemoglobin α; HBB, hemoglobin β major; SP-B, surfactant protein B; SP-C, surfactant protein C; HIF, hypoxia-inducible factor; ACT, β-actin; NFE2, nuclear factor (erythroid-derived 2, p45); KLF, Kruppel-like factor 1 (erythroid); CP2, alpha-globin transcription factor CP2; ALAS2, δ-aminolevulinate synthase 2; TTF-1, thyroid transcription factor (also known as Nkx2-1); SLC4A1, solute carrier family 4 (anion exchanger), member 1 (also known as band 3). Oligonucleotides were designed according to murine sequences found at the GenBank accession numbers listed in the right column.
Experimental results by qPCR were verified by repetition with RNA extracted from different aliquots of cells (at least three independent reactions were performed per template/primer combination). For relative quantification in qPCR, a mathematical model was used, incorporating the effects of the efficiency of amplification for each primer pair over a 104 range of template dilutions (36), and starting template concentrations were normalized by comparison with β-actin amplification. Semiquantitative RT-PCR was similarly run, using the protocol already described, followed by polyacrylamide gel electrophoresis (PAGE) and ethidium bromide staining. Densitometric comparison was normalized against β-actin.
Western Blotting
Rinsed cells were lysed in low salt buffer (10 mM Hepes, 10 mM KCl, 0.1 mM EDTA, and 1 mM DTT) containing 0.5% NP40 (Sigma), nuclei were pelleted by centrifugation, and the supernatant was recovered as a cytosolic extract. Nuclear extracts were obtained by resuspending in high salt buffer (20 mM Hepes, 400 mM NaCl, 1 mM EDTA, and 1 mM DTT) and centrifugation. Proteins were separated by SDS-PAGE (NuPAGE gels; Invitrogen), transferred onto nitrocellulose, and probed with antibodies to HBA, HBB (both from Santa Cruz Biotechnology), HIF-1α, proSP-C, preprocessed surfactant protein B, or proSP-B (all from Chemicon/Millipore), or HIF-2α (Novus Biologicals, Littleton, CO). Peroxidase-conjugated secondary antibodies and a SuperSignal West Dura chemiluminescence kit (both from Pierce Biotechnology, Rockford, IL) were used for detection with a FluorChem 8900 Imager (Alpha Innotech, San Leandro, CA). Purified human hemoglobin (Sigma) was used as a standard. Semiquantification was performed by normalizing the optical density to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH; antibody from Chemicon/Millipore). HIF-2α nuclear quantification was performed by normalizing against TATA-binding protein (TBP; Abcam, Cambridge, MA).
RESULTS
Hemoglobin Is Expressed by Primary ATII Cells In Vivo
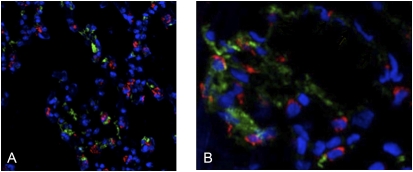
Our previous results demonstrated that hemoglobin mRNAs and polypeptides are expressed by several cultured ATII cell lines and primary ATII cells isolated from rat lungs, as determined by RT-PCR, immunoblotting, and protein sequencing (5). To determine further whether hemoglobin is expressed by pulmonary alveolar cells in vivo, immunohistochemistry was performed, using sections from perfused mouse lungs. Figure 1A shows that hemoglobin (green) and the ATII-specific marker proSP-C (red) proteins colocalize within ATII cells in the alveoli, although in distinct subcellular compartments. With the exception of a few residual erythrocytes, the only cells in the lung tissue sections that stained positive for hemoglobin were alveolar epithelial cells (determined by antibody staining for other cell markers, including the endothelial cell-specific VE-cadherin; data not shown).
Frozen sections of normal human lung tissue containing alveoli were also subjected to immunohistochemical analyses, using a variety of antibodies. In Figure 1B, hemoglobin protein (green) is localized within cells that stain positively for the ATII cell–specific marker proSP-C (red; nuclei of all cell types stained blue). Within ATII cells, the patterns of hemoglobin and proSP-C visualization are consistent with cytoplasmic and Golgi/endosomal compartment staining, respectively. Hemoglobin staining that appears to be unassociated with ATII cells is evident within the lumen of capillaries. Anti-hemoglobin antibodies also did not stain Clara cells, as identified using the anti-CC10 antibody (data not shown).
For obvious reasons, human lung sections, unlike mouse tissue, could not be perfused before removal, a technical problem that previously complicated this type of in vivo human analysis. However, for clarity, we selected lung-tissue sections for Figure 1 that contained very little residual blood, which is located primarily within alveolar capillaries (identified by costaining with the VE-cadherin antibody, a marker for endothelial cells, the other most abundant cell type in alveoli; data not shown). The only nucleated cells to display clearly defined hemoglobin staining in our analyses of human lung sections (which contained a variety of tissues) were ATII cells.
Figure 1.
Hemoglobin protein is expressed by alveolar Type II cells in vivo. Immunohistochemical staining of normal murine lung tissue (A) and a normal human alveolar section (B) was paraformaldehyde-fixed and costained with anti-hemoglobin (green) and anti–proSP-C (red) antibodies (colors are attributable to fluorescent secondary antibodies). Cell nuclei were stained with TO-PRO-3 (blue).
ATII Cells Express Erythroid and Heme-Specific Factors
Hematopoietic development and heme biosynthesis are associated with a complex pattern of coordinated gene expression and transcriptional activity (37–39). To examine whether hemoglobin expression in ATII cell may involve some of the same genes that are critical in erythroid lineages, the relative expressions of these factors between ATII cells and erythroid precursors were evaluated (Table 2). Using semiquantitative RT-PCR and gel densitometry, both primary ATII and MLE-15 cells were shown to express mRNA for the highly specific transcription factors GATA1, GATA2, and p45 NFE2 (nuclear factor erythroid-derived 2, 45kDa). These factors are closely associated with globin expression in erythroid precursors, and are required for hematopoietic development (40–42). Moreover, ATII primary cells expressed moderate levels of aminolevulinic acid synthase 2 (ALAS2), an isoform of the rate-limiting porphyrin biosynthetic enzyme previously shown to be expressed solely in erythroid cells (43). To demonstrate the identity and purity of these cell populations, the expressions of SP-B, SP-C, the surfactant protein-regulating thyroid transcription factor-1 (TTF-1/Nkx2-1), and the erythroid membrane protein SCL4A1 were also measured in murine MH-S macrophage and BB88 erythroid cell lines (before and after induction). Neither control expressed SP-B, SP-C, or TTF-1. Similarly, MLE-15 and primary ATII cells did not express SCL4A1. BB88 erythroid cells expressed HBA and HBB, as well as GATA1 and ALAS2 (Table 2).
TABLE 2.
ALVEOLAR TYPE II CELLS EXPRESS ERYTHROID-SPECIFIC GENES
| MLE-15 | ATII Primary Cells | MH-S Macrophage | BB88 Erythroid Noninduced | BB88 Erythroid Induced | |
|---|---|---|---|---|---|
| HBA | + | ++ | Negative | + | +++ |
| HBB | + | ++ | Negative | + | +++ |
| GATA1 | + | + | Negative | ++ | +++ |
| GATA2 | ++ | + | Negative | +++ | ++ |
| NFE2 | + | + | Negative | + | + |
| ALAS2 | ± | ++ | Negative | ++ | +++ |
| SP-B | +++ | +++ | Negative | Negative | Negative |
| SP-C | +++ | +++ | Negative | Negative | Negative |
| TTF-1/Nkx2-1 | ++ | ++ | Negative | Negative | Negative |
| SLC4A1/band3 | Negative | Negative | Negative | + | +++ |
Definition of abbreviations: HBA, hemoglobin α; HBB, hemoglobin β major; NFE2, nuclear factor (erythroid-derived 2); ALAS2, δ-aminolevulinate synthase 2; SP-B, surfactant protein B; SP-C, surfactant protein; TTF-1, thyroid transcription factor cells, also known as Nkx2-1); SLC4A1, solute carrier family 4 (anion exchanger), member 1, also known as band 3. “±” indicates extremely low concentrations. MH-S and BB88 are ATCC cell designations as described in Methods.
Exposure of MLE-15 Cells to Hypoxia or Mimics of Hypoxia Increases Hemoglobin and Decreases Surfactant Protein Expression
MLE-15 cells maintain a number of ATII-associated characteristics, including phospholipid secretion and surfactant protein synthesis/secretion (44). Both new immunohistoochemical staining (data not shown) and our previously published results (5) demonstrated the expression of hemoglobin mRNA and protein in these cells. Thus, given the role of hemoglobin in oxygen-binding, MLE-15 cells were used in our subsequent experiments to determine the effects of hypoxia on hemoglobin expression in ATII cells.
Because ATII was reported to be surprisingly tolerant to acute hypoxia (45), but to show increased apoptosis during prolonged hypoxic exposure (15), the viability of MLE-15 cells in hypoxia was measured, to design appropriate experiments that would minimize cell death. Cellular viability, as measured by trypan blue exclusion and caspase induction, showed no significant effects after up to 20 hours of hypoxic exposure (data not shown). Thus, further experiments were limited to 20-hour courses of hypoxia.
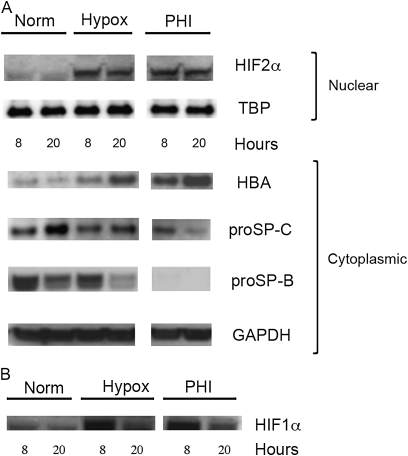
MLE-15 cells, grown under normoxic (21% O2) or hypoxic (1.0% O2) conditions, were harvested after 8 or 20 hours, and Western blot analyses were performed using cellular lysates. As expected, hypoxic exposure resulted in a significant increase in HIF-2α protein concentrations, compared with control cells (Figure 2A). Surprisingly, a dramatic increase in HBA protein concentrations was also detected with increasing hypoxic exposure.
Figure 2.
Hypoxia affects hemoglobin and surfactant protein (SP) concentrations in alveolar Type II (ATII) cells. Protein lysates were prepared from murine ATII-like cell line MLE-15 exposed to normoxia (Norm) or 1.5% O2 (hypoxia; Hypox) for 8 or 20 hours (or to 800 μM of the prolyl-4-hydroxylase inhibitor L-mimosine; PHI). Western blots were prepared from SDS-PAGE of 25 μg of total protein per sample (or 40 μg nuclear protein for hypoxic-inducible factor [HIF]–1α and HIF2α detection), and probed with the antibodies HIF1α, HIF2α, hemoglobin (HBA), proSP-C, proSP-B, TATA binding protein (TBP), or GAPDH.
To correlate these observed changes further in hemoglobin expression during hypoxia with increased cellular levels of HIF, cells were exposed in parallel to the PHI L-mim under normoxic conditions. This PHI was shown to mimic exposure to hypoxia, dramatically increasing nuclear accumulation of the active HIF transcription factor (31). As shown in Figure 2A, the exposure of MLE-15 cells to this PHI for 8 and 20 hours resulted in increases in both HIF and hemoglobin proteins that closely paralleled those of cells exposed to hypoxia. These results suggest that the increase in hemoglobin expression in these cells under hypoxia is associated with increased cellular concentrations of HIF transcription factors. Because of the function of ATII cells in surfactant production, the effect of hypoxic exposure on SP-B and SP-C production was also evaluated. Unexpectedly, by 20 hours, the concentrations of pre-processed forms of SP-B and SP-C protein dramatically decreased in cells exposed to either hypoxia or PHI (Figure 2A).
HIF-1α and HIF-2α share 48% amino-acid identity, and previous studies implicated roles for both in ATII cells. HIF-1α protein was also detectable in MLE-15 nuclear extracts at the time points measured, and was similarly induced by hypoxia and PHI after 8 hours (Figure 2B). However, by 20 hours of hypoxic exposure, concentrations of HIF-1α protein had dramatically decreased. These results are consistent with studies suggesting that HIF-related changes in response to (prolonged) 20 hours of hypoxia in ATII cells are attributable to HIF-2α (14, 46). However, these results may also support a possible role for HIF-1α in an earlier hypoxic response. Reports that link HIF-1α to the regulation of HIF-2α protein expression support a hypothesis that HIF-1α may be required for the expression of HIF-2α observed during prolonged hypoxic exposure.
Exposure of ATII Cells to Hypoxia or Mimics of Hypoxia Increases Hemoglobin Gene Expression
To determine if increased hemoglobin protein concentrations in ATII cells exposed to hypoxia were correlated with similar changes in gene expression, parallel aliquots of MLE-15 cells exposed to the same experimental conditions were analyzed by real-time qPCR. After 20 hours of exposure to hypoxia, steady-state levels of HBA and HBB mRNAs increased approximately 10-fold and fivefold, respectively, whereas SP-B mRNA did not change significantly, and SP-C concentrations decreased slightly (Table 2). Even larger increases in globin mRNA concentrations were evident in cells exposed to the PHI L-mim. Treatment with PHI similarly enhanced the decrease of SP-C mRNA. Both HIF-1α and HIF-2α mRNAs were highly expressed in MLE-15 cells, but no changes were evident in HIF gene expression under hypoxia. These results suggest that HIF-2α acts as a transcription factor affecting hemoglobin gene expression.
As expected because of stabilization, HIF-2α protein concentrations dramatically increased over the hypoxic time course (Figure 2), without significant change in steady-state mRNA levels (Table 3). Similarly, levels of hemoglobin protein (α-globin antibody) increased at least 20-fold after 20 hours of exposure to hypoxia or PHI, most likely because of a parallel increase in HBA mRNA levels. Although SP-B protein concentrations also decreased under hypoxia in these cells, steady-state concentrations of SP-B mRNA did not significantly change. Conversely, concentrations of SP-C mRNA decreased moderately (1.5-fold) after 20 hours of exposure to hypoxia, consistent with reduced concentrations of proSP-C protein. As observed for protein analyses, PHI treatment with L-mim amplified the observed changes in mRNA concentrations.
TABLE 3.
HYPOXIA INCREASES HEMOGLOBIN EXPRESSION AND DECREASES SP-C EXPRESSION IN ATII CELLS
| Gene | Experimental Condition | Change as Compared with Control |
|---|---|---|
| HBA | Hypoxia | 10-fold increase (± 1.4) |
| HBA | PHI | 24-fold increase (± 4.2) |
| HBB | Hypoxia | 5-fold increase (± 1.0) |
| HBB | PHI | 7-fold increase (± 1.3) |
| SP-B | Hypoxia | No significant change |
| SP-B | PHI | No significant change |
| SP-C | Hypoxia | 1.5-fold decrease (± 0.4) |
| SP-C | PHI | 3-fold decrease (± 1.2) |
| HIF1α | Hypoxia | No significant change |
| HIF1α | PHI | No significant change |
| HIF2α | Hypoxia | No significant change |
| HIF2α | PHI | No significant change |
Definition of abbreviations: PHI, 800 μm L-mimosine; HBA, hemoglobin α; HBB, hemoglobin β; SP-B, surfactant protein B; SP-C, surfactant protein C; HIF, hypoxia-inducible factor.
Prolyl-4-Hydroxylase Inhibitors Mimic the Hypoxic Response of MLE-15 Cells
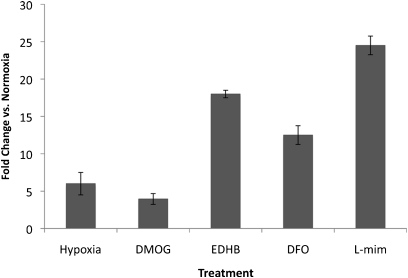
To correlate the observed changes in hemoglobin gene expression during hypoxia more specifically to increased cellular concentrations of HIF, cells were exposed to several additional agents commonly used to mimic hypoxia, by stabilizing the HIFα subunit through an inhibition of the same prolyl-4-hydroxylases. These PHIs included the selective agents DMOG, EDHB, and L-mim, as well as the iron-chelating, nonselective agent deferoxamine (EDHB also has iron-chelating properties) (47). In all cases, the exposure of MLE-15 cells to hypoxia or PHIs significantly increased the cellular concentrations of HBA mRNA, as measured by real-time qPCR (Figure 3). L-mim was chosen as a standard PHI for further studies because of its combination of effectiveness, solubility, and low toxicity.
Figure 3.
Prolyl-4-hydroxylase inhibitors up-regulate hemoglobin expression in ATII cells. Real-time quantitative PCR (qPCR) was used to measure changes in HBA mRNA concentrations in murine MLE-15 cells exposed to either hypoxia (1.5% O2) or different prolyl-4-hydroxylase inhibitors: L-mimosine (L-mim), ethyl-3,4-dihydroxybenzoate (EDHB), dimethyloxallyl glycine (DMOG), or deferoxamine (DFO) for 20 hours. Control cells were grown in parallel and exposed to normoxia with no inhibitors for 20 hours. For obtaining X-fold increases over control cells, starting cDNA template concentrations were normalized by comparison with β-actin amplification. Data are presented as mean ± SEM; n = 3.
Hypoxia Increases the Expression of Globin-Specific Transcription Factors
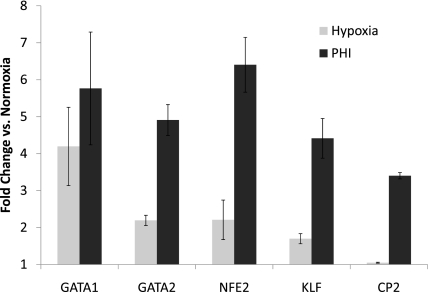
The identification of erythroid-specific transcription factors in MLE-15 and ATII cells (Table 3) suggests that these factors may play similar roles in globin expression in ATII cells. Also, because hemoglobin mRNA and protein are dramatically up-regulated during hypoxia, we investigated whether globin-associated transcription factor expression is similarly affected by hypoxic exposure in MLE-15 cells. As shown in Figure 4, both hypoxia and PHI treatment resulted in significant increases in steady-state mRNA concentrations of several well-characterized globin transcription factors. Also, hypoxia and PHI treatment increased the mRNA expression of other globin-associated genes, including those of the rate-limiting porphyrin biosynthetic enzymes ALAS1 and coproporphyrinogen oxidase (CPOX). No effect was evident on concentrations of the erythroid-specific ALAS2; data not shown. These results are consistent with our hypothesis that hemoglobin expression in erythroid cells and ATII shares important regulatory similarities, and that hemoglobin may play a role in the oxygen-sensing pathway in alveolar epithelial cells.
Figure 4.
RNA levels of many transcription factors associated with globin gene expression increase in MLE cells exposed to hypoxia. Real-time qPCR was used to measure steady-state concentrations of mRNAs extracted from MLE cells exposed to different experimental conditions: control (normoxia), 20-hour exposure to 1.5% O2 (Hypoxia), or 20-hour exposure to the prolyl-4-hydroxylase inhibitor (PHI) L-mim. Fold-change in specific mRNA concentrations, compared with those in control cells, is depicted (0 = no change from concentrations in control cells). Levels of initial cDNA templates between samples were normalized by comparison with β-actin amplification. Target genes and primers used are as shown in Table 1. GATA1 and GATA2, globin associated transcription factor isoforms 1 and 2, respectively; NFE2, nuclear factor (erythroid-derived 2, p45); KLF, Kruppel-like factor 1 (erythroid); CP2, alpha-globin transcription factor CP2. Data are presented as mean ± SEM; n = 3.
GATA1 Is Required for Hemoglobin Expression in ATII Cells
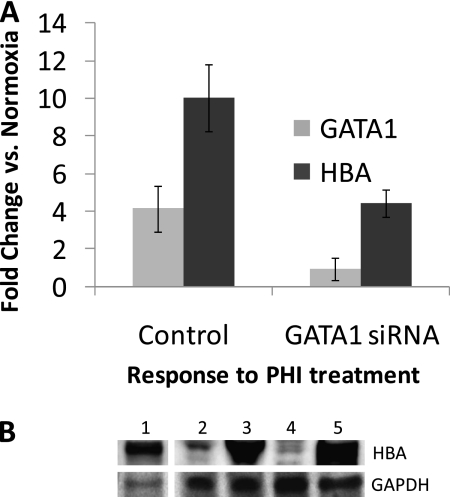
Because most globin-associated genes contain GATA1-binding sites, we hypothesized that GATA1 would be a prime candidate for a further investigation of GATA1's effects on globin expression in MLE cells. The results of transient transfections of MLE cells with GATA1 siRNA indicated that modest reductions in GATA1 mRNA concentrations (∼30%) did not appear to affect the steady-state concentrations of existing HBA mRNA, most likely because of insufficient GATA1 knockdown or HBA mRNA stability over this time frame (data not shown). However, GATA1 knockdown did dramatically reduce the up-regulation of HBA mRNA in response to PHI treatment (Figure 5A).
Figure 5.
GATA1 is required for globin gene expression in ATII cells. (A) Transient small, interfering RNA (siRNA) transfection. Real-time qPCR was used to measure steady-state mRNA levels from MLE lysates after 3 days of transfection with GATA1 siRNA (as normalized to β-actin amplification). Effect of transfection on the up-regulation of both GATA1 and HBA mRNAs is shown after treatment of transfected cells for 20 hours with the hypoxia mimic L-mim (PHI), compared with that seen in nontreated control cells (transfection of scrambled siRNA). Three experimental replicates were performed (n = 3) and presented as mean ± SEM. (B) Inducible GATA1 shRNA transfection. Protein lysates were prepared from stable MLE-15 transfectants (antibiotic-selectable cell lines) that express GATA1-specific shRNA under the control of a tetracycline-inducible promoter. Western blots were prepared from SDS-PAGE of 25 μg of total protein per sample (or 4 μg from the murine erythroleukemia cell line BB88), and probed with antibodies for hemoglobin (HBA) or GAPDH (control). Lane 1, BB88 cells; lane 2, MLE transfectant in normoxia (control, not tetracycline-induced); lane 3, MLE transfectant cultured for 20 hours with PHI (not tetracycline-induced); lane 4, MLE transfectant induced to express GATA1 short hairpin RNA (shRNA) by 2 weeks of exposure to tetracycline; lane 5, shRNA-induced transfectant cultured for 20 hours with PHI.
To achieve a long-term knockdown of GATA1 mRNA, we generated stably transfected MLE clones containing a GATA1 shRNA construct under the control of a tetracycline-inducible promoter. Clones induced to express the GATA1 shRNA for 2 weeks demonstrated a 35% reduction in GATA1 mRNA concentrations, and a 70% reduction in HBA mRNA concentrations. The effects of this GATA1 knockdown were also apparent in the concentrations of hemoglobin protein (Figure 5B, lanes 2 and 4). These results strongly suggest that GATA1 is necessary for hemoglobin expression in MLE cells. Despite the moderate knockdown of GATA1 in this clone, PHI treatment increased concentrations of both GATA1 and HBA mRNAs (twofold and fourfold increases, respectively), as well as globin protein concentrations (Figure 5B, lane 5), although the total concentrations of both mRNAs were still significantly lower than those concentrations in noninduced control transfectants treated with PHI (55% of control GATA1 concentrations, and 43% of HBA concentrations).
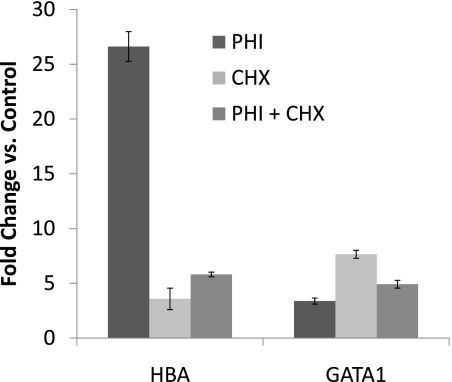
Protein synthesis was inhibited by CHX to determine whether the rise in hypoxia-induced or PHI-induced up-regulation of HBA mRNA concentrations required de novo protein synthesis. Surprisingly, CHX significantly increased the concentrations of both GATA1 and HBA mRNAs, even in the absence of hypoxic treatment (Figure 6), strongly suggesting that GATA1 gene expression (and possibly HBA expression) may be normally suppressed by one or more inhibitors in these cells. Conversely, treatment with CHX dramatically abrogated the up-regulation of HBA mRNA by the hypoxia mimic, indicating that the hypoxia-induced up-regulation of hemoglobin in ATII cells requires de novo protein synthesis (e.g., of GATA1 or other transcription factors), and is not attributable solely to direct transcriptional activation through HIF stabilization.
Figure 6.
Up-regulation of globin gene expression during hypoxic responses in ATII cells requires de novo protein synthesis, and may involve removal of transcriptional inhibition. MLE-15 cells were exposed to 20-hour treatment with L-mim (PHI), cycloheximide (CHX), or both. Real-time qPCR was used to compare fold-changes in steady-state mRNA concentrations, compared with those in control cells (normoxia, not exposed to PHI or CHX). Levels of initial cDNA templates between samples were normalized by comparison with β-actin amplification. Data are presented as mean ± SEM; n = 3.
DISCUSSION
In this study, we provide evidence implicating hemoglobin as an oxygen-sensitive gene in alveolar epithelial cells. Previously, we discovered that hemoglobin RNA and protein are expressed in several alveolar cell lines, including A549 and MLE-15, as well as in primary ATII cells purified from murine and rat lungs (5). Here, we demonstrate that hemoglobin mRNA and protein are up-regulated during hypoxic exposure in the murine ATII-like cell line MLE-15.
In vivo analyses of human and murine lung sections for hemoglobin and cell-specific markers strengthen our previous study, and further confirm the presence of hemoglobin polypeptides in the cytosol of ATII cells (as identified by staining for the ATII-specific marker proSP-C). These results are in agreement with another report in which hemoglobin localized to the corners of the alveoli in rat lung sections, an area occupied primarily by ATII cells (7).
Erythroid differentiation and the hemoglobin biosynthetic pathway are highly coordinated and tightly regulated processes involving a number of specific transcription factors and enzymes. A number of these factors, including p45 NFE2 and ALAS2, are traditionally defined as erythroid-specific. However, semiquantitative RT-PCR demonstrated low concentrations of both mRNAs in alveolar epithelial cells (Table 2). Furthermore, the transcription factors GATA2 and GATA1, which are necessary for early and late hematopoietic development, respectively, as well as the expression of erythroid-specific target genes including those for the α-globin and β-globin (41, 42, 48), were also expressed by both MLE-15 and ATII primary cell lines. The concentrations of HBA, HBB, and ALAS2 were noticeably greater in ATII primary cells compared with MLE-15 cells. Because primary cells more closely represent in vivo ATII, these data may be more representative of the true expression levels of these genes. The expression of GATA1 in a nonhematopoietic cell line, HeLa, was shown to induce the transcription of several erythroid-specific genes, including α-globin and β-globin, suggesting that the presence of GATA1 is sufficient to induce an erythroid pattern of gene expression (49).
Hemoglobin expression by cells of the pulmonary epithelium may have considerable implications in the physiology and pathology of the lung because of the many defined roles inherent to the structures of the hemoglobin molecule and its derived peptides, including gas exchange, nitric oxide homeostasis, blood pressure regulation, and protection against oxidative and nitrosative stress (50–55). The protein may function in oxygen or carbon dioxide transport across the blood–airway interface, or may have a simpler, myoglobin-like role in oxygen storage, or may even trap toxins such as carbon monoxide. Furthermore, recent data establish both myoglobin and hemoglobin as intrinsic nitrite reductases, with critical roles in regulating hypoxic nitric oxide signaling (56–58). Hemoglobin was shown to function as a nitric oxide dioxygenase by controlling O2 binding, further emphasizing a critical role in nitric oxide metabolism and detoxification (59). Other cellular hemoglobins, and specifically neuroglobin, were shown to be cytoprotective during hypoxia and ischemia, potentially by modulating the mRNA expression of hypoxic response genes (60, 61). During hypoxia, oxygen homeostasis is maintained by stimulating erythropoiesis and the synthesis of hemoglobin. These studies all support a role for globin genes in protecting against hypoxic injury. Recently, the discovery of hemoglobin in the lens and cornea of prehematopoietic murine embryos suggests a developmental role for hemoglobin, independent of oxygen metabolism (11). The presence of hemoglobin in pulmonary epithelial cells could play a major role in protection against the stress of functioning in the most highly oxidative cellular environment within the mammalian body.
HIFs control numerous genes involved in adaptations to low oxygen tension. We postulate that globin gene induction in ATII cells may constitute a response that is coordinated with HIF stabilization during hypoxia. Both HIF-1α and HIF-2α protein were u-regulated in response to hypoxia. However, after 20 hours, protein concentrations of HIF-1α dramatically decreased, suggesting that HIF-1α and HIF-2α are differentially regulated by prolonged hypoxia. This result is consistent with previous studies indicating that, in lung epithelial cells, the up-regulation of HIF-1α protein occurs during short-term hypoxia, but not with mild and prolonged hypoxic exposure. Feedback inhibition of HIF-1α by HIF-2α was implicated during chronic hypoxia (14, 46). No change in mRNA concentrations of either HIF was detected. These results were expected, because the cellular concentrations of these transcription factors are regulated posttranslationally by protein stabilization. HIF-2α is of particular interest as an oxygen-sensitive gene in ATII cells, because HIF-2α mRNA transcripts are expressed in murine alveolar epithelium at a higher concentration than HIF-1α mRNA (62), and studies indicate that HIF-2α has a role in ATII function, especially during late gestation (2, 23, 27, 32). Hypoxia was shown to increase ATII vascular endothelial growth factor expression and the glucose transporter isoform 1 (GLUT-1) through HIF-2α–mediated transcriptional activation (45, 62, 63). Our results confirm previous reports that implicated HIF-2α as the key HIF in prolonged hypoxia in ATII cells, and also indicate a possible role for HIF-1α in early hypoxic gene regulation. Furthermore, our results with PHIs, which stabilize the HIFα subunit, further support our hypothesis that the activation of HIFs is associated with increased hemoglobin expression in ATII cells exposed to hypoxia, though not through the direct transcriptional activation of globin genes. HIF-targeted knockout or overexpression studies would allow for a further elucidation of the roles of specific HIFs in ATII cells.
As already discussed, the identification of erythroid-specific genes in MLE-15 and ATII cells suggests that, as in erythroid cells, these factors may have a similar role in globin expression in ATII cells. Hypoxia is widely known for directly up-regulating a number of HIF target genes involved in erythropoiesis, including erythropoietin and those involved in iron metabolism. ALAS2 contains a sequence similar to that found in HIFs that directly confers oxygen-dependent degradation (64). These data suggest that HIFs may play a key role in the activation of genes associated with the induction of globin transcription. The significant increases in mRNA concentrations of several globin-associated transcription factors in response to hypoxia or PHI treatment suggest a role for these factors in hypoxia-induced hemoglobin expression in ATII cells through an HIF-mediated pathway. Small alterations in the expression of upstream genes, such as transcription factors during hypoxia, have the potential to result in dramatic changes in globin gene expression.
Our results strongly suggest that GATA1 is required for normal and hypoxia-induced globin expression in ATII cells. Because of the significant complexity of globin gene regulation and the many unknown details and promiscuous pathways involved in the hypoxic response, GATA1 activation alone is highly unlikely to regulate the dramatic up-regulation of globins during hypoxia. For instance, the GATA1 promoter lacks a known hypoxia-response element for HIF, although HIFα does contain a GATA1 site. The hypoxia-induced HBA mRNA increase appears to require de novo protein synthesis, and therefore the HIF-associated up-regulation of globin expression in ATII cells is most likely indirect (upstream), rather than directly mediated through globin transcriptional activation by stabilized HIF. This suggests the involvement of additional factors, some of which may regulate GATA1. The observed increase in GATA1 mRNA expression after CHX treatment, similar to that seen with hypoxia, is consistent with the existence of a transcriptional inhibitor that may actively suppress ATII globin expression in the absence of hypoxic stimulation. Cycloheximide also moderately increases HBA gene expression (although to much lower extent than hypoxia or PHI), which may indicate the removal of an inhibitor or upstream GATA1 activation by CHX.
In addition to increases in hemoglobin mRNA and protein, hypoxic exposure resulted in marked decreases in the protein of the alveolar epithelial–associated genes proSP-B and proSP-C in MLE-15 cells. Because real-time qPCR analyses indicated no change in steady-state mRNA concentrations of SP-B (despite dramatic decreases in proSP-B protein), the observed decreases in protein concentrations during hypoxia may be the result of increases in RNA stability or decreased protein stability or translation, a phenomenon that was observed to be enhanced with the use of hypoxia mimics. Because of the defined time course of our experiments and the known stability of SP-B mRNA, quantitative PCR protocols may be biased toward the identification of gene transcription increases rather than decreases. Prolonged hypoxic exposure did not affect MLE-15 cell viability, indicating that the inhibition of surfactant protein gene expression is not attributable to cell toxicity. Studies indicate that ATII cells are tolerant of acute hypoxia and can maintain their ATP content close to normoxic conditions (45). However, hypoxic insult often results in a disruption of expression of tissue-specific genes. TTF- 1/Nkx2.1 is currently the only transcription factor known to regulate all four surfactant protein promoter activities (65–67). MLE-12 cells treated with cobalt chloride (which simulates hypoxia via the activation of HIF), although indicating the repression of TTF-1 expression as well as the down-regulation of SP-B promoter activity, did not alter SP-C promoter activity (68). These data suggest that the SP-B and SP-C promoters are differentially regulated under oxidative stress.
The elucidation of oxygen-sensitive genes in ATII cells is of particular interest because epithelial dysfunction is often associated with increased edema and subsequent hypoxia. Our results confirm that HIF-2α is a key hypoxia-inducible transcription factor in ATII cells, and suggest that hypoxia triggers a coordinated pathway in ATII cells that leads to an increase in functional hemoglobin protein expression as an effector response. Because HIF-2α was directly implicated in the late stages of alveolar development, hemoglobin may play an additional or coordinate role in ATII cells, independent of oxygen metabolism. The identification of several erythroid-specific factors in ATII cells suggests a possible role in globin gene regulation. Further studies are underway to identify the specific mechanisms and proteins involved in hemoglobin up-regulation and surfactant protein disruption.
Acknowledgments
We thank Dr. Jennifer Isaacs (Department of Pharmacology, Medical University of South Carolina, Charleston, SC) for her assistance with the hypoxia experiments, and Steve Glasser (Children's Hospital Research Foundation, Cincinnati, OH) for providing primary mouse ATII cells.
This work was supported by the National Heart, Lung, and Blood Institute.
Originally Published in Press as DOI: 10.1165/rcmb.2009-0307OC on May 27, 2010
Author Disclosure: J.E.B. received a sponsored grant (5R01HL85738) from the National Institutes of Health (more than $100,000). None of the other authors have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.van Tuyl M, Liu J, Wang J, Kuliszewski M, Tibboel D, Post M. Role of oxygen and vascular development in epithelial branching morphogenesis of the developing mouse lung. Am J Physiol Lung Cell Mol Physiol 2005;288:L167–L178. [DOI] [PubMed] [Google Scholar]
- 2.Ahmad S, Ahmad A, Gerasimovskaya E, Stenmark KR, Allen CB, White CW. Hypoxia protects human lung microvascular endothelial and epithelial-like cells against oxygen toxicity: role of phosphatidylinositol 3-kinase. Am J Respir Cell Mol Biol 2003;28:179–187. [DOI] [PubMed] [Google Scholar]
- 3.Stenmark KR, Gebb SA. Lung vascular development: breathing new life into an old problem. Am J Respir Cell Mol Biol 2003;28:133–137. [DOI] [PubMed] [Google Scholar]
- 4.Crapo JD, Barry BE, Gehr P, Bachofen M, Weibel ER. Cell number and cell characteristics of the normal human lung. Am Rev Respir Dis 1982;126:332–337. [DOI] [PubMed] [Google Scholar]
- 5.Newton DA, Rao KM, Dluhy RA, Baatz JE. Hemoglobin is expressed by alveolar epithelial cells. J Biol Chem 2006;281:5668–5676. [DOI] [PubMed] [Google Scholar]
- 6.Giardina B, Messana I, Scatena R, Castagnola M. The multiple functions of hemoglobin. Crit Rev Biochem Mol Biol 1995;30:165–196. [DOI] [PubMed] [Google Scholar]
- 7.Bhaskaran M, Chen H, Chen Z, Liu L. Hemoglobin is expressed in alveolar epithelial Type II cells. Biochem Biophys Res Commun 2005;333:1348–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu L, Zeng M, Stamler JS. Hemoglobin induction in mouse macrophages. Proc Natl Acad Sci USA 1999;96:6643–6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wride MA, Mansergh FC, Adams S, Everitt R, Minnema SE, Rancourt DE, Evans MJ. Expression profiling and gene discovery in the mouse lens. Mol Vis 2003;9:360–396. [PubMed] [Google Scholar]
- 10.Ohyagi Y, Yamada T, Goto I. Hemoglobin as a novel protein developmentally regulated in neurons. Brain Res 1994;635:323–327. [DOI] [PubMed] [Google Scholar]
- 11.Mansergh FC, Hunter SM, Geatrell JC, Jarrin M, Powell K, Evans MJ, Wride MA. Developmentally regulated expression of hemoglobin subunits in avascular tissues. Int J Dev Biol 2008;52:873–886. [DOI] [PubMed] [Google Scholar]
- 12.Semenza GL. Oxygen-regulated transcription factors and their role in pulmonary disease. Respir Res 2000;1:159–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jean JC, Rich CB, Joyce-Brady M. Hypoxia results in an HIF-1–dependent induction of brain-specific aldolase C in lung epithelial cells. Am J Physiol Lung Cell Mol Physiol 2006;291:L950–L956. [DOI] [PubMed] [Google Scholar]
- 14.Uchida T, Rossignol F, Matthay MA, Mounier R, Couette S, Clottes E, Clerici C. Prolonged hypoxia differentially regulates hypoxia-inducible factor (HIF)–1alpha and HIF-2alpha expression in lung epithelial cells: implication of natural antisense HIF-1alpha. J Biol Chem 2004;279:14871–14878. [DOI] [PubMed] [Google Scholar]
- 15.Krick S, Eul BG, Hanze J, Savai R, Grimminger F, Seeger W, Rose F. Role of hypoxia-inducible factor–1alpha in hypoxia-induced apoptosis of primary alveolar epithelial type II cells. Am J Respir Cell Mol Biol 2005;32:395–403. [DOI] [PubMed] [Google Scholar]
- 16.Safran M, Kaelin WG Jr. HIF hydroxylation and the mammalian oxygen-sensing pathway. J Clin Invest 2003;111:779–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fedele AO, Whitelaw ML, Peet DJ. Regulation of gene expression by the hypoxia-inducible factors. Mol Interv 2002;2:229–243. [DOI] [PubMed] [Google Scholar]
- 18.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic–helix–loop–helix–PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA 1995;92:5510–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG Jr. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 2001;292:464–468. [DOI] [PubMed] [Google Scholar]
- 20.Patel SA, Simon MC. Biology of hypoxia-inducible factor–2alpha in development and disease. Cell Death Differ 2008;15:628–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu AY, Frid MG, Shimoda LA, Wiener CM, Stenmark K, Semenza GL. Temporal, spatial, and oxygen-regulated expression of hypoxia-inducible factor–1 in the lung. Am J Physiol 1998;275:L818–L826. [DOI] [PubMed] [Google Scholar]
- 22.Wiesener MS, Jurgensen JS, Rosenberger C, Scholze CK, Horstrup JH, Warnecke C, Mandriota S, Bechmann I, Frei UA, Pugh CW, et al. Widespread hypoxia-inducible expression of HIF-2alpha in distinct cell populations of different organs. FASEB J 2003;17:271–273. [DOI] [PubMed] [Google Scholar]
- 23.Compernolle V, Brusselmans K, Acker T, Hoet P, Tjwa M, Beck H, Plaisance S, Dor Y, Keshet E, Lupu F, et al. Loss of HIF-2alpha and inhibition of VEGF impair fetal lung maturation, whereas treatment with VEGF prevents fatal respiratory distress in premature mice. Nat Med 2002;8:702–710. [DOI] [PubMed] [Google Scholar]
- 24.Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol Cell Biol 2003;23:9361–9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiesener MS, Turley H, Allen WE, Willam C, Eckardt KU, Talks KL, Wood SM, Gatter KC, Harris AL, Pugh CW, et al. Induction of endothelial PAS domain protein–1 by hypoxia: characterization and comparison with hypoxia-inducible factor–1alpha. Blood 1998;92:2260–2268. [PubMed] [Google Scholar]
- 26.Zhang Q, Moe OW, Garcia JA, Hsia CC. Regulated expression of hypoxia-inducible factors during postnatal and postpneumonectomy lung growth. Am J Physiol Lung Cell Mol Physiol 2006;290:L880–L889. [DOI] [PubMed] [Google Scholar]
- 27.Hosford GE, Olson DM. Effects of hyperoxia on VEGF, its receptors, and HIF-2alpha in the newborn rat lung. Am J Physiol Lung Cell Mol Physiol 2003;285:L161–L168. [DOI] [PubMed] [Google Scholar]
- 28.Wikenheiser KA, Vorbroker DK, Rice WR, Clark JC, Bachurski CJ, Oie HK, Whitsett JA. Production of immortalized distal respiratory epithelial cell lines from surfactant protein C/simian virus 40 large tumor antigen transgenic mice. Proc Natl Acad Sci USA 1993;90:11029–11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chesebro B, Wehrly K, Housman D. Lack of erythroid characteristics in Ia-positive leukemia cell lines induced by Friend murine leukemia virus: brief communication. J Natl Cancer Inst 1978;60:239–242. [DOI] [PubMed] [Google Scholar]
- 30.Mbawuike IN, Herscowitz HBMH-S. A murine alveolar macrophage cell line: morphological, cytochemical, and functional characteristics. J Leukoc Biol 1989;46:119–127. [DOI] [PubMed] [Google Scholar]
- 31.Warnecke C, Griethe W, Weidemann A, Jurgensen JS, Willam C, Bachmann S, Ivashchenko Y, Wagner I, Frei U, Wiesener M, et al. Activation of the hypoxia-inducible factor–pathway and stimulation of angiogenesis by application of prolyl hydroxylase inhibitors. FASEB J 2003;17:1186–1188. [DOI] [PubMed] [Google Scholar]
- 32.Asikainen TM, Ahmad A, Schneider BK, Ho WB, Arend M, Brenner M, Gunzler V, White CW. Stimulation of HIF-1alpha, HIF-2alpha, and VEGF by prolyl 4-hydroxylase inhibition in human lung endothelial and epithelial cells. Free Radic Biol Med 2005;38:1002–1013. [DOI] [PubMed] [Google Scholar]
- 33.Vorbroker DK, Profitt SA, Nogee LM, Whitsett JA. Aberrant processing of surfactant protein C in hereditary SP-B deficiency. Am J Physiol 1995;268:L647–L656. [DOI] [PubMed] [Google Scholar]
- 34.Vorbroker DK, Voorhout WF, Weaver TE, Whitsett JA. Posttranslational processing of surfactant protein C in rat Type II cells. Am J Physiol 1995;269:L727–L733. [DOI] [PubMed] [Google Scholar]
- 35.Stahlman MT, Besnard V, Wert SE, Weaver TE, Dingle S, Xu Y, von Zychlin K, Olson SJ, Whitsett JA. Expression of ABCA3 in developing lung and other tissues. J Histochem Cytochem 2007;55:71–83. [DOI] [PubMed] [Google Scholar]
- 36.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001;29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao A, Moi P. Regulation of the globin genes. Pediatr Res 2002;51:415–421. [DOI] [PubMed] [Google Scholar]
- 38.Ney PA. Gene expression during terminal erythroid differentiation. Curr Opin Hematol 2006;13:203–208. [DOI] [PubMed] [Google Scholar]
- 39.Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene 2002;21:3368–3376. [DOI] [PubMed] [Google Scholar]
- 40.Orkin SH. GATA-binding transcription factors in hematopoietic cells. Blood 1992;80:575–581. [PubMed] [Google Scholar]
- 41.Tsai FY, Keller G, Kuo FC, Weiss M, Chen J, Rosenblatt M, Alt FW, Orkin SH. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 1994;371:221–226. [DOI] [PubMed] [Google Scholar]
- 42.Weiss MJ, Keller G, Orkin SH. Novel insights into erythroid development revealed through in vitro differentiation of GATA-1 embryonic stem cells. Genes Dev 1994;8:1184–1197. [DOI] [PubMed] [Google Scholar]
- 43.Bishop DF. Two different genes encode delta-aminolevulinate synthase in humans: nucleotide sequences of cDNAs for the housekeeping and erythroid genes. Nucleic Acids Res 1990;18:7187–7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wikenheiser KA, Clark JC, Linnoila RI, Stahlman MT, Whitsett JA. Simian virus 40 large T antigen directed by transcriptional elements of the human surfactant protein C gene produces pulmonary adenocarcinomas in transgenic mice. Cancer Res 1992;52:5342–5352. [PubMed] [Google Scholar]
- 45.Ouiddir A, Planes C, Fernandes I, VanHesse A, Clerici C. Hypoxia upregulates activity and expression of the glucose transporter GLUT1 in alveolar epithelial cells. Am J Respir Cell Mol Biol 1999;21:710–718. [DOI] [PubMed] [Google Scholar]
- 46.Stroka DM, Burkhardt T, Desbaillets I, Wenger RH, Neil DA, Bauer C, Gassmann M, Candinas D. HIF-1 is expressed in normoxic tissue and displays an organ-specific regulation under systemic hypoxia. FASEB J 2001;15:2445–2453. [DOI] [PubMed] [Google Scholar]
- 47.Wang J, Buss JL, Chen G, Ponka P, Pantopoulos K. The prolyl 4-hydroxylase inhibitor ethyl-3,4-dihydroxybenzoate generates effective iron deficiency in cultured cells. FEBS Lett 2002;529:309–312. [DOI] [PubMed] [Google Scholar]
- 48.Pevny L, Simon MC, Robertson E, Klein WH, Tsai SF, D'Agati V, Orkin SH, Costantini F. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature 1991;349:257–260. [DOI] [PubMed] [Google Scholar]
- 49.Layon ME, Ackley CJ, West RJ, Lowrey CH. Expression of GATA-1 in a non-hematopoietic cell line induces beta-globin locus control region chromatin structure remodeling and an erythroid pattern of gene expression. J Mol Biol 2007;366:737–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crawford MJ, Goldberg DE. Role for the salmonella flavohemoglobin in protection from nitric oxide. J Biol Chem 1998;273:12543–12547. [DOI] [PubMed] [Google Scholar]
- 51.Gross SS, Lane P. Physiological reactions of nitric oxide and hemoglobin: a radical rethink. Proc Natl Acad Sci USA 1999;96:9967–9969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mallory GB Jr. Surfactant proteins: role in lung physiology and disease in early life. Paediatr Respir Rev 2001;2:151–158. [DOI] [PubMed] [Google Scholar]
- 53.Wright JR. Pulmonary surfactant: a front line of lung host defense. J Clin Invest 2003;111:1453–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bishop AE. Pulmonary epithelial stem cells. Cell Prolif 2004;37:89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poynter SE, LeVine AM. Surfactant biology and clinical application. Crit Care Clin 2003;19:459–472. [DOI] [PubMed] [Google Scholar]
- 56.Hendgen-Cotta UB, Merx MW, Shiva S, Schmitz J, Becher S, Klare JP, Steinhoff HJ, Goedecke A, Schrader J, Gladwin MT, et al. Nitrite reductase activity of myoglobin regulates respiration and cellular viability in myocardial ischemia–reperfusion injury. Proc Natl Acad Sci USA 2008;105:10256–10261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Crawford JH, Isbell TS, Huang Z, Shiva S, Chacko BK, Schechter AN, Darley-Usmar VM, Kerby JD, Lang JD Jr, Kraus D, et al. Hypoxia, red blood cells, and nitrite regulate NO-dependent hypoxic vasodilation. Blood 2006;107:566–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rassaf T, Flogel U, Drexhage C, Hendgen-Cotta U, Kelm M, Schrader J. Nitrite reductase function of deoxymyoglobin: oxygen sensor and regulator of cardiac energetics and function. Circ Res 2007;100:1749–1754. [DOI] [PubMed] [Google Scholar]
- 59.Gardner PR, Gardner AM, Brashear WT, Suzuki T, Hvitved AN, Setchell KD, Olson JS. Hemoglobins dioxygenate nitric oxide with high fidelity. J Inorg Biochem 2006;100:542–550. [DOI] [PubMed] [Google Scholar]
- 60.Khan AA, Wang Y, Sun Y, Mao XO, Xie L, Miles E, Graboski J, Chen S, Ellerby LM, Jin K, et al. Neuroglobin-overexpressing transgenic mice are resistant to cerebral and myocardial ischemia. Proc Natl Acad Sci USA 2006;103:17944–17948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu Z, Liu J, Guo S, Xing C, Fan X, Ning M, Yuan JC, Lo EH, Wang X. Neuroglobin-overexpression alters hypoxic response gene expression in primary neuron culture following oxygen glucose deprivation. Neuroscience 2009;162:396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ema M, Taya S, Yokotani N, Sogawa K, Matsuda Y, Fujii-Kuriyama Y. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1alpha regulates the VEGF expression and is potentially involved in lung and vascular development. Proc Natl Acad Sci USA 1997;94:4273–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boussat S, Eddahibi S, Coste A, Fataccioli V, Gouge M, Housset B, Adnot S, Maitre B. Expression and regulation of vascular endothelial growth factor in human pulmonary epithelial cells. Am J Physiol Lung Cell Mol Physiol 2000;279:L371–L378. [DOI] [PubMed] [Google Scholar]
- 64.Abu-Farha M, Niles J, Willmore WG. Erythroid-specific 5–aminolevulinate synthase protein is stabilized by low oxygen and proteasomal inhibition. Biochem Cell Biol 2005;83:620–630. [DOI] [PubMed] [Google Scholar]
- 65.Ikeda K, Clark JC, Shaw-White JR, Stahlman MT, Boutell CJ, Whitsett JA. Gene structure and expression of human thyroid transcription factor–1 in respiratory epithelial cells. J Biol Chem 1995;270:8108–8114. [DOI] [PubMed] [Google Scholar]
- 66.Kelly SE, Bachurski CJ, Burhans MS, Glasser SW. Transcription of the lung-specific surfactant protein C gene is mediated by thyroid transcription factor 1. J Biol Chem 1996;271:6881–6888. [DOI] [PubMed] [Google Scholar]
- 67.Korfhagen TR, Whitsett JA. Transcriptional control in the developing lung. The Parker B. Francis lectureship. Chest 1997;111:83S–88S. [DOI] [PubMed] [Google Scholar]
- 68.Guo X, Lin HM, Lin Z, Montano M, Sansores R, Wang G, DiAngelo S, Pardo A, Selman M, Floros J. Surfactant protein gene A, B, and D marker alleles in chronic obstructive pulmonary disease of a Mexican population. Eur Respir J 2001;18:482–490. [DOI] [PubMed] [Google Scholar]