Abstract
Human airway epithelial cells cultured in vitro at the air–liquid interface (ALI) form a pseudostratified epithelium that forms tight junctions and cilia, and produces mucin. These cells are widely used in models of differentiation, injury, and repair. To assess how closely the transcriptome of ALI epithelium matches that of in vivo airway epithelial cells, we used microarrays to compare the transcriptome of human large airway epithelial cells cultured at the ALI with the transcriptome of large airway epithelium obtained via bronchoscopy and brushing. Gene expression profiling showed that global gene expression correlated well between ALI cells and brushed cells, but with some differences. Gene expression patterns mirrored differences in proportions of cell types (ALIs have higher percentages of basal cells, whereas brushed cells have higher percentages of ciliated cells), that is, ALI cells expressed higher levels of basal cell–related genes, and brushed cells expressed higher levels of cilia-related genes. Pathway analysis showed that ALI cells had increased expression of cell cycle and proliferation genes, whereas brushed cells had increased expression of cytoskeletal organization and humoral immune response genes. Overall, ALI cells provide a good representation of the in vivo airway epithelial transcriptome, but for some biologic questions, the differences between in vitro and in vivo environments need to be considered.
Keywords: gene expression, airway epithelium, air–liquid interface
CLINICAL RELEVANCE.
Air–liquid interface cultures are useful tools for studying the airway epithelium in vitro. This study compares the transcriptomes of these cultures with cells directly brushed from the airways.
The human airway epithelium is a layer of ciliated, secretory, undifferentiated columnar and basal cells, pseudostratified in the large airways and becoming columnar and cuboidal in the small airways. The epithelium functions to clear mucus, maintain the homeostasis of fluid and electrolytes, and provide a critical barrier against inhaled pathogens, toxic gases, xenobiotics, and particulates (1–3). In healthy individuals, airway epithelial repair is a continual process in response to cell senescence and injury, characterized by cell proliferation and differentiation to maintain the normal epithelial layer (4–7).
The most commonly used models to study the biology of human airways are so-called “air–liquid interface” (ALI) cultures, where human airway epithelium obtained by biopsy, brushing, surgery, or lung transplant, or postmortem, is cultured on a collagen substratum in the presence of growth factors and retinoic acid, with the apical surface of the cells exposed to air (8–11). Over 2–3 weeks, the cells differentiate and form a pseudostratified epithelium that forms tight junctions, develops cilia, and produces mucin (12). ALI cultured cells have been used extensively to study the biology of the airway epithelium, and as models for injury and repair of the airway epithelium in health and disease (11, 13–18).
As in any in vitro cell culture model, in vitro conditions impose a milieu different from that encountered in vivo, with stresses that are undoubtedly different than those in vivo. To assess the similarities and differences between the biology of human ALI airway epithelial cultures and the biology of human airway epithelial cells immediately sampled in vivo, we used microarrays to compare the transcriptome of human large airway epithelium cultured on the ALI to the transcriptome of human airway epithelium brushed from the large airways of healthy nonsmokers. We hypothesized that the transcriptome of cultured ALI cells would overlap to a large extent with that of the brushed large airway epithelial cells, but that some differences imposed by the ALI model would exist. The data show that although a strong overall correlation in gene expression exists between cells grown in culture at the ALI and cells obtained directly from the large airway epithelium, differences are also evident, explained in part by altered differential cell counts, and by differences introduced by the biology of the cell culture environment. Some of the results of these studies were previously reported in the form of an abstract (19).
MATERIALS AND METHODS
In Vivo Samples of Large Airway Epithelia
Large airway epithelia from healthy nonsmokers (n = 12) were sampled via flexible bronchoscopy and brushing (as described in full detail in the online supplement).
Air–Liquid Interface Cell Cultures
Large airway epithelium from a healthy nonsmoker was cultured (n = 12) at the ALI for 16–18 days, according to the supplier's specifications (EpiAirway; MatTek, Ashland, MA). Cells were derived from healthy human primary explant tissue of a 23-year-old white male nonsmoker. The basal culture medium consisted of Dulbecco's modified Eagle's medium plus growth factors. Tissue was cultured at 37°C in 5.0% CO2 (for full details of the immunohistochemistry, see the online supplement). The HG-U133 Plus 2.0 array (Affymetrix, Santa Clara, CA), which includes 54,675 probe sets, was used for transcriptome analysis. Details of RNA extraction, microarray processing, and analysis are reported in the online supplement.
Analysis of Gene Expression Relating to Differential Cell Type
A list of experimentally validated ciliary and basal body–related genes (157 probe sets, representing 75 genes) was used to evaluate ciliated cells (20) (Table E1). A list of genes with enriched expression in mouse tracheal basal cells (184 probe sets, representing 73 genes) was used to evaluate basal cell–related gene expression (Table E2). A list of airway epithelium secretory cell–related genes (17 probe sets, representing 10 genes) was compiled from a review of the literature (Table E3). These lists were used to compare genes for individual cell types in large airway epithelium and ALI cultured cells. The mean normalized expression values were plotted for genome-wide as well as cilia-related, basal body–related, and secretory cell–related genes. The relative mean levels of expression in ALI cells versus brushed cells were plotted for examples of genes from each group.
Classification of Biologic Function of Differentially Expressed Genes
Gene ontology analysis (GO) was performed for all differentially expressed genes, using Genespring software (Genespring version 7.3.1; Agilent Technologies, Inc., Santa Clara, CA). Pathway analysis was performed using Ingenuity Pathways Analysis (www.ingenuity.com; for full details, see the online supplement).
Statistical Analysis
For details of the statistical analysis, see the online supplement.
Web Deposition of Data
All microarray data were deposited in the Gene Expression Omnibus site (http://www.ncbi.nlm.nih.gov/geo), which is curated by the National Center for Bioinformatics. The accession number is GSE18637. Subsets of the brushed airway samples used for microarray analysis in this study were included in previous publications, none of which involved comparisons with samples grown at the ALI (21–23).
RESULTS
Sampling the Large Airway Epithelium In Vivo
Large airway samples were collected from 12 healthy nonsmokers (Table 1). All individuals had no significant medical history and a normal general physical examination. All were HIV-negative, with blood and urinary parameters within normal range and a normal chest x-ray. Urine nicotine, urine cotinine, and venous blood carboxyhemoglobin concentrations confirmed the nonsmoking status of all individuals. Pulmonary function testing revealed normal spirometry, lung volumes, and diffusing capacity. Large airway epithelial cells were obtained by flexible bronchoscopy and by brushing the large (third-order to fourth-order bronchi) airways. The number of epithelial cells recovered ranged from 4.2–15 × 106, with an average of 99% ± 1% epithelial cells recovered. Microarray analysis showed the expression of CD45, consistent with a low level of inflammatory-cell contamination. No expression of CD11C was evident, consistent with an absence of dendritic cells in the samples.
TABLE 1.
CHARACTERISTICS OF STUDY POPULATION USED FOR IN VIVO BRUSHED LARGE AIRWAY EPITHELIAL SAMPLES*
| Parameter | Healthy Nonsmokers |
|---|---|
| n | 12 |
| Sex (male/female) | 7/5 |
| Age (years) | 42 ± 7 |
| Race (black/white/other) | 3/5/4 |
| Smoking history (pack-years) | 0 |
| Venous carboxyhemoglobin (%)† | 0.38 ± 0.59 |
| Urine nicotine | Undetectable |
| Urine cotinine | Undetectable |
| Pulmonary function parameters‡ | |
| FVC | 104 ± 13 |
| FEV1 | 97 ± 32 |
| FEV1/FVC | 82 ± 4 |
| TLC | 99 ± 13 |
| DLCO | 97 ± 10 |
| Number of large airway epithelial cells recovered x 106 | 7.6 ± 3.0 |
| Percent epithelial cells | 99 ± 1 |
| Percent inflammatory cells | 1.0 ± 0.3 |
Definition of abbreviations: FVC, forced vital capacity; FEV1, forced expiratory volume in 1 second; TLC, total lung capacity; DLCO, diffusing capacity.
Data are presented as mean ± SD.
Venous carboxyhemoglobin is a secondary marker of current smoking; nonsmokers, normal value < 1.5%.
Pulmonary function testing parameters are given as percentages of predicted value, with the exception of FEV1/FVC, which is reported as percent observed.
Comparison of Cell Populations
The cell populations of large airway epithelia recovered by brushing were similar to those observed in previous studies (Table 2) (21). The histology of ALI cells is depicted in Figure E1. On average, ALI cultured cells included fewer ciliated cells (P < 10−8), a similar number of secretory cells (P > 0.6), more undifferentiated columnar cells (P < 10−8), and more basal cells (P < 10−4).
TABLE 2.
CELL DIFFERENTIALS OF LARGE AIRWAY EPITHELIA RECOVERED IN VIVO VERSUS LARGE AIRWAY EPITHELIA DERIVED FROM AIR–LIQUID INTERFACE CULTURES*
| Cell Type | In Vivo Brush† | Coefficient of Variation (Brush, %) | Air–Liquid Interface‡ | Coefficient of Variation (ALI, %) | P Value |
|---|---|---|---|---|---|
| Ciliated (%) | 57 ± 2 | 9.9 | 27 ± 1 | 10.0 | 1.82 × 10−9 |
| Secretory (%) | 11 ± 1 | 43.0 | 12 ± 2 | 33.4 | > 0.7 |
| Undifferentiated (%) | 11 ± 1 | 24.3 | 29 ± 2 | 12.3 | 1.72 × 10−9 |
| Basal (%) | 21 ± 1 | 16.7 | 31 ± 1 | 8.3 | 2.46 × 10−5 |
As percent total epithelial cells (mean ± SEM).
n = 12.
n = 6.
Overall Comparison of Gene Expression in Cultured versus Brushed Cells
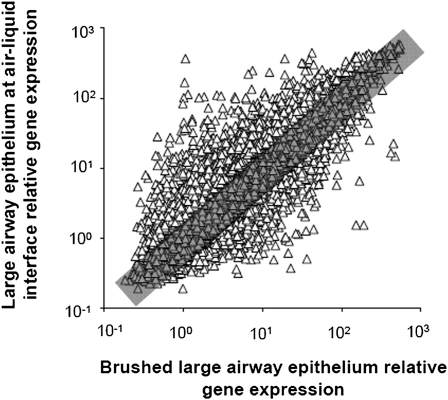
To examine differences in gene expression between cultured ALI cells and brushed cells obtained from large airway epithelia, a transcriptional analysis was performed using the Affymetrix Human Genome U133 Plus 2.0 array. A comparison of global gene expression demonstrated that, based on a P call of “present” in at least 20% of samples from both groups, cultured cells expressed 30,053 probe sets, and the large airway epithelia expressed 31,379 probe sets, with 27,579 probe sets expressed in both cell types. Eighty-one percent of those sets expressed in both cell types (22,261 probe sets) showed similar (within twofold) expression between cultured and brushed cells. Five thousand three hundred and eighteen probe sets were differentially expressed. Of those 5,318, 3,509 (66%) showed relatively increased expression in ALI cells, and 1,809 (34%) showed relatively increased expression in brushed large airway epithelial cells. Computation of the Spearman rank correlation coefficient for the probe sets expressed in both cell types showed that overall, gene expression in ALI cultures correlated well with gene expression in brushed airway cells (Spearman rank correlation coefficient, 0.89; P < 0.001; Figure 1). Similar results were seen when the analysis was repeated using eight independent brushed large airway samples (Spearman rank correlation coefficient, 0.81; P < 0.001; results not shown). An unsupervised hierarchical cluster analysis of the gene expression profiles for the 12 ALI samples and 12 large airway brushed cell samples was performed using a Spearman correlation. This analysis showed that ALI and brushed cells clustered separately. (Figure E2). This was true whether genome-wide expression values were used in the cluster analysis (Figure E2A), or cluster analysis was performed on cilia-related genes (Figure E2B), secretory cell–related genes (Figure E2C), or basal cell–related genes (Figure E2D).
Figure 1.
Correlation of overall gene expression of large airway epithelium air–liquid interface (ALI; n = 12) with brushed large airway epithelia of healthy nonsmokers (n = 12). Ordinate indicates relative gene expression in large airway epithelium ALI cultured cells. Abscissa indicates relative gene expression in brushed cells from large airway epithelia. Each is plotted on a logarithmic scale. Triangles represent each probe set that was present in at least 20% of samples. Gray rectangle indicates an up or down twofold change. Tight clustering indicates a good correlation between cultured cells and brushed cells (Spearman rank correlation coefficient, 0.89; P < 0.001). Overall, 81% of probe sets were similarly expressed within twofold.
To evaluate for variability among the brushed samples because of age or sex, two subgroup analyses were performed. Analyses of genome-wide expression among brushed cells from men (n = 7) versus women (n = 5) revealed 16 differentially expressed probe sets, representing 12 genes. Of these genes, five were Y-linked and two were X-linked, leaving five other genes differentially expressed between the two groups (Table E4). Similarly, brushed cells were divided into older and younger groups based on the median age (n = 6 in each), and evaluated for genome-wide differences in expression. In this analysis, no differences were found.
Cilia-Related Genes
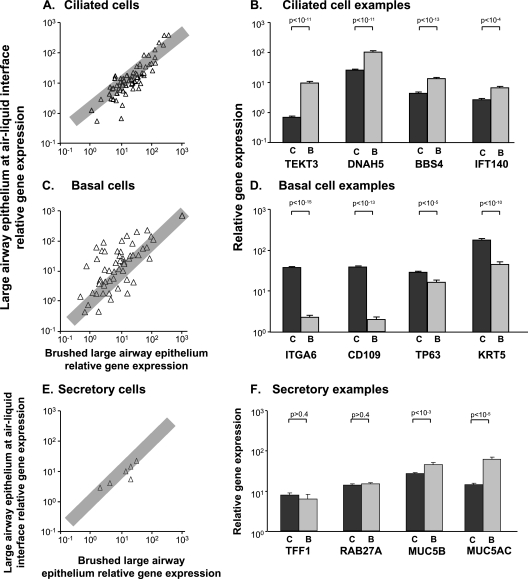
Because the brushed large airway epithelia contained a higher proportion of ciliated cells than did ALI cultures (Table 2), we assessed whether the expression of genes related to cilia structure, function, and maintenance was different in the in vitro versus in vivo samples. A list of 157 probe sets representing 75 cilia-related genes was examined (20) (Table E1 contains a list of the genes). Of these, 123 probe sets representing 70 genes demonstrated a P call of “Present” in at least 20% of both cultured and brushed cells, and 67% (47 of 70) of the genes showed similar expression (within twofold) between cultured and brushed cells. The Spearman rank correlation coefficient was 0.85 (P < 0.001) (Figure 2A). When the analysis was repeated with eight independent large airway epithelial samples, results were similar (Spearman rank correlation coefficient, 0.91; P < 0.001; data not shown). Examination of 24 cilia-related genes that were differentially expressed by more than twofold showed that 92% (23 of 25) exhibited relatively increased expression in brushed cells relative to ALI cells. Specific cilia-related genes that demonstrated relatively increased expression in large airway brushed cells included the structural proteins TEKT3, DNAH5, BBS4, and IFT140 (Figure 2B).
Figure 2.
Correlation of expression values of ciliated, basal, and secretory cell-related genes in large airway epithelium ALI cultured cells and brushed large airway epithelia. (A, C, and E) Ordinate indicates relative gene expression in ALI cells, whereas abscissa indicates relative gene expression in brushed cells, plotted on a logarithmic scale. Triangles represent each gene present in at least 20% of samples. When multiple probe sets represent a gene, the probe set with the highest expression values was selected. Gray rectangle indicates an up or down twofold change. (B, D, and F) Ordinate indicates relative gene expression in ALI and brushed cells. Abscissa indicates examples of markers of each cell type. Dark gray bars, ALI cells; light gray bars, brushed cells from large airway epithelia. Each bar represents relative mean expression with standard error. P values are represented in brackets above bars. (A) Correlation of cilia-related genes (Spearman rank correlation coefficient, 0.85; P < 0.001). Overall, 67% of genes were similarly expressed within twofold. Of genes that were differentially expressed, 92% showed increased expression in brushed cells relative to ALI cultures. (B) Relative expression in examples of cilia-related genes (TEKT3, DNAH5, BBS4, and IFT40). Each was more highly expressed in brushed cells relative to ALI. (C) Correlation of basal cell–related genes (Spearman rank correlation coefficient, 0.64: P < 0.001). Overall, 42% of genes were similarly expressed within twofold. Of genes that were differentially expressed, 78% showed increased expression in ALI relative to brushed cells. (D) Relative expression of examples of basal cell markers (integrin A6, CD109, TP63, and keratin 5). Each was more highly expressed in ALI cells relative to brushed cells. (E) Correlation of secretory cell–related genes (Spearman rank correlation coefficient, 0.90; P < 0.02). Overall, 83% of genes were similarly expressed within twofold. (F) Relative expression levels of examples of secretory genes. TFF1, RAB27A, and MUC5B all showed similar expression in ALI cells relative to brushed cells, whereas MUC5AC was more highly expressed in brushed cells relative to ALI. Although the P value for MUC5B was significant, the fold change of −1.6 did not meet our criteria for differential expression of more than twofold. (B, D, and F) C, large airway epithelium ALI cultures; B, large airway epithelium brushed cells.
Basal Cell–Related Genes
Because higher percentages of basal cells in the ALI cultured cells occurred relative to brushed large airway epithelia (Table 2), we examined the expression of basal cell–related genes (see Table E2 for a list of the genes). Of 73 genes analyzed, 56 demonstrated a P call of “Present” in at least 20% of both cell types; 42% (24 of 56) demonstrated similar expression (within twofold) between cultured and brushed cells. The Spearman rank correlation coefficient was 0.64 (P < 0.001) (Figure 2C). When the analysis was repeated with eight independent large airway epithelial samples, the results were similar (Spearman rank correlation coefficient, 0.89; P < 0.001; results not shown). Of 32 basal cell–related genes that were differentially expressed, 78% (25 of 32) showed relatively increased expression in ALI cells compared with brushed large airway epithelia. Examples of basal cell markers with increased expression in the ALI included integrin A6, CD109, tumor protein 63, and cytokeratin 5 (Figure 2D).
Secretory Cell–Related Genes
A similar number of secretory cells were observed in the ALI cultured cells relative to brushed large airway epithelial cells (Table 2). We examined the expression of secretory cell–related genes to determine whether similar expression was observed. The expression of 17 secretory cell–related probe sets representing 10 genes was examined (see Table E3 for a list of the genes). Six of these genes were expressed with a P call of “Present” in at least 20% of both ALI cells and brushed cells. Mucin 2 (MUC2) was expressed by brushed large airway epithelial cells, but not by ALI cells. MUC7, MUC8, and MUC19 were not expressed in either brushed large airway cells or cultured ALI cells. The Spearman rank correlation coefficient was 0.90 (P < 0.02) (Figure 2E). When the analysis was repeated with eight independent large airway epithelial samples, the results were similar (Spearman rank correlation coefficient, 0.89; P < 0.001). MUC5AC demonstrated relatively increased expression in large airway brushed cells (Figure 2F). The remaining 83%, that is, five of six genes (MUC5B, MUC16, TFF1, TFF3, and RAB27A), were similarly expressed (within twofold) in both cell types.
Gene Ontology Analysis of Differentially Expressed Genes
GO was performed, using Genespring software, on all differentially expressed genes to determine their biologic functions (Table 3). The top 15 biologic functional categories, selected on the basis of significance (according to P values), are shown. Categories of interest with relatively increased expression in ALI relative to brushed cells included cell cycle (score, 9.34; P < 2 × 10−7) and cell proliferation (score, 6.82; P < 3 × 10−6), as expected from cultured versus in vivo cells. Categories of interest that showed relatively increased expression in brushed cells obtained from the large airway epithelia included cytoskeleton organization (score, 6.85; P < 3 × 10−6) and humoral immune response (score, 2.58; P < 3 × 10−4).
TABLE 3.
COMMON CATEGORIES OF GENES DIFFERENTIALLY EXPRESSED IN LARGE AIRWAY EPITHELIUM CELLS CULTURED AT AIR–LIQUID INTERFACE VERSUS CELLS BRUSHED FROM LARGE AIRWAY EPITHELIA*
| Expression Relatively Increased in Air–Liquid Interface Relative to Brushed Epithelia |
Expression Relatively Increased in Brushed Epithelia Relative to Air–Liquid Interface |
||||
|---|---|---|---|---|---|
| GO Biologic Function | Score† | P Value‡ | GO Biologic Function | Score† | P Value‡ |
| Mitotic cell cycle | 4.22 | 1.02 × 10−14 | Cytoskeleton-dependent intracellular transport | 3.12 | 5.19 × 10−12 |
| M phase of mitotic cell cycle | 3.17 | 4.17 × 10−12 | Microtubule-based movement | 3.12 | 5.19 × 10−12 |
| Mitosis | 3.08 | 1.35 × 10−11 | Microtubule-based process | 3.73 | 2.59 × 10−9 |
| Cell cycle | 9.98 | 1.38 × 10−10 | Cytoskeleton organization and biogenesis | 6.55 | 4.22 × 10−6 |
| Regulation of progression through cell cycle | 7.25 | 1.99 × 10−10 | Antigen presentation, exogenous antigen | 0.91 | 1.00 × 10−5 |
| M phase | 3.43 | 2.24 × 10−9 | Antigen processing, exogenous antigen via major histocompatibility antigen Class II | 0.91 | 1.22 × 10−5 |
| Cell division | 3.03 | 4.71 × 10−9 | Fibril organization and biogenesis | 0.30 | 3.70 × 10−5 |
| Cell cycle checkpoint | 1.32 | 4.91 × 10−9 | Response to cold | 0.40 | 7.73 × 10−5 |
| Development of epidermis | 1.50 | 7.31 × 10−9 | Humoral immune response | 2.62 | 8.11 × 10−5 |
| Development of ectoderm | 1.67 | 1.76 × 10−8 | Detection of biotic stimulus | 0.71 | 8.98 × 10−5 |
| Cell proliferation | 7.17 | 4.97 × 10−8 | Neutral amino-acid transport | 0.50 | 3.21 × 10−4 |
| Monosaccharide metabolism | 2.37 | 2.39 × 10−7 | Oligosaccharide metabolism | 0.71 | 3.20 × 10−4 |
| Glucose catabolism | 1.45 | 1.15 × 10−7 | Cytoskeletal anchoring | 0.71 | 1.93 × 10−4 |
| Glucose metabolism | 1.85 | 1.25 × 10−7 | Nitric oxide transport | 0.30 | 1.44 × 10−4 |
| Hexose metabolism | 2.37 | 1.46 × 10−7 | Detection of pest, pathogen, or parasite | 0.60 | 1.09 × 10−4 |
Gene ontology analysis (GO) was performed, using Genespring software, on all differentially expressed genes (magnitude of fold-change ALI cells compared with brushed cells, > 2; P < 0.01 after Benjamini-Hochberg multiple test correction). Biologic functional categories were selected on the basis of significance. Top 15 categories (based on P values) in each group are shown.
Score refers to the percentage of differentially expressed genes belonging to each GO category.
P value represents the probability that the group of genes falls within a given GO category by chance, ranked in descending order of P values.
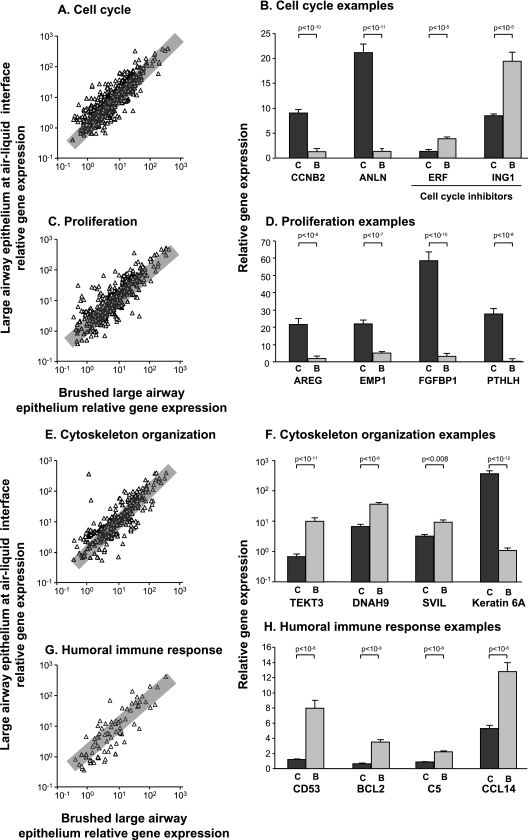
To characterize the expression of each of these categories of genes within ALI cells and brushed large airway cells, a Spearman rank correlation coefficient was calculated. Within the gene ontology (GO) category “cell cycle” (GO, 7,049), 1,260 probe sets corresponding to 590 genes of known function were expressed with a P call of “Present” in at least 20%. The Spearman rank correlation coefficient was 0.86 (P < 0.001) (Figure 3A). Examples of genes demonstrating increased expression in ALI cells included cyclin B2 and anillin, whereas the tumor suppressors v-ets erythroblastosis virus E26 oncogene homolog 2 (avian) repressor factor and inhibitor of growth family member 1 were more highly expressed by brushed cells (Figure 3B). In the GO category “cell proliferation” (GO, 8,283), 785 probe sets were expressed by both cell types, with a P call of “Present” in at least 20%, corresponding to 366 genes of known function. The Spearman rank correlation coefficient was 0.83 (P < 0.001) (Figure 3C). Amphiregulin, epithelial membrane protein 1, fibroblast growth factor binding protein, and parathyroid-like hormone are examples of genes that showed relatively increased expression in ALI (Figure 3D). Within the GO category “cytoskeletal organization and biogenesis” (GO, 7,010), 593 probe sets corresponding to 260 genes of known function were expressed, with a P call of “Present” in at least 20%. The Spearman rank correlation coefficient was 0.84 (P < 0.001) (Figure 3E). Tetkin 3, dynein heavy chain 9, and supervillin are examples of genes that showed relatively decreased expression in ALI cells (Figure 3F). Notably, of 34 genes that showed relatively decreased expression in ALI cells, 38% (13 of 34) were in the dynein family, suggesting that their decreased expression was likely related to the lower percentage of ciliated cells in cultures. Another gene in this category, keratin 6A, demonstrated expression that was markedly increased in cultured cells relative to brushed cells (Figure 3F). Because karatin 6A was shown to be a marker of basal cells that are actively proliferating (24), this is an example of a gene whose expression is increased because of both the increased presence of basal cells and the biology of proliferation during differentiation in ALI cultures. Within the GO category “humoral immune response” (GO, 6,959), 138 probe sets corresponding to 77 genes of known function were expressed, with a P call of “Present” in at least 20%. The Spearman rank correlation coefficient was 0.84 (P < 0.001) (Figure 3G). CD53, B-cell CLL/lymphoma 2 (BCL2), complement 5 (C5), and chemokine (C-C motif) ligand 14 (CCL14) are examples of genes that showed relatively decreased expression in ALI cultured cells (Figure 3H).
Figure 3.
Expression of the gene ontology categories cell cycle, cell proliferation, cytoskeleton organization, and humoral immune response genes in large airway epithelium ALI compared with brushed large airway epithelia. (A, C, E, and G) Ordinate indicates relative gene expression in ALI, whereas abscissa indicates relative gene expression in brushed cells, plotted on a logarithmic scale. Triangles represent each gene present in at least 20% of samples. When multiple probe sets represented a gene, the probe set with the highest expression values was used. Gray rectangle indicates twofold change. (B, D, F, and H) Ordinate indicates relative gene expression in ALI and brushed cells. Abscissa indicates examples of genes from each category. Dark gray bars, ALI; light gray bars, brushed cells. Each bar represents relative mean expression with standard error. P values are represented in brackets above bars. (A) Correlation of cell cycle genes (Spearman rank correlation coefficient, 0.86; P < 0.001). Overall, 72% of genes were similarly expressed. (B) Expression of cell cycle genes. Cyclin B2 (CCNB2) and anillin were more highly expressed in ALI relative to brushed cells, whereas tumor-suppressor genes ERF and ING1 were more highly expressed in brushed cells relative to ALI. (C) Correlation of cell proliferation genes. Overall, 66% of genes were similarly expressed within twofold (Spearman rank correlation coefficient, 0.83; P < 0.001). (D) Expression of cell proliferation genes. Amphiregulin (AREG), EMP1, fibroblast growth factor BP1 (FGFBP1), and PTH-like hormone (PTHLH) were more highly expressed in ALI relative to brushed large airway epithelial cells. (E) Correlation of cytoskeleton organization and biogenesis (Spearman rank correlation coefficient, 0.84; P < 0.001). Overall, 61% of genes were similarly expressed within twofold. (F) Expression of cytoskeleton organization genes. Tetkin 3 (TEKT3), dynein heavy chain 9 (DNAH9), and supervillin (SVIL) were more highly expressed in brushed cells relative to ALI. In addition to other keratins and tubulins within this category, keratin 6A was more highly expressed in ALI relative to brushed cells. (G) Correlation of humoral immune response genes (Spearman rank correlation coefficient, 0.84; P < 0.001). Overall, 56% of genes were similarly expressed within twofold. (H) Expression of humoral immune response genes. CD53, B-cell CLL/lymphoma 2 (BCL2), complement factor 5 (C5), and chemokine (C-C motif) ligand 14 (CCL14) were more highly expressed in brushed cells relative to ALI. (B, D, F, and H) C, large airway epithelium ALI cultures; B, large airway epithelium brushed cells.
Assessment of Functional Pathways
To assess differences between ALI cultures and brushed cells further, all differentially expressed genes were examined using Ingenuity Pathway Analysis (www.ingenuity.com) on differentially expressed genes (magnitude of fold-change in ALI cells compared with brushed cells, > 2; P < 0.01 after Benjamini-Hochberg multiple-test correction). Canonical pathways were selected on the basis of significance and ratio. In total, 3,509 genes demonstrated higher expression in ALI cultured cells, and 1,809 genes demonstrated higher expression in brushed cells from large airway epithelia. Eighty-one pathways were more highly expressed in ALI cultured cells, and 22 pathways were more highly expressed in brushed cells. The main canonical pathways whose expression was relatively increased in ALI cells included integrin signaling (Figure E3A) (ratio, i.e., number of pathway genes in the differentially expressed dataset compared with the total number of genes in the pathway, 0.26; P < 0.001), ephrin receptor signaling (ratio, 0.24; P < 0.001), IL-8 signaling (ratio, 0.24; P < 0.001), p53 signaling (Figure E3B; ratio, 0.32; P < 0.001), and cell cycle checkpoint regulation (ratio, 0.31; P < 0.001). The main canonical pathways with relatively increased expression in brushed cells relative to ALI cultured cells included the metabolism of xenobiotics by cytochrome p450 (Figure E3C; ratio, 0.09; P < 0.001), fatty-acid metabolism (ratio, 0.08; P < 0.001), peroxisome proliferator activator receptor α/retinoid X receptor α (ratio, 0.11; P < 0.001), the complement system (Figure E3D; ratio, 0.14; P < 0.001), and the antigen presentation pathway (ratio, 0.13; P < 0.001; Table 4).
TABLE 4.
PATHWAY ANALYSIS OF GENES DIFFERENTIALLY EXPRESSED IN LARGE AIRWAY EPITHELIAL CELLS GROWN AT AIR–LIQUID INTERFACE VERSUS LARGE AIRWAY BRUSHED CELLS*
| Pathways with Genes with Increased Expression in Air–Liquid Interface Cells Relative to Brushed Cells |
Pathways with Genes with Increased Expression in Brushed Cells Relative to Air–Liquid Interface Cells |
||||
|---|---|---|---|---|---|
| Functional Pathway | Ratio† | P Value‡ | Functional Pathway | Ratio† | P Value‡ |
| Integrin signaling | 0.26 | 3.18 × 10−9 | Metabolism of xenobiotics by cytochrome p450 | 0.09 | 5.53 × 10−7 |
| Ephrin receptor signaling | 0.24 | 5.05 × 10−8 | Fatty acid metabolism | 0.08 | 2.57 × 10−4 |
| IL-8 signaling | 0.24 | 1.85 × 10−7 | PXR/RXR activation | 0.11 | 5.85 × 10−3 |
| p53 signaling | 0.32 | 6.02 × 10−7 | Complement system | 0.14 | 2.91 × 10−2 |
| Cell cycle checkpoint regulation | 0.31 | 3.53 × 10−5 | Antigen presentation pathway | 0.13 | 1.70 × 10−2 |
Definition of abbreviation: PXR/RXR, peroxisome proliferator activator receptor α/retinoid X receptor α.
Functional pathway analysis was carried out using Ingenuity Pathway Analysis (www.ingenuity.com) on all differentially expressed genes (magnitude of fold-change ALI cells compared to brushed cells > 2, P < 0.01, after Benjamini-Hochberg multiple test correction). Canonical pathways were selected on the basis of significance and ratio.
Ratio refers to the number of differentially expressed pathway genes compared to the total number of genes in the curated pathway.
P value represents the likelihood that the association between the genes in the pathway is attributable to chance, calculated using the right-tailed Fisher exact test.
DISCUSSION
ALI cultures are a powerful tool for studying human airway epithelium in vitro, and are widely used to model airway epithelial differentiation, injury, and repair, and the function of specific genes and biologic pathways, and to assess gene transfers (8–10, 12, 13, 15, 17, 25, 26). Using gene expression profiling, we assessed how well ALI cultures represent the transcriptome of the airway epithelium in vivo. The data show a large and significant overlap between the transcriptomes of human airway epithelia cultured in vitro at the ALI and brushed cells obtained directly from the large airway epithelium of healthy nonsmokers. Overall, 81% of expressed genes showed similar expression profiles. However, differences are evident. The majority of those genes with differential expression showed relatively increased expression in ALI cultured cells. When genes relating specifically to ciliated and secretory cells were examined, the majority showed a similar expression within twofold, whereas the expression of basal cell–related genes showed less similarity. Analyses of the functional categories of differentially expressed genes revealed that categories relating to proliferative processes had increased expression in ALI cells, whereas categories relating to the cytoskeleton and immune response had a relatively higher expression in brushed cells directly obtained from large airway epithelia. Unsupervised hierarchical clustering analysis showed that, as might be expected, ALI and brushed large airway samples cluster separately. These observed disparities in gene expression are likely attributable, in part, to differences in cell populations, with ALI containing more basal cells, and brushed airway epithelia containing increased percentages of ciliated cells. Moreover, differences were likely to result from the milieu of the in vitro environment. This study used a single method to culture cells at the ALI. Additional differences might be discovered if different culture methods were used. It would also be interesting to compare gene expression in vitro versus in vivo with paired samples from the same individual. However, the present study shows that even when independent samples from distinct individuals are used, the correlation is good. Among most categories with observed differences, the majority of genes still showed similar expression, that is, ALI cultures provide a good representation of the airway epithelium in vivo, but for specific genes or pathways, attention should be paid to whether differences exist.
Changes in Gene Expression Relating to Different Cell Populations
To assess changes in gene expression secondary to differences in cell populations between ALI cultured cells and large airway brushed cells, we analyzed the expression of genes relating to ciliated, basal, and secretory cells. The data demonstrated that, of the differentially expressed genes, the majority of cilia and secretory genes were similarly expressed by the ALI and brushed cells. In contrast, the expression of basal cell–related genes was relatively increased in ALI cultures relative to brushed cells, consistent with the increased percentage of basal cells in ALI cultures. The mucins 7, 8, and 19 were not expressed in either sample, likely because their expression in normal lung tissue is restricted to the submucosal glands (27). MUC5B is also expressed in the submucosal glands of normal tissue, but was previously shown to be expressed by goblet cells in vitro (27). MUC2 was not expressed by ALI cells; it is found in very low concentrations in normal airway epithelium (27). MUC5AC showed a relatively higher expression in brushed cells. The majority of the remaining secretory cell–related genes showed similar expression between brushed and cultured cells. The identification of ciliated cells in this study relied on visual inspection, using phase microscopy. The number of ciliated cells may have been underestimated, because visual examination could have missed some cells undergoing differentiation. However, among the differentially expressed cilia-related genes, most showed relatively decreased expression in ALI cultures, making it less likely that we underestimated the number of ciliated cells in cultures.
Functional Categories of Differentially Expressed Genes
To analyze the functional categories of genes that were differentially expressed, GO was used. When analyzing the group of genes showing relatively increased expression in ALI cells, a large number of these genes fell into categories relating to cell cycle/proliferation, mitosis, and the metabolism or catabolism of glucose. This finding is consistent with growth in an in vitro environment, where cells are rapidly proliferating and have high metabolic demands. An examination of the major categories of genes that showed relatively higher expression in large airway epithelium brushed cells demonstrated that these genes fell into two main categories. One group related to the cytoskeleton and microtubule organization. Within these GO categories were many genes relating to cilia structure and function, likely reflecting the lower percentage of ciliated cells in the ALI culture. Another group of categories related to the immune response, including antigen presentation, the humoral immune response, and the recognition of pathogens. Pulmonary epithelial cells have the ability to express major histocompatibility antigen (MHC) Class I and II receptors (2), and many of the genes in these categories relate to MHC genes. The brushed cells had an opportunity to interact in vivo with components of the immune system, which may provide an explanation for the increased expression of these genes in brushed cells.
Assessment of Functional Pathways
The functional pathway analysis of genes showing relatively higher expression in ALI cultured cells revealed that the dominant canonical pathways were related to signaling and communication between cells. In contrast, the dominant canonical pathways in brushed cells from large airway epithelia were related to the immune response and xenobiotic and fatty-acid metabolism. Many components of the integrin signaling pathway showed relatively increased expression in ALI cultured cells, including the ligands of the integrins laminin and fibronectin (28). Collagens were shown to activate the integrin signaling pathway, and being grown on a collagen matrix likely contributed to the activation of this pathway in ALI cells (28). Ephrins and their receptors mediate bidirectional signaling between neighboring cells, and orchestrate developmental patterning by modulating adhesive or repulsive cell properties (29). Components of the ephrin signaling pathway demonstrated increased expression in ALI cells, which was likely a product of the culture environment. Cells were cultured in the presence of epidermal growth factor; its activated receptor, EGFR, was shown to activate the ephrin signaling pathway independently (30). The p53 pathway, which had relatively higher expression in large airway brushed cells, plays well-documented roles in responding to stress, regulating cell cycle progression, DNA repair, and apoptosis (31). Many of the components showing increased expression in this pathway specifically relate to cell cycle progression and apoptosis, and were likely increased because of cell proliferation in culture.
Interestingly, fewer pathways exhibited relatively increased expression in brushed cells. Of the pathways that were increased in brushed cells, the most significant relate to the metabolism of xenobiotics, the immune response, and inflammation. The airway epithelium is constantly in contact with oxidants in the environment, and even healthy nonsmokers are exposed to some low concentrations of xenobiotics (32). Brushed cells likely exhibit the basal expression of the pathway to metabolize xenobiotics. Cultured ALI cells in our study were not exposed to an oxidant stress, but ALI cells were shown to activate this pathway in response to cigarette smoke (26), that is, the response to oxidants is not a limitation of ALI cells. The complement system demonstrated increased expression in brushed cells. Airway epithelial cells were shown to synthesize many components of the complement system as part of their host defense function, and they interact with and modulate inflammatory cells (7, 33). In vivo brushed cells are more likely to have ongoing exposures that activate these pathways.
In Vitro Culture of Cells at the Air–Liquid Interface
Wu and colleagues (8) and Whitcutt and colleagues (9) first described the method of using a biphasic culture system to expose the apical surface of cells to ALI and promote mucociliary differentiation, using cultured primary tracheal hamster cells. The technique was later expanded to human airway epithelial cells (10). The cells were able to form tight junctions, produce mucin, and differentiate to form cilia; an addition of retinoic acid to the culture medium prevented squamous metaplasia (12). Subsequent studies using pseudostratified, differentiated human airway cells at the ALI were used to create models of specific disease states such as asthma and cystic fibrosis, to examine cytokine production and as models of differentiation (15, 34). Controversy remains concerning whether these model systems represent intrinsic genetic differences attributable to these disease processes, or they are a product of culture conditions and environment. ALI cultures incubated with various antibacterials were shown to have a reduced secretion of inflammatory cytokines and to maintain epithelial integrity, indicating that antibacterial agents may modulate the anti-inflammatory process mediated by epithelial cells (35). ALI cells were exposed to cigarette smoke, which induced oxidant-related gene expression, up-regulated cytokines IL-6 and IL-8, and reduced vascular endothelial growth factor A (VEGF) (36, 37). A variety of pathogens were studied in vitro using ALI, including mycoplasma, Pseudomonas aeruginosa, influenza, parainfluenza, and respiratory syncytial virus, to understand the factors affecting infectivity, the maintenance of barrier function, electrolyte transport, and mucus production (38–42).
ALI cultures were also useful in studying the proliferation and differentiation of human airway epithelial cells. For example, IL-13 is known to induce airway remodeling, and was shown in ALI cells to decrease the expression of forkhead box J1 (FOXJ1), induce the loss of cilia, promote goblet-cell metaplasia, and decrease ciliary beat frequency (43, 44). Many of these effects were reversed when IL-13 was inhibited (45). Goblet-cell metaplasia was also shown to be promoted by E-cadherin (which is induced by oxidative stress via the activation of EGFR) and by IL-9 during airway repair after injury (16, 46, 47). Trefoil factor family 3 peptide induces ciliogenesis, and is able to promote ciliated cell differentiation in vitro, partly via an EGFR-dependent pathway (48). Blocking the tyrosine kinase receptor erbB2 (v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog [avian]) was shown to decrease the number of ciliated cells in ALI culture, whereas treatment with its ligand, heregulin-α, stimulated differentiation.
Global gene expression was used to assess the differentiation of human airway epithelium (17), as well as the response of human airway epithelium to an exogenous stimulus. Maunders and colleagues (36) exposed ALI cultured cells to cigarette smoke in vitro, and used a microarray to show that antioxidant and mitogen-activated protein kinase pathways were activated. ALI cells were exposed to isolated components of cigarette smoke, and gene expression was analyzed to identify the mechanisms of toxicity (49). Primary brushed cells, obtained from smokers with COPD, were grown in culture at the ALI by Pierrou and colleagues (26), and were used to confirm the in vitro response of oxidant genes to cigarette smoke exposure in chronic obstructive pulmonary disease. Woodruff and colleagues (50) used a microarray to study the response of patients with asthma to inhaled corticosteroids, and used ALI cells to confirm that similar effects were observed in vitro. Ross and colleagues (17) described changes in gene expression over time associated with the mucociliary differentiation process, and identified candidate genes that may be involved in ciliogenesis, as well as signaling networks involved in differentiation.
Using the genome-wide transcriptome as the phenotype, the present study shows that for the majority of genes, ALI cultured cells serve as a good model for the gene expression of brushed cells obtained directly from the large airway epithelia of normal nonsmokers. Some differences, however, were evident, which may be explained in part by differences in proportions of cell types, and in part by the in vitro versus in vivo milieu. In that context, the use of ALI should always take into consideration variations, if any, in differences among the genes and pathways between the ALI versus airway epithelium in vivo.
Supplementary Material
Acknowledgments
We thank B. Ferris for expert technical assistance, Y. Strulovici-Barel for assistance with data analysis, T. O'Connor and A. Krause for helpful discussions, and N. Mohamed and T. Virgin-Bryan for help in preparing the manuscript.
This work was supported in part by National Institutes of Health grants P01 HL51746 (R.G.C.), R01 HL074326 (R.G.C.), P50 HL084936 (R.G.C.), UL1-RR024996, and K12 RR024997 (A.E.T.), and by the Starr Foundation/Starr Cancer Consortium.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2009-0453OC on June 4, 2010
Author Disclosure: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Breeze RG, Wheeldon EB. The cells of the pulmonary airways. Am Rev Respir Dis 1977;116:705–777. [DOI] [PubMed] [Google Scholar]
- 2.Robbins RA, Rennard SI. Biology of airway epithelial cells. In: Crystal RG, West JB, Weibel ER, Barnes PJ, editors. The lung, 2nd ed. Philadelphia: Lippincott Raven Publishers; 1997. p. 445–458.
- 3.Knight DA, Holgate ST. The airway epithelium: structural and functional properties in health and disease. Respirology 2003;8:432–446. [DOI] [PubMed] [Google Scholar]
- 4.Bishop AE. Pulmonary epithelial stem cells. Cell Prolif 2004;37:89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rawlins EL, Hogan BL. Epithelial stem cells of the lung: privileged few or opportunities for many? Development 2006;133:2455–2465. [DOI] [PubMed] [Google Scholar]
- 6.Crystal RG, Randell SH, Engelhardt JF, Voynow J, Sunday ME. Airway epithelial cells: current concepts and challenges. Proc Am Thorac Soc 2008;5:772–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnes PJ. Immunology of asthma and chronic obstructive pulmonary disease. Nat Rev Immunol 2008;8:183–192. [DOI] [PubMed] [Google Scholar]
- 8.Wu R, Sato GH, Whitcutt MJ. Developing differentiated epithelial cell cultures: airway epithelial cells. Fundam Appl Toxicol 1986;6:580–590. [DOI] [PubMed] [Google Scholar]
- 9.Whitcutt MJ, Adler KB, Wu R. A biphasic chamber system for maintaining polarity of differentiation of cultured respiratory tract epithelial cells. In Vitro Cell Dev Biol 1988;24:420–428. [DOI] [PubMed] [Google Scholar]
- 10.de Jong PM, van Sterkenburg MA, Kempenaar JA, Dijkman JH, Ponec M. Serial culturing of human bronchial epithelial cells derived from biopsies. In Vitro Cell Dev Biol Anim 1993;29A:379–387. [DOI] [PubMed] [Google Scholar]
- 11.Wu R, Zhao YH, Chang MM. Growth and differentiation of conducting airway epithelial cells in culture. Eur Respir J 1997;10:2398–2403. [DOI] [PubMed] [Google Scholar]
- 12.Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, Randell SH. Well-differentiated human airway epithelial cell cultures. Methods Mol Med 2005;107:183–206. [DOI] [PubMed] [Google Scholar]
- 13.Gruenert DC, Basbaum CB, Widdicombe JH. Long-term culture of normal and cystic fibrosis epithelial cells grown under serum-free conditions. In Vitro Cell Dev Biol 1990;26:411–418. [DOI] [PubMed] [Google Scholar]
- 14.Vermeer PD, Einwalter LA, Moninger TO, Rokhlina T, Kern JA, Zabner J, Welsh MJ. Segregation of receptor and ligand regulates activation of epithelial growth factor receptor. Nature 2003;422:322–326. [DOI] [PubMed] [Google Scholar]
- 15.Zabner J, Scheetz TE, Almabrazi HG, Casavant TL, Huang J, Keshavjee S, McCray PB Jr. CFTR DeltaF508 mutation has minimal effect on the gene expression profile of differentiated human airway epithelia. Am J Physiol Lung Cell Mol Physiol 2005;289:L545–L553. [DOI] [PubMed] [Google Scholar]
- 16.Casalino-Matsuda SM, Monzon ME, Forteza RM. Epidermal growth factor receptor activation by epidermal growth factor mediates oxidant-induced goblet cell metaplasia in human airway epithelium. Am J Respir Cell Mol Biol 2006;34:581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross AJ, Dailey LA, Brighton LE, Devlin RB. Transcriptional profiling of mucociliary differentiation in human airway epithelial cells. Am J Respir Cell Mol Biol 2007;37:169–185. [DOI] [PubMed] [Google Scholar]
- 18.Arold SP, Malavia N, George SC. Mechanical compression attenuates normal human bronchial epithelial wound healing. Respir Res 2009;10:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dvorak AK, Tilley AE, O'Connor TP, Crystal RG. Do air–liquid interface cultures of normal human airway epithelium provide a true representation of gene expression in the normal human airway epithelium? Am J Respir Crit Care Med 2009;179:A2393. [Google Scholar]
- 20.Gherman A, Davis EE, Katsanis N. The Ciliary Proteome Database: an integrated community resource for the genetic and functional dissection of cilia. Nat Genet 2006;38:961–962. [DOI] [PubMed] [Google Scholar]
- 21.Carolan BJ, Heguy A, Harvey BG, Leopold PL, Ferris B, Crystal RG. Up-regulation of expression of the ubiquitin carboxyl–terminal hydrolase L1 gene in human airway epithelium of cigarette smokers. Cancer Res 2006;66:10729–10740. [DOI] [PubMed] [Google Scholar]
- 22.Carolan BJ, Harvey BG, De BP, Vanni H, Crystal RG. Decreased expression of intelectin 1 in the human airway epithelium of smokers compared to nonsmokers. J Immunol 2008;181:5760–5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanni H, Kazeros A, Wang R, Harvey BG, Ferris B, De BP, Carolan BJ, Hubner RH, O'Connor TP, Crystal RG. Cigarette smoking induces overexpression of a fat-depleting gene AZGP1 in the human. Chest 2009;135:1197–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moll R, Divo M, Langbein L. The human keratins: biology and pathology. Histochem Cell Biol 2008;129:705–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pickles RJ, McCarty D, Matsui H, Hart PJ, Randell SH, Boucher RC. Limited entry of adenovirus vectors into well-differentiated airway epithelium is responsible for inefficient gene transfer. J Virol 1998;72:6014–6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pierrou S, Broberg P, O'Donnell RA, Pawlowski K, Virtala R, Lindqvist E, Richter A, Wilson SJ, Angco G, Moller S, et al. Expression of genes involved in oxidative stress responses in airway epithelial cells of smokers with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2007;175:577–586. [DOI] [PubMed] [Google Scholar]
- 27.Rose MC, Voynow JA. Respiratory tract mucin genes and mucin glycoproteins in health and disease. Physiol Rev 2006;86:245–278. [DOI] [PubMed] [Google Scholar]
- 28.van der FA, Sonnenberg A. Function and interactions of integrins. Cell Tissue Res 2001;305:285–298. [DOI] [PubMed] [Google Scholar]
- 29.Lackmann M, Boyd AW. EPH, a protein family coming of age: more confusion, insight, or complexity? Sci Signal 2008;1:re2. [DOI] [PubMed] [Google Scholar]
- 30.Larsen AB, Pedersen MW, Stockhausen MT, Grandal MV, van Deurs DB, Poulsen HS. Activation of the EGFR gene target EPHA2 inhibits epidermal growth factor-induced cancer cell motility. Mol Cancer Res 2007;5:283–293. [DOI] [PubMed] [Google Scholar]
- 31.Vousden KH, Prives C. Blinded by the light: the growing complexity of P53. Cell 2009;137:413–431. [DOI] [PubMed] [Google Scholar]
- 32.Heffner JE, Repine JE. Pulmonary strategies of antioxidant defense. Am Rev Respir Dis 1989;140:531–554. [DOI] [PubMed] [Google Scholar]
- 33.Thompson AB, Robbins RA, Romberger DJ, Sisson JH, Spurzem JR, Teschler H, Rennard SI. Immunological functions of the pulmonary epithelium. Eur Respir J 1995;8:127–149. [DOI] [PubMed] [Google Scholar]
- 34.Hackett TL, Warner SM, Stefanowicz D, Shaheen F, Pechkovsky DV, Murray LA, Argentieri R, Kicic A, Stick SM, Bai TR, et al. Induction of epithelial–mesenchymal transition in primary airway epithelial cells from patients with asthma by transforming growth factor–beta1. Am J Respir Crit Care Med 2009;180:122–133. [DOI] [PubMed] [Google Scholar]
- 35.Zimmermann GS, Neurohr C, Villena-Hermoza H, Hatz R, Behr J. Anti-inflammatory effects of antibacterials on human bronchial epithelial cells. Respir Res 2009;10:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maunders H, Patwardhan S, Phillips J, Clack A, Richter A. Human bronchial epithelial cell transcriptome: gene expression changes following acute exposure to whole cigarette smoke in vitro. Am J Physiol Lung Cell Mol Physiol 2007;292:L1248–L1256. [DOI] [PubMed] [Google Scholar]
- 37.Thaikoottathil JV, Martin RJ, Zdunek J, Weinberger A, Rino JG, Chu HW. Cigarette smoke extract reduces VEGF in primary human airway epithelial cells. Eur Respir J 2009;33:835–843. [DOI] [PubMed] [Google Scholar]
- 38.Slepushkin VA, Staber PD, Wang G, McCray PB Jr, Davidson BL. Infection of human airway epithelia with H1N1, H2N2, and H3N2 influenza A virus strains. Mol Ther 2001;3:395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krunkosky TM, Jordan JL, Chambers E, Krause DC. Mycoplasma pneumoniae host–pathogen studies in an air–liquid culture of differentiated human airway epithelial cells. Microb Pathog 2007;42:98–103. [DOI] [PubMed] [Google Scholar]
- 40.Chen L, Song W, Davis IC, Shrestha K, Schwiebert E, Sullender WM, Matalon S. Inhibition of Na+ transport in lung epithelial cells by respiratory syncytial virus infection. Am J Respir Cell Mol Biol 2009;40:588–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halldorsson S, Gudjonsson T, Gottfredsson M, Singh PK, Gudmundsson GH, Baldursson O. Azithromycin maintains airway epithelial integrity during Pseudomonas aeruginosa infection. Am J Respir Cell Mol Biol 2010;42:62–68. [DOI] [PubMed] [Google Scholar]
- 42.Palermo LM, Porotto M, Yokoyama CC, Palmer SG, Mungall BA, Greengard O, Niewiesk S, Moscona A. Human parainfluenza virus infection of the airway epithelium: viral hemagglutinin–neuraminidase regulates fusion protein activation and modulates infectivity. J Virol 2009;83:6900–6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Booth BW, Sandifer T, Martin EL, Martin LD. IL-13–induced proliferation of airway epithelial cells: mediation by intracellular growth factor mobilization and ADAM17. Respir Res 2007;8:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gomperts BN, Kim LJ, Flaherty SA, Hackett BP. IL-13 regulates cilia loss and FOXJ1 expression in human airway epithelium. Am J Respir Cell Mol Biol 2007;37:339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laoukili J, Perret E, Willems T, Minty A, Parthoens E, Houcine O, Coste A, Jorissen M, Marano F, Caput D, et al. IL-13 alters mucociliary differentiation and ciliary beating of human respiratory epithelial cells. J Clin Invest 2001;108:1817–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim S, Schein AJ, Nadel JA. E-cadherin promotes EGFR-mediated cell differentiation and MUC5AC mucin expression in cultured human airway epithelial cells. Am J Physiol Lung Cell Mol Physiol 2005;289:L1049–L1060. [DOI] [PubMed] [Google Scholar]
- 47.Vermeer PD, Harson R, Einwalter LA, Moninger T, Zabner J. Interleukin-9 induces goblet cell hyperplasia during repair of human airway epithelia. Am J Respir Cell Mol Biol 2003;28:286–295. [DOI] [PubMed] [Google Scholar]
- 48.Lesimple P, van Seuningen SI, Buisine MP, Copin MC, Hinz M, Hoffmann W, Hajj R, Brody SL, Coraux C, Puchelle E. Trefoil factor family 3 peptide promotes human airway epithelial ciliated cell differentiation. Am J Respir Cell Mol Biol 2007;36:296–303. [DOI] [PubMed] [Google Scholar]
- 49.Sexton K, Balharry D, Berube KA. Genomic biomarkers of pulmonary exposure to tobacco smoke components. Pharmacogenet Genomics 2008;18:853–860. [DOI] [PubMed] [Google Scholar]
- 50.Woodruff PG, Boushey HA, Dolganov GM, Barker CS, Yang YH, Donnelly S, Ellwanger A, Sidhu SS, Dao-Pick TP, Pantoja C, et al. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci USA 2007;104:15858–15863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.