Abstract
Transforming growth factor (TGF)–β1 is a key mediator of lung remodeling and fibrosis. Epithelial cells are both a source of and can respond to TGF-β1 with epithelial-to-mesenchymal transition (EMT). We recently determined that TGF-β1–induced EMT in lung epithelial cells requires the presence of c-Jun N-terminal kinase (JNK) 1. Because TGF-β1 signals via Smad complexes, the goal of the present study was to determine the impact of JNK1 on phosphorylation of Smad3 and Smad3-dependent transcriptional responses in lung epithelial cells. Evaluation of JNK1-deficient lung epithelial cells demonstrated that TGF-β1–induced terminal phosphorylation of Smad3 was similar, whereas phosphorylation of mitogen-activated protein kinase sites in the linker regions of Smad3 was diminished, in JNK1-deficient cells compared with wild-type cells. In comparison to wild-type Smad3, expression of a mutant Smad3 in which linker mitogen-activated protein kinase sites were ablated caused a marked attenuation in JNK1 or TGF-β1–induced Smad-binding element transcriptional activity, and expression of plasminogen activator inhibitor–1, fibronectin-1, high-mobility group A2, CArG box–binding factor–A, and fibroblast-specific protein–1, genes critical in the process of EMT. JNK1 enhanced the interaction between Smad3 and Smad4, which depended on linker phosphorylation of Smad3. Conversely, Smad3 with phosphomimetic mutations in the linker domain further enhanced EMT-related genes and proteins, even in the absence of JNK1. Finally, we demonstrated a TGF-β1–induced interaction between Smad3 and JNK1. Collectively, these results demonstrate that Smad3 phosphorylation in the linker region and Smad transcriptional activity are directly or indirectly controlled by JNK1, and provide a putative mechanism whereby JNK1 promotes TGF-β1–induced EMT.
Keywords: transforming growth factor–β1, c-Jun N-terminal kinase 1, Smad3, epithelial-to-mesenchymal transition
CLINICAL RELEVANCE.
These findings provides new insights into the potential mechanism whereby c-Jun N-terminal kinase (JNK) 1 promotes fibrosis in the lung and suggest that JNK1 is a relevant therapeutic target in patients with tissue fibrosis.
The cytokine, transforming growth factor (TGF)–β, has been identified as one of the “master regulators” in induction of fibrosis in many tissues, including the lung (1). TGF-β can act upon many different cell types, including epithelial cells, wherein it was demonstrated to induce epithelial-to-mesenchymal transition (EMT). EMT represents the process whereby fully differentiated epithelial cells undergo transition to a mesenchymal phenotype, giving rise to fibroblasts and myofibroblasts, and has recently emerged as a process that is potentially critical in the development of fibrosis, including pulmonary fibrosis (2, 3).
TGF-β signals through two types of transmembrane serine/threonine (Ser/Thr) kinase receptors. Upon TGF-β binding, the type II receptor phosphorylates the type I receptor (4, 5). Subsequently, the receptor-activated Smads (R-Smads), Smad2 and -3, are phosphorylated by the activated TGF-β type I receptor on the C-terminal SSXS motif (6). The activated R-Smads form a complex with the common partner, Smad4, and are translocated into the nucleus (4), where they interact with other transcription factors to regulate the transcription of TGF-β–responsive genes, including connective tissue growth factor, α–smooth muscle actin (α-SMA), collagen1A, and plasminogen activator inhibitor (PAI)–1 (7, 8).
c-Jun N-terminal kinases (JNKs) are a member of a larger group of Ser/Thr protein kinases known as the mitogen-activated protein (MAP) kinase (MAPK) family, which can be activated by TGF-β through TGF-β–activated kinase 1 (9, 10). Besides a C-terminal phosphorylation region, Smad2 and Smad3 contain a proline- and serine-rich linker region, which contains phosphorylation sites for the MAPKs extracellular signal–regulated kinase (ERK) (PX(S/T)P), and the non-ERK MAPK sites (XX(S/T)P) for JNK and p38 MAPK (11). The functional implications of phosphorylation of the Smad2/3 in linker region by MAPKs, and the consequences for Smad transcriptional activity and gene expression remain unclear. For example ERK-mediated Smad2 linker phosphorylation resulted in nuclear export and subsequent reduced transcriptional activity (11–13). On the other hand, JNK-mediated phosphorylation in the linker region of Smad3 resulted in an enhanced hetero-oligomerization with Smad4, and enhanced nuclear translocation of the complex (14, 15).
Recent work in our laboratory identified JNK1 as a crucial mediator of TGF-β1–induced EMT (16) and lung fibrosis (17), and the importance of Smad3 in the development of tissue fibrosis is well established (18, 19). These collective findings led us to hypothesize that JNK activation induced by TGF-β causes Smad3 linker phosphorylation, and that this promotes Smad3/4-dependent transcriptional activation of genes associated with EMT.
MATERIALS AND METHODS
Cell Culture
Primary mouse tracheal epithelial (MTE) cells were isolated and cultured according to previously published methods (16, 20). A line of spontaneously transformed mouse alveolar type II epithelial cells (C10) (21) was cultured in CRML-1066 medium, containing 50 U/ml penicillin–50 μg/ml streptomycin, 2 mM L-glutamine, and 10% FBS, all from GIBCO-BRL (Carlsbad, CA). When applicable, in both culture systems, recombinant human TGF-β1 (R&D Systems, Minneapolis, MN) was added for the indicated time points. All chemicals used were purchased from Sigma-Aldrich (St. Louis, MO), unless otherwise noted.
Western Blotting
MTE or C10 whole-cell lysates were prepared by the addition of lysis buffer (20 mM Tris, 150 mM NaCl, 1% [vol/vol] Nonidet P-40, 1 mM DTT, 1% [vol/vol] Protease Inhibitor Cocktail, 1% [vol/vol] Phosphatase Inhibitor Cocktail). Total protein was assessed by the Bio-Rad DC Protein Assay kit (Bio-Rad, Hercules, CA), according to manufacturer's instructions and 5–30 μg of protein was loaded. α-SMA antibody was purchased from Sigma. Full-length JNK1/2, Smad3, C-terminal phospho-Smad3, histone H3 (all from Cell Signaling Technology, Danvers, MA) S208Linker Pospho-Smad3 (kindly provided by Dr. F. Liu), E-cadherin (Invitrogen, Carlsbad, CA), Smad4, α-tubulin, fibroblast-specific protein (FSP)–1, and β-actin (all Santa Cruz biotechnology, Santa Cruz, CA) protein abundance was evaluated by western blotting.
α-Smad3, 4 and α-Phospho-Serine Immunoprecipitation
Wild-type (WT) or JNK1-deficient (−/−) cells were lysed in 50 mM Tris, 150 mM NaCl, 10% (vol/vol) glycerol 0.5% (vol/vol) Nonidet P-40, 1 mM EDTA, 1 mM DTT, 1% (vol/vol) Protease Inhibitor Cocktail, 1% (vol/vol) Phosphatase Inhibitor Cocktail, 500 μM NaF, 100 μM β-glycerophosphate, 100 μM sodium–pyro-PO4, 10 μg/ml leupeptin, and 1% (vol/vol) aprotinin. An input of 100 μg of total protein was precleared with protein G-sepharose (Invitrogen), and subsequently incubated with 1 μg anti-Smad3, 2 μg of anti-Smad4, 1 μg of anti–phospho-serine (Biocompare, San Francisco, CA) or IgG control (Jackson ImmunoResearch Laboratories, Westgrove, PA), followed by the addition of protein G-sepharose. Western blotting was used as described previously here to assess Smad3, S208 linker pospho-Smad3, and Smad4 levels.
JNK1 and Smad3 siRNA
C10 cells were incubated with Dharmacon SMARTpool control nontargeting small interfering (si)RNA (100 nM) or Dharmacon SMARTpool siRNA specific against JNK1, Smad2, or Smad3 (100 nM) (Dharmacon, Lafayette, CO), and subsequently transfected, and exposed to TGF-β1 for the indicated time points for evaluation by Western blotting or Taqman analysis.
Transfections and Plasmids
Transient transfections were performed using Nanofectin (PAA, Pasching, Austria) according to manufacturer's instructions. Smad-binding element (SBE) 4–luciferase (0.25 μg) was kindly provided by Dr. B. Vogelstein (Johns Hopkins Kimmel Cancer Center, Baltimore, MD). Plasmids encoding Smad3, Smad4, and ΔSmad4 (1–514) were kindly provided by Dr. J. Massague (Memorial Sloan-Kettering Cancer Center, New York, NY). Plasmids encoding Smad3–Erk/Pro-directed kinase site mutant (EPSM), and Smad3-phosphomimetic (PM; containing mutations of T179E, S204D and S208D) were kindly provided by Dr. K. Matsuzaki (Kansai Medical University, Osaka, Japan) and Dr. F. Liu (Center for Advanced Biotechnology and Medicine, State University of New Jersey, Piscataway, NJ), respectively. pSV–β-gal (0.25 μg) was employed to correct for transfection efficiency. Luciferase (Promega, Madison, WI) and β-galactosidase (Tropix, Bedford, MA) were measured according to manufacturer's instructions.
Gene Expression
Total RNA was isolated from C10 cells using the RNeasy mini-kit (Qiagen, Valencia, CA), subjected to reverse transcription and DNase treatment to produce cDNA for Taqman gene analysis using SYBR green or Assays on Demand for the individual target genes (Applied Biosystems, Foster City, CA). Primer sequences are provided in Table 1. Sequences were taken from Genbank. All accession numbers are denoted.
TABLE 1.
PRIMER SEQUENCES
| Gene | Accession No. | Sequences (5′ → 3′) | Amplicon (bp) | |
|---|---|---|---|---|
| Hmga2 | NM_010441 | Forward | AAGGCAGCAAAAACAAGAGC | 121 |
| Reverse | GCAGGCTTCTTCTGAACGAC | |||
| CBF-A | L36663 | Forward | GGGAAAAATGTTCGTTGGTG | 130 |
| Reverse | CCCTCTTGATCGTCCAGTGT | |||
| Fsp-1 | NM_0113111 | Forward | CTGGGGAAAAGGACAGATGA | 109 |
| Reverse | TGCAGGACAGGAAGACACAG | |||
| E-cadherin | NM_009864.2 | Forward | AGCCATTGCCAAGTACATCC | 133 |
| Reverse | AAAGACCGGCTGGGTAAACT | |||
| PAI-1 | NM_008871 | Forward | AGTCTTTCCGACCAAGAGCA | 105 |
| Reverse | GACAAAGGCTGTGTGGAGGAAG | |||
| Fn1 | NM_010233 | Forward | GTGTAGCACAACTTCCAATTACGAA | 90 |
| Reverse | GGAATTTCCGCCTCGAGTCT |
Primer sequences for quantitative PCR cycling conditions with similar efficiencies to obtain simultaneous amplification in the same run. Sequences were taken from GeneBank; all accession numbers are denoted.
Statistical Analysis
Data were evaluated by one-way ANOVA using the Tukey Honestly Significant Difference test to adjust for multiple comparisons. Results with a P value less than 0.05 or smaller were considered statistically significant.
RESULTS
Smad3 Linker Phosphorylation in Lung Epithelial Cells Stimulated with TGF-β1 Requires JNK1
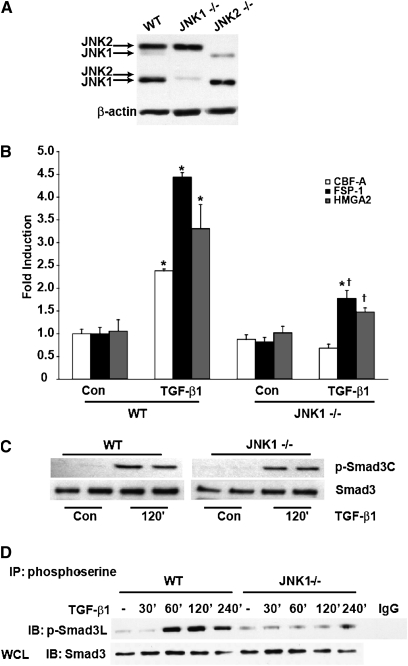
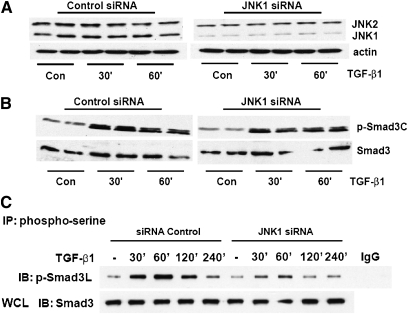
We recently elucidated the importance of JNK1 in TGF-β1–induced EMT (16). Specifically, we demonstrated that TGF-β1–induced Smad DNA-binding activity and induction of mesenchymal genes associated with EMT were diminished in JNK1−/− MTE cells compared with WT cells. We first sought to extend upon those observations in WT and JNK1−/– MTE cells. Data in Figure 1A confirm the JNK1−/− genotype. Note that two isoforms of JNK1 and JNK2 are expressed in WT MTE cells and C10 cells (data not shown). Hereafter, only the major bands of JNK1 and -2 will be indicated. We next analyzed mRNA expression of the EMT transcriptional regulators, high-mobility group A2 (HMGA2) (22), and CArG box-binding factor-A (CBF-A) (23), and the EMT marker, FSP-1 (23), comparatively in WT and JNK1−/− MTE cells. Results in Figure 1B demonstrate that TGF-β1–induced expression of HMGA2 was substantially decreased in JNK1−/− MTE cells compared with WT counterparts, consistent with our previous data (16). Evaluation of mRNA expression of the EMT regulator, CBF-A, and the EMT marker protein, FSP-1, also demonstrated significant attenuations in response to TGF-β1 in JNK1−/− cells as compared with WT groups (Figure 1B). These findings demonstrate that, in cells lacking JNK1, expression of EMT regulatory genes is markedly attenuated. To address further the mechanisms whereby JNK1 exerts its role in promoting EMT, we evaluated the impact of Smad3 phosphorylation in WT cells, or cells lacking JNK1. After stimulation with TGF-β1, Smad3 terminal phosphorylation was equal in JNK1−/− MTE cells compared with WT cells (Figure 1C), in agreement with our previous work (16). Because TGF-β1 can also result in phosphorylation of R-Smads at their linker regions, which was reported to depend on JNK (14), we examined the magnitude of the linker phosphorylation of Smad3 in WT and JNK1−/− MTE cells. Immunoprecipitation of serine-phosphorylated proteins, followed by assessment of Smad3 linker phosphorylation by Western blot analysis, revealed that TGF-β1–induced phosphorylation of Smad3 in the linker region was significantly attenuated in JNK1−/− MTE cells compared with cells isolated from WT mice, which showed robust linker phosphorylation of Smad3 in response to TGF-β1 (Figure 1D). To exclude the possibility that the observed reduction of Smad3 linker phosphorylation in JNK1−/− MTE cells originated from a compensatory mechanism due to a chronic lack of JNK1, we ablated JNK1 acutely in a line of lung alveolar epithelial cells using an siRNA-mediated approach (Figure 2A). Similar to the findings in MTE cells, TGF-β1–induced Smad3 terminal phosphorylation was largely unaffected in cells treated with siRNA for JNK1 compared with siRNA control cells (Figure 2B). In contrast, SiRNA-mediated knockdown of JNK1 diminished TGF-β1–induced Smad3 linker phosphorylation significantly compared with control siRNA groups (Figure 2C), in agreement with the observations in JNK1−/− MTE cells (Figure 1D). These results demonstrate that Smad3 phosphorylation in the linker region is increased in response to TGF-β1 and requires the presence of JNK1.
Figure 1.
Epithelial-to-mesenchymal transition (EMT) gene expression, Smad 3 terminal and linker phosphorylation in wild-type (WT) and c-Jun N-terminal kinase (JNK) 1–deficient (−/−) mouse tracheal epithelial (MTE) cells after exposure to transforming growth factor (TGF)–β1. (A) Confirmation of the JNK1−/− genotype in MTE cells by Western blot analysis of total JNK. Lysates were prepared from WT, JNK1−/−, and JNK2−/− MTE cells for comparative evaluation of JNK1 and -2 isoforms; β-actin was used as loading control. (B) Assessment of TGF-β1–mediated increases of EMT regulatory genes in WT and JNK1−/− MTE cells. WT or JNK1−/− MTE cells were cultured in the presence or absence of 5 ng/ml of TGF-β1 for 24 hours, and total RNA was isolated for evaluation of CArG box–binding factor-A (CBF-A), fibroblast-specific protein (FSP)–1, and high-mobility group A2 (HMGA2) by real-time PCR analysis. mRNA abundance was normalized to β-actin. Results are expressed as fold change compared with WT sham controls, and reflect mean values (±SEM) (n = 3). *P < 0.05 (ANOVA) compared with WT sham control groups; †P < 0.05 compared with TGF-β1–exposed WT cells. (C) Assessment of Smad3 C-terminal phosphorylation induced by TGF-β1 in WT or JNK1−/− MTE cells. Cells were exposed to 5 ng/ml TGF-β1 for the indicated time points, cell lysates were prepared, and equal amounts of protein (20 μg) were separated by SDS-PAGE and subjected to Western blot analysis to assess C-terminal phosphorylated and total Smad3 content. (D) Evaluation of Smad3 linker phosphorylation in WT or JNK1−/− MTE cells exposed to TGF-β1 (5 ng/ml) for the indicated times. Phospho-serine–containing proteins were immunoprecipated from cells at the indicated times, and immunoprecipitates subjected to Western blot analysis to assess Smad3 phosphorylation of serine 208 in the linker region. IgG: control sample wherein preimmune IgG was used in the immunoprecipitation. Total Smad3 content in whole-cell lysates (WCL) was used as a loading control. Shown is a representative example of three independent experiments. con, control; IB, Immunoblot; IP, immunoprecipitated.
Figure 2.
Smad3 terminal and linker phosphorylation after acute ablation of JNK1 in C10 cells. C10 cells transfected with control or JNK1-specific small interfering (si)RNA (100 nM) were exposed to 5 ng/ml of TGF-β1 for the indicated time points. (A) Confirmation of JNK1 knockdown evaluated by Western blotting. β-actin was used as loading control. (B) C10 cell lysates were prepared and equal amounts of protein (20 μg) were separated by SDS-PAGE and subjected to Western blot analysis to assess C-terminal phosphorylated Smad3 and total Smad 3 content. (C) Assessment of serine 208-phosphorylated Smad3 content in cells treated with control or JNK1-specific siRNAs after stimulation with TGF-β1 for the indicated time points. Phospho-serine containing proteins were immunoprecipitated from cells at the indicated times, and immunoprecipitates subjected to Western blot analysis to assess Smad3 phosphorylation of serine 208 in the linker region. IgG: control sample wherein preimmune IgG was used in the immunoprecipitation. Total Smad3 content in whole-cell lysates (WCL) was used as a control. Shown are representative data of three independent experiments.
Expression of a MKK7/JNK1 Fusion Protein Enhances SBE Transcriptional Activity in a Smad4-Dependent Manner
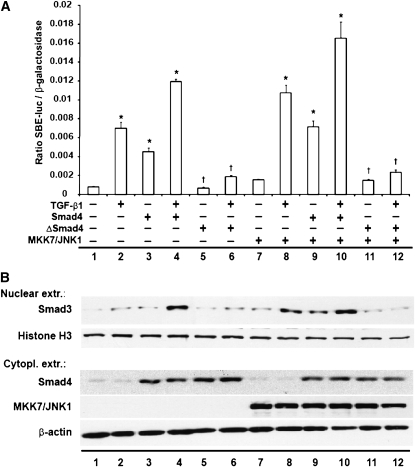
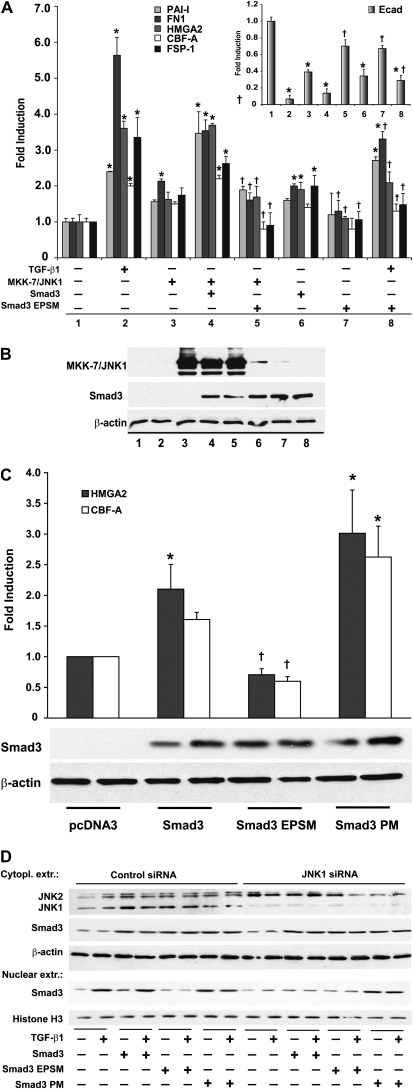
Mitogen-activated protein kinase kinase 7 (MKK7) is a specific activator of JNK (24), and fusion of MKK7 to JNK1 causes constitutive activation of JNK. Indeed, overexpression of an MKK7/JNK1 fusion protein in various cells, including lung epithelial cells, results in a significant increase of c-jun transcriptional activity (Ref. 25, and data not shown). To assess the role of JNK1 in the Smad signaling pathway, transactivation of an SBE promoter reporter construct, which preferentially recognizes Smad3/4 heterodimers (26), was evaluated. Expression of MKK7/JNK1 alone slightly increased SBE promoter transactivation compared with control transfected cells (Figure 3A). Expression of MKK7/JNK1, together with Smad4, significantly increased SBE transcriptional activity, which was further increased in the presence of TGF-β1. In contrast, expression of MKK7/JNK1, together with a transcriptionally inactive variant of Smad4 (ΔSmad4), failed to enhance further SBE transcriptional activity over levels observed after expression of MKK7/JNK1, and attenuated the ability of TGF-β1 to increase SBE transcriptional activity. Similar results were obtained using an artificial PAI-1 promoter luciferase construct (data not shown). Collectively, these results show that active JNK1 controls Smad3/4-dependent transcriptional activity. To evaluate further the role of JNK1 in the regulation of Smad3/4, we evaluated, in the same experiment, nuclear translocation of the Smad4 binding partner, Smad3 (Figure 3B). Expression of MKK7/JNK1 (lane 7) or Smad4 alone (lane 3) led to a moderate increase in nuclear Smad3, compared with sham controls (lane 1), as assessed via Western blotting of nuclear fractions (Figure 3B). However, expression of MKK7/JNK1 together with Smad4 resulted in a significant increase of nuclear Smad3 (lane 9), which was further increased in the presence of TGF-β1 (lane 10). In contrast, expression of MKK7/JNK1 together with ΔSmad4 failed to enhance Smad3 nuclear translocation (lane 11), and, in the presence of TGF-β1, no further increases in nuclear Smad3 were observed (lane 12). On the contrary, the increases in nuclear Smad3 observed in MKK7/JNK1-expressing cells that were stimulated with TGF-β1 (lane 8) were not observed in the presence of ΔSmad4 (lane 12). Bottom panels of Figure 3B confirm expression of the various constructs. In aggregate, these results demonstrate that active JNK1 regulates Smad3/4 nuclear translocation, and enhances the stimulatory effects on Smad3/4 transcriptional responses induced by TGF-β1.
Figure 3.
Expression of Mitogen-activated protein kinase kinase 7 (MKK7)/JNK1 fusion protein enhances Smad-binding element (SBE) transcriptional activity in a Smad4-dependent manner. (A) SBE promoter–luciferase reporter and β-gal (0.15 μg each) constructs were transfected in C10 cells together with plasmids encoding MKK7/JNK1 (1 μg), Smad4 (1 μg), or ΔSmad4 (1 μg). After 24 hours, cells were stimulated with TGF-β1 (5 ng/ml). At 48 hours after transfection, cells were lysed to measure luciferase and β-gal activities. Shown are representative graphs of three independent experiments. Data are expressed as luciferase units corrected for β-gal, and values are expressed as means (±SEM) (n = 3). *P < 0.05 (ANOVA) compared with control (lane 1). †P < 0.05 compared with respective Smad4 groups (lane 5 compared with lane 3, lane 6 compared with lane 4, lane 11 compared with lane 9, and lane 12 compared with lane 10). (B) Assessment of Smad3 in nuclear extracts via Western blot analysis. Histone H3 was used as loading control. MKK7/JNK1 and Smad4 content was assessed in cytoplasmic extracts to confirm expression of constructs after transfection. Shown are representative data of three independent experiments.
Expression of a Smad3 Linker Mutant Ablates JNK1-Dependent SBE Transcriptional Activity
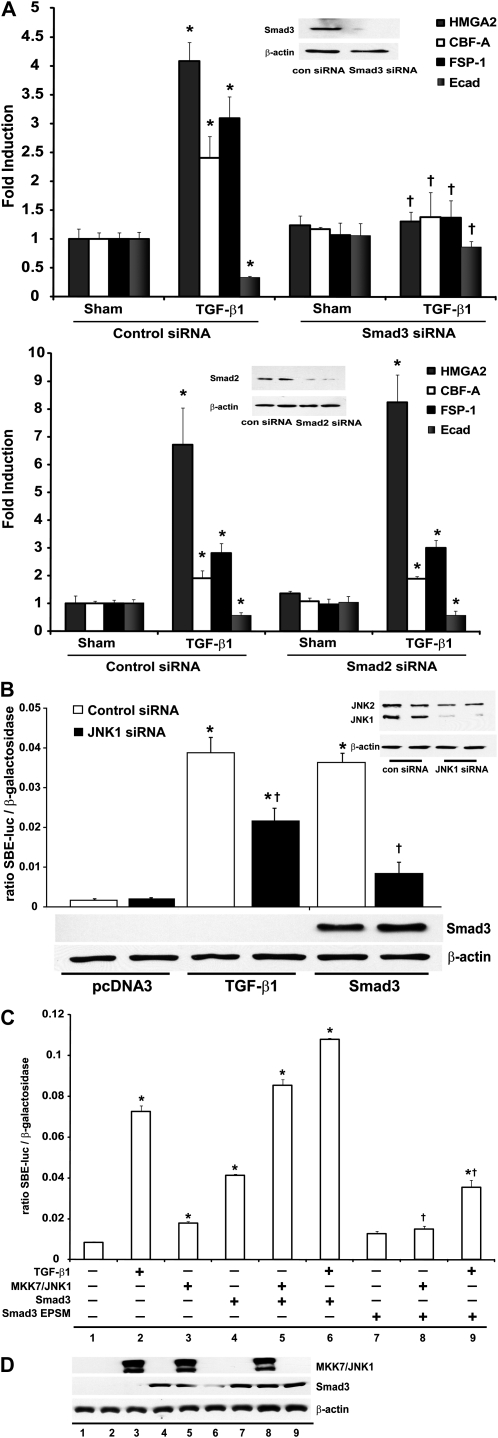
The respective roles of Smad2 and -3 in TGF-β1–induced EMT remain to be fully elucidated. Primarily Smad3 has been associated with regulating EMT and tissue fibrosis (19). However, Smad2 has also been linked to EMT (27). To examine the relative importance of the different receptor Smads, Smad2 or -3 were ablated using an siRNA approach. Results in Figure 4A demonstrate that ablation of Smad3 (top), but not Smad2 (bottom), attenuated the TGF-β1–induced mRNA expression of the EMT regulators HMGA2, CBF-A, and FSP-1. Furthermore, siRNA-mediated ablation of Smad3, but not Smad2, attenuated the TGF-β1–induced down-regulation of E-cadherin mRNA compared with cells transfected with control siRNA. These findings confirm the functional importance of Smad3, but not Smad2, in TGF-β–induced EMT transcriptional program in C10 lung epithelial cells. Based upon our findings so far, demonstrating the importance of Smad3 and JNK1 in EMT, the requirement of JNK1 in augmenting Smad3 linker phosphorylation after TGF-β1, the stimulatory effect of active MKK7/JNK1 on Smad4-dependent transcription, and nuclear translocation of Smad3, we chose to evaluate further the role of JNK1 in regulating Smad3-dependent transcriptional activity and EMT. Results in Figure 4B demonstrate that TGF-β–induced SBE transcriptional activity was significantly attenuated by ablation of JNK1 in lung epithelial cells using an siRNA compared with control siRNA transfected cells. Interestingly, the ability of Smad3 to induce SBE transcriptional activity was also markedly decreased after siRNA-mediated ablation of JNK1 (Figure 4B), highlighting the role of endogenous JNK1 in regulating Smad3-dependent transcriptional responses. To investigate further the role of JNK1 in this context, we expressed MKK7/JNK1 together with Smad3. Coexpression of MKK7/JNK1 and Smad3 resulted in a synergistic increase of SBE transcriptional activity compared with expression of individual constructs (Figure 4C). However, coexpression of MKK7/JNK1 and a mutant form of Smad3 (Smad3-EPSM), which cannot be phosphorylated in the linker region (14), failed to activate SBE transcriptional activity robustly compared with vector control transfected cells. Moreover, TGF-β1–induced SBE transcriptional activity was also markedly attenuated after overexpression of Smad3-EPSM (Figure 4C), and similar results were obtained using a PAI-1 promoter construct (data not shown). Western blot analyses verified depletion of Smad2, Smad3, and JNK1 after siRNA (Figures 4A and 4B, insets), expression of MKK7/JNK1, and Smad3 constructs (Figure 4D). Collectively, these data suggest that JNK1 plays a role in TGF-β1–induced SBE transcriptional activity through Smad3 linker phosphorylation.
Figure 4.
Expression of a Smad3 linker mutant cDNA ablates JNK1-dependent SBE transcriptional activity (A) C10 cells transfected with control, Smad3-, or Smad2-specific siRNAs (100 nM) were treated with TGF-β1 (5 ng/ml, 24 h). Total RNA was isolated to assess mRNA abundance of HMGA2, CBF-A, FSP-1, and E-cadherin by quantitative PCR (Q-PCR). mRNA abundance was normalized to β-actin. Results are expressed as fold change compared with sham controls and reflect means (±SEM) (n = 3). Inset: confirmation of Smad3 and Smad2 knockdown via Western blot analysis of Smad3 or Smad2. β-actin was used as loading control. *P < 0.05 (ANOVA) compared with sham control siRNA groups; †P < 0.05 compared with TGF-β1–exposed control siRNA transfected groups. (B) C10 cells transfected with control or JNK1-specific siRNAs (100 nM), followed by transfection of a SBE-sensitive promoter–luciferase reporter, β-gal (0.15 μg), in the presence or absence of Smad3 (1 μg), were lysed to measure luciferase and β-gal activity. Data are expressed as luciferase units corrected for β-gal, and values are expressed as means (±SEM) (n = 3). *P < 0.05 compared with pcDNA3 control; †P < 0.05 compared with respective control siRNA groups, as determined by one-way ANOVA. Lower panel represents confirmation of expression of Smad3 via Western blot analysis. Inset represents confirmation of JNK1 knockdown via Western Blot analysis. (C) SBE-sensitive promoter–luciferase reporter and β-gal (0.15 μg each) constructs were transfected in C10 cells, together with plasmids encoding MKK7/JNK1 (1 μg), Smad3 (1 μg), or Smad3–extracellular signal–regulated kinase (ERK)/Pro-directed kinase site mutant (EPSM) (1 μg). At 36 hours after transfection, cells were lysed to measure luciferase and β-gal activity. Shown are representative graphs of three independent experiments. Data are expressed as described in (A). *P < 0.05 compared with the control (lane 1); †P < 0.05 (lane 8 compared with lane 5 and lane 9 compared with lane 6, as determined by one-way ANOVA). (D) Confirmation of MKK7/JNK1, Smad3, and Smad3-EPSM protein content in the treatment groups shown in (B) via Western blot analysis.
Requirement for Smad3 Linker Phosphorylation in the Induction of TGF-β1–Induced Expression of Genes Important in EMT
Several transcriptional regulators involved in the induction of EMT have been identified (23, 28). Because we recently demonstrated the requirement of JNK1 in the induction of EMT (16), and that JNK1 controls Smad3-dependent transcriptional activity, we next verified the requirement of Smad3 linker phosphorylation in the induction of EMT regulators in lung epithelial cells. We expressed a WT Smad3, Smad3-EPSM, or a Smad3 construct with PM mutations in the linker region (Smad3-PM) in the presence or absence of MKK-7/JNK1 and stimulation with TGF-β1. Results in Figure 5A demonstrate slight increases in mRNA expression of PAI-1, fibronectin (FN)-1, HMGA2, CBF-A, and FSP-1 in cells expressing MMK7/JNK1, and similar increases were seen after overexpression of Smad3. Coexpression of MKK7/JNK1 and Smad3 led to significant increases in mRNA levels of PAI-1, FN-1, HMGA2, CBF-A, and FSP-1, which were completely abrogated in cells coexpressing MKK7/JNK1 and the Smad3 linker mutant, Smad3-EPSM. Importantly, expression of Smad3-EPSM also significantly attenuated TGF-β1–induced mRNA expression of FN-1, HMGA2, CBF-A, and FSP-1 (Figure 5A). As expected, mRNA content of the epithelial marker, E-cadherin, was markedly decreased in response to TGF-β1, and decreases in E-cadherin mRNA were also apparent in cells expressing MKK7/JNK1. Expression of the Smad3 linker mutant, Smad3-EPSM, partially restored E-cadherin levels (Figure 5A inset; compare lane 8 to lane 2 and lane 5 to lane 3), demonstrating the importance of Smad3 linker phosphorylation in the loss of the epithelial phenotype in response to TGF-β1, or expression of MKK7/JNK1, respectively. Results in Figure 5B confirm expression of various constructs. To address whether Smad3 linker phosphorylation is sufficient in the induction of EMT transcriptional regulators, we expressed a Smad3 construct with PM mutations in the linker region (Smad3-PM). Results in Figure 5C demonstrate that mRNA levels of HMGA2 and CBF-A were increased in cells expressing Smad3-PM compared with WT Smad3, whereas expression of Smad3-EPSM failed to increase mRNA expression of these EMT regulators above control levels, consistent with earlier observations. These findings demonstrate a direct link between Smad3 linker phosphorylation status and activation of an EMT transcriptional program.
Figure 5.
Expression of Smad3 linker mutant that lacks mitogen-activated protein kinase (MAPK) phosporylation sites reduces TGF-β1–induced Smad3 nuclear translocation and attenuates JNK1-dependent expression of EMT transcriptional regulators and mesenchymal genes. (A) Total RNA was isolated from C10 cells treated with TGF-β1 (24 h) or transfected with MKK7/JNK1 (1 μg), Smad3 (1 μg), or Smad3 linker-mutant (EPSM) (1 μg). mRNA abundance of plasminogen activator inhibitor (PAI)–1, fibronectin (FN)-1, HMGA2, CBF-A, FSP-1, and E-cadherin (Ecad; inset) was assessed by Q-PCR. mRNA abundance was normalized to β-actin content. Results are expressed as fold change compared with sham control (lane 1) and reflect mean values (±SEM) (n = 3). *P < 0.05 (ANOVA) compared with lane 1; †P < 0.05 lane 5 compared with lane 4, lane 7 compared with lane 6, and lane 8 compared with lane 2. (B) Confirmation of MKK7/JNK1, Smad3, and Smad3-EPSM protein content by Western blot analysis. β-actin was used as loading control. (C) Assessment of mRNA abundance of HMGA2 and CBF-A by Q-PCR in C10 cells transfected with Smad3, Smad3-EPSM, or Smad3-phosphomimetic (PM; 1 μg each). mRNA abundance was normalized to β-actin. Results are expressed as fold change compared with pcDNA3 transfected controls, and reflect means (±SEM) (n = 3). *P < 0.05 (ANOVA) compared with pcDNA3 controls; †P < 0.05 compared with Smad3. Lower panel: confirmation of expression of Smad3 constructs by Western blot analysis. β-actin was used as loading control. (D) Assessment of Smad3 in nuclear extracts via Western Blot analysis. C10 cells transfected with control or JNK1-specific siRNAs (100 nM), followed by transfection with plasmids encoding Smad3, Smad3 linker-mutant (EPSM), or Smad3-PM (1 μg each). After 24 hours, cells were stimulated with TGFβ-1 (5 ng/ml) for an additional 24 hours before the isolation of nuclear extracts. Bottom panels: nuclear content of Smad3, and Histone H3 as a loading control. Top panels: confirmation of JNK1 knockdown via Western Blot analysis. β-actin was used as loading control.
We next evaluated the role of Smad3 linker phosphorylation and JNK1 in TGF-β1–induced nuclear translocation of Smad3. As expected, clear increases in nuclear Smad3 content were observed after stimulation of cells with TGF-β1, and similar findings were observed in cells expressing Smad3 (Figure 5D). In cells expressing Smad3-EPSM, TGF-β1–induced increases in nuclear Smad3 were abolished. Conversely, in cells expressing Smad3-PM, nuclear presence of Smad3 was markedly enhanced even in the absence of TGF-β1. To confirm the role of JNK1 in Smad3 nuclear translocation, JNK1 was ablated via siRNA. Indeed, loss of JNK1 resulted in decreased TGF-β1–induced nuclear translocation of Smad3, and similar results were apparent in cells expressing WT Smad3 that were stimulated with TGF-β1. In contrast, in cells expressing Smad3-PM, nuclear content of Smad3 remained elevated, even in the absence of JNK1. Collectively, these results demonstrate that the presence of JNK1 is required for nuclear translocation of Smad3, and that this requires linker phosphorylation of Smad3.
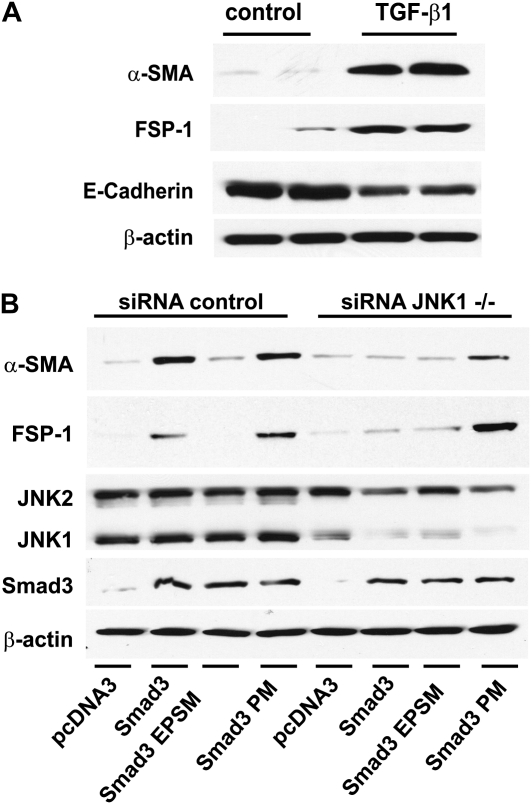
We next confirmed that TGF-β1 induces EMT in the C10 lung epithelial cell line by monitoring the content of mesenchymal and epithelial proteins. Indeed, results in Figure 6A demonstrate increases in α-SMA and FSP-1 content, and decreased expression of E-Cadherin in response to exposure to TGF-β1, consistent with EMT. We next assessed whether expression of Smad3 would be sufficient to drive EMT-specific protein expression. Indeed, after overexpressing of Smad3 or Smad3-PM, α-SMA and FSP-1 protein abundance was increased (Figure 6B). In contrast, expression of Smad3-EPSM at equivalent levels failed to increase α-SMA and FSP-1 protein content, consistent with the differential abilities of these constructs to induce EMT-linked gene expression (Figure 5D).
Figure 6.
Expression of EMT proteins in C10 cells exposed to TGF-β1, and the requirement of JNK1, and Smad3 linker phosphorylation, in Smad3-induced EMT. (A) C10 cells were treated with 5 ng/ml TGF-β1 for 48 hours. Cell lysates were prepared for assessment of α–smooth muscle actin (SMA), FSP-1, and E-cadherin content by Western blotting. β-actin was used as loading control. (B) C10 cells were transfected with control or JNK1-specific siRNAs (100 nM), and 24 hours later transfected with pcDNA3, Smad3, Smad3-EPSM, or Smad3-PM (all 1 μg). After 24 hours, lysates were prepared, and equal amounts of protein (20 μg) were separated by SDS-PAGE and subjected to Western blot analysis to assess α-SMA, FSP-1, JNK1/2, and Smad3 content. β-actin was used as loading control. Shown are data representative of three independent experiments.
To address the functional importance of JNK1 in regulating Smad3-induced EMT, we ablated JNK1 with siRNA. Results in Figure 6B demonstrate that the ability of Smad3 to increase α-SMA and FSP-1 content was almost completely abrogated after siRNA-mediated ablation of JNK1. In contrast, ablation of JNK1 did not affect the ability of Smad3-PM to increase levels of α-SMA and FSP-1, suggesting that PM mutations in the linker domain of Smad3 bypass the requirement of JNK1 in the induction of these parameters of EMT. Collectively, these results demonstrate that JNK1 can regulate mRNA expression and content of EMT regulators and mesenchymal markers through linker phosphorylation of Smad3.
JNK Associates with Smad3 and Increases Binding of Smad3 and Smad4 through Phosphorylation of Smad3 in the Linker Region
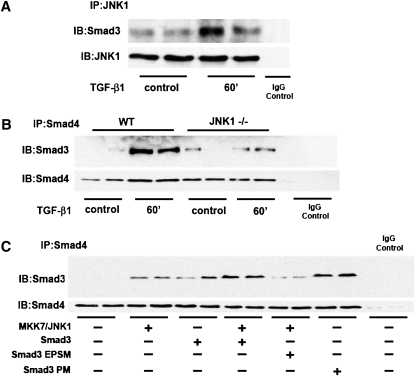
Given our findings that JNK1 controls Smad3/4 transcriptional responses via Smad3 linker phosphorylation, we next determined whether JNK1 interacted with Smad3. Results in Figure 7A demonstrate that immunoprecipitation of JNK in WT MTE cells resulted in pulldown of Smad3, and that after exposure to TGF-β1, the association between Smad3 and JNK1 was enhanced. Immunoprecipitation of Smad4, followed by Western blot analysis of Smad3, demonstrated marked increases in Smad3 and Smad4 association in WT cells exposed to TGF-β1. In contrast, the TGF-β1–induced association between Smad3 and Smad4 was largely abrogated in MTE cells lacking JNK1 (Figure 7B), suggesting that the Smad3 and Smad4 complex formation requires JNK1. Furthermore, expression of MKK7/JNK1 or Smad3 alone increased Smad3 association with Smad4 (Figure 7C). Coexpression of MKK7/JNK1 with Smad3 further increased this association, whereas coexpression with the Smad3 linker mutant, Smad3-EPSM, reduced Smad3/Smad4 association to basal levels. Finally, expression of Smad3-PM alone increased Smad3/Smad4 association, confirming that Smad3 linker phosphorylation is sufficient to enhance the interaction between Smad3 and Smad4 (Figure 7C). Collectively, these findings show that JNK1 can control phosphorylation of Smad3 in the linker region, which subsequently promotes association of Smad3 with Smad4, leading to increased expression of genes important in the regulation of EMT.
Figure 7.
JNK1 associates with Smad3 and increases Smad3/Smad4 binding through phosphorylation of the linker region of Smad3. MTE cells were cultured with or without TGF-β1 for indicated time points, and total cell lysates were prepared. (A) JNK1 was immunoprecipitated, and immunoprecipitated samples were subjected to Western blot analysis to assess Smad3. JNK1 abundance was assessed to ensure immunoprecipitation of JNK1. IgG control lane reflects sample that was incubated with preimmune IgG control antibody. (B). WT or JNK1−/− MTE cells were incubated with 5 ng/ml of TGF-β1 for 60 minutes. Smad4 was immunoprecipitated, and immunoprecipitated samples were subjected to Western blot analysis to assess Smad3 abundance. IgG control lane reflects sample that was incubated with preimmune IgG control antibody. Lower panel: Smad4 content was assessed to verify immunoprecipitation of Smad4. (C) C10 cells were transfected with combinations of MKK7/JNK1, Smad3, Smad3-EPSM, or Smad3-PM (1 μg each). Smad4 was immunoprecipitated, and immunoprecipitated samples were subjected to Western blot analysis to assess Smad3 abundance. Lower panel: Smad4 content was assessed to verify immunoprecipitation of Smad4. IgG control lane reflects sample that was incubated with preimmune IgG control antibody.
DISCUSSION
A number of processes and cell types have been linked to the development of lung fibrosis, and evidence for a role for EMT herein is accumulating (29). The profibrotic cytokine, TGF-β1, plays a major role in fibrogenesis, and has been demonstrated to be one of the cardinal cytokines that induces EMT (2, 3). The molecular details whereby TGF-β regulates the EMT process remain enigmatic. A substantial number of TGF-β1 effecter functions require signaling via Smads, and TGF-β1–induced tissue fibrosis requires Smad3 (18). In the present study, we demonstrate that presence of Smad3 is required for the induction of the EMT-linked genes, HMGA2, CBF-A, and FSP-1, by TGF-β1, consistent with previous observations demonstrating the requirement of Smad3 in EMT in the eye, kidney, and lung (30–32). Our findings do not support a causal role for Smad2 in TGF-β1–induced EMT, which is in agreement with another report in proximal tubular epithelial cells (33). Nonetheless, in alveolar type II epithelial cells, TGF-β was shown to induce formation of stable complexes between the tyrosine 654 phosphorylated version of β-catenin, and phosphorylated Smad2, in association with EMT-related protein expression (27). In contrast, other studies demonstrate that loss of Smad2 is in fact permissive for EMT (34, 35). Overall, these findings clearly demonstrate that Smad2 and -3 clearly have nonredundant roles in the regulation of EMT, findings that warrant further study.
Recently, our laboratory demonstrated the pivotal importance of the JNK1 signaling pathway in augmenting the expression of profibrotic mediators in lung fibrosis, and identified JNK1 as a crucial amplifier of TGF-β1–induced EMT (16, 17). The results presented in the current study extend those observations, and demonstrate that JNK1 is critical in Smad3-induced EMT, and that JNK1 promotes Smad3-dependent transcriptional activation of genes important in the regulation of EMT by promoting phosphorylation in the Smad3 linker regions. These findings provide new insights into the potential mechanisms whereby JNK1 promotes TGF-β1–induced EMT. Numerous studies have demonstrated evidence for crosstalk between JNK and TGF-β1 signaling pathways. For example, TGF-β–activated kinase 1 can mediate the activation of JNK1 (14, 36). Indeed, it has been reported that JNK activation by TGF-β occurs rapidly, and that JNK phosphorylates Smad3 outside its −SSXS motif, which facilitates Smad3 activation by the TGF-β receptor complex, leading to enhanced transcriptional responses (9). Phosphorylation of the Smad3 linker domain by other kinases, such as p38 and Rho-associated protein kinase (ROCK), in addition to JNK, also has been associated with enhanced Smad3 transcriptional activity, and suggests that phosphorylation of R-Smads in the linker regions plays an important role in transmitting signals from serine receptor kinases and receptor tyrosine kinases (9, 14, 37). Indeed, it has been reported that the Smad3 linker region contains a transcriptional activation domain that cooperates with the C-terminal domain of Smad3 to enhance TGF-β1–induced transcriptional activation (15). Evidence to support the enhanced association of Smad3 with Smad4, after Smad3 linker phosphorylation by JNK, has been previously reported, based upon the use of SP600125 as an inhibitor or JNK (14). Our present studies extend those prior observations in demonstrating that cells that lack JNK1 display lowered association between Smad3 and Smad4 after administration of TGF-β1. Furthermore, we demonstrate here that expression of MKK7/JNK1 is sufficient to enhance the interaction between Smad3 and Smad4, and enhances SBE transcriptional activity in a Smad4-dependent manner, which requires linker phosphorylation of Smad3. Our findings, demonstrating that EMT-regulatory genes were also enhanced after expression of MKK7/JNK1, and were attenuated in cells expressing a Smad3 construct refractory to linker phosphorylation, strongly suggest that JNK-dependent phosphorylation of Smad3 and subsequently enhanced transcriptional activity of Smad3/4 complexes are important in the regulation of EMT.
Intriguingly, Ras causes phosphorylation of Smad2 and Smad3 in the linker domains via ERK MAPKs, which has been demonstrated to repress Smad signaling (11, 38). Furthermore, dephosphorylation of Smad2/3 in the linker regions by small C-terminal domain phosphatase 1 has been demonstrated to result in enhanced TGF-β–dependent transcriptional responses (39), suggesting that linker phosphorylation had an inhibitory role. These disparate effects of linker phosphorylation on Smad signaling demonstrate that the functional outcome of linker phosphorylation is highly context dependent, and additionally may depend on the combinations of linker Ser/Thr sites that are phosphorylated. Recently, these disparate effects were further emphasized by the lack of Smad3 208/213 linker phosphorylation after TGF-β1 in human lung fibroblasts. Intriguingly however, this study revealed an important role of JNK/Smad3 signaling pathways in TGF-β–induced up-regulation of coagulation factor XII by regulating Smad3 binding to the SBE site in the factor XII promoter (40).
Despite a growing number of studies that demonstrate a stimulatory role of JNK in TGF-β1–induced signaling and EMT, numerous other studies have demonstrated evidence that JNK can abrogate the effects of TGF-β. JNK-dependent phosphorylation of the activator protein 1 (AP-1) family member, c-Jun, has been shown to inhibit Smad3-dependent transcription via the formation of a functional complex between Smad3/c-Jun, which represses transcription (41). Furthermore, JNK-dependent phosphorylation of c-Jun and JunB has been demonstrated to mediate the antagonistic effects of inflammatory cytokines on TGF-β/Smad signaling (36). These disparate results seem to suggest that the functional outcome of JNK may be cell type and context specific, and could depend on the target that is phosphorylated by JNK. Despite the aforementioned studies that demonstrate an inhibitory role of the AP-1 family members, c-Jun and JunB in TGF-β1/Smad signaling, an intriguing recent report demonstrates that expression of the AP-1 family member, Fra2, is sufficient to cause pulmonary fibrosis, and that expression of Fra2 is enhanced in patients with idiopathic pulmonary fibrosis (42). Furthermore, IL-13–induced fibrosis was shown to involve signaling through IL-13Rα2 leading to activation of the TGF-β1 promoter via a c-Jun/Fra2 site (43). Further studies to determine whether Fra2 exerts these effects via regulation of Smads and/or an EMT transcriptional program remain to be conducted.
Smad3 plays an important role in the transcriptional control of a large number of diverse target genes. The SBE contains a repeated AGAC sequence (44), found in many promoters of TGF-β target genes (45). In contrast to Smad2, Smad3 and Smad4 can bind directly to the SBE. It has been unraveled that these interactions are not sufficient to convey promoter selectivity (5). Therefore, it is likely that Smad3–Smad4 complexes synergize with other transcription factors and/or coactivators to induce transcription of distinct target genes. Two recently described transcription factors important in EMT have been identified as HMGA2 and CBF-A. TGF-β1 has been shown to cause induction of the HMGA2 gene in a Smad-dependent manner, and ectopic expression of HMGA2 has been demonstrated to be sufficient to induce a mesenchymal phenotype characterized by a strong down-regulation of E-cadherin (22). Similarly, overexpression of CBF-A also is sufficient to drive EMT (23). Our present work demonstrates that TGF-β1–induced expression of HMGA2 and CBF-A mRNAs requires JNK1, and depends on the phosphorylation status of the Smad3 linker region. Furthermore, we demonstrate here that expression of a PM Smad3 linker mutant is sufficient to enhance expression of CBF-A and HMGA2. These findings, combined with our observations that JNK1, in cooperation with Smad3, also increases expression of these critical EMT regulators, strongly suggest a role of JNK1 in regulating EMT via phosphorylation of Smad3 in the linker domain.
Although the present study demonstrates that Smad3 linker phosphorylation is important in the augmentation of transcriptional expression of EMT regulators, the exact mechanisms responsible for these events remain to be unraveled. A plausible scenario is that Smad3 phosphorylation promotes the association with transcriptional coactivators to augment transcription of EMT genes. In this regard, cooperation between HMGA2 and Smads in the regulation of expression of Snail-1 has been demonstrated during the induction of EMT, which involved a physical interaction between Smad3 and HMGA2 (28, 46). Whether such interaction is enhanced after phosphorylation of Smad3 in the linker domain remains to be tested further. Whether HMGA2 or CBF-A are direct targets of phosphorylation by JNK1, and the impact of phosphorylation on their activity as regulators of the EMT transcriptome, also remains to be investigated. A recent study suggests that the role of Smad3 in epithelial transitions may be more subtle, based upon findings that, in epithelial–myofibroblast transition, a Smad3-dependent role exists in the early mesenchymal phase of this process, whereas the late myogenic phase was Smad3 independent, and that this involved the regulation of myocardin-related transcription factor by Smad3 (47).
In summary, our present study demonstrates that the stimulatory role of JNK1 in TGF-β1–induced EMT requires phosphorylation of Smad3 in the linker region, and provides new insights into the potential mechanism whereby JNK1 promotes fibrosis in the lung. Additional studies will be required to address formally the role of JNK1 and Smad3 signaling in cells of epithelial origin in the pathogenesis of tissue fibrosis. The results of the present study, coupled to findings demonstrating that JNK is activated in lung tissue from patients with idiopathic pulmonary fibrosis (48, 49), and in fibroblasts derived from patients with pulmonary fibrosis (50), strongly suggest that JNK1 is a relevant therapeutic target in patients with tissue fibrosis.
Acknowledgments
The authors are indebted to Drs. J. Massague (Memorial Sloan-Kettering Cancer Center, New York, NY), B. Vogelstein (Johns Hopkins Kimmel Cancer Center, Baltimore, MD), F. Liu (Center for Advanced Biotechnology and Medicine, State University of New Jersey, Piscataway, NJ), and K. Matsuzaki (Kansai Medical University, Osaka, Japan) for kindly providing valuable plasmids and antibodies.
This work was supported by grant T32 HL076122 and R01 HL085464 from the National Institutes of Health.
Originally Published in Press as DOI: 10.1165/rcmb.2009-0282OC on June 25, 2010
Author Disclosure: J.F.A. received a sponsored grant from Centocor R&D for $50,001–$100,000, the American Thoracic Society for $50,001–$100,000 and Parker B. Francis Foundation for more than $100,001; Y.M.W.J.-H. received sponsored grants from the National Institutes of Health (NIH) one grant for $10,001–$50,000 and three grants for more than $100,001. J.L.J.v.d.V. received a sponsored grant from NIH for $10,001–$50,000; E.C.H.L.B. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript; A.S.G. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Strieter RM, Mehrad B. New mechanisms of pulmonary fibrosis. Chest 2009;136:1364–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim KK, Kugler MC, Wolters PJ, Robillard L, Galvez MG, Brumwell AN, Sheppard D, Chapman HA. Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix. Proc Natl Acad Sci USA 2006;103:13180–13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willis BC, Liebler JM, Luby-Phelps K, Nicholson AG, Crandall ED, du Bois RM, Borok Z. Induction of epithelial–mesenchymal transition in alveolar epithelial cells by transforming growth factor–beta1: potential role in idiopathic pulmonary fibrosis. Am J Pathol 2005;166:1321–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heldin CH, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through Smad proteins. Nature 1997;390:465–471. [DOI] [PubMed] [Google Scholar]
- 5.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 2003;113:685–700. [DOI] [PubMed] [Google Scholar]
- 6.Macias-Silva M, Abdollah S, Hoodless PA, Pirone R, Attisano L, Wrana JL. MADR2 is a substrate of the TGFbeta receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell 1996;87:1215–1224. [DOI] [PubMed] [Google Scholar]
- 7.Dennler S, Goumans MJ, ten Dijke P. Transforming growth factor beta signal transduction. J Leukoc Biol 2002;71:731–740. [PubMed] [Google Scholar]
- 8.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 2003;425:577–584. [DOI] [PubMed] [Google Scholar]
- 9.Engel ME, McDonnell MA, Law BK, Moses HL. Interdependent Smad and JNK signaling in transforming growth factor–beta–mediated transcription. J Biol Chem 1999;274:37413–37420. [DOI] [PubMed] [Google Scholar]
- 10.Hocevar BA, Prunier C, Howe PH. Disabled-2 (dab2) mediates transforming growth factor beta (TGFbeta)–stimulated fibronectin synthesis through TGFbeta-activated kinase 1 and activation of the JNK pathway. J Biol Chem 2005;280:25920–25927. [DOI] [PubMed] [Google Scholar]
- 11.Kretzschmar M, Doody J, Timokhina I, Massague J. A mechanism of repression of TGFbeta/Smad signaling by oncogenic ras. Genes Dev 1999;13:804–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Caestecker MP, Yahata T, Wang D, Parks WT, Huang S, Hill CS, Shioda T, Roberts AB, Lechleider RJ. The Smad4 activation domain (SAD) is a proline-rich, p300-dependent transcriptional activation domain. J Biol Chem 2000;275:2115–2122. [DOI] [PubMed] [Google Scholar]
- 13.de Caestecker MP, Parks WT, Frank CJ, Castagnino P, Bottaro DP, Roberts AB, Lechleider RJ. Smad2 transduces common signals from receptor serine–threonine and tyrosine kinases. Genes Dev 1998;12:1587–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mori S, Matsuzaki K, Yoshida K, Furukawa F, Tahashi Y, Yamagata H, Sekimoto G, Seki T, Matsui H, Nishizawa M, et al. TGF-beta and HGF transmit the signals through JNK-dependent Smad2/3 phosphorylation at the linker regions. Oncogene 2004;23:7416–7429. [DOI] [PubMed] [Google Scholar]
- 15.Wang G, Long J, Matsuura I, He D, Liu F. The Smad3 linker region contains a transcriptional activation domain. Biochem J 2005;386:29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alcorn JF, Guala AS, van der Velden J, McElhinney B, Irvin CG, Davis RJ, Janssen-Heininger YM. Jun n-terminal kinase 1 regulates epithelial-to-mesenchymal transition induced by TGF-beta1. J Cell Sci 2008;121:1036–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alcorn JF, van der Velden J, Brown AL, McElhinney B, Irvin CG, Janssen-Heininger YM. c-Jun N-terminal kinase 1 is required for the development of pulmonary fibrosis. Am J Respir Cell Mol Biol 2009;40:422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonniaud P, Kolb M, Galt T, Robertson J, Robbins C, Stampfli M, Lavery C, Margetts PJ, Roberts AB, Gauldie J. Smad3 null mice develop airspace enlargement and are resistant to TGF-beta–mediated pulmonary fibrosis. J Immunol 2004;173:2099–2108. [DOI] [PubMed] [Google Scholar]
- 19.Roberts AB, Tian F, Byfield SD, Stuelten C, Ooshima A, Saika S, Flanders KC. Smad3 is key to TGF-beta–mediated epithelial-to-mesenchymal transition, fibrosis, tumor suppression and metastasis. Cytokine Growth Factor Rev 2006;17:19–27. [DOI] [PubMed] [Google Scholar]
- 20.Wu R, Smith D. Continuous multiplication of rabbit tracheal epithelial cells in a defined, hormone-supplemented medium. In Vitro 1982;18:800–812. [DOI] [PubMed] [Google Scholar]
- 21.Malkinson AM, Dwyer-Nield LD, Rice PL, Dinsdale D. Mouse lung epithelial cell lines—tools for the study of differentiation and the neoplastic phenotype. Toxicology 1997;123:53–100. [DOI] [PubMed] [Google Scholar]
- 22.Thuault S, Valcourt U, Petersen M, Manfioletti G, Heldin CH, Moustakas A. Transforming growth factor–beta employs HMGA2 to elicit epithelial–mesenchymal transition. J Cell Biol 2006;174:175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venkov CD, Link AJ, Jennings JL, Plieth D, Inoue T, Nagai K, Xu C, Dimitrova YN, Rauscher FJ, Neilson EG. A proximal activator of transcription in epithelial–mesenchymal transition. J Clin Invest 2007;117:482–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tournier C, Whitmarsh AJ, Cavanagh J, Barrett T, Davis RJ. Mitogen-activated protein kinase kinase 7 is an activator of the c-Jun NH2-terminal kinase. Proc Natl Acad Sci USA 1997;94:7337–7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lei K, Nimnual A, Zong WX, Kennedy NJ, Flavell RA, Thompson CB, Bar-Sagi D, Davis RJ. The bax subfamily of Bcl2-related proteins is essential for apoptotic signal transduction by c-Jun NH(2)–terminal kinase. Mol Cell Biol 2002;22:4929–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Massague J, Wotton D. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J 2000;19:1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim KK, Wei Y, Szekeres C, Kugler MC, Wolters PJ, Hill ML, Frank JA, Brumwell AN, Wheeler SE, Kreidberg JA, et al. Epithelial cell alpha3beta1 integrin links beta-catenin and Smad signaling to promote myofibroblast formation and pulmonary fibrosis. J Clin Invest 2009;119:213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thuault S, Tan EJ, Peinado H, Cano A, Heldin CH, Moustakas A. HMGA2 and Smads co-regulate Snail1 expression during induction of epithelial-to-mesenchymal transition. J Biol Chem 2008;283:33437–33446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willis BC, Borok Z. TGF-beta–induced EMT: mechanisms and implications for fibrotic lung disease. Am J Physiol Lung Cell Mol Physiol 2007;293:L525–L534. [DOI] [PubMed] [Google Scholar]
- 30.Saika S, Kono-Saika S, Ohnishi Y, Sato M, Muragaki Y, Ooshima A, Flanders KC, Yoo J, Anzano M, Liu CY, et al. Smad3 signaling is required for epithelial–mesenchymal transition of lens epithelium after injury. Am J Pathol 2004;164:651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato M, Muragaki Y, Saika S, Roberts AB, Ooshima A. Targeted disruption of TGF-beta1/Smad3 signaling protects against renal tubulointerstitial fibrosis induced by unilateral ureteral obstruction. J Clin Invest 2003;112:1486–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hackett TL, Warner SM, Stefanowicz D, Shaheen F, Pechkovsky DV, Murray LA, Argentieri R, Kicic A, Stick SM, Bai TR, et al. Induction of epithelial–mesenchymal transition in primary airway epithelial cells from patients with asthma by transforming growth factor–β1. Am J Respir Crit Care Med 2009;180:122–133. [DOI] [PubMed] [Google Scholar]
- 33.Phanish MK, Wahab NA, Colville-Nash P, Hendry BM, Dockrell ME. The differential role of Smad2 and Smad3 in the regulation of pro-fibrotic TGFbeta1 responses in human proximal-tubule epithelial cells. Biochem J 2006;393:601–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoot KE, Lighthall J, Han G, Lu SL, Li A, Ju W, Kulesz-Martin M, Bottinger E, Wang XJ. Keratinocyte-specific Smad2 ablation results in increased epithelial–mesenchymal transition during skin cancer formation and progression. J Clin Invest 2008;118:2722–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Runyan CE, Hayashida T, Hubchak S, Curley JF, Schnaper HW. Role of SARA (Smad anchor for receptor activation) in maintenance of epithelial cell phenotype. J Biol Chem 2009;284:25181–25189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verrecchia F, Tacheau C, Wagner EF, Mauviel A. A central role for the JNK pathway in mediating the antagonistic activity of pro-inflammatory cytokines against transforming growth factor–beta–driven Smad3/4-specific gene expression. J Biol Chem 2003;278:1585–1593. [DOI] [PubMed] [Google Scholar]
- 37.Kamaraju AK, Roberts AB. Role of Rho/ROCK and p38 map kinase pathways in transforming growth factor–beta–mediated Smad-dependent growth inhibition of human breast carcinoma cells in vivo. J Biol Chem 2005;280:1024–1036. [DOI] [PubMed] [Google Scholar]
- 38.Matsuura I, Wang G, He D, Liu F. Identification and characterization of ERK MAP kinase phosphorylation sites in Smad3. Biochemistry 2005;44:12546–12553. [DOI] [PubMed] [Google Scholar]
- 39.Wrighton KH, Willis D, Long J, Liu F, Lin X, Feng XH. Small C-terminal domain phosphatases dephosphorylate the regulatory linker regions of Smad2 and Smad3 to enhance transforming growth factor–beta signaling. J Biol Chem 2006;281:38365–38375. [DOI] [PubMed] [Google Scholar]
- 40.Jablonska E, Markart P, Zakrzewicz D, Preissner KT, Wygrecka M. Transforming growth factor–beta1 induces expression of human coagulation factor XII via Smad3 and JNK signaling pathways in human lung fibroblasts. J Biol Chem 2010;285:11638–11651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dennler S, Prunier C, Ferrand N, Gauthier JM, Atfi A. c-Jun inhibits transforming growth factor beta–mediated transcription by repressing Smad3 transcriptional activity. J Biol Chem 2000;275:28858–28865. [DOI] [PubMed] [Google Scholar]
- 42.Eferl R, Hasselblatt P, Rath M, Popper H, Zenz R, Komnenovic V, Idarraga MH, Kenner L, Wagner EF. Development of pulmonary fibrosis through a pathway involving the transcription factor FRA-2/AP-1. Proc Natl Acad Sci USA 2008;105:10525–10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fichtner-Feigl S, Strober W, Kawakami K, Puri RK, Kitani A. IL-13 signaling through the IL-13alpha2 receptor is involved in induction of TGF-beta1 production and fibrosis. Nat Med 2006;12:99–106. [DOI] [PubMed] [Google Scholar]
- 44.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGF beta–inducible elements in the promoter of human plasminogen activator inhibitor–type 1 gene. EMBO J 1998;17:3091–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ross S, Hill CS. How the Smads regulate transcription. Int J Biochem Cell Biol 2008;40:383–408. [DOI] [PubMed] [Google Scholar]
- 46.Vincent T, Neve EP, Johnson JR, Kukalev A, Rojo F, Albanell J, Pietras K, Virtanen I, Philipson L, Leopold PL, et al. A Snail1–Smad3/4 transcriptional repressor complex promotes TGF-beta mediated epithelial–mesenchymal transition. Nat Cell Biol 2009;11:943–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Masszi A, Speight P, Charbonney E, Lodyga M, Nakano H, Szaszi K, Kapus A. Fate-determining mechanisms in epithelial–myofibroblast transition: major inhibitory role for Smad3. J Cell Biol 2010;188:383–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang XM, Zhang Y, Kim HP, Zhou Z, Feghali-Bostwick CA, Liu F, Ifedigbo E, Xu X, Oury TD, Kaminski N, et al. Caveolin-1: a critical regulator of lung fibrosis in idiopathic pulmonary fibrosis. J Exp Med 2006;203:2895–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshida K, Kuwano K, Hagimoto N, Watanabe K, Matsuba T, Fujita M, Inoshima I, Hara N. MAP kinase activation and apoptosis in lung tissues from patients with idiopathic pulmonary fibrosis. J Pathol 2002;198:388–396. [DOI] [PubMed] [Google Scholar]
- 50.Shi-Wen X, Rodriguez-Pascual F, Lamas S, Holmes A, Howat S, Pearson JD, Dashwood MR, du Bois RM, Denton CP, Black CM, et al. Constitutive Alk5-independent c-Jun N-terminal kinase activation contributes to endothelin-1 overexpression in pulmonary fibrosis: evidence of an autocrine endothelin loop operating through the endothelin A and B receptors. Mol Cell Biol 2006;26:5518–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]