Abstract
Epidemiological studies indicate an increased risk of subsequent primary ovarian cancer from women with breast cancer. We have recently identified a 28-gene expression signature that predicts, with high accuracy, the clinical course in a large population of breast cancer patients. This prognostic gene signature also accurately predicts response to chemotherapy commonly used for treating breast cancer, including CMF, Tamoxifen, Paclitaxel, Docetaxel, and Doxorubicin (Adriamycin), in a panel of 60 cancer cell lines of nine different tissue origins. This prompted us to investigate whether this prognostic gene signature could also predict clinical outcome in other cancer types of epithelial origins, including ovarian cancer (n = 124), colon tumors (n = 74), and lung adenocarcinomas (n = 442). The results show that the gene expression signature contributes significantly more accurate (P < 0.05; compared with random prediction) prognostic information in multiple cancer types independent of established clinical parameters. Furthermore, the functional pathway analysis with curated database delineated a biological network with tight connections between the signature genes and numerous well established cancer hallmarks, indicating important roles of this prognostic gene signature in tumor genesis and progression.
Keywords: prognostic gene signature, breast cancer, ovarian cancer, colon cancer, lung adenocarcinoma
Introduction
For women with breast cancer, an increased risk of primary ovarian cancer has been observed from epidemiological studies (1). This risk is highest for women with early-onset breast cancer (younger than age 50 years at diagnosis). To date, two major genes, BRCA1 and BRCA2, have been identified to be associated with susceptibility to breast and ovarian cancer. However, mutations in these two genes only account for 2 to 3 percent of all breast cancers (2). It has been proposed that additional genes that are associated with susceptibility to breast and ovarian cancer exist (2). Identification of other susceptibility genes could provide crucial information to guide clinicians to assess the risk of subsequent ovarian cancer in breast cancer patients.
Previously, we identified a 28-gene breast cancer prognostic signature in a population-based study (3). A unified classification scheme was later developed for patient stratification based on the expression patterns of the 28-gene signature. The prognostic categorization system was validated with more than 2,000 breast cancer patient samples quantified with heterogeneous DNA microarray platforms (4). This prognostic gene signature was also found to predict response to chemotherapy commonly used for treating breast cancer, including CMF, Tamoxifen, Paclitaxel, Docetaxel, and Doxorubicin (Adriamycin), in a panel of 60 cancer cell lines (NCI-60) of nine different tissue origins (4). Based on these results, we hypothesize that the 28-gene prognostic signature reveals molecular characteristics important to tumor genesis and progression. To test this hypothesis, we first sought to investigate whether the 28-gene signature reveals common biological processes involved in recurrence and metastases of breast and ovarian cancer. Next, we sought to explore whether the 28-gene prognostic signature could also predicts clinical outcome in other cancer types with epithelial origin, including colon cancer and non-small cell lung cancer.
Material and Methods
Patients and Samples
Ovarian Cancer
The ovarian cancer cohort (n = 124) was retrieved from Bild et al (5). 94.4% (117/124) of these ovarian cancer patients had advanced stages (III and IV).
Colon Cancer
The first cohort contained 50 patients with stage II colon adenocarcinoma (6). None of the patients had emergency surgery or received any adjuvant chemotherapy. Twenty-five patients developed a distant metastasis (liver in 22 patients; lung in five patients) within 52 months. The other 25 patients remained disease-free for at least 60 months, with mean follow-up of 79 months. The second cohort contained 24 patients with stage II colon adenocarcinoma (7). None of these patients received adjuvant chemotherapy. Ten patients developed a liver metastasis within 55 months. The other 14 patients remained disease-free for at least 60 months, with mean follow-up of 72.2 months.
Non-small Cell Lung Cancer
The cohort from Shedden et al (8) contained 442 lung adenocarcinomas collected from multiple cancer centers and institutes. Two hundred and seventy-six patients were in stage I, 94 in stage II, and 68 in stage III and four patients with undefined stage.
DNA Microarray Analysis
The RNA extraction and cDNA preparation in these studies was described in their original publications. The ovarian cancer dataset from Bild et al (5) were assayed with Affymetrix U133A (retrieved with record GSE3149 from Gene Expression Omnibus). Two colon cancer datasets were all generated with Affymetrix U133A arrays (7,6). The lung adenocarcinoma datasets from Shedden et al (8) were generated with Affymetrix U133A.
Patient Stratification in Ovarian Cancer
The ovarian cancer cohort (n = 124) from Bild et al. (5) was used to explore whether the 28-gene signature reveals molecular portraits common in breast cancer and ovarian cancer. To avoid over-fitting in the validation, the data set was randomly partitioned into a training set (n = 82) and a test set (n = 42). The 28 gene predictors were fitted in a Cox hazard proportional model on the training set, and a survival risk score was generated for each patient. A high risk score represents a high probability of post-operative treatment failure, and similarly for a low risk score. The median of the survival risk scores in the training set was used as the cutoff point to stratify patients into different prognostic groups. A patient with a risk score higher than median risk score was classified into poor-prognosis group; whereas a patient with a lower risk score was classified into good-prognosis group. The same cutoff value and prognostic model were applied to patient stratification in the test set.
Prognostic Prediction of Recurrence in Colon Cancer
The matching genes in the 28-gene signature were identified with Affymetrix IDs. Twenty-five common genes were found in each colon cancer cohort. If a gene has multiple probes, the average expression of multiple probes was used in the classification. The patient cohort from Barrier et al (6) was used as training set (n = 50), while the cohort from another study by Barrier et al (7) was used as an independent validation set (n = 24). A training model was built with the 25 signature genes to classify recurrence in colon cancer patients using a Linear Discriminant Analysis function in SAS 9.1. A 10-fold cross validation was used to evaluate the performance of the training model. This training model was used to predict tumor recurrence in each patient in the validation set.
Prognostic Categorization of Non-Small Cell Lung Cancer
The patient samples collected from the University of Michigan Cancer Center (UM) and Moffitt Cancer Center (HLM) form the training set (n = 256), whereas the samples obtained from Memorial Sloan-Kettering Cancer Center (MSK, n = 104) and the Dana-Farber Cancer Institute (DFCI, n = 82) constitute an independent validation set. Gene symbols were used to find the matching genes in the signature. In the training set (UM and HLM cohorts), a Cox proportional hazard model was constructed by using the matching genes as covariates to predict lung cancer survival after the initial treatment. A risk score was generated for each patient in this cohort. Based on the distribution of the risk scores in the training set, a cutoff point representing the peak value in the histogram was identified to stratify patients into high- or low-risk groups. This cutoff risk score and the training model were applied in prognostic categorization in the validation set (MSK and DFCI cohorts).
Statistical analysis
Patient survival rates were assessed with Kaplan-Meier analysis using log-rank tests. Cox hazard proportional model was used to generate a risk score for each ovarian cancer patient based on the 28-gene signature. All statistical analyses were performed with software package R (9).
Biological Pathway Analysis
Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Redwood City, CA) is a proprietary web-based curated database which provides contents of gene and protein interactions reported in the literature. In this study, we used IPA to delineate molecular networks of genes interacting with the 28-gene signature. Core analysis identified the most significant biological functions and processes from the merged network generated for the 28-gene signature.
Results
Recent studies showed that a prognostic gene signature identified from breast cancer cells might be able to predict clinical outcome in multiple tumor types (10,5,11,12). A set of 28 genes predicted recurrence-free survival (including metastasis and relapse) and overall survival in multiple independent breast cancer cohorts (3,4). In the present study, we sought to investigate whether this breast cancer prognostic gene signature also predicts clinical outcomes in other cancer types of epithelial origins, including ovarian cancer (n = 124), colon cancer (n = 74), and non-small cell lung cancer (n = 442).
28-gene prognostic signature predicts ovarian cancer outcome
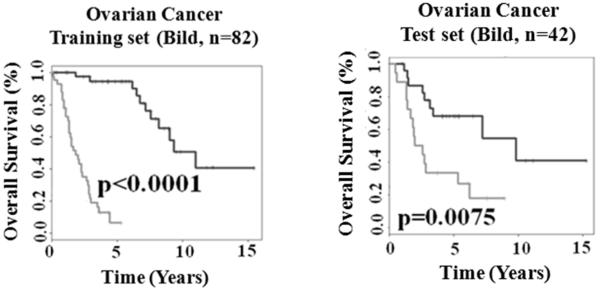
Ovarian cancer is a common malignancy in women, whose prognosis is bleak due to a usually advanced disease stage at the time of diagnosis. Common genetic risk factors of susceptibility to breast and ovarian cancer have recently been proposed (2). To explore whether the 28-gene signature reveals common molecular features affecting breast and ovarian cancer survival, an ovarian cancer cohort from Bild et al (5) was analyzed. This ovarian cancer cohort (n = 124) was randomly split into a training set (n = 82) and a test set (n = 42). A Cox model was built on the training set using the signature genes as covariates. A survival risk score was generated for each patient. The median of the risk scores in the training set was identified as a cutoff point for patient stratification. Patients with a risk score greater than the cutoff were stratified into the high-risk group, and otherwise, into the low-risk group. In the prognostic model evaluation, the high- and low-risk groups had significantly (log-rank P < 0.0001) different relapse-free survival in the training cohort in Kaplan-Meier analysis (Fig. 1A). This training model and stratification scheme were applied to the test set, and generated significant prognostic stratification (log-rank P = 0.0075) in Kaplan-Meier analysis (Fig. 1B). The details of the prognostic Cox model were provided in http://www.hsc.wvu.edu/mbrcc/fs/GuoLab/pdfs/Shruti_Rathnagiriswaran_Thesis.pdf. These results indicate that the 28-gene signature reflects common biological processes involved in breast and ovarian cancer metastases and relapse. The 28-gene signature could indentify more aggressive ovarian cancers that were more likely to develop recurrence after surgical resections and initial treatment. Therefore, the high risk patients defined with this gene signature might benefit from second line chemotherapy.
Figure 1.
The 28-gene prognostic signature predicts overall survival in ovarian cancer. Kaplan-Meier analyses of the training cohort (A) and test cohort from Bild et al. (5). The upper curves represent the gene expression-define low risk group, and the lower curve represent the high risk group. The median of the risk scores with a value of 0.301 generated by fitting the Cox proportional hazard model on the training set was taken as the cut-off in both training and test sets.
28-gene prognostic signature is an independent predictor of colon cancer recurrence
In order to extent the potential usefulness of the 28-gene prognostic signature, we explored its value for predicting clinical outcome in patients with stage II colon cancer. To construct a molecular classifier to predict colon cancer recurrence, 50 patients with stage II colon adenocarcinoma (6) were used as training cohort. Twenty-five genes within the 28-gene signature were identified from the DNA microarray data. These signature genes were used to classify recurrence in each patient with the Linear Discriminant Analysis algorithm. The performance of the classifier was evaluated in a 10-fold cross validation on the training set (Table 1). The prognostic signature correctly predicted recurrence in 94% (47/50) of patients, with a sensitivity of 100% (25/25) and a specificity of 88% (22/25). The model identified in the training cohort was applied to predict recurrence in each patient in the validation set (n = 24) with a patient cohort retrieved from Barrier et al (7). In the validation, the prognostic signature correctly predicted recurrence in 75% (18/24) patients, with a sensitivity of 80% (8/10) and a specificity of 71.43% (10/14). Both cohorts contained only stage II lymph node negative colon adenocarcinomas. These results indicate that the 28-gene prognostic signature provides independent prognostic information in addition to tumor stage. Once validated in larger, independent cohorts this signature could be potentially used to select lymph node-negative patients for receiving adjuvant chemotherapy.
Table 1.
Prediction accuracy of colon cancer recurrence using the 28-gene prognostic signature. All patients were with tumor stage II at diagnosis.
| Patients | Sensitivity (Recurrence within 5-y) |
Specificity (no recurrence within 5-y) |
Overall Accuracy |
P-Value* |
|---|---|---|---|---|
| Training set (n=50) (5) |
100% (25/25) | 88% (22/25) | 94% (47/50) | 4.8e-7 |
| Validation set (n=24) (6) |
80% (8/10) | 71.43% (10/14) | 75% (18/24) | 0.04 |
P<0.05 represents the overall accuracy is significantly higher than that of random prediction (two-sided Z-tests).
28-gene prognostic signature predicts lung cancer survival
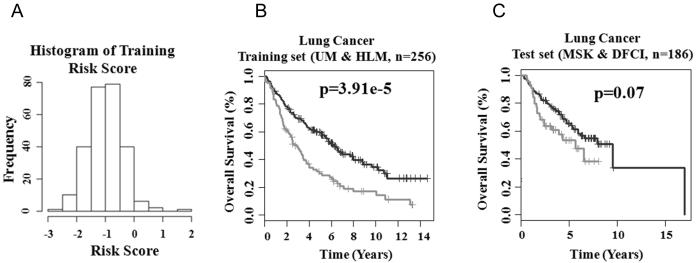
To explore the clinical relevance of the 28-gene prognostic signature for the prognostication of patients with non-small cell lung cancer, the lung adenocarcinoma cohorts (UM and HLM) retrieved from Shedden et al (8) were used as a training set (n = 256). A Cox model of overall survival was constructed based on the 28-gene signature, with each gene variable as a covariate. A survival risk score was generated for every patient, with a higher risk score representing a greater probability of treatment failure (i.e., death). Based on the histogram representing distribution of gene expression-defined risk scores in this cohort (Fig. 2A), a cutoff value of −0.75, the peak value in the histogram, was used to stratify patients into high- and low-risk groups. This cutoff value represents the linear additive expression levels of all the signature genes in lung cancer patients. This stratification separated patients into two groups with distinct overall survival (log-rank P < 3.91e-5) in Kaplan-Meier analysis (Fig. 2B). This cutoff risk score and training model were applied to the validation set (MSK and DFCI, n = 186). The 28-gene signature generated borderline significant prognostic categorization in the validation set (log-rank P = 0.07; Fig. 2C) in Kaplan-Meier analysis. In all studied lung adenocarcinoma cohorts, the low-risk groups had 73.54% to 82.15% of 2.5-year postoperative survival rate, representing a significantly better prognosis compared with the corresponding high-risk groups for which the 2.5-year survival was ranging from 53.76% to 63.51%. As the majority of non-small cell lung cancer recurrence occurs within 2 years after surgery (13), these results indicate that the 28-gene prognostic signature could be used to predict postoperative survival in non-small cell lung cancer patients.
Figure 2.
The 28-gene prognostic signature predicts overall survival in lung cancer. (A) Histogram of gene expression-defined risk scores in the training cohort from Shedden et al (8). The peak value with risk score of −0.75 in the histogram was defined as the cutoff in prognostic categorization. Gene expression-defined high- (lower curves) and low-risk groups (upper curves) had remarkably different post-operative lung cancer survival in both training (B) and test cohorts (C).
Functional pathway analysis
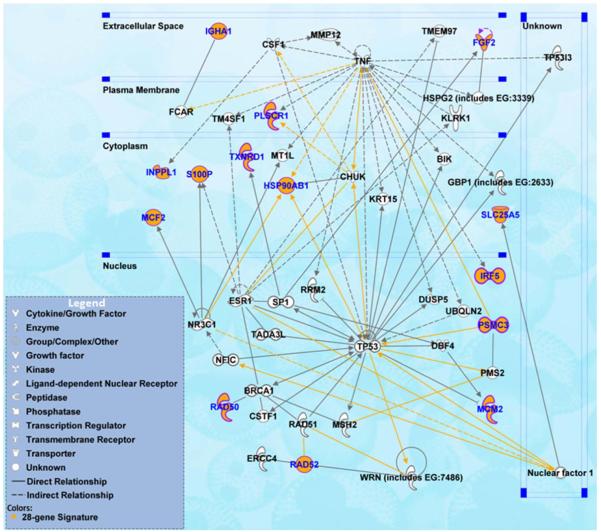
The 28-gene prognostic signature was able to distinguish more aggressive tumors in multiple cancer types, indicating that this signature might be involved in important mechanisms of tumor genesis and progression. Functional pathway analysis was performed based on curated database of molecular interactions reported in the literature using Ingenuity Pathway Analysis. The results show that the signature genes interact with multiple prominent cancer signaling pathways, including TP53, TNF and ER, the BRCA1 breast cancer and ovarian cancer risk gene, the KRT15 stem cell marker, as well as DNA repair proteins RAD51 and ERCC4 (Fig. 3).
Figure 3.
Functional pathway analysis of the 28-gene prognostic signature using Ingenuity Pathway Analysis. The biological network showed genes interacting with the signature genes as reported in the literature.
Discussion
Genome-wide association studies utilizing human tissue samples have enhanced the prognostic capacity of cancer outcomes. Four breast cancer signatures, including intrinsic subtypes (14), poor prognosis signature (MammaPrint®) (15), recurrence score (Oncotype DX®) (16), and wound response (17), represent largely the same prognostic space (18). Our identified 28-gene breast cancer prognostic signature predicted disease-free survival and overall survival in a large population of more than 2,000 breast cancer patient with heterogeneous disease stage, including both early stage and advanced breast cancers (3,4). In the evaluation, the 28-gene prognostic signature is comparable as Oncotype DX® and could potentially be more accurate than the other above mentioned signatures in terms of predicting disease-free survival and overall survival in van de Vijver’s cohort (15). More importantly, the 28-gene breast cancer signature showed prognostic ability beyond early-stage breast cancer. The 28-gene prognostic signature quantified disease-free survival and overall survival in a broad patient population including those with advanced stage (T3/T4), tumor grade III, lymph node metastasis, or negative estrogen receptor status (ER−) (4). These results indicate that the 28-gene signature might extend the prognostic space defined by MammaPrint® and Oncotype DX® that primarily target early stage breast cancer. To confirm this conjecture, this study investigated whether the 28-gene prognostic signature could predict clinical outcomes in other tumor types of epithelial origin, including ovarian cancer (n = 124), colon cancer (n = 74), and lung adenocarcinoma (n = 442).
In each studied cancer type, a patient stratification scheme was developed based on the expression of the 28-gene prognostic signature, and was validated on independent patient cohorts. Based on the clinical outcome provided in two colon cancer cohorts, a machine learning algorithm Linear Discriminant Analysis was used in the model construction on the training set (n = 50) with stage II colon carcinoma to predict patients’ recurrence after surgery. The model accuracy was 94% on the training cohort in a 10-fold cross validation. This prognostic model was applied to a test set (n = 24) and achieved an overall accuracy of 75% in the independent validation. These results are more accurate (P < 0.04) compared with random predictions. In the prognostic validation of lung adenocarcinoma, a prognostic model was built with Cox model using the gene expression profiles as covariates. The cutoff point for prognostic categorization was defined based on histogram of gene expression defined-risk scores on the training cohort (n = 256). This stratification scheme was applied to an independent validate set (n = 186). The gene signature separated patients into different prognostic groups with different (log-rank P = 0.07) clinical outcomes in Kaplan-Meier analysis. Similarly, the Cox model was used in the prognostic validation on ovarian cancer. In both training and test cohorts (n = 124), the gene expression defined-model provided significant (log-rank P < 0.0075) post-operative prognostic stratification in Kaplan-Meier analyses.
Epidemiological studies strongly indicate that an association exists between breast cancer and the risk of subsequent ovarian cancer (1). Begfeldt’s group found that a primary breast cancer patient has a two-fold increased risk of a primary ovarian cancer. Several genes have been identified to be associated with susceptibility to breast cancer and ovarian cancer, including BRCA1, BRCA2, TP53, PTEN, and STK11/LKB1. However, mutations in these genes only account for very limited portions of breast cancer and ovarian cancer (2). Identification of other susceptibility genes could provide essential information to guide clinicians to assess the risk of subsequent ovarian cancer in breast cancer patients. The 28-gene signature was shown to be predictive of clinical outcomes in both breast cancer and ovarian cancer. Furthermore, the signature genes were shown to interact with TP53 and BRCA1 in the biological association network (Fig. 3). Together, this signature might reveal essential genomic information for estimating the risk of consequent ovarian cancer in breast cancer patients.
This study confirmed that the identified 28-gene prognostic signature could predict clinical outcomes in multiple cancer types with epithelial origins. Thus, this 28-gene signature could extend breast cancer prognostic space defined by MammaPrint® and Oncotype DX®, among other breast cancer signatures with potential clinical utility (5,11,10,12). The functional pathway analysis with curated IPA database delineated a biological network with tight connections between the signature genes and numerous well established cancer hallmarks, indicating important roles of this prognostic gene signature in tumor genesis and progression.
Acknowledgements
We thank Dr. Jame Abrahim at West Virginia University for thoughtful discussions. This research is supported by National Library of Medicine R01LM009500 (Guo) and NCRR P20 RR16440 Supplement (Guo) from the NIH.
Footnotes
Disclaimer
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the views of the National Institute for Occupational Safety and Health.
References
- 1.Bergfeldt K, Rydh B, Granath F, Gronberg H, Thalib L, Adami HO, Hall P. Risk of ovarian cancer in breast-cancer patients with a family history of breast or ovarian cancer: a population-based cohort study. Lancet. 2002;360:891–894. doi: 10.1016/S0140-6736(02)11023-3. [DOI] [PubMed] [Google Scholar]
- 2.Wooster R, Weber BL. Breast and Ovarian Cancer. N Engl J Med. 2003;348:2339–2347. doi: 10.1056/NEJMra012284. [DOI] [PubMed] [Google Scholar]
- 3.Ma Y, Qian Y, Wei L, et al. Population-Based Molecular Prognosis of Breast Cancer by Transcriptional Profiling. Clin Cancer Res. 2007;13:2014–2022. doi: 10.1158/1078-0432.CCR-06-2222. [DOI] [PubMed] [Google Scholar]
- 4.Rathnagiriswaran S, Wan YW, Abraham J, Castranova V, Qian Y, Guo NL. A population-based gene signature is predictive of breast cancer survival and chemoresponse. Int J Oncol. 2010;36:607–616. doi: 10.3892/ijo_00000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 6.Barrier A, Boelle PY, Roser F, et al. Stage II Colon Cancer Prognosis Prediction by Tumor Gene Expression Profiling. J Clin Oncol. 2006;24:4685–4691. doi: 10.1200/JCO.2005.05.0229. [DOI] [PubMed] [Google Scholar]
- 7.Barrier A, Roser F, Boelle PY, et al. Prognosis of stage II colon cancer by non-neoplastic mucosa gene expression profiling. Oncogene. 2006;26:2642–2648. doi: 10.1038/sj.onc.1210060. [DOI] [PubMed] [Google Scholar]
- 8.Shedden K, Taylor JM, Enkemann SA, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everitt B, Hothorn T. A Handbook of Statistical Analyses Using R. Chapman & Hall/CRC; Boca Raton, FL: 2006. [Google Scholar]
- 10.Liu R, Wang X, Chen GY, et al. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356:217–226. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 11.Chi JT, Wang Z, Nuyten DS, et al. Gene expression programs in response to hypoxia: cell type specificity and prognostic significance in human cancers. PLoS Med. 2006;3:e47. doi: 10.1371/journal.pmed.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minn AJ, Gupta GP, Siegel PM, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cibas E, Ducatman B. In: Cytology: Diagnostic Principles and Clinical Correlates. 2nd edition Cibas E, Ducatman B, editors. W.B. Saunders; Edinburgh: 2006. [Google Scholar]
- 14.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van de Vijver MJ, He YD, van ’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 16.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 17.Chang HY, Nuyten DS, Sneddon JB, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci U S A. 2005;102:3738–3743. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Massague J. Sorting Out Breast-Cancer Gene Signatures. N Engl J Med. 2007;356:294–297. doi: 10.1056/NEJMe068292. [DOI] [PubMed] [Google Scholar]