Abstract
Microarray-based comparative genomic hybridizations (CGH) interrogate genomic DNA to identify structural differences such as amplifications and deletions that are easily detected as large signal aberrations. Subtle signal deviations caused by single nucleotide polymorphisms (SNPs) can also be detected but is challenged by a high AT content (81%) in P. falciparum. We compared genome-wide CGH signal to sequence polymorphisms between parasite strains 3D7, HB3, and Dd2 using NimbleGen microarrays. From 23,191 SNPs (excluding var/rif/stevor genes), our CGH probe set detected SNPs with > 99.9% specificity but low (< 10%) sensitivity. Probe length, melting temperature, GC content, SNP location in the probe, mutation type, and hairpin structures affected SNP sensitivity. Previously unrecognized variable number tandem repeats (VNTRs) also were detected by this method. These findings will guide the redesign of a probe set to optimize an openly available CGH microarray platform for high resolution genotyping suitable for population genomics studies.
Keywords: Comparative genomic hybridization, malaria genomics, genotyping, single nucleotide polymorphism, variable number tandem repeats
Despite a campaign by the World Health Organization (WHO) to eradicate malaria and years of research dedicated to studying the disease, malaria still exacts a large burden on some of the most impoverished regions of the world. Four species of Plasmodium cause malaria in humans with Plasmodium falciparum causing the most severe and deadly form of the disease in humans. There are an estimated 1–2 million deaths and 515 million clinical episodes of falciparum malaria annually [1]. Completed sequencing projects for humans, the mosquito vector, and the causative parasite promise breakthroughs in combating malaria awaiting only the development of creative applications that tap this wealth of information.
The P. falciparum genome spans nearly 23 megabases (Mb) on 14 chromosomes with a high AT content of 81% [2]. The original genome sequencing project focused on a single parasite strain, 3D7, and recent efforts by the Broad Institute have generated extensive comparative sequence from many strains, including genome assemblies of strains HB3 and Dd2 [3]. The majority of the parasite life cycle is spent in haploid form including the asexual, erythrocytic stages responsible for disease morbidity. The haploid nature of the parasite is useful for genome-wide studies; however the genome's AT content requires optimization of molecular techniques.
Microarrays traditionally have been used to measure gene expression levels, but they also allow direct interrogation of genomic DNA (gDNA), i.e. comparative genomic hybridizations (CGH), to reveal structural variation at high resolution. CGH microarrays readily detect copy number polymorphisms (CNPs), such as segmental amplifications or deletions and also have potential for direct allelic variation scanning as first demonstrated in yeast [4]. Sequence polymorphisms in target gDNA cause less efficient hybridization to the microarray probe (relative to a perfectly matched reference sample) creating a detectable difference in hybridization signal intensity. This signal intensity is affected by the number and location of mismatches between the target and probe sequence. These signal variations point to locations in the genome associated with polymorphisms purely on the basis of hybridization kinetics. Hybridization signal-based detection of “single feature polymorphisms” (SFPs) can indicate the presence of a variant but can not explicitly identify the polymorphism without using probes targeted to specific alleles.
Investigations of metronidazole resistance in Helicobacter pylori demonstrated the detection of SFPs corresponding to deletions and individual SNPs using a microarray [5]. SFPs were characterized further using a redesigned chip that effectively resequenced the polymorphic positions. This approach was sensitive enough to identify specific point mutations that occurred during several rounds of increasing metronidazole pressure which were associated with hyper-resistance to this bactericidal compound. Yeast tiling microarrays have been used for genome-wide SNP discovery without relying on allele-specific probes [6]. The tiled probes provide redundant coverage for any given nucleotide allowing the location of the polymorphism to be reliably mapped to a range of a few nucleotides. This approach allowed the detection of > 90% of the 30,303 known SNPs between two yeast strains. Furthermore, chip-based genotyping can be powerful for genome-wide association studies; for example two Mendelian traits were mapped in canines to regions < 1 Mb using a genotyping array and only twenty individuals [7], and a Wellcome Trust Case Control Consortium human study identified disease susceptibility factors using a 500k SNP typing array [8].
Plasmodium studies relying on CGH microarrays have demonstrated the ability to identify CNPs including segmental duplications and deletions [9]. A P. falciparum CGH microarray covering approximately 50% of the genome's coding regions measured genetic variation between 14 field and laboratory parasite strains [10] and identified 23,653 SFPs genome-wide (including var/rif/stevor genes) across all strains and also pointed to 500 genes that are evolving at a higher than neutral rate. Recently, Jiang et al. conducted an analysis of hybridization-based detection of SFP and found a large number associated with var/rif/stevor genes [11]; they also specifically evaluated SNP detection for 2651 preselected SNPs identified in an earlier comparative sequencing study [12].
Understanding variation in var genes is particularly interesting because they encode for the P. falciparum erythrocyte membrane protein 1 (PfEMP1) which mediates cytoadherence through binding to host receptors and is the major cell-surface variant antigen [13–15]. The nearly 100 var gene family members are distributed across the 14 chromosomes in both subtelomeric clusters and internal clusters[15–17] where their highly polymorphic nature make accurate SNP detection through hybridization especially challenging. Furthermore, individual parasite clones have unique var gene repertoires with minimal overlap limited to just several family members [18, 19], meaning that probes designed from a reference genome will not be able to effectively interrogate the majority of var genes in other parasite genomes. Similarly, there is limited utility to hybridization-based SNP detection in the subtelomeric rif and stevor multi-gene families which also encode highly polymorphic proteins that are exported to the host cell surface [20, 21].
The NimbleGen CGH platform uses a two-dye system to competitively hybridize test (cy5) and reference (cy3) samples labeled with fluorescent dyes. Log2 ratios of test to reference signal indicate a hybridization bias if one sample better matches the chip design sequence. Multiple contiguous probes that exhibit a hybridization bias indicates a segmental duplication or deletion event has occurred while individual probes that exhibit a bias correspond to SNPs or small indel events. A signal ratio cutoff is used to identify probes that correspond to SFPs.
The microarrays used here were designed to detect CNPs in large segments of the genome and consequently, were not targeted or specifically designed for SNP detection. Population genomic studies will require optimal identification of CNPs that could be important determinants of emerging drug resistances; however a powerful corollary to this approach will be to encode a set of high-resolution SNP markers that can be genotyped on the same chip. We observed that some known SNPs sufficiently influenced hybridization to be detected. Here, we assess the overall capabilities of a first generation P. falciparum probe set designed for the NimbleGen microarray platform to correctly identify SNPs genome-wide. A comprehensive set of sequence polymorphisms between 3D7, HB3, and Dd2 was compiled using available sequence data [3], and was compared to CGH data from 3D7-HB3, 3D7-Dd2, HB3-Dd2 hybridizations. We determined SNP sensitivity and SNP specificity in a completely unbiased, whole-genome method. Sensitivity with respect to probe length, melting temperature, GC content, SNP location, and base mutation revealed how these parameters affect SNP detection, and will facilitate the production of an openly available platform designed for unbiased, genome-wide surveys of polymorphisms optimized for SNP detection. The custom design capabilities of this high-resolution microarray platform can be cost-effectively optimized for individual experiments and will facilitate community access to a powerful genomics tool.
Results
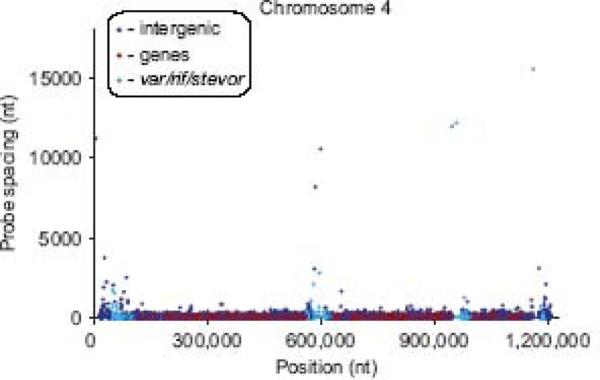
385,585 probes designed from the P. falciparum 3D7 genome were synthesized on microarrays through maskless photolithography [22, 23]. The probe set consisted of variable length probes ranging from 45 to 83-mers designed to all regions of the genome, including genes and intergenic regions. This probe size range is optimal for detection of CNPs but is considerably longer than has been described for SFP genotyping. Each probe corresponded to a unique sequence that occurred only once in the entire reference genome with a 48 bp median probe spacing and an 8 bp median probe overlap. Probe coverage on this array represented 69.4% of the nucleotides in the complete reference genome and 77.7% of nucleotides in genes. Coding regions are well-represented in the probe set by virtue of their sequence characteristics, not because they were targeted by this design as has been the case for other expression-focused chips. Regions of the telomeres and subtelomeres, as well as other highly repetitive regions, were covered less densely, including some large gaps that will be inaccessible to microarray-based approaches because sufficiently unique probes cannot be designed. Regardless, our design procedure provides an unbiased assessment of the uniqueness of genome regions that will be most amenable to chip-based genotyping as well as other shotgun and `next-gen' technologies. Genome wide probe distribution (Fig. 1) illustrates the wider probe spacing in the subtelomeric regions and in areas of polymorphic multigene families. Unique probes were found in these regions but at reduced density. In intergenic regions, there was no median probe overlap, but in genes – excluding var/rif/stevor genes – there was a 17 bp median probe overlap. On average, genes had probes with 16 nt overlap, intergenic regions had no probe overlap, and genome-wide – including all repetitive regions – probes had an 8 nt overlap.
Figure 1. Microarray probe spacing on chromosome 4.
Spacing between adjacent probes is plotted by probe location, as measured in nucleotides. Probes are colored according to gene location as indicated in the legend. Gaps are visible where unique probes could not be designed due to low sequence complexity or repeated sequences.
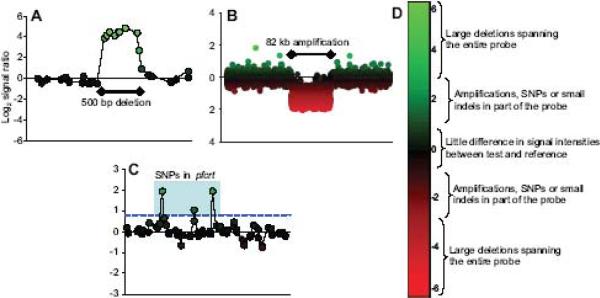
A variety of polymorphisms was detected between 3D7 and test samples, including amplifications, deletions, and possible SNPs or other SFPs (Fig. 2; Supplemental Fig. S1). SNP loci were detected when a base change overlapped a probe in the test gDNA in comparison to the reference gDNA, impacting the relative hybridization of the test and reference gDNAs that led to a signal displacement towards the reference. This was illustrated in a 3D7-Dd2 hybridization for which a signal bias towards 3D7 was observed for probes that interrogated known polymorphisms in Dd2 pfcrt, the P. falciparum chloroquine resistance transporter gene (Fig. 2C). Similarly, hybridization signals were displaced for various structural features; extreme signal bias was associated with deletions while smaller biases were associated with amplifications, SNPs, or indels (Fig. 2D). Large, segmental features were easily recognized because of their affect on multiple, contiguous probes allowing accurate breakpoint identification within 300 nt of the exact breakpoint (Supplemental Fig. S2).
Figure 2. Feature detection by CGH microarrays.
Hybridization signals on CGH microarrays identify various polymorphisms. Each data point is a microarray probe and its log2 signal ratio (y-axis) is plotted by genome position (x-axis). (A) A 500 bp segment is deleted in HB3 and corresponds to a set of probes exhibiting a very strong hybridization bias towards 3D7. (B) An 82 kb amplification is detected in Dd2 showing a moderate hybridization bias towards Dd2. (C) SNPs in Dd2 pfcrt are detected by individual probes exhibiting a hybridization bias towards 3D7 (light blue box). A blue dashed line indicates the 0.8 cutoff used to indicate the presence of SFPs. (D) Signal ratio ranges are associated with different types of polymorphisms.
Overall, 17,966 SFPs were detected between HB3-3D7 or Dd2-3D7 using a stringent 0.8 signal ratio cutoff (segmental amplifications and deletions were omitted from this analysis due to the confounding effect on hybridization signal: 24,779 SFPs were detected in the entire genome when including SFPs in amplifications or deletions). The majority of these probes were in highly polymorphic, multigene family members (var/rif/stevor genes); because individual parasite genomes have unique complements of these multigene families, signal bias often is due to factors other than point mutations so we have also defined our optimizing parameters by excluding the multigene families. The total number of SFPs detected was 3235 when polymorphic multigene families were excluded (Supplemental Table S1). As a first unbiased pass with no attempt to specifically target SNPs and using longer oligonucleotide probes, this number of SFPs suggests a strong potential for tiled CGH chips to include dense SNP marker coverage. However it is necessary to understand the signal to noise relationship in these hybridization ratios.
CGH detects SNPs with high specificity
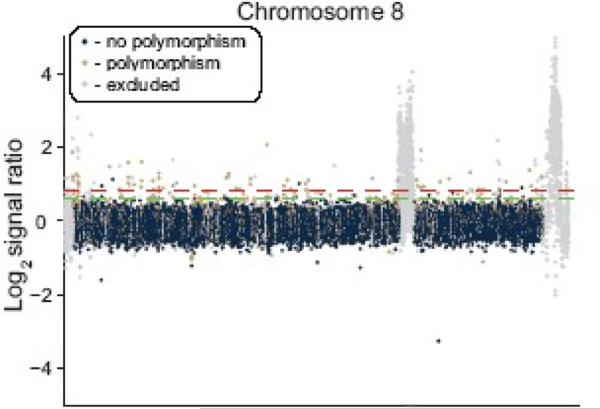
CGH signal indicates the presence of various sequence polymorphisms when an individual probe's signal exceeds a threshold that separates signal from noise (Fig. 3). This SNP detection was confounded at deletions, amplifications, and polymorphic multigene families; i.e. signal deviation reflected more than point mutations. Therefore probes in these regions were omitted from SNP analysis. Additional probes were omitted because homologous sequence could not be identified in the genome assemblies due to incomplete assemblies, or polymorphic sequences where there was not sufficient HB3/Dd2 sequence identity to map to 3D7 (light gray probes in Fig. 3). Approximately 11% and 29% of all probes were excluded from the HB3 and Dd2 hybridizations, respectively. With the exception of removing these regions of potentially confounding signals and inaccuracies, our analysis is the first in P. falciparum to comprehensively assess genome-wide SNPs without pre-selection based on specific characteristics.
Figure 3. SNP detection between HB3 and 3D7.
CGH data for chromosome 8 from a 3D7-HB3 hybridization. Probes that fall above a signal ratio cutoff are classified as SFPs. The signal ratio may be adjusted for more aggressive or conservative identification of polymorphic loci − 0.8 and 0.6 cutoffs are indicated by red and green dashed lines, respectively. Probes are colored according to sequence predictions made through the HB3 genome assembly as indicated in the legend. Probes in deletions, amplifications, and multigene families were excluded from subsequent SNP analyses. The internal region depicted on this chromosome spans a var cluster.
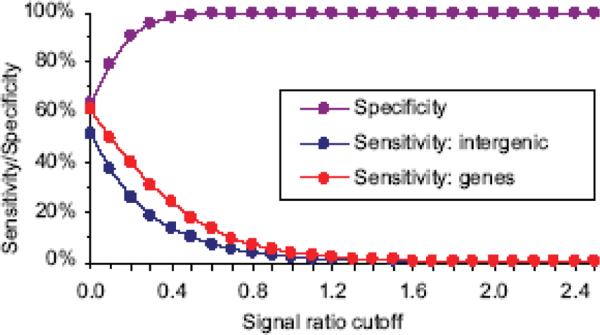
Quantitative error rates of genome-wide SNP detection for the CGH platform were calculated as sensitivity and specificity over a range of signal ratio cutoffs as shown in Fig. 4. Sensitivity is the portion of gold probes that exceeded the signal ratio cutoff in Fig. 3; i.e. the proportion of polymorphisms correctly recognized as SFPs. Specificity is the portion of blue colored probes that did not exceed the signal ratio cutoff in Fig. 3. Sensitivity and specificity analyses across the range of signal ratios characterized the relationship between false positives and false negatives. This identifies the point at which we achieve high specificity (i.e. few false positives were observed) while maximizing sensitivity (Fig. 4) leading us to chose the 0.8 signal ratio cutoff. Sensitivity was low in this dataset and a subtle but noticeable difference in sensitivity was observed between genes and intergenic regions. Specificity did not vary between genes and intergenic regions and was very high overall. At a 0.8 cutoff, specificity surpassed 99.9%. A key observation here is that false positives were extraordinarily rare in this dataset. Given the large number of true negatives (probes that did not span a polymorphism), a 0.8 cutoff yielding 99.94% specificity and 6.9% sensitivity in genes was used for all analyses. Significantly, our data indicate that cost-effective, single replication hybridization genotyping does not risk false positives, however it is certain that probe optimization for parameters affecting SNP detection and/or replication will provide increased sensitivity.
Figure 4. Genome-wide sensitivity and specificity for SNP detection.
Sensitivity and specificity are shown across a range of signal ratio cutoffs. Differences in specificity between genes and intergenic regions are negligible so they are not displayed separately.
Parameters affecting SNP detection
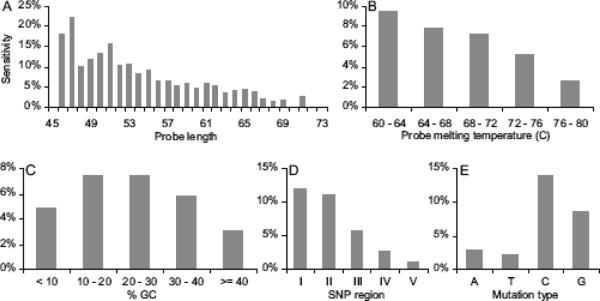
Probe and polymorphism characteristics including probe length and specific base variations affected the ability to detect SNPs. Fig. 5 illustrates the variable sensitivity to SNPs in genes according to probe length, melting temperature, GC content, SNP location in the probe, and point mutation type. Probe length analysis determined that shorter probes were more sensitive to SNPs (Fig. 5A). A 47 nt probe length had the greatest sensitivity at 22.2%. Probe lengths ranged from 45 – 83mers however none of the 45-mers encompassed SNPs while there were 141 probes longer than 71 nt that encompassed SNPs, but none of these detected polymorphisms. Notably, probe length is a key distinguishing feature of platforms dedicated to CNP vs. SNP detection. Probe melting temperatures ranged from 60 – 80°C and analysis showed that probes with lower melting temperatures were more sensitive (Fig. 5B). The lowest melting temperature group had the greatest sensitivity at 9.5% while the highest melting temperature group had 2.6% sensitivity. Probe GC content ranged from 4 – 52% and intermediate GC content showed the highest sensitivity at 7.4%. Probes with the most extreme GC content had the worst sensitivity; no probes with GC content greater than 45% detected SNPs (Fig. 5C). A SNP region analysis examining the effect of SNP location divided the probe into five equal regions: region I was the 5' section, region V was the 3' section, and region III occupied the center of the probe. Region I had the highest SNP sensitivity (12.0%), and this degraded when the SNP was closer to the 3' end reaching as low as 1.2% sensitivity in region V (Fig. 5D). Finer scale analysis demonstrated that there was degraded sensitivity at the extreme 5' end and that maximum sensitivity was near the junction of regions I and II (Supplemental Fig. S3). Mutation type analysis grouped SNPs by original base (in the 3D7 reference genome) synthesized into the microarray probe. As shown in Fig. 5E, SNP sensitivity was drastically worse for mutations that diverged from A's and T's (2.9% and 2.2% respectively) compared with detecting mutations that diverged from C's and G's (14.0% and 8.6% respectively). The highest sensitivity was observed for the C to A mutation (18.0%).
Figure 5. The effects of probe and polymorphism parameters on sensitivity to SNPs.
All graphs display sensitivity on the y-axis. Calculations are for probes in genes. SNP sensitivity is measured for (A) Probe length. (B) Probe melting temperature. (C) Probe GC content. (D) SNP region: location of the SNP in the probe where region I is 5' and region V is the 3' end. (E) Mutation type: base changes classified by the nucleotide in the reference genome; e.g. the C column represents sensitivity to C→A, C→T, or C→G point mutations.
Secondary structure affects polymorphism detection
CGH probes or the hybridized gDNA could contain sequences with inverted, self-complementary stretches that were prone to forming hairpin structures. The base composition bias of the P. falciparum genome enhanced the likelihood of such secondary structure formations. These features could interfere with polymorphism detection, especially when the hairpin structure occurred in a region of the probe that targeted polymorphic nucleotides, or the hairpin structure was stable enough to affect general probe-target hybridization.
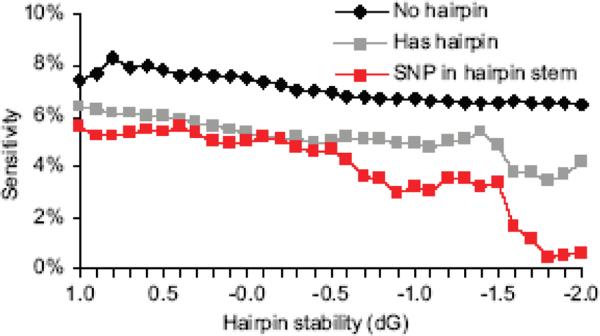
Polymorphisms in pfcrt were detected by individual probes as shown in Fig. 2C. However, other polymorphisms in pfcrt did not surpass the 0.8 signal ratio cutoff. A comprehensive list of polymorphisms in the pfcrt Dd2 allele was taken from PlasmoDB [24] and mapped to the probe set revealing that three SNPs were not detected by probes. Secondary structure predictions of these probe sequences revealed relatively stable hairpin structures, or hairpins in the probe region that queried the polymorphism (Supplemental Table S2; Supplemental Fig. S4). Probe hairpin analysis relied on UNAFold [25] for secondary structure and change in free energy (dG) predictions where dG was used as a measure of hairpin stability; structures with a lower dG were more stable. It was possible for a sequence to have multiple folds and the analysis took all predicted structures into account. Reduced sensitivity was observed for probes with hairpin structures, and this reduced sensitivity was more pronounced when the SNP occurred in the stem of a hairpin structure (Fig. 6). With increasing hairpin structure stability, SNP sensitivity was further reduced.
Figure 6. Hairpin structures reduce SNP sensitivity.
All probe sequences were examined for potential to form secondary structures. In some cases, multiple structures of varying stability are predicted for a single sequence. The change in free energy (dG) indicates relative hairpin stability. Probes were classified as containing a hairpin, or containing no hairpins across a range of free energy cutoffs. The probes that span a SNP in the hairpin stem give the lowest sensitivity at all free energies.
CGH detects other polymorphisms
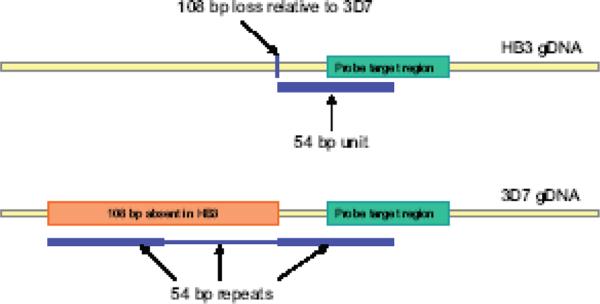
Rare false positives for presumed underlying SNPs were observed; at the 0.8 signal ratio cutoff used in this study, approximately 10% of the SFPs corresponded to areas where sequencing data indicated no polymorphisms. Possible explanations include inconsistency due to hybridization noise or aberrations, incorrect sequence data, or that different mutations had accumulated in parasites cultivated from the same initial clones over time. We chose 10 false positives from HB3 for resequencing where available sequence information indicated the sequences were identical to 3D7. Each of these positives was in a gene and multiple CGH experiments corroborated the presence of HB3 polymorphisms (data not shown). Suprisingly, resequencing revealed no sequence polymorphisms at any of the probe target sites in the DNA. Further inspection of sequencing data revealed other, unexpected polymorphisms (Supplemental Table S3). Each of the resequenced loci had polymorphisms from one of two classes: 1) SNP adjacent to the probe target site; 2) variable number tandem repeat polymorphisms (Fig. 7). In cases where a SNP was found adjacent to the probe target site, secondary structure predictions revealed that the SNP gave rise to new hairpin folds. These structures were predicted to be more stable and form in regions of the gDNA targeted by the probes. In the second polymorphism class, tandem repeat units ranged in size from 3 – 54 bp. At each of these loci, 3D7 and HB3 differed in the number of times the sequence was repeated, but both strains carried the repeat. This repeat copy polymorphism ranged from a difference of 1 to 5 units. The repeats partially overlapped parts of the nucleotide sequence represented in the probes, however, the differing number of repeats caused no sequence polymorphism at the probe target sequence.
Figure 7. CGH detects VNTR polymorphisms between 3D7 and HB3.
Schematic of the 3D7 and HB3 alleles for an SFP locus on chromosome 1. At this locus, a 54 bp unit is repeated three times in 3D7 but is present only once in the HB3 genome. This 54 bp repeat partially overlaps with the probe target area in the gDNA.
CGH SNP sensitivity limits
A focused analysis of a 200 kb region of chromosome 8 (nucleotides 80,000 – 280,000) was chosen for having relatively high probe density (median probe overlap of 12 nucleotides including genes and intergenic regions). At a 0.8 signal ratio cutoff, 12.8% of all SNPs in genes were detected. In some cases, a single SNP was queried by two or three different probes; if one of these probes exceeded the signal ratio cutoff, then a SNP was considered to be detected. For SNPs covered by multiple probes in this 200 kb region, 21.6% were detected.
Discussion
It is possible to detect SNPs with high specificity in the P. falciparum genome using a CGH design optimized for copy number variation. Previous investigations of probe design relied on limited sets of probes to examine individual parameters and often hybridize artificial samples such as PCR products [26–30]. The results from the current study were derived from more then 300,000 probes using complex biological samples and a global SNP set for HB3/Dd2/3D7 derived from their genome sequences. We observe very high specificity, however our chip optimized for CNP was not sensitive for whole-genome level SNP detection. Because our study comprehensively evaluated SNPs, without pre-selecting for favorable characteristics, it was possible to assess the probe features that dictate sensitivity and specificity. We define the probe parameters that define the degree to which point mutations can be assayed reliably. Our analysis differs from, but compliments the recent study by Jiang et al. which used the Affymetrix PFSANGER microarray with over 2.2 million P. falciparum probes to focus on SNP detection in a set of 2651 SNPs reporting 81% sensitivity [11]. Together, these studies demonstrate that gene chips can carefully target “high quality SNPs” to provide very high sensitivity and specificity that will be necessary for high-resolution genome-wide association studies.
Sensitivity for SNP genotyping by hybridization is compounded by the high AT content of the P. falciparum genome. For example, sensitivity was better in coding regions, reflecting the relatively low sequence complexity and repetitive stretches in intergenic regions. Decreased probe density in subtelomeric regions and polymorphic multigenic families illustrate the challenge of assaying these regions using tiling and resequencing arrays, just as they are not amenable to PCR applications or assembly following shotgun sequencing. Probe performance parameters have been evaluated for gene expression applications [26], however in gene expression studies, sensitivity refers to the ability to detect the relative abundance of transcripts to minimize the impact of sequence polymorphisms on hybridization signals. Conversely in CGH applications used to identify SFPs, sensitivity refers to the ability to detect small polymorphisms through their impact on hybridization signal. This distinction is important because relatively long probes increase transcript detection sensitivity in gene expression arrays, and also CNP detection in gDNA, but decrease polymorphism detection sensitivity in CGH arrays.
The customizable microarray synthesis outlined here provides a cost-effective approach to generate microarrays with variable length probes, allowing for iterative design to incorporate new data to optimize between experiments or for particular targets. Other studies have reported that longer probes are prone to cross-hybridization or hybridizing to targets with one or more polymorphisms [26, 30]. For P. falciparum SNP detection, the optimal probe length or melting temperature has not been defined. Our data indicate a steady improvement in performance with shorter probe lengths down to 45 nt. However, at some point shorter probes should become detrimental to SNP detection and it is not established that commonly used 25-mers are the optimal length, especially in the AT rich P. falciparum genome. A study of probe length effect on mismatch discrimination provides some insight into this issue [30]. Perfect match (PM): mismatch (MM) signal ratio comparison for 25, 30, and 35-mers finds that 30-mers have a greater ability to distinguish PM/MM probes with a signal ratio > 2. Although 25-mers have a greater average PM:MM ratio, 30-mers were able to distinguish a greater percentage of PM/MM pairs with a 2-fold signal difference. Another study estimates that 50-mers are less prone to cross-hybridization than 25-mers [26]. For SNP calling, probes longer than 25-mers may be able to correctly type more loci and probes designed at variable lengths for each individual target may provide even greater discrimination. Probes longer than uniform 25-mers employed by Affymetrix, likely will give greater specificity in an AT rich genome while enhancing CNP analysis. Studies comparing variable probe length performance to fixed length performance will be helpful in optimizing probe design.
Probes with the highest melting temperatures and highest GC content have greater thermodynamic stability leading to lower SNP sensitivity. This observation agrees with Letowski et al. who demonstrate that 56% GC 50-mers hybridize to targets carrying five central mismatches with no appreciable loss of signal relative to perfect match targets under 42°C and 47°C hybridization conditions [28]. They find, however, that when the five mismatches are distributed across the probe or the probe is 42% GC, there is an appreciable decrease in signal intensity. A SARS resequencing chip also shows reduced polymorphism recognition associated with high probe GC content [31].
Greater sensitivity to base changes from G or C in the reference 3D7 sequence can be explained by the loss of the stronger G-C triple bond as being more disruptive to probe hybridization than losing the weaker A-T double bond. In addition to hydrogen bonding, base size can also allow specific mutations to be more readily detected; this can account for why the microarray is most sensitive to C to A mutations. In a C-G base pair, a C to A mutation would lead to an A-G mismatch composed of two large bases ultimately causing greater disruption of double helix stability than other base pairs.
SNPs that reside nearer to the 5' end of the probe are more efficiently detected indicating that steric hindrance is occurring where the DNA fragments are physically unable to access the 3' end of the probes; the NimbleGen CGH platform uses probes that are 3' anchored. The Affymetrix platform uses probe pairs consisting of a perfect match and mismatch (MM) such that a mismatched nucleotide is centrally located in the MM probe. The underlying assumption is that a central mismatch is most disruptive to hybridization so the MM signal is a measure of non-specific hybridization [32]. Future designs can account for this by including probes which are 5' anchored or by using spacer molecules to distance the oligonucleotides from the solid support [33]. By targeting optimal SNP defined by our parameters, sensitivity will be drastically improved if the probes are designed such that the polymorphic nucleotide resides nearer to the 5' end of the probe.
Stable hairpins interfere with SNP detection and this is most evident when SNPs occur in predicted hairpin stems. Hairpin stems are less accessible for target sequence hybridization, and relatively stable hairpins can affect general hybridization to the probe. In cases where SNPs in the DNA cause new or more stable hairpins, the secondary structure may cause those gDNA fragments to be less competitive in hybridizing to the microarray probes meaning the SNPs are easier to detect. In addition to self hybridization in the probes and gDNA, it is likely there is also some interstrand hybridization of the gDNA fragments.
Detailed investigation of false positive SFP led us to uncover additional polymorphisms of two different types. The minority class consisted of SNPs adjacent to the probe target area which allowed more stable hairpin structures encompassing the target sequence. These are not false positives because there is an underlying polymorphism. However, it is important to note from this observation that not only is the probe sequence important, but qualities of the gDNA fragments themselves including target flanking regions may affect probe-target hybridization. The second class consisted of previously unrecognized VNTRs which are currently being analyzed through comparative genomics studies. None of the false positives we investigated were true false positives because they identified the presence of previously unrecognized polymorphisms.
Our sensitivity and specificity was derived from a global analysis of 3D7/HB3/Dd2 sequence polymorphisms. Specifically, we did not target high-quality SNPs, nor selectively exclude challenging SNPs as is often done when reporting genotyping parameters. Additionally, we treated the var/rif/stevor and amplification/deletion SFPs separately because they would artificially inflate our apparent capacity to detect SNPs based purely on threshold levels. When analyzing a region of relatively high probe density there was greater SNP sensitivity: 6.9% genome-wide compared to 12.8% in the high probe density region (Table 1). Furthermore, and not surprisingly, multi-probe coverage of a SNP was associated with increased sensitivity (21.6%). This performance difference reflected SNP location in the probe and illustrates the improvement of a new design that incorporates this feature to target SNPs. For a SNP, an ideal probe can be designed to optimally target that SNP, however, in the case of SNPs with multi-probe coverage, it was not expected that all SNPs were interrogated by the ideal probe, so allele-specific probes designed to optimally target individual SNPs would be expected to give performance surpassing 21.6% SNP sensitivity. Our data indicate that cost-effective, single replicate hybridization genotyping does not risk false positives and that SNPs can be typed genome-wide at less then 1 SNP per 4 kb. Probe optimization will further increase this resolution.
Table 1.
CGH SNP sensitivity in probe subsets
| Probe subset | SNP sensitivity |
|---|---|
| Genome-wide | 6.9% |
| High probe density | 12.8% |
| High probe density, multiprobe coverage | 21.6% |
SNP sensitivity in genes is low throughout the genome. Regions of higher probe density exhibit greater SNP sensitivity. With replicated data where statistical tests can identify more subtle but consistent signal deviations, SNP sensitivity is even greater.
Optimization for SNP detection will include optimizing probe parameters identified in this study: shorten probe lengths to 50-mers or lower, restrict probe sequences to a moderate GC content, eliminate probes that form stable hairpin formations, target GC point mutations, place SNPs off-center in the probe towards the 5' end. Additional P. falciparum studies provide insights [10, 11] and other developments can contribute to SNP sensitivity. For example, gDNA fragment length significantly affects probe performance [29] and it is likely that standard 42°C hybridization conditions could be varied, especially to accommodate the AT rich genome [28]. The use of nucleotide analogues – such as peptide nucleic acids – could also be adapted to photolithography [34] to take advantage of their greater binding affinities to compensate for the presence of secondary structure or allow greater mismatch discrimination [35]. More extensive use of artificial mismatches using base analogs, especially in resequencing or SNP applications, should be considered in light of the findings that they enhance SNP detection [36], that distributed mismatches are more disruptive to hybridization [28], and that there are unique interactions for different mismatched base pairs that are still not fully understood [37, 38].
Microarrays offer vast opportunities for genome-wide surveys of genetic polymorphisms. However, many polymorphisms will not be accessible to any hybridization-based approaches. Using current technology, we estimate that a redesigned probe set may be able to detect up to 18% of all P. falciparum SNPs; this is a genome-wide sensitivity rate and not one for a limited set of high quality SNPs. Sensitivity will improve with the use of allele specific probes where we estimate a genome-wide SNP sensitivity rate that exceeds 22%. Significantly, we show that single replicate hybridization allows genotyping without risking false positives. As chips advance to greater probe densities, increased probe numbers will allow increased resolution and redundant coverage for more accurate and precise mapping of polymorphisms. It is apparent from human studies that genome structural variation may be an important contributor to polymorphism that must be interrogated in addition to SNPs [39]. The information from this study will lead to an openly available, refined probe set that measures genome structural variations and has been optimized for SNP detection in P. falciparum. These technologies will ultimately allow quicker and more cost effective identification of all polymorphisms present in a sample which can be linked to phenotypes or used in evolutionary studies.
Methods
Fresh cultures of 3D7, HB3, Dd2, and clones progeny lines derived from genotyped stock material were grown at 37°C and 5% hematocrit in O+ human red blood cells (Indiana regional blood center, Indianapolis, IN) using RPMI 1640 with L-glutamine (Invitrogen, Carlsbad, CA) supplemented with 50 mg/L hypoxanthine, 25 mM HEPES, 0.5% Albumax I (Invitrogen), 0.25% sodium bicarbonate and 0.01 mg/ml gentamicin under an atmosphere of 90% nitrogen, 5% oxygen, 5% carbon dioxide. Culture media was changed every 1–2 days and parasitemia was maintained below 6%.
Microarray probes were selected using NimbleGen's standard CGH probe design protocol, modified for the P. falciparum genome [40]. Briefly, probes were tiled through the genome at a 4 bp interval spacing using filtering criteria of Tm 60–80°C and length 45–85 bp. The resulting probes were clustered with nearest neighbors and sorted to remove probes with extensive sequence identity to any other probe. Regardless of length, any probe with more than one 45-mer exact match in the genome was discarded. The most unique 385,585 probes, based on average 15-mer frequency and base pair composition score, were synthesized on the chip using maskless photolithography [22, 23]. The current CGH design, “080222_Plasmodium_3D7_WG_CGH” can be ordered from NimbleGen. All raw data and information concerning our optimization and ongoing designs will be updated and available at: http://www.nd.edu/~ferdilab/.
Labeling and hybridization were conducted using standard procedures for CGH [40]. Genomic DNA was sonicated to achieve random 500 – 2000 bp fragments. DNA was denatured at 98°C for 10 minutes in the presence of 1 O.D. Cy3 or Cy5 labeled random 9mers (TriLink Biotechnologies, San Diego, CA). The denatured sample was quick chilled in ice water and incubated with 100 units Klenow fragment (NEB, Ipswich, MA), and dNTP mix (6 mM each in Tris, EDTA (TE), Sigma Aldrich, St. Louis, MO) for 2 hours at 37°C. Reactions were terminated by addition of 0.5 M EDTA, precipitated with isopropanol, and resuspended in water. The test and reference samples were combined (13 μl each), dried in a Speed-Vac on low heat, and resuspended in hybridization buffer (NimbleGen Systems, Inc., Madison, WI). The combined sample was denatured at 95°C for 5 minutes, and cooled prior to hybridization overnight (16–20 hours) at 42°C. The microarrays were hybridized in a MAUI Hybridization Station (BioMicro Systems, Salt Lake City, UT). Microarrays were sequentially washed in Wash buffer I (15 seconds at 42°C; 2 minutes at room temperature), Wash buffer II (1 minute at room temperature), Wash buffer III (15 seconds at room temperature; NimbleGen Systems, Inc.), and dried in an Array-Go-Round (NimbleGen Systems, Inc.) for 1 minute.
Microarrays were scanned at 5 μm resolution using a GenePix4000B scanner (Axon Instruments, Molecular Devices Corp., Sunnyvale, CA). Data was extracted from scanned images using NimbleScan extraction software (NimbleGen Systems, Inc.). The Cy3 and Cy5 signal intensities were normalized through the Qspline implementation in the affy Bioconductor package (www.bioconductor.org) [41].
Contigs from the HB3 and Dd2 genome assemblies were downloaded from the Broad Institute's website (http://www.broad.mit.edu). BLAST databases for HB3 and Dd2 were created from the genome assemblies using formatdb from the NCBI toolkit. A BLAST search was conducted using each probe sequence against the HB3 and Dd2 assemblies using blastall from the NCBI toolkit with low complexity filtering turned off and an e-value cutoff of 1e-16. The BLAST results were parsed with a custom perl script using the bioperl Bio:SearchIO module and a custom perl module. Only blast hsp alignments spanning the entire probe length were considered. This script tallied the number of perfect matches, number of mismatches, and specific mutations that occurred in mismatches. Parsed results were stored in a database to allow cross-referencing with probe information and hybridization signals.
Regions of amplification or deletion were identified using Spotfire DecisionSite visualization software (www.spotfire.com). These segments were not included in subsequent SNP analysis. Probes aligning with annotated polymorphic, multigene families (var/rif/stevor) also were excluded from SNP analysis. Custom perl and bash scripts were written to evaluate individual polymorphisms to determine if the particular probe overlapping a polymorphism surpassed the hybridization signal ratio cutoff under investigation. Because the polymorphism was known, it was possible to classify the result for each probe – as True/False Positive/Negative for HB3 and Dd2 – which were tallied and binned by parameter of interest and sensitivity and specificity values calculated. In the SNP region, mutation type, and secondary structure analyses, only probes that spanned a single SNP were analyzed; all other analyses included probes that spanned multiple SNPs and/or small indels.
Sensitivity and specificity were calculated by:
where the condition is determined from the genome assemblies and the test outcome is determined from the CGH data.
SNP sensitivity analysis of HB3×Dd2 progeny used Dd2 SNP loci where HB3 was identical to the 3D7 reference genome. Microsatellite markers were mapped to physical locations in the reference genome. SNPs between microsatellite markers at the same genetic distance were assigned parental alleles. SNPs between markers with a different genetic distance were not assigned parental alleles. Regions in intergenic regions, multigene families, amplification, and deletion were not included in this analysis. A t-test was used to test for significant differences in CGH signal at each SNP locus by allele type.
Secondary structure predictions and associated dG were generated by UNAFold [25] with the parameters:
All secondary structures and dGs predicted for each probe were parsed from the results and converted to a string with custom perl scripts for database storage and analysis. These results were cross-referenced with microarray data in database software and analyzed with a custom perl script. Some probe sequences had multiple potential folds and all predicted folds were taken into account during data analysis.
Oligonucleotide primers were designed using VectorNTI (Invitrogen) to amplify and sequence PCR fragments. Primers were obtained from IDT (Integrated DNA Technologies). PCR reactions were setup with 50 ng DNA, 2 μl of each primer (0.4 μM) and 46 μl of PCR master mix. All amplifications were performed at 94°C for 2 m; 35 cycles of 94°C for 30 s, 48°C for 30 s, 62°C for 1.5 min; and 62°C for 5 min. Five μl of the PCR products were run on a 1% agarose gel with ethidium bromide to check for quality of amplification. If a single band was observed with no extensive primer dimers, 5 μl of PCR product were treated with 2 μl ExoSAP-IT (USB, Cleveland, OH) at 37°C for 30 min and 80°C for 15 min. Cycle sequencing was performed on the treated PCR product using the Big Dye Terminator Sequence Reaction mix (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. Sequencing reactions were run on an ABI 3700XL DNA analyzer (Applied Biosystems). Sequencing data were assembled and analyzed using the Contig Express module of Vector NTI.
Supplementary Material
Acknowledgements
The authors wish to acknowledge L. Checkley for excellent technical support. We are grateful to the Broad Institute for making HB3 and Dd2 genome sequence data publicly available prior to publication. This research was supported by NIH grants A1055035 and AI072517 to MTF. J. Tan was supported by an Arthur J. Schmitt Presidential Fellowship from the University of Notre Dame.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–7. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Volkman SK, et al. A genome-wide map of diversity in Plasmodium falciparum. Nat. Genet. 2007;39:113–9. doi: 10.1038/ng1930. [DOI] [PubMed] [Google Scholar]
- [4].Winzeler EA, et al. Direct allelic variation scanning of the yeast genome. Science. 1998;281:1194–7. doi: 10.1126/science.281.5380.1194. [DOI] [PubMed] [Google Scholar]
- [5].Albert TJ, et al. Mutation discovery in bacterial genomes: metronidazole resistance in Helicobacter pylori. Nat. Methods. 2005;2:951–3. doi: 10.1038/nmeth805. [DOI] [PubMed] [Google Scholar]
- [6].Gresham D, et al. Genome-wide detection of polymorphisms at nucleotide resolution with a single DNA microarray. Science. 2006;311:1932–6. doi: 10.1126/science.1123726. [DOI] [PubMed] [Google Scholar]
- [7].Karlsson EK, et al. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat. Genet. 2007;39:1321–8. doi: 10.1038/ng.2007.10. [DOI] [PubMed] [Google Scholar]
- [8].Wellcome, Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Carret CK, et al. Microarray-based comparative genomic analyses of the human malaria parasite Plasmodium falciparum using Affymetrix arrays. Mol. Biochem. Parasitol. 2005;144:177–86. doi: 10.1016/j.molbiopara.2005.08.010. [DOI] [PubMed] [Google Scholar]
- [10].Kidgell C, et al. A systematic map of genetic variation in Plasmodium falciparum. PLoS Pathog. 2006;2:e57. doi: 10.1371/journal.ppat.0020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jiang H, et al. Detection of genome-wide polymorphisms in the AT-rich Plasmodium falciparum genome using a high-density microarray. BMC Genomics. 2008;9:398. doi: 10.1186/1471-2164-9-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mu J, et al. Genome-wide variation and identification of vaccine targets in the Plasmodium falciparum genome. Nat. Genet. 2007;39:126–30. doi: 10.1038/ng1924. [DOI] [PubMed] [Google Scholar]
- [13].Baruch DI, et al. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- [14].Smith JD, et al. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell. 1995;82:101–10. doi: 10.1016/0092-8674(95)90056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Su XZ, et al. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- [16].Aurrecoechea C, et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539–43. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rubio JP, Thompson JK, Cowman AF. The var genes of Plasmodium falciparum are located in the subtelomeric region of most chromosomes. EMBO J. 1996;15:4069–77. [PMC free article] [PubMed] [Google Scholar]
- [18].Junior L. H. Freitas, et al. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature. 2000;407:1018–22. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- [19].Kraemer SM, et al. Patterns of gene recombination shape var gene repertoires in Plasmodium falciparum: comparisons of geographically diverse isolates. BMC Genomics. 2007;8:45. doi: 10.1186/1471-2164-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cheng Q, et al. stevor and rif are Plasmodium falciparum multicopy gene families which potentially encode variant antigens. Mol. Biochem. Parasitol. 1998;97:161–76. doi: 10.1016/s0166-6851(98)00144-3. [DOI] [PubMed] [Google Scholar]
- [21].Kyes SA, Rowe JA, Kriek N, Newbold CI. Rifins: a second family of clonally variant proteins expressed on the surface of red cells infected with Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9333–8. doi: 10.1073/pnas.96.16.9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nuwaysir EF, et al. Gene expression analysis using oligonucleotide arrays produced by maskless photolithography. Genome Res. 2002;12:1749–55. doi: 10.1101/gr.362402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Singh-Gasson S, et al. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat. Biotechnol. 1999;17:974–8. doi: 10.1038/13664. [DOI] [PubMed] [Google Scholar]
- [24].Bahl A, et al. PlasmoDB: the Plasmodium genome resource. A database integrating experimental and computational data. Nucleic Acids Res. 2003;31:212–5. doi: 10.1093/nar/gkg081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–81. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Chou CC, Chen CH, Lee TT, Peck K. Optimization of probe length and the number of probes per gene for optimal microarray analysis of gene expression. Nucleic Acids Res. 2004;32:e99. doi: 10.1093/nar/gnh099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lane S, Evermann J, Loge F, Call DR. Amplicon secondary structure prevents target hybridization to oligonucleotide microarrays. Biosens Bioelectron. 2004;20:728–35. doi: 10.1016/j.bios.2004.04.014. [DOI] [PubMed] [Google Scholar]
- [28].Letowski J, Brousseau R, Masson L. Designing better probes: effect of probe size, mismatch position and number on hybridization in DNA oligonucleotide microarrays. J. Microbiol. Methods. 2004;57:269–78. doi: 10.1016/j.mimet.2004.02.002. [DOI] [PubMed] [Google Scholar]
- [29].Liu WT, Guo H, Wu JH. Effects of target length on the hybridization efficiency and specificity of rRNA-based oligonucleotide microarrays. Appl. Environ. Microbiol. 2007;73:73–82. doi: 10.1128/AEM.01468-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Relogio A, Schwager C, Richter A, Ansorge W, Valcarcel J. Optimization of oligonucleotide-based DNA microarrays. Nucleic Acids Res. 2002;30:e51. doi: 10.1093/nar/30.11.e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wong CW, et al. Tracking the evolution of the SARS coronavirus using high-throughput, high-density resequencing arrays. Genome Res. 2004;14:398–405. doi: 10.1101/gr.2141004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Selinger DW, et al. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat. Biotechnol. 2000;18:1262–8. doi: 10.1038/82367. [DOI] [PubMed] [Google Scholar]
- [33].Shchepinov MS, Case-Green SC, Southern EM. Steric factors influencing hybridisation of nucleic acids to oligonucleotide arrays. Nucleic Acids Res. 1997;25:1155–61. doi: 10.1093/nar/25.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Liu ZC, et al. Light-directed synthesis of peptide nucleic acids (PNAs) chips. Biosens Bioelectron. 2007;22:2891–7. doi: 10.1016/j.bios.2006.12.005. [DOI] [PubMed] [Google Scholar]
- [35].Armitage BA. The impact of nucleic acid secondary structure on PNA hybridization. Drug Discov. Today. 2003;8:222–8. doi: 10.1016/s1359-6446(03)02611-4. [DOI] [PubMed] [Google Scholar]
- [36].Guo Z, Liu Q, Smith LM. Enhanced discrimination of single nucleotide polymorphisms by artificial mismatch hybridization. Nat. Biotechnol. 1997;15:331–5. doi: 10.1038/nbt0497-331. [DOI] [PubMed] [Google Scholar]
- [37].Naef F, Lim DA, Patil N, Magnasco M. DNA hybridization to mismatched templates: a chip study. Phys Rev E Stat Nonlin Soft Matter Phys. 2002;65:040902. doi: 10.1103/PhysRevE.65.040902. [DOI] [PubMed] [Google Scholar]
- [38].Wang Y, Miao ZH, Pommier Y, Kawasaki ES, Player A. Characterization of mismatch and high-signal intensity probes associated with Affymetrix genechips. Bioinformatics. 2007;23:2088–95. doi: 10.1093/bioinformatics/btm306. [DOI] [PubMed] [Google Scholar]
- [39].Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Selzer RR, et al. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer. 2005;44:305–19. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]
- [41].Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.