Abstract
Smad6 and Smad7 are inhibitory Smads induced by transforming growth factor β-Smad signal transduction pathways in a negative-feedback mechanism. Previously it has been thought that inhibitory Smads bind to the type I receptor and block the phosphorylation of receptor-activated Smads, thereby inhibiting the initiation of Smad signaling. Conversely, few studies have suggested the possible nuclear functions of inhibitory Smads. Here, we present compelling evidence demonstrating that Smad6 repressed bone morphogenetic protein-induced Id1 transcription through recruiting transcriptional corepressor C-terminal binding protein (CtBP). A consensus CtBP-binding motif, PLDLS, was identified in the linker region of Smad6. Our findings show that mutation in the motif abolished the Smad6 binding to CtBP and subsequently its repressor activity of transcription. We conclude that the nuclear functions and physical interaction of Smad6 and CtBP provide a novel mechanism for the transcriptional regulation by inhibitory Smads.
Members of the transforming growth factor β (TGF-β) proteins are secreted multifunctional proteins that exhibit a diverse set of cellular responses. TGF-βs induce the expression of a variety of genes involved in cell proliferation and differentiation (8, 10, 25, 29, 31, 36). During development, TGF-β and related factors regulate cell and tissue differentiation, morphogenetic processes, and embryonic organization (33, 45). TGF-β expression and responsiveness also regulate human disease development (5, 9, 26, 35, 44).
TGF-βs activate type II and type I transmembrane serine/threonine kinase receptors (for recent reviews, see references 25 and 36). Ligand-activated receptors, in turn, activate intracellular effectors, Smads (for recent reviews, see references 8, 25, 29, 31, and 46). Smads form a family of proteins that have been evolutionarily conserved from Drosophila and Caenorhabditis elegans to vertebrates. Smad proteins are classified into three groups: receptor-activated Smad (R-Smad), common Smad (co-Smad, i.e., Smad4) and inhibitory Smad (I-Smad). Upon ligand stimulation, R-Smads are phosphorylated by the activated type I receptor and then form complexes with Smad4. The heteromeric complexes of R-Smads and Smad4 are then translocated into the nucleus, where they mediated ligand-induced changes in the transcription of a variety of genes (for reviews, see references 8, 11, 25, 29, 31, 46, and 47). The heteromeric Smad complex activates transcription through its ability to functionally cooperate with several promoter-specific transcription factors and/or to bind specific DNA sequences (11, 27).
I-Smads are induced by TGF-β/Smad signal transduction pathways in a negative-feedback mechanism (1, 17, 18, 41). Specifically, activin/TGF-β signaling induces Smad7 expression and bone morphogenetic proteins (BMPs) induce Smad6 expression, thus providing a ligand-induced negative-feedback loop for Smad signaling (1, 41). It is generally thought that I-Smads competitively interfere with the binding of the R-Smads to the type I receptor, thus preventing their phosphorylation (1, 18), and that Smad6 also interferes with the BMP-induced formation of the heteromeric Smad1-Smad4 complex (17). However, recent studies indicate potential functions of I-Smads in the nucleus. Smad6 physically links with Hoxc-8 and histone deacetylases(HDACs) to repress BMP-induced osteopontin gene transcription (3, 4). However, the precise mechanism of how Smad6 acts as a transcriptional (co)repressor remains to be elucidated.
Transcriptional repression, like transcriptional activation, has emerged as a common mechanism of transcriptional regulation (37). DNA sequence-specific repressors mediate their effect by recruiting corepressors to the target promoter. Corepressors are non-DNA-binding proteins, including mSin3A, SMRT/NCoR, Groucho, and C-terminal binding protein (CtBP). One mechanism of transcriptional repression is the recruitment of HDACs, which can enzymatically remove acetyl group from histones as well as nonhistone proteins, which is thought to cause a condensed, transcriptionally inactive chromatin. CtBP is a cellular protein that binds to the C-terminal region of the human adenovirus E1A proteins via the highly conserved PLDLS motif (6, 38). Two members of the CtBP family have been identified in humans, CtBP1 (the original E1A-interacting CtBP) and CtBP2 (7). CtBPs repress transcription in both an HDAC-dependent and HDAC-independent manner, depending on the promoter context. CtBP1 has been reported to be associated with HDAC1 and HDAC2 as well as class II HDACs (e.g., HDAC5) for the potential histone deacetylation on a target promoter (7).
In this study we provide compelling evidence for the association of CtBP1 with inhibitory Smad6. The physical interaction between CtBP1 and Smad6 was both direct and physiologically relevant, as demonstrated by using glutathione S-transferase (GST) in vitro binding assays and coimmunoprecipitation experiments. The CtBP binding was through the PLDLS motif in the linker region of Smad6. Notably, mutations in the PLDLS motif abolished the ability of Smad6 to bind CtBP and consequently decreased its capacity for inhibiting BMP signaling. The potential nuclear repressor functions of Smad6 are consistent with the presence of Smad6 protein in the nucleus and are further supported by the observation that Smad6 repressed transcription when tethered to Gal4-binding DNA. Thus, recruitment of CtBP by Smad6 provides a unique mechanism to directly inhibit BMP signaling in the nucleus.
MATERIALS AND METHODS
Plasmids.
Mammalian expression plasmids for Flag-tagged Smad6 and its HA-tagged Smad6 deletion mutants, the Smad6-Smad7 chimera (gift of Kohei Miyazono), were previously described (16). HA-tagged full-length Smad6 and Gal4-Smad6 were made by transferring the Smad6 coding region (EcoRI-XhoI) from Flag-Smad6 into the EcoRI and SalI sites of the pXF3H vector and pXF2Gal, respectively. A mutation in the CtBP-binding motif 290PLDLS294 in Smad6 was generated by PCR, and the mutated motif was cloned into pXF3H vector. HA-tagged Smad7 was similarly made in the pXF3H vector. NLS-Smad6 was made by transferring the Smad6 coding region (EcoRI-XhoI) into the EcoRI and SalI sites of the pXF1N vector, which contains an in-frame nuclear localization signal (NLS) sequence (PKKKRK) of simian virus 40 (SV40) large T antigen before the EcoRI site (unpublished data). Flag-CtBP (a gift of Richard Goodman) (50) and myc-CtBP (a gift of Eric Olson) (49) were previously described.
Northern blot hybridization.
Exponentially growing P19 cells were infected for 1 h with adenoviruses expressing Smad6 (104 PFU/μl; a gift of Jingsong Zhao [51]). After 24 h, the cells were treated with 25 ng of BMP2 per ml for 4 h. Total RNA was prepared using the TriZol kit (Invitrogen) as specified by the manufacturer. Equal amounts of RNA were then separated in an agarose gel and stained with ethidium bromide for visualization of RNA. RNA was then transferred to a nitrocellulose membrane. Hybridization to a 32P-labelled Id1 gene probe (human Id1 coding region [a gift of Xiao-Hong Sun]) was carried out at 65°C in a solution containing 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt's reagent, and 0.1% sodium dodecyl sulfate (SDS). Hybridized bands were then visualized by autoradiography.
Immunoprecipitation and Western blot analysis.
Immunoprecipitations using anti-Flag or anti-HA antibody were carried out essentially as described previously (14, 15). For detection of Smad6-bound CtBP as described in the legend to Fig. 4A, 293T cells were transfected with expression plasmids for HA-tagged Smad6 and Myc-tagged CtBP1. After 48 h, the cells were harvested in Flag lysis buffer (0.01 M Tris · Cl [pH 8], 300 mM NaCl, 1% Triton X-100). HA-Smad6 was immunoprecipitated using anti-HA antibody (12CA5 [Roche] and HA1.1 [Babco]) conjugated to protein A-agarose beads, and after extensive washing it was eluted in SDS sample loading buffer (Bio-Rad). Eluted proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE), transferred to nitrocellulose, and detected in an immunoblot with anti-HA antibody for Smad6 protein and anti-Myc antibody (9E10 [Santa Cruz Biotech]) for Smad6-bound CtBP protein. Antibody-bound proteins were visualized by horseradish peroxidase-conjugated secondary antibody followed by chemiluminescence (Pierce). Immunoprecipitation and Western blotting shown in Fig. 4B, 5B, and 6C were similarly conducted with the antibodies specified in the figure legends.
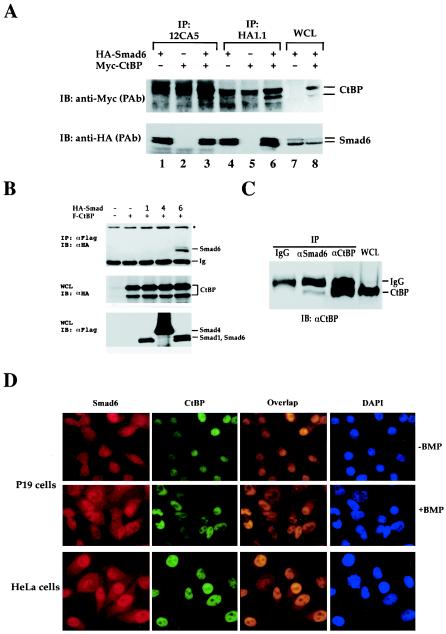
FIG. 4.
Smad6 interacts with the transcription corepressor CtBP in vivo. (A) Smad6 coimmunoprecipitates with CtBP. 293T cells were transfected with HA-tagged Smad6 and myc-tagged CtBP. Cell lysates were immunoprecipitated (IP) with anti-HA antibodies (12CA5 or HA1.1)and then immunoblotted (IB) with an anti-Myc polyclonal antibody to detect Smad6-bound CtBP (upper panel) and anti-HA to determine the level of immunoprecipitated Smad6 (bottom). Whole-cell lysates (WCL) were also directly immunoblotted with anti-Myc or anti-HA antibodies to demonstrate the expression of transfected CtBP and Smad6 (lanes 7 and 8). (B) Smad6 interaction with CtBP is specific. Immunoprecipitation-Western analysis was conducted as for panel A, except that anti-Flag and anti-HA antibodies were used for IP and IB, respectively. (C) Endogenous interaction between Smad6 and CtBP. The IP-Western blot procedure was carried out similarly to that in panel A, with the antibodies indicated. (D) Smad6 is colocalized with CtBP. Immunofluorescence was performed as in Fig. 2A.
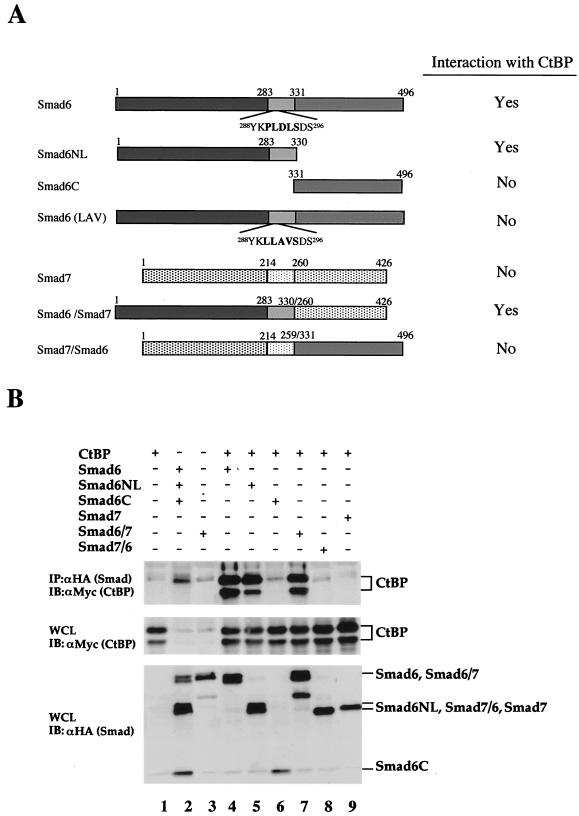
FIG. 5.
The Smad6 NL domain mediates its interaction with CtBP. (A) Schematic diagram of Smad6 constructs. Deletion mutants of Smad6 and Smad6-Smad7 chimera were shown by different shadings: dark gray, MH1; light gray, linker; medium gray, MH2. Their interaction with CtBP from immunoprecipitation experiments (B; see also Fig. 6C) is summarized. (B) Mapping of the CtBP-interacting domain on Smad6. 293T cells were transfected with HA-tagged Smad constructs and myc-tagged CtBP. Smad-bound CtBP was detected by anti-HA immunoprecipitation (IP) coupled with anti-Myc immunoblotting (IB) (upper panel). The levels of transfected protein in whole-cell lysates (WCL) are shown (bottom panels).
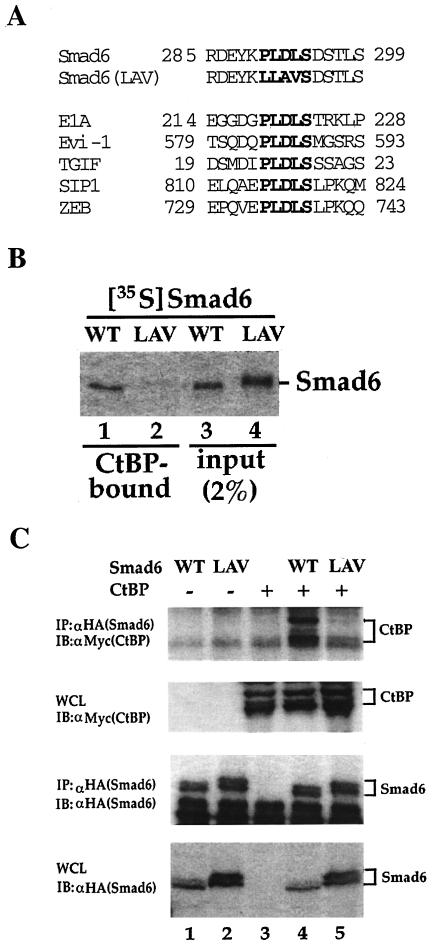
FIG. 6.
The PLDLS motif is essential for Smad6 interaction with CtBP. (A) Alignment of PLDLS motifs. The PLDLS motif of Smad6 and the mutated version are shown together with other CtBP-binding proteins. (B) The PLDLS→LLAVS mutation abrogates the binding of Smad6 to CtBP. Equal amounts (1 μg) of GST-CtBP fusion protein on glutathione-Sepharose beads were incubated with 35S-labeled Smad6 in vitro. After extensive washing, CtBP-Smad6 complex was resolved by SDS-PAGE and CtBP-bound Smad6 was visualized by autoradiography. WT, wild-type Smad6; LAV, Smad6(LAV) mutant. (C) The PLDLS mutant of Smad6 fails to interact with CtBP in vivo. 293T cells were transfected with HA-tagged Smad6 and myc-tagged CtBP. Smad6-bound CtBP was detected by anti-HA IP coupled with anti-Myc IB (upper panel). The levels of transfected protein in whole-cell lysates are shown. WT, wild-type Smad6; LAV, Smad6(LAV) mutant.
For endogenous Smad6-CtBP interaction (Fig. 4C), 293T cells were lysed in Flag buffer and immunoprecipitation was carried out using anti-Smad6, anti-CtBP, or a control antibody. The immunocomplex was then blotted using anti-CtBP antibody (Santa Cruz Biotech).
Transcription reporter assays.
Plasmid Id1-Luc, which contains the luciferase gene under control of the human Id1 promoter −1147/+88 (a gift of Tetsuya Taga with permission from Robert Benezra [32]), was used to measure BMP-induced transcription. Transfections and reporter assays were carried out as described previously (12, 13). P19 cells were transfected by using Lipofectamine (Invitrogen). Generally, exponentially grown cells at 25 to 30% confluency were transfected with expression plasmids for Smad6 and/or reporter plasmids. The amount of transfected DNA was always made the same by adding vector DNA when needed. At 36 h after transfection, the cells were treated with 25 ng of BMP2 (R&D System) per ml for 12 h. They were then harvested for measurement of luciferase and β-galactosidase activities. All assays were performed in triplicate, and all values were normalized for transfection efficiency against β-galactosidase activity.
For the Gal4 transrepression assay, a plasmid encoding Gal4-Smad6 (Smad6 fused to the Gal4 DNA-binding domain) was cotransfected with the Gal4-luciferase reporter plasmid Gal4-TK-Luc into P19 cells. Transfected cells were treated for 12 h with or 25 ng of BMP2 per ml or left untreated. The ability of Gal4-Smad6 to repress the heterologous Gal4-binding promoter was quantitated by measuring the luciferase expression from the Gal4-binding promoter.
Biotinylated DNA oligonucleotide precipitation.
Biotinylated Id1 promoter DNA oligonucleotide, corresponding to nt −919 to −889 (5′-Biotin-CAG GCC TGG CGT CTA ACG GTC TGA GCC GCT G -3′) was synthesized. Immobilization of biotinylated DNA and adsorption of cellular proteins to the DNA were carried out using Dynabeads (Dynal) as specified by the manufacturer. Briefly, 400 μg of whole-cell lysates from 293T cells was incubated with 40 pmol of biotinylated DNA at 4°C for 1 h in BW buffer containing 10 mM Tris · HCl [pH 7.5], 1 mM EDTA, and 2 M NaCl, followed by extensive washing in S1 buffer (10 mM Tris · HCl [pH 7.5], 10% glycerol, 150 mM NaCl, 0.5 mM EDTA, 0.5 mM dithiothreitol, 1 mM MgCl 2, 0.1% NP-40). DNA-bound protein was then subjected to SDS-PAGE followed by Western blotting using antibodies (see Fig. 7A). The cell lysates were immunoblotted with different antibodies to demonstrate the expression levels of transfected proteins.
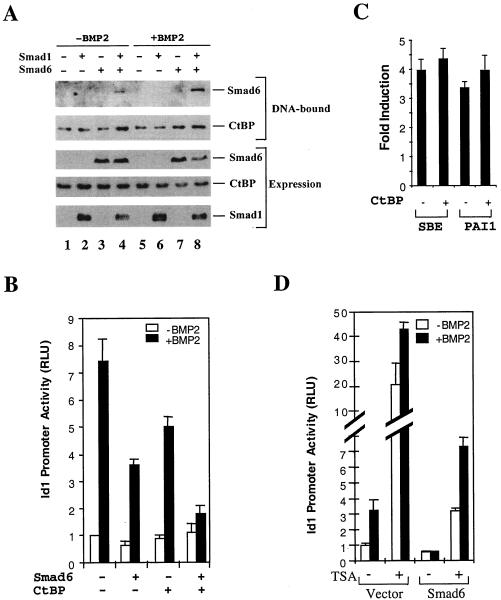
FIG. 7.
Smad6 and CtBP are recruited to the Id1 promoter, and they cooperate to repress the BMP-induced Id1 transcription. (A) Smad6 and CtBP coexist on the Id1 promoter. 293T cells were transfected with expression plasmids for Smad1 and Smad6 and treated with BMP2 for 4 h as indicated. (B) Smad6 cooperates with CtBP to inhibit BMP-dependent Id1 induction. P19 cells were transfected with the indicated expression plasmids. BMP2 treatment and the Id1-luciferase assay were done as in Fig. 1B. (C) Overexpression of CtBP has no effects on TGF-β-induced promoter activity. TGF-β-responsive HaCaT cells were transfected with SBE-luc or PAI1-luc, with or without CtBP, and treated with 5 ng of TGF-β per ml for 24 h. The y-axis value represents the induction by TGF-β, i.e., the ratio of Id1 promoter activity in the presence of TGF-β to that in the absence of TGF-β. (D) TSA inhibits the ability of Smad6 to repress BMP-induced Id1 induction. HeLa cells were transfected with the indicated Smad6 expression plasmids and treated with 10 ng of TSA per ml for 18 h. BMP2 treatment and the Id1-luciferase assay were done as in Fig. 1B. RLU, relative light units.
Immunofluorescence.
HeLa and P19 cells, untransfected or transfected as specified in the text and figure legends (see Fig. 2A, 3B, and 4D), were grown on coverslips, fixed with cold methanol, and blocked with 2% bovine serum albumin in phosphate-buffered saline (pH 7). The cells were then stained with anti-Smad6 polyclonal or anti-CtBP monoclonal antibodies, followed by Texas Red-conjugated anti-rabbit or fluorescein isothiocyanate (FITC)-conjugated anti-mouse antibody, and examined using a Zeiss Axioplan II microscope.
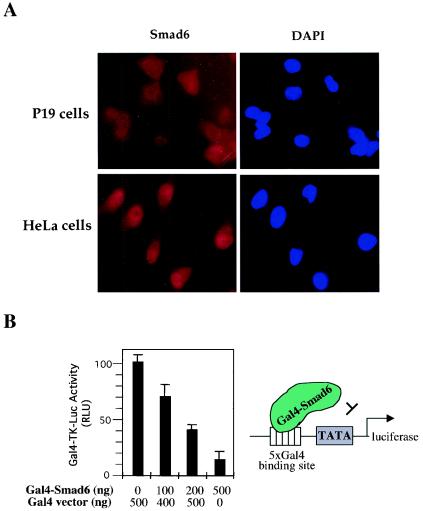
FIG. 2.
Smad6 has repressor activity. (A) Smad6 is localized in the nucleus and cytoplasm. Endogenous Smad6 was detected using anti-Smad6 staining followed by FITC-conjugated secondary antibody and visualized under a fluorescence microscope. DAPI indicates nuclear staining. (B) Smad6 has transcriptional repressor activity. P19 cells were cotransfected with Gal4-Smad6 and the luciferase reporter plasmid pGal4-TK-luc. The basal transactivation of the Gal4-TK-luc reporter is scored as 100 in the absence of Gal4-Smad6. Note that increasing levels of Gal4-Smad6 have an inhibitory activity on the Gal4-binding promoter. RLU, relative light units.
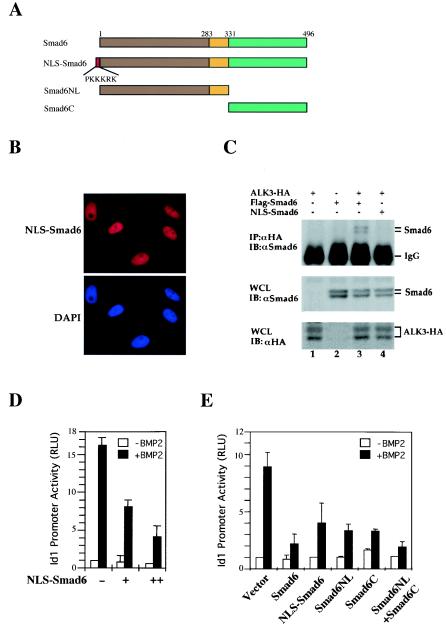
FIG. 3.
Nucleus-targeted Smad6 retains the ability to repress the BMP-induced Id1 response. (A) Schematic diagram of the structure of Smad6 and mutant versions of it. NLS-Smad6 contains an N-terminally fused NLS from SV40 large T antigen. (B) NLS-Smad6 is localized exclusively in the nucleus. NLS-Smad6 was transfected into P19 cells, and its expression was detected using anti-Smad6 staining followed by FITC-conjugated secondary antibody.DAPI indicates nuclear staining. (C) NLS-Smad6 does not bind to ALK3. 293T cells were transfected with HA-tagged ALK3 and Smad6 constructs. Cell lysates were immunoprecipitated (IP) with anti-HA antibodies (12CA5) and then immunoblotted (IB) with an anti-Smad6 polyclonal antibody to detect receptor-bound Smad6 (upper panel). Whole-cell lysates (WCL) were also directly immunoblotted with anti-HA or anti-Smad6 antibodies to demonstrate the expression of ALK3 and Smad6 (bottom panels). (D) NLS-Smad6 is capable of repressing the Id1 promoter. NLS-Smad6 was transfected into P19 cells together with Id1-luciferase reporter. BMP treatment and luciferase measurement were carried out as described in the legend to Fig. 1B. (E) Smad6NL and Smad6C synergize to repress the Id1 promoter. Various Smad6 constructs were transfected into P19 cells together with Id1-luciferase reporter. BMP treatment and luciferase measurement were carried out as described in the legend to Fig. 1B. Note that Smad6NL alone retains ability to repress the Id1 promoter. RLU, relative light units.
RESULTS
Smad6 inhibits BMP2-induced Id1 transcription.
Smad6 is an inhibitory Smad that is induced by TGF-β ligands (1, 17, 18). Several studies have shown that Smad6 forms nonregulatable associations with the BMP type I receptor and to a lesser extent with the TGF-β receptor (18). The Smad6-receptor interaction blocks the association of R-Smad to the receptor and subsequent phosphorylation (18). Meanwhile, Smad6 interacts with Smad1 to inhibit the receptor-mediated formation of Smad1-Smad4 heteromeric complex (17). Recent studies suggest that Smad6 has nuclear functions (3, 4).
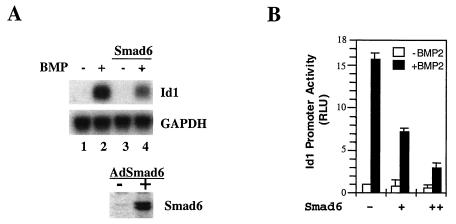
Smad6 may function as a repressor in many pathways, since its expression can be induced by different conditions such as growth factors (1, 18, 42). We first attempted to investigate whether Smad6 represses BMP-induced Id1 gene transcription. Id1 is an immediately-early response gene to BMPs, strongly induced in both human and mice (20, 21, 23, 30, 32, 43); it was therefore chosen for our analysis of the BMP response. To recapitulate this BMP response in mouse embryonic carcinoma P19 cells, we first analyzed the mRNA accumulation of Id1 in response to BMP2. As shown in Fig. 1A, Id1 mRNA was induced by BMP2 after a 4-h treatment (compare lane 2 with lane 1). Notably, overexpression of Smad6 inhibited BMP2-induced Id1 transcription. Adenovirus infection-mediated expression of Smad6 significantly inhibited BMP2-induced Id1 mRNA accumulation (compare lane 4 with lane 2).
FIG. 1.
Smad6 inhibits BMP-induced Id1 transcription. (A) Smad6 inhibits BMP2-induced Id1 mRNA accumulation. Exponentially growing mouse embryonic P19 cells were infected with adenovirus Smad6, and then treated with 25 ng of BMP2 per ml, as indicated. Id1 mRNA was detected by Northern hybridization. Equal levels of RNA were loaded per lane. GAPDH, glyceraldehyde 3-phosphate dehydrogenase. (B) Smad6 inhibits BMP-induced activation of the Id1 promoter in P19 cells. Cells were transfected with the Id1-luc reporter plasmid and, at 36 h after transfection, treated with BMP for 12 h; luciferase values were measured. RLU, relative light units.
We next examined the BMP2-dependent Id1 induction by using a BMP-responsive reporter gene Id1-luc, which contains the −1147/+88 upstream regulatory sequence of the Id1 gene linked to the luciferase reporter gene (32). Corresponding to the induction of Id1 mRNA (Fig. 1A), the promoter activity of Id1 was highly responsive to BMP treatment, with a 15-fold increase in the Id1 promoter activity in P19 cells (Fig. 1B). Transfection of cytomegalovirus-driven Smad6 significantly inhibited the Id1 promoter activity in response to BMP2 (Fig. 1B). Inhibitory function could be attributed to two separate functions of Smad6, i.e., its inhibition of R-Smad activation and/or its repressor activity in the nucleus.
Smad6 had transcription repressor activity.
To define the inhibitory role of Smad6 in BMP signaling in the nucleus versus the cytoplasm, we first examined the subcellular localization of endogenous Smad6. For this goal, we used human P19 and HeLa cells. As shown in Fig. 2A, Smad6 was localized in both the cytoplasm and nucleus of P19 cells, while it was located predominantly in the nucleus of HeLa cells.
The nuclear localization of Smad6 suggests that it may possess nuclear functions. Using the Gal4 transcription assay, we tested whether Smad6 had intrinsic repressor activity. Smad6 was fused to the GAL4 DNA-binding domain (Gal4-Smad6). The ability of Gal4-Smad6 to activate or repress the basal activity from a heterologous Gal4-TK promoter was assessed in P19 cells. We normalized the basal Gal4-TK promoter activity without Smad6 as 100%. As shown in Fig. 2B, Gal4-Smad6 decreased the Gal4-TK-luc promoter activity in a dose-dependent manner, with Gal4-Smad6 at high doses inhibiting the Gal4-TK-luc reporter to 15% (Fig. 2B). While studies have shown that BMP can induce transcription of the Smad6 gene (1, 18), it has no effects in regulating directly the repressing activity of Smad6 protein (data not shown). Thus, our findings suggest that Smad6 possesses intrinsic repressor activity and may be directly involved in the repression of gene transcription in the nucleus.
Smad6 inhibits BMP signaling independent of its binding to receptor.
To elucidate the nuclear function of Smad6, it was necessary to exclude the inhibitory effects of Smad6 at the BMP receptor level. To distinguish the inhibitory effect of Smad6 at the receptor level or the nuclear level, we engineered a Smad6 variant that contained the NLS sequence (PKKKRK) of SV40 large T antigen at the N terminus of Smad6, designated NLS-Smad6 (Fig. 3A). To test whether NLS-Smad6 was truly localized in the nucleus, we transfected this variant into P19 cells. Immunofluorescence examination of NLS-Smad6 showed its localization exclusively in the nucleus (Fig. 3B). In addition, in transfected cells, NLS-Smad6 did not coimmunoprecipitate with the BMP type I receptor ALK3, while wild-type Smad6 physically interacted with ALK3, strongly suggesting the exclusion of NLS-Smad6 outside of the nucleus (Fig. 3C). We next determined whether NLS-Smad6 could still inhibit BMP2-induced Id1 promoter activity. As shown in Fig. 3D, we observed that the ability of Smad6 to inhibit Id1-luc expression was maintained in NLS-Smad6. These findings suggest that the nucleus-localized NLS-Smad6 may directly repress the Id1 transcription without interfering BMP receptors.
The Smad-receptor association is determined primarily by the contact between the L3 loop in the MH2 domain of R-Smads and the L45 loop in the type I receptor. It is reasonable to speculate that the MH2 domain of Smad6 similarly contacts BMP type I receptors. To distinguish further between the direct nuclear function of Smad6 from its functions at the receptor level, we compared the inhibitory effects of the NL domain (i.e., the MH1-linker) and C domain (MH2) of Smad6 on Id1 promoter activity. As shown in Fig. 3E, the NL and C domains could inhibit BMP2-induced Id1 promoter activity in an additive manner. While Smad6NL and Smad6C exhibited moderate inhibitory effects on Id1 promoter activity, the combination of the two resulted in an inhibition as effectively as the full-length Smad6 on the Id1 promoter. These data further support the notion that in addition to the interference with the type I receptor (through MH2 domain), Smad6 inhibits BMP signaling through the MH1-linker region.
Smad6 interacts with the corepressor CtBP in vivo.
Transcription repression is controlled by HDAC and corepressors such as mSin3 and CtBP. HDACs associate with the TGF-β corepressors TGIF (46), c-Ski (2, 24, 40, 48), inhibitory Smads (3), and even directly with Smad3 (22), suggesting an important role of chromatin modulation in TGF-β-mediated gene transcription. In a search for candidate proteins that mediate the nuclear functions of Smad6, we found that Smad6 physically interacted with the corepressor CtBP1. CtBP1 is a transcriptional corepressor that was identified as an E1A-interacting protein (6, 38). Many transcriptional repressors are associated with CtBP proteins (7). To examine the Smad6-CtBP interaction in vivo, we performed a coimmunoprecipitation experiment. HA-tagged Smad6 and myc-tagged CtBP1 were transfected into 293T cells, and the Smad6-CtBP1 interaction was analyzed by anti-HA immunoprecipitation followed by anti-Myc Western blotting. Figure 4A shows that Smad6 and CtBP1 were present in the same anti-HA immunoprecipitated complex (lanes 3 and 6). The Smad6-CtBP interaction was not observed in cells expressing only HA-Smad6 or Myc-CtBP1. Immunoprecipitation using two different anti-HA antibodies gave similar results (Fig. 4A).
To determine whether CtBP1 binding to Smad6 is specific, we included Smad1 (R-Smad) and Smad4 (co-Smad) as controls. Significantly, neither Smad1 nor Smad4 could interact with CtBP1 in a coimmunoprecipitation experiment (Fig. 4B). Furthermore, another inhibitory Smad, Smad7, also failed to interact with CtBP1 (Fig. 5B). These results demonstrate the specific association between Smad6 and CtBP1.
Since Smad6 was specifically associated with CtBP1, we next evaluated the ability of Smad6 to interact with CtBP1 at endogenous levels. Interaction of endogenous Smad6 with CtBP1 was determined using coimmunoprecipitation analysis. As shown in Fig. 4C, endogenous CtBP1 was present in the Smad6 immunoprecipitated complex. As a control, unrelated antibody immunoglobulin G did not pull down CtBP1 (Fig. 4C). We also examined whether Smad6 and CtBP are similarly subcellularly localized. As shown in Fig. 4D, in both P19 and HeLa cells, endogenous Smad6 showed similar colocalization with CtBP in the nucleus. In HeLa cells the colocalization was more apparent since Smad6 is more strongly distributed in the nucleus (Fig. 4D and 2A). The localization of Smad6 and CtBP was not regulated by BMP2 in P19 cells (Fig. 4D), in agreement with the lack of BMP regulation of Smad6-CtBP coimmunoprecipitation (data not shown). Therefore, Smad6 interacts with CtBP1 at physiological levels.
Direct Smad6-CtBP association is mediated by the PLDLS motif of Smad6.
To map the Smad6 domain that interacts with CtBP1, we performed a transfection experiment using deletion mutants of Smad6 and Smad6-Smad7 chimera (Fig. 5A). The interaction of these mutants with CtBP1 is shown in Fig. 5B. While Smad6NL bound to CtBP1 (Fig. 5B, lane 5), deletion of NL domains (as in Smad6C) abolished the ability to interact with CtBP1 (lane 6). Similarly, the Smad6/7 chimera that contained the Smad6 NL domains interacted with CtBP1 (lane 7). In contrast to Smad6, inhibitory Smad7 could not interact with CtBP1 (lane 7), and likewise, the reverse Smad7/6 chimera failed to bind to CtBP1 (lane 9). Therefore, Smad6 interacted directly and specifically through its MH1-linker region with CtBP1.
CtBP binds to E1A or most repressors through a conserved PLDLS motif (7). Examination of the Smad6 sequence identified a consensus PLDLS motif in the linker region of Smad6, located at amino acids 290 to 294 (Fig. 6A). To determine whether this motif is essential for Smad6-CtBP interaction, we generated a Smad6 mutant that contained point mutations in the motif by changing PLDLS into LLAVS, named the Smad6(LAV) mutant (Fig. 6A). The GST in vitro binding assay was used to assess the binding of radiolabeled Smad6 (wild type or LAV mutant) to GST-CtBP (Fig. 6B). Results of this analysis showed that wild-type Smad6 directly interacted with CtBP but that this interaction was lost in the mutant (Fig. 6B). The failure of Smad6(LAV) to bind to CtBP was also confirmed by a coimmunoprecipitation experiment (Fig. 6C). Furthermore, Smad7 has no PLDLS motif, which may explain the absence of Smad7 binding to CtBP (Fig. 5B). These results suggest that Smad6-CtBP interaction is mediated by the PLDLS motif in Smad6.
Smad6 and CtBP coexist on the Id-1 promoter and cooperate to repress Id1 transcription.
Having established the interaction between Smad6 and CtBP, we carried out oligonucleotide DNA precipitation experiments to test whether Smad6 and CtBP were in the same nucleoprotein complex on the promoter of the Id1 gene. A 31-bp biotinylated oligonucleotide probe containing the GC-rich region and two GTCT elements in the human Id1 promoter (nucleotides nt −919 to −889 relative to the transcription initiation site) was used to retrieve the nucleoprotein complex from cell lysates. In transfected BMP-responsive 293T cells, Smad6 bound to the DNA only in the presence of Smad1 (Fig. 7A, lanes 4 and 8) and BMP treatment increased the association of Smad6 with the Id1 promoter DNA. This suggests that Smad6 binding to DNA may be through Smad1. CtBP was also present in this nucleoprotein complex, which seemed to be independent of BMP treatment. Interestingly, the presence of Smad6 (lane 7) or the combination of Smad6 and Smad1 (lanes 4 and 8) enhanced the binding of CtBP to DNA.
Having established the interaction between Smad6 and CtBP on DNA, we next explored the functional role of this interaction in the BMP-induced Id1 transcription. While transfection of the expression plasmid for Smad6 could repress the Id1 promoter by 50% in P19 cells, CtBP1 alone had a minimal effect in repressing the Id1 promoter activity. Coexpression of CtBP1 with Smad6 significantly repressed the transcriptional activation from the Id1 promoter (Fig. 7B), suggesting that cooperation of Smad6 and CtBP1 is required for maximal repressing activity towards the Id1 promoter. As a control, we tested whether overexpression of CtBP also repressed the TGF-β-induced promoter activity. We used both the synthetic Smad-binding element promoter and the natural 800-bp plasminogen activator inhibitor 1 promoter, both of which are inducible by TGF-β. As shown in Fig. 7C, overexpression of CtBP had little effect on the TGF-β-induced activity of these promoters; in contrast, we often observed a slight increase in the TGF-β-induced promoter activity.
Both Smad6 and CtBP are associated with HDACs. We therefore examined whether HDACs were involved in the Smad6-mediated repression of Id1 promoter by using the HDAC inhibitor trichostatin A (TSA). As shown in Fig. 7D, TSA at 10 ng/ml significantly enhanced the promoter activity of Id1 in the absence of Smad6. Notably, while Smad6 expression completely shut down the activity of the Id1 promoter, TSA treatment could partly relieve the repressing activity of Smad6, and thus BMP2-induced Id1 promoter activity was observed. These data suggest that Smad6 repressed BMP signaling partly in a HDAC-dependent manner.
CtBP is required for the repressing activity of Smad6.
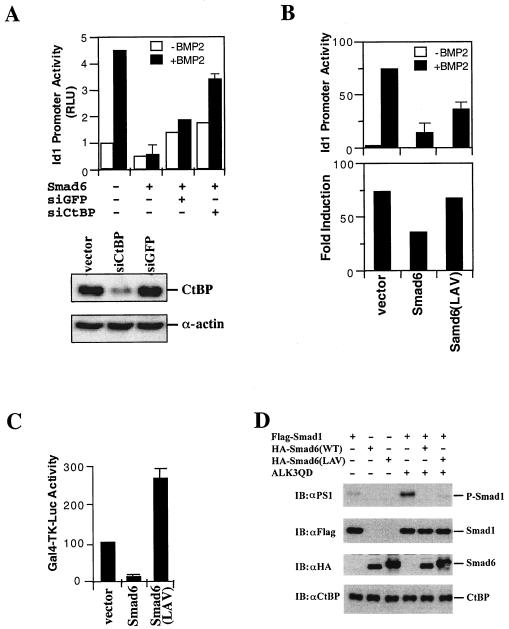
Since Smad6 physically interacted and functionally cooperated with CtBP, we sought to determine whether CtBP is essential in the Smad6-mediated repression of BMP signaling. We first tested the effect of loss of CtBP expression on the Smad6-mediated repression of the Id1 promoter. An expression vector carrying small interfering RNA (siRNA) for CtBP, called siCtBP (39), was transfected into HeLa cells and could reduce the expression of CtBP by 90% (Fig. 8A, bottom). We found that siCtBP could partly restore the BMP-induced Id1 promoter activity (Fig. 8A, bar graph). In sharp contrast, siGFP, an irrelevant siRNA vector, had little effects on the BMP response (Fig. 8A, bar graph).
FIG. 8.
CtBP binding is required for maximal inhibitory function of Smad6 in BMP-induced Id1 transcription. (A) siRNA of CtBP blocks Smad6-mediatd repression on the BMP-dependent Id1 promoter activity. HeLa cells were transfected with the indicated siRNA expression plasmids. BMP2 treatment and the Id1-luciferase assay were done as in Fig. 1B. The bottom panel shows the expression levels of CtBP knocked down by siCtBP but not by siGFP. (B) Smad6 mutant defective in CtBP binding has reduced activity to inhibit Id1 induction. P19 cells were transfected with the indicated expression plasmids. BMP2 treatment and the Id1-luciferase assay were done as in Fig. 1B. In the top panel, the y axis indicates relative light units (RLU) with or without BMP2; in bottom panel, the y axis represents BMP-mediated induction, i.e., the ratio of Id1 promoter activity in the presence of BMP2 to that in the absence of BMP2. (C) The Smad6 mutant defective in CtBP binding has lost its repressor activity. P19 cells were transfected and the Gal4-TK-luc reporter was scored as described in the legend to Fig. 2B. Smad6(LAV), the PLDLS mutant of Smad6 in Gal4 vector. Note that Gal4-Smad6(LAV) was converted from a repressor to an activator. (D) Loss of CtBP binding has no effect on the ability of Smad6 to inhibit receptor-mediated Smad1 phosphorylation. P19 cells were transfected with indicated expression plasmids, and after 48 h the cell lysates were subjected to Western blot analysis using the indicated antibodies. αPS1, antibody against phospho-Smad1.
We then tested whether loss of CtBP1 binding impaired the ability of Smad6 to repress Id1 promoter activity. To address this issue, we compared Smad6 and its CtBP-binding-defective LAV mutant for their effects on BMP2-induced Id1-lucifease activity. The results are shown in Fig. 8B. In comparing the absolute induction peak by BMP2, it appears that the Smad6(LAV) mutant only partially inhibited the BMP2-induced response whereas wild-type Smad6 strongly inhibited BMP2-induced Id1 reporter activity. Significantly, when we compared their effects on the fold induction of Id1 promoter activity by BMP, we found that the mutant completely lost its repressing function on Id1-luciferase response.
To further confirm that the mutation in PLDLS motif affects the repressor function of Smad6, we analyzed the repressor activity of Smad6 in a Gal4-based transcription assay. We found that unlike wild-type Smad6, this mutant Smad6 could not repress the heterologous Gal4 promoter (Fig. 8C). On the contrary, Gal4-Smad6(LAV) exhibited transactivation activity. We also excluded the possibility that the mutant may have lost its inhibition on receptor signaling. As shown in Fig. 8D, Smad6(LAV) retained the ability to block the phosphorylation of Smad1 as potently as wild-type Smad6 did. Taken together, these data in Fig. 8 clearly suggest that the CtBP binding is essential for the nuclear repressor activity of Smad6.
DISCUSSION
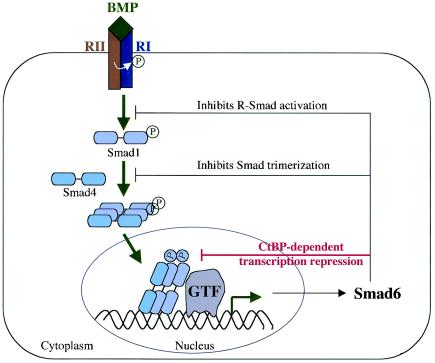
TGF-β signaling is tightly regulated through both positive and negative mechanisms. One of the negative-feedback mechanisms during TGF-β signaling is the production of Inhibitory Smads, which are generally thought to inhibit TGF-β signaling at the level of Smad activation in the cytoplasm (1, 17, 18, 41). In this study, we demonstrated for the first time that Smad6 binds to the nuclear corepressor CtBP to repress BMP-induced transcription, representing a novel mechanism for how Smad6 inhibits BMP signaling (Fig. 9).
FIG. 9.
Working model for Smad6 functions in BMP signaling. The BMP signaling pathway is illustrated with points of Smad6 action. Smad6, an immediate-early gene product of BMP signaling, inhibits R-Smad activation and R-Smad-Smad4 heteromerization and directly represses transcription in the nucleus. RI, BMP type I receptor; RII, BMP type II receptor; GTF, general transcription factors such as TATA-binding proteins and associated factors.
Several lines of evidence support the transcriptional repressor role of Smad6. First, Smad6 has intrinsic repressor activity, as demonstrated by the Gal4-based transcription assay. The Gal4-Smad6 fusion protein, when tethered to the Gal4-binding promoter elements, inhibits the activation of the heterologous promoter containing Gal4-binding sites. This is in contrast to the activity of Smad7, the other I-Smad in vertebrates, which has transactivation capacity (34). These observations are consistent with the observation of their differential ability to bind to corepressor CtBP. Unlike Smad6, Smad7 does not have the PLDLS motif and is clearly unable to bind to CtBP. Most notably, a point mutation in the PLDLS motif switched Smad6 from a repressor to a transcription activator.
Second, CtBP1 physically interacts and functionally cooperates with Smad6 in the regulation of BMP-induced Id1 transcription. This not only identifies CtBP1 as a new physiological partner for Smad6 but also assigns a new function to Smad6 in directly controlling transcription, since the majority of the previously identified CtBP-interacting proteins are transcription repressors (7). For example, oncogenic Evi-1 represses Smad-induced transcription of TGF-β responsive genes through its CtBP-binding motifs (19). Similarly, mutations in the CtBP-binding motif of TGIF abolish its function in repressing TGF-β target genes and may be linked to holoprosencephaly disease (28). In this study, Smad6-CtBP interaction occurs under physiological conditions, and the interaction depends on the integrity of the CtBP-binding PLDLS motif in Smad6. Most importantly, mutation in the PLDLS motif markedly decreases the inhibitory function of Smad6 in BMP-induced Id1 promoter activity. Therefore, our findings directly connect the inhibitory role of Smad6 in BMP signaling with transcription corepressor CtBP. This may also provide a logical explanation of how Smad6, when recruited to the BMP-responsive promoter through interaction with Hoxc-8, repressed OPN transcription (4). Further studies are needed to determine whether CtBP participate in the repression of the OPN gene and other BMP-dependent target genes.
Finally, the subcellular localization profile of Smad6 also supports the nuclear function of Smad6. Although its localization varies among cell types, Smad6 can be found in the nucleus or even predominantly located in the nucleus. It appears that the inhibitory function of Smad6 in the nucleus, where it colocalizes with CtBP, is responsible for its repressor activity. We carried out experiments using NLS-Smad6 and the Smad6NL variant to further distinguish the nuclear function of Smad6 from its antireceptor mechanism. NLS-Smad6 is an engineered Smad6 variant that failed to bind to the type I receptor ALK3 and was exclusively nucleus-localized in BMP-responsive P19 cells. We found that nucleus-localized NLS-Smad6 potently repressed the BMP2-induced Id1 transcriptional activation, suggesting that nuclear Smad6 suffices to confer the repression to the Id1 promoter. Consistent with our model, the Smad6NL (containing the DNA-binding MH1 domain and the CtBP-binding linker region, but lacking the MH2 domain) could also repress transcription from the Id1 promoter, although this variant lacks the ability to bind to the type I receptor.
Our finding expands our understanding of the molecular mechanisms involved in the negative regulation of BMP signaling by Smad6 and highlights its physiological function as a transcription repressor in the nucleus. To date, the precise timing for how Smad6 represses BMP target genes is not known. Considering that Smad6 is induced by BMPs, Smad6 may serve as a mechanism to effectively shut off the BMP-induced transcription through both inhibiting R-Smad activation in the cytoplasm and repressing (deactivating) the BMP target genes. It is possible that Smad6 selectively inhibits a subset of BMP-regulated genes. The nuclear function of Smad6 may depend on its DNA-binding activity (4) or its interaction with other DNA-binding transcription factors, including Hoxc-8 (4) and Smad1 (17). Ligand-inducible Smad1-Smad6 interaction may occur in the nucleus and may recruit CtBP to a Smad1-responsive promoter. Further investigations to determine the existence and regulation of Smad1-Smad6-CtBP assembly will provide insights into the precise mechanism underlying the repressor functions of Smad6.
Acknowledgments
We thank Rik Derynck for critically reading the manuscript; Xiao-Fan Wang for advice, Kohei Miyazono for providing numerous reagents including Smad6 cDNA, deletion mutants, and Smad6/7 chimeras; Richard Goodman for providing Flag-CtBP; Eric Olson for providing myc-CtBP; Xiao-Hong Sun for providing Id1 cDNA; and Tetsuya Taga and Robert Benezra for providing the Id1-luc reporter. Smad6 adenovirus was kindly provided by Jingsong Zhao with permission from Kohei Miyazono. Sincere thanks also go to Irene Harrison for her critical reading and editing.
This work was supported by grants from American Cancer Society (RSG-02-145-01-CCG to X.L. and RSG-00-214-01-CCG to X.-H.F.) and the National Institutes of Health (F32 GM70690 to Y.J.S., R01 CA95731 to F.C.B., R01 GM53874 to Y.S., and R01 GM63773 to X.-H.F.). X.-H.F. is a Leukemia & Lymphoma Society Scholar.
REFERENCES
- 1.Afrakhte, M., A. Moren, S. Jossan, S. Itoh, K. Sampath, B. Westermark, C. H. Heldin, N. E. Heldin, and P. ten Dijke. 1998. Induction of inhibitory Smad6 and Smad7 mRNA by TGF-β family members. Biochem. Biophys. Res. Commun. 249:505-511. [DOI] [PubMed] [Google Scholar]
- 2.Akiyoshi, S., H. Inoue, J. Hanai, K. Kusanagi, N. Nemoto, K. Miyazono, and M. Kawabata. 1999. c-Ski acts as a transcriptional co-repressor in transforming growth factor-β signaling through interaction with Smads. J. Biol. Chem. 274:35269-35277. [DOI] [PubMed] [Google Scholar]
- 3.Bai, S., and X. Cao. 2002. A nuclear antagonistic mechanism of inhibitory Smads in transforming growth factor-β signaling. J. Biol. Chem. 277:4176-4182. [DOI] [PubMed] [Google Scholar]
- 4.Bai, S., X. Shi, X. Yang, and X. Cao. 2000. Smad6 as a transcriptional corepressor. J. Biol. Chem. 275:8267-8270. [DOI] [PubMed] [Google Scholar]
- 5.Blobe, G. C., W. P. Schiemann, and H. F. Lodish. 2000. Role of transforming growth factor β in human disease. N. Engl. J. Med. 342:1350-1358. [DOI] [PubMed] [Google Scholar]
- 6.Boyd, J. M., T. Subramanian, U. Schaeper, M. La Regina, S. Bayley, and G. Chinnadurai. 1993. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J. 12:469-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chinnadurai, G. 2002. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell 9:213-224. [DOI] [PubMed] [Google Scholar]
- 8.Dennler, S., M. J. Goumans, and P. Ten Dijke. 2002. Transforming growth factor β signal transduction. J. Leukoc. Biol. 71:731-740. [PubMed] [Google Scholar]
- 9.Derynck, R., R. J. Akhurst, and A. Balmain. 2001. TGF-β signaling in tumor suppression and cancer progression. Nat. Genet. 29:117-129. [DOI] [PubMed] [Google Scholar]
- 10.Derynck, R., and X.-H. Feng. 1997. TGF-β receptor signaling. Biochim. Biophys. Acta 1333:F105-F150. [DOI] [PubMed] [Google Scholar]
- 11.Derynck, R., Y. Zhang, and X. H. Feng. 1998. Smads: transcriptional activators of TGF-β responses. Cell 95:737-740. [DOI] [PubMed] [Google Scholar]
- 12.Feng, X.-H., Y.-Y. Liang, M. Liang, W. Zhai, and X. Lin. 2002. Direct interaction of c-Myc with Smad2 and Smad3 to inhibit TGF-β-mediated induction of the CDK inhibitor p15INK4B. Mol. Cell 9:133-143. [DOI] [PubMed] [Google Scholar]
- 13.Feng, X.-H., X. Lin, and R. Derynck. 2000. Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15Ink4B transcription in response to TGF-β. EMBO J. 19:5178-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng, X.-H., Y. Zhang, R.-Y. Wu, and R. Derynck. 1998. The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for Smad3 in TGF-β-induced transcriptional activation. Genes Dev. 12:2153-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng, X. H., E. H. Filvaroff, and R. Derynck. 1995. Transforming growth factor-β (TGF-β)-induced down-regulation of cyclin A expression requires a functional TGF-β receptor complex. Characterization of chimeric and truncated type I and type II receptors. J. Biol. Chem. 270:24237-24245. [DOI] [PubMed] [Google Scholar]
- 16.Hanyu, A., Y. Ishidou, T. Ebisawa, T. Shimanuki, T. Imamura, and K. Miyazono. 2001. The N domain of Smad7 is essential for specific inhibition of transforming growth factor-β signaling. J. Cell Biol. 155:1017-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hata, A., G. Lagna, J. Massague, and A. Hemmati-Brivanlou. 1998. Smad6 inhibits BMP/Smad1 signaling by specifically competing with the Smad4 tumor suppressor. Genes Dev. 12:186-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imamura, T., M. Takase, A. Nishihara, E. Oeda, J. Hanai, M. Kawabata, and K. Miyazono. 1997. Smad6 inhibits signalling by the TGF-β superfamily. Nature 389:622-626. [DOI] [PubMed] [Google Scholar]
- 19.Izutsu, K., M. Kurokawa, Y. Imai, K. Maki, K. Mitani, and H. Hirai. 2001. The corepressor CtBP interacts with Evi-1 to repress transforming growth factor β signaling. Blood 97:2815-2822. [DOI] [PubMed] [Google Scholar]
- 20.Katagiri, T., M. Imada, T. Yanai, T. Suda, N. Takahashi, and R. Kamijo. 2002. Identification of a BMP-responsive element in Id1, the gene for inhibition of myogenesis. Genes Cells 7:949-960. [DOI] [PubMed] [Google Scholar]
- 21.Korchynskyi, O., and P. ten Dijke. 2002. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J. Biol. Chem. 277:4883-4891. [DOI] [PubMed] [Google Scholar]
- 22.Liberati, N. T., M. Moniwa, A. J. Borton, J. R. Davie, and X. F. Wang. 2001. An essential role for Mad homology domain 1 in the association of Smad3 with histone deacetylase activity. J. Biol. Chem. 276:22595-22603. [DOI] [PubMed] [Google Scholar]
- 23.Lopez-Rovira, T., E. Chalaux, J. Massague, J. L. Rosa, and F. Ventura. 2002. Direct binding of Smad1 and Smad4 to two distinct motifs mediates bone morphogenetic protein-specific transcriptional activation of Id1 gene. J. Biol. Chem. 277:3176-3185. [DOI] [PubMed] [Google Scholar]
- 24.Luo, K., S. L. Stroschein, W. Wang, D. Chen, E. Martens, S. Zhou, and Q. Zhou. 1999. The Ski oncoprotein interacts with the Smad proteins to repress TGF-β signaling. Genes Dev. 13:2196-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Massagué, J. 2000. How cells read TGF-beta signals. Nat. Rev. Mol. Cell. Biol. 1:169-178. [DOI] [PubMed] [Google Scholar]
- 26.Massagué, J., S. Blain, and R. Lo. 2000. TGF-β signaling in growth control, cancer, and heritable disorders. Cell 103:295-309. [DOI] [PubMed] [Google Scholar]
- 27.Massagué, J., and D. Wotton. 2000. Transcriptional control by the TGF-β/Smad signaling system. EMBO J. 19:1745-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melhuish, T. A., and D. Wotton. 2000. The interaction of the carboxyl terminus-binding protein with the Smad corepressor TGIF is disrupted by a holoprosencephaly mutation in TGIF. J. Biol. Chem. 275:39762-39766. [DOI] [PubMed] [Google Scholar]
- 29.Miyazono, K., K. Kusanagi, and H. Inoue. 2001. Divergence and convergence of TGF-β/BMP signaling. J. Cell. Physiol. 187:265-276. [DOI] [PubMed] [Google Scholar]
- 30.Miyazono, K., and K. Miyazawa. 2002. Id: a target of BMP signaling. Sci. STKE 2002:E40. [DOI] [PubMed] [Google Scholar]
- 31.Moustakas, A., S. Souchelnytskyi, and C. H. Heldin. 2001. Smad regulation in TGF-β signal transduction. J. Cell Sci. 114:4359-4369. [DOI] [PubMed] [Google Scholar]
- 32.Nakashima, K., T. Takizawa, W. Ochiai, M. Yanagisawa, T. Hisatsune, M. Nakafuku, K. Miyazono, T. Kishimoto, R. Kageyama, and T. Taga. 2001. BMP2-mediated alteration in the developmental pathway of fetal mouse brain cells from neurogenesis to astrocytogenesis. Proc. Natl. Acad. Sci. USA 98:5868-5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Padgett, R., and G. Patterson. 2001. New developments for TGF-β. Dev. Cell 1:343-349. [DOI] [PubMed] [Google Scholar]
- 34.Pulaski, L., M. Landstrom, C. H. Heldin, and S. Souchelnytskyi. 2001. Phosphorylation of Smad7 at Ser-249 does not interfere with its inhibitory role in transforming growth factor-β-dependent signaling but affects Smad7-dependent transcriptional activation. J. Biol. Chem. 276:14344-4349. [DOI] [PubMed] [Google Scholar]
- 35.Rich, J., A. Borton, and X. Wang. 2001. Transforming growth factor-β signaling in cancer. Microsc. Res. Tech. 52:363-373. [DOI] [PubMed] [Google Scholar]
- 36.Roberts, A. B., and R. Derynck. 2001. Meeting report: signaling schemes for TGF-β. Sci. STKE 2001:PE43. [DOI] [PubMed]
- 37.Roberts, S. G. 2000. Mechanisms of action of transcription activation and repression domains. Cell. Mol. Life Sci. 57:1149-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schaeper, U., J. M. Boyd, S. Verma, E. Uhlmann, T. Subramanian, and G. Chinnadurai. 1995. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc. Natl. Acad. Sci. USA 92:10467-10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi Y, J. Sawada, G. Sui, El B. Affar, J.R. Whetstine, F. Lan, H. Ogawa, M.P. Luke, Y. Nakatani, and Y. Shi. 2003. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422:735-738. [DOI] [PubMed] [Google Scholar]
- 40.Sun, Y., X. Liu, E. Ng-Eaton, H. F. Lodish, and R. A. Weinberg. 1999. SnoN and Ski protooncoproteins are rapidly degraded in response to transforming growth factor β signaling. Proc. Natl. Acad. Sci. USA 96:12442-12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takase, M., T. Imamura, T. K. Sampath, K. Takeda, H. Ichijo, K. Miyazono, and M. Kawabata. 1998. Induction of Smad6 mRNA by bone morphogenetic proteins. Biochem. Biophys. Res. Commun. 244:26-29. [DOI] [PubMed] [Google Scholar]
- 42.Topper, J., J. Cai, Y. Qiu, K. Anderson, Y.-Y. Xu, J. Deeds, R. Feeley, C. Gimeno, E. Woolf, O. Tayber, G. Mays, B. Sampson, F. Schoen, J. Gimbrone, and D. Falb. 1997. Vascular MADs: two novel MAD-related genes selectively inducible by flow in human vascular endothelium. Proc. Natl. Acad. Sci. USA 94:9314-9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valdimarsdottir, G., M. J. Goumans, A. Rosendahl, M. Brugman, S. Itoh, F. Lebrin, P. Sideras, and P. Ten Dijke. 2002. Stimulation of Id1 expression by bone morphogenetic protein is sufficient and necessary for bone morphogenetic protein-induced activation of endothelial cells. Circulation 106:2263-2270. [DOI] [PubMed] [Google Scholar]
- 44.Wakefield, L. M., and A. B. Roberts. 2002. TGF-β signaling: positive and negative effects on tumorigenesis. Curr. Opin. Genet. Dev. 12:22-29. [DOI] [PubMed] [Google Scholar]
- 45.Whitman, M., and M. Mercola. 2001. TGF-β superfamily signaling and left-right asymmetry. Sci. STKE 2001:RE1. [DOI] [PubMed]
- 46.Wotton, D., R. S. Lo, S. Lee, and J. Massague. 1999. A Smad transcriptional corepressor. Cell 97:29-39. [DOI] [PubMed] [Google Scholar]
- 47.Wrana, J. L. 2000. Regulation of Smad activity. Cell 100:189-192. [DOI] [PubMed] [Google Scholar]
- 48.Xu, W., K. Angelis, D. Danielpour, M. M. Haddad, O. Bischof, J. Campisi, E. Stavnezer, and E. E. Medrano. 2000. Ski acts as a co-repressor with Smad2 and Smad3 to regulate the response to type β transforming growth factor. Proc. Natl. Acad. Sci. USA 97:5924-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, C. L., T. A. McKinsey, J. R. Lu, and E. N. Olson. 2001. Association of COOH-terminal-binding protein and MEF2-interacting transcription repressor (MITR) contributes to transcriptional repression of the MEF2 transcription factor. J. Biol. Chem. 276:35-39. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, Q., D. W. Piston, and R. H. Goodman. 2002. Regulation of corepressor function by nuclear NADH. Science 295:1895-1897. [DOI] [PubMed] [Google Scholar]
- 51.Zhao, J., W. Shi, H. Chen, and D. Warburton. 2000. Smad7 and Smad6 differentially modulate transforming growth factor β-induced inhibition of embryonic lung morphogenesis. J. Biol. Chem. 275:23992-23997. [DOI] [PubMed] [Google Scholar]