Summary
The infusion of natural killer (NK) cells is a promising therapy for patients with advanced malignancies. Clinical expanded NK cell products were compared to freshly isolated NK cells. Autologous peripheral blood mononuclear cells were collected by apheresis from 8 patients. NK cells were isolated by anti-CD3 negative selection followed by anti-CD56 positive selection. They were then expanded by coculture with IL-2 and an irradiated EBV-transformed lymphoblastoid cell line (EBV-TM-LCL) to produce 14 NK products. Molecular changes in the 14 NK cell products were characterized using gene and microRNA expression microarrays. EBV-TM-LCL feeder cells from 3 lots were also analyzed since they were expanded for over 90 days and each lot was used for multiple NK cell expansions. The gene expression profiles among the 3 EBV-TM-LCL lots used showed no differences and were not affected by their time in culture. Freshly isolated and expanded NK cells had distinct gene and microRNA expression profiles. Compared to fresh NK cells, expanded NK cells overexpressed 1,098 genes and 28 human microRNAs. Genes in the crosstalk between dendritic cells and NK cells and metabolic pathways were up-regulated in expanded NK cells, while genes in a number of immune function pathways were down-regulated. Among all the most up-regulated genes were the NK cell activating receptor natural cytotoxicity triggering receptor 3 (NCR3), myxovirus restistance 1 (MX1), lymphotoxin β (LTB) and BCL2-associated X protein (BAX) Although some expanded NK cell product variability was observed, perhaps related to patient factors, further studies on larger numbers of products will be needed to determine the impact of these differences on clinical outcomes.
Keywords: consistency, expansion, gene and microRNA, natural killer cells, potency
INTRODUCTION
Natural killer (NK) cells are an important component of the innate immune system. Immune therapy, utilizing adoptive NK cell infusions, is a promising cell-based anti-tumor therapy for patients with advanced malignancies. Both expanded and unstimulated NK cells collected from autologous and allogeneic donors are being used in clinical trials to treat patients with cancer and hematologic malignancies (1–3). In allogeneic transplantation, NK cells can mediate potent anti-cancer activity without causing graft-versus-host disease (4).
Expanded tumor infiltrating lymphocytes (TIL) have been effectively used to treat patients with metastatic melanoma (5;6), however, the isolation and expansion of TIL is difficult and these cells cannot be obtained from all patients. The ability of NK cells to kill tumor cells without the need to recognize tumor-specific antigens and their ease of collection and isolation makes them appealing as potential effectors for cancer immunotherapy(2). NK cell recognition of targets is regulated through a balance of activating and inhibitory signals (7). NK cells have the ability to kill target cells directly as well as mediate antibody-dependent cellular cytotoxicity. They can also induce tumor apoptosis via the perforin/granzyme pathway or through death receptor ligands. Such ligands include tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) or Fas ligand (2;8).
NK cells can be isolated from peripheral blood mononuclear cells (PBMCs) by selection with anti-CD56 and paramagnetic beads or by using a combination of negative selection of CD3+ cells with anti-CD3 and positive selection with anti-CD56 (1;9;10). For autologous NK cell immunotherapy, the administration of large quantities of functional NK cells to achieve a threshold to induce tumor suppression is desirable. It has been previously shown that a pure population of NK cells can be expanded approximately 500-fold by co-culturing NK cells with an irradiated Epstein-Barr virus-transformed lymphoblastoid cell line (EMV-TM-LCL) in interleukin (IL)-2 containing media (10). The goal of this study was to compare freshly isolated and ex vivo expanded NK cells from cancer patients.
We used global gene and microRNA expression analysis to assess the expanded NK cell concentrates produced in our clinical cellular therapy laboratory to treat 8 cancer patients at our institution on an NK cell clinical trial. We compared NK cells freshly isolated from each patient to the expanded NK cell products in order to characterize their differences and to identify biomarkers in addition to TRAIL that might be useful in assessing the potency of expanded NK cell products. The protocol allowed for multiple expanded NK cell treatments for patients who had stable or responsive disease following initial therapy. Three patients were treated with multiple infusions, allowing us to perform a preliminary assessment of variability in the products due to both inter-patient and manufacturing factors. The NK cell expansion process involved the co-culture of NK cells with an irradiated EBV-TM-LCL. Since differences related to the culture duration of EBV-TM-LCL feeders could potentially impact NK cell expansion, we also analyzed EBV-TM-LCL with global gene expression profiling.
MATERIALS AND METHODS
Study Design
PBMCs were collected from patients with metastatic cancer using a blood cell separator (Cobe Spectra, Gambro BCT, Lakewood, CO, USA). These subjects were not treated with cytokines or growth factors before the collection procedure. NK cells were purified from PBMCs through CD3-positive T-cell depletion and subsequent enrichment of CD56-positive cells on CliniMACS system (Miltenyi Biotec, Auburn, CA, USA). The isolated NK cells were divided into 4 to 6 aliquots with 1 to 100 × 106 cells in each aliquot. The NK cell aliquots were either expanded immediately or cryopreserved and expanded later. The NK cells were cryopreserved in Plasma-Lyte A (Baxter Healthcare, Deerfield, IL, USA) supplemented with 4% human serum albumin (Talecris Biotherapeutics, Research Triangle Park, NC, USA), 5% dimethyl sulfoxide (DMSO) and 6% pentastarch using a controlled rate freezer and stored in liquid nitrogen. Fresh or cryopreserved NK cells were expanded for 14 to 16 days in media containing 500 IU/mL of recombinant human IL-2 (rHuIL-2, Hoffmann-La Roche Inc., Nutley, NJ, USA) co-cultured with irradiated EBV-TM-LCL cells at a 10:1 feeder to NK-cell ratio. These studies were approved by an institutional review board (IRB) of National Heart, Lung and Blood Institute (NHLBI).
Culture of EBV-TM-LCL Feeders
The good manufacturing practice (GMP)-certified human EBV-TM-LCL was obtained from the master cell bank generated at Fred Hutchinson Cancer Research Center (FHCRC, Seattle, WA, USA) (10;11). EBV-TM-LCL cells were maintained in 180-cm2 300-mL bags (Baxter Healthcare Corporation, Deerfield, IL, USA) at 0.2–1.0 × 106/mL in RPMI-1640 supplemented with 10% heat-inactivated human AB plasma. The culture bags were placed into an incubator at 37±1°C, 5±1% CO2, and 90±5% humidity. Cell doubling time was 15–24 hours 1 week after culture initiation. The cultures were split every 3 to 4 days thereafter.
Our NK cell expansion protocol allowed each lot of EBV-TM-LCLs to be maintained in culture for up to 90 days and during the culture period multiple aliquots from each lot of cultured EBV-TM-LCLs were used for several separate NK cell expansions.
Expansion of NK Cells
Isolated NK cells were combined with irradiated EBV-TM-LCL cells (1:10 ratio) in X-VIVO 20 supplemented with 10% heat-inactivated human AB plasma and 500 IU/mL of rHuIL-2 (Hoffmann-La Roche Inc.). The cells were cultured in 180-cm2 300-mL bags (Baxter Healthcare Corporation) in an incubator at 37±1°C, 5±1% CO2, and 90±5% humidity. Four days after initiation of culture, half of the medium was replaced with fresh medium. Two days later, the concentration of NK cells was adjusted to 1 × 106 cells/mL using the same medium. Thereafter, the expanded cells were counted and diluted every 48 hours to keep the expanded NK cells at the concentration of 1 × 106 cells/mL.
Gene Expression Profiling
Total RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA, USA) and the RNA concentration was assessed using ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Total RNA (3 μg) was amplified from 0.5 × 106 to 107 cells into anti-sense RNA (aRNA). Total RNA from PBMCs pooled from 6 normal donors was also amplified into aRNA to serve as the reference. Both reference and test sample aRNA were directly labeled using Universal Linkage System aRNA Fluorescent Labeling Kit (Kreatech Biotechnology, Amsterdam, Netherlands) with Cy3 (green) for reference and Cy5 (red) for test sample, respectively. Whole-genome human 36K microarrays were printed in the Infectious Disease and Immunogenetics Section of Department of Transfusion Medicine using a commercial probe set which contains 35,035 probes, representing approximately 25,100 unique genes and 39,600 transcripts excluding control oligonucleotides (Operon Human Genome Array-Ready Oligo Set version 4.0, Huntsville, AL, USA). The complete list of genes in the microarray is online at http://nciarray.nci.nih.gov/gal_files/Hs-CCDTM36k-1px.gal. Hybridization was carried out at 42°C for 18 hours. The arrays were then washed and scanned on a GenePix 4000B (Molecular Devices, Sunnyvale, CA, USA).
MicroRNA Expression Profiling
Total RNA was extracted using TRIzol (Invitrogen) and the RNA concentration was assessed using ND-1000 Spectrophotometer (NanoDrop Technologies). Total RNA isolated (2 μg) was directly labeled with miRCURY LNA Array Power Labeling Kit (Exiqon, Woburn, MA, USA) according to the manufacturer’s instructions. The total RNA from the EBV cell line was used as the reference. The test sample was labeled with Hy5 and the reference with Hy3. A microRNA probe set was designed using mature antisense microRNA sequences consisting of 1,564 unique microRNAs from human, mouse, rat, and virus plus 14 control probes. The probes were 5′ amine modified and printed in duplicates on CodeLink activated slides (General Electric, GE Health, NJ, USA) via covalent bonding at the Infectious Disease and Immunogenetics Section of the Department of Transfusion Medicine (Clinical Center, NIH, Bethesda, MD, USA). After labeling, the sample and the reference were co-hybridized to the microRNA array at room temperature over night as previously described (12;13). The slides were then washed and scanned on a GenePix 4000B (Molecular Devices).
Data Processing and Analysis
Resulting data files were uploaded to the mAdb (http://madb.nci.nih.gov)and the raw microarray data were retrieved and analyzed by BRB-ArrayTools (http://linus.nci.nih.gov/BRB-ArrayTools.html) developed at the Division of Cancer Treatment and Diagnosis, Biometric Research Branch, National Cancer Institute. The data were filtered according to a general procedure to exclude spots with the minimum intensity that was arbitrarily set to 100 in both fluorescence channels. If one-channel intensity is below the minimum while the other was above, the lower intensity was arbitrarily set to the minimum. Flagged spots and spots with diameters less than 10 μm were also excluded from the analyses. Afterwards, the spot-filtered data were normalized using Lowess Smoother. We selected genes that were expressed in more than 50 percent of samples for analyses. We then conducted hierarchical cluster analysis on those genes with Cluster and TreeView software (14). Differentially expressed genes or microRNAs in freshly isolated versus expanded NK cells were identified using t test or F tests with a P value cutoff of 0.01 (gene) or 0.05 (miRNA). The data were adjusted for class comparisons by False Discovery Rate (FDR) < 0.10. BRB-ArrayTools was used for multidimensional scaling, analysis of variance between groups, and gene set expression comparison. Ingenuity Pathway Analysis (http://www.ingenuity.com, Ingenuity Systems Inc., Redwood City, CA, USA) was used for analysis of functional pathways. Gene ontology selection was performed through mAdb. Analysis of MicroRNA target and cluster was done using BRB-ArrayTools, Target Scan (http://www.targetscan.org), and miRGen (http://www.diana.pcbi.upenn.edu/miRGen.html).
Gene Expression Analysis by Quantitative PCR
To validate the results of the microarray analysis, 5 genes were selected for analysis by quantitive real-time/reverse-transcription polymerase chain reaction (RT-PCR). Gene expressions for LTB (Assay ID Hs00242737_m1), TUBB (Assay ID Hs00742828_s1), JUN (Assay ID Hs99999141_s1), NCR3(Assay ID Hs00394809_m1) and FOSB (Assay ID Hs01547109_m1) were quantified by TaqMan Gene Expression Assays (Applied Biosystems) according to manufacturer’s protocol and normalized by 18s rRNA(Assay ID Hs99999901_s1). The resulting data showed in the heat maps and fold changein expression data was obtained by using DataAssist™ v2.0 software (Applied Biosystems).
RESULTS
Expanded NK Cell Products and EBV-TM-LCLs
Freshly isolated NK cells and expanded NK cells from 8 patients were studied. A total of 14 expanded NK cell products were manufactured from the 8 patients and 3 lots of EBV-TM-LCLs were used as feeder cells to produce the expanded NK cells. Each lot of EBV-TM-LCLs was maintained in culture for up to 90 days and aliquots were removed as needed for NK cell expansion. As a result, each lot of EBV-TM-LCLs was used for several NK cell expansions. Some patients were treated with multiple doses of expanded NK cells and since each NK cell expansion was performed at a different time, expanded NK cells produced for the same patient but at different times may have been co-cultured with different EBV-TM-LCL lots. The relation between specific expanded NK cell products with specific EBV-TM-LCL lots and their duration in culture is summarized in Table 1.
TABLE 1.
Expanded NK Cells: Culture Duration, TRAIL Expression, Viability and LCL Cells used as Feeder Cells.
| Patient | Expanded NK Cells | Days in Culture | Feeder Cells* | CD56 TRAIL (MFI) | Viability |
|---|---|---|---|---|---|
| 1 | eNK-1-1 | 14 | LCL-1-1-D16 | 1,741 | 90 |
|
| |||||
| 2 | eNK-2-1 | 14 | LCL-1-2-D28 | 1,008 | 86 |
| eNK-2-2 | 14 | LCL-1-5-D75 | 1,779 | 86 | |
| eNK-2-3 | 14 | LCL-2-2-D29 | 838 | 84 | |
| eNK-2-4 | 14 | LCL-3-4-D46 | 1,104 | 81 | |
|
| |||||
| 3 | eNK-3-1 | 15 | LCL-1-2-D28 | 860 | 89 |
| eNK-3-2 | 14 | LCL-1-5-D75 | 563 | 86 | |
| eNK-3-3 | 15 | LCL-3-3-D43 | 354 | 89 | |
|
| |||||
| 4 | eNK-4-1 | 16 | LCL-2-1-D15 | 1,573 | 86 |
|
| |||||
| 5 | eNK-5-1 | 15 | LCL-2-1-D15 | 1,460 | 88 |
| eNK-5-2 | 15 | LCL-3-2-D24 | 1,500 | 90 | |
|
| |||||
| 6 | eNK-6-1 | 16 | LCL-3-2-D24 | 921 | 91 |
|
| |||||
| 7 | eNK-7-1 | 14 | LCL-3-5-D64 | 2,330 | 85 |
|
| |||||
| 8 | eNK-8-1 | 14 | LCL-4-1-D69 | 1,394 | 71 |
LCL indicates lymphoblastoid cell line; eNK, expanded NK; D, days.
Corresponding feeder cells of which specimens were available.
Gene Expression Profiles of EBV-TM-LCL Feeders
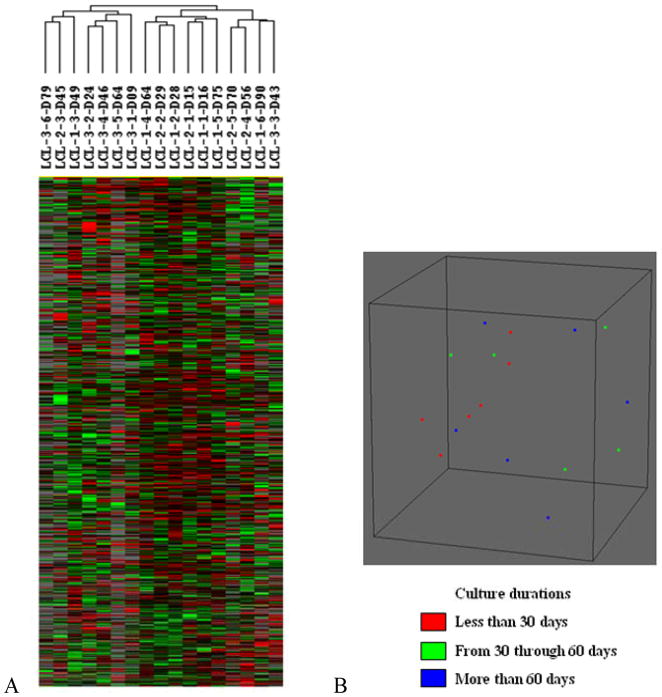
In order to determine if differences might exist in the EBV-TM-LCLs used as feeder cells for NK cell expansion, we analyzed the 3 separate lots of EBV-TM-LCL used in this study over their approximately 90 days in culture. Each lot was sampled approximately once every 15 days and a total of 17 EBV-TM-LCL samples were analyzed by global gene expression profiling. Unsupervised hierarchical clustering of the 12,283 genes remaining after filtering revealed no differences between the cultured LCL cell lines (Figure 1A). No significant differences were shown in class comparisons including analysis of variance, based on LCL lot and culture duration. In addition, analysis of the 17 samples with multidimensional scaling using BRB-ArrayTools also found no significant difference due to lot or culture duration (Figure 1B).
FIGURE 1. Global gene expression analysis and multidimensional scaling of EBV-transformed lymphoblastoid cell lines (EBV-TM-LCL) used as feeder cells for NK-cell expansion.
(A) Global gene expression analysis: 3 EBV-TM-LCL lots cultured for up to 90 days were analyzed (n = 17). The 12,283 genes remaining after filtering were analyzed by unsupervised hierarchical clustering analysis. No differences were noted between the cultured cell lines. The sample label LCL-3-6-D79 indicates the sixth cell line from EBV-TM-LCL lot 3 with culture duration of 79 days. (B) Unsupervised multidimensional scaling analysis of gene expression data using centered correlation: each spot represents one EBV-TM-LCL sample. No effects of culture duration on EBV-TM-LCL were noted.
Gene Expression Profiles of Freshly Isolated and Expanded NK Cells
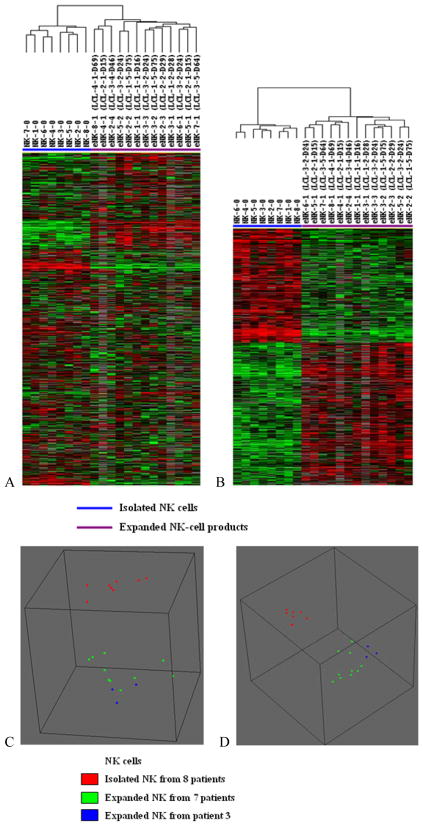
The freshly isolated NK cells from each of the 8 patients and 13 expanded NK cell products were analyzed by gene expression profiling. One of the expanded NK cell products from patient 2 was not available for analysis. Unsupervised hierarchical clustering analysis of 9,634 genes that remained after filtering separated the 21 samples into two groups, the isolated NK-cells and the expanded NK-cell products (Figure 2A). A comparison of isolated NK cells and expanded NK cell products using paired t tests (P < 0.01, FDR < 0.10) revealed that 1,997 genes were differentially expressed. The expression of 1,098 genes was up-regulated in expanded NK-cell products and 879 genes were down-regulated. Supervised hierarchical clustering analysis of the 1,997 differentially expressed genes showed obvious discrimination between the isolated NK-cells and the expanded NK-cell products (Figure 2B). Among the expanded NK cell products, all three products from patient 3 were found in one cluster, but 3 expanded NK cell products from patient 2 and 2 expanded products from patient 5 did not cluster together.
FIGURE 2. Global gene expression and multidimensional scaling analysis of NK-cell products.
(A) The 9,634 genes remaining after filtering were analyzed by unsupervised hierarchical clustering. Significant differences were found between the freshly isolated NK cells and the expanded NK-cell products. The sample label NK-7-0 indicates freshly isolated NK cells from patient 7 and eNK-8-1 indicates the first expanded NK-cell product from patient 8. (B) Supervised hierarchical clustering analysis of the expression of 1,997 differentially expressed genes showed obvious discrimination between the freshly isolated NK cells and the expanded NK-cell products. (C) Unsupervised multidimensional scaling analysis using centered correlation of the 9,634 genes. Each spot represents one NK-cell sample. The expanded NK-cell products were distinguished from the isolated NK cells. (D) Supervised multidimensional scaling analysis using centered correlation of the 1,997 differentially expressed genes separated the samples into the expanded and the freshly isolated NK cells. Among the expanded NK cell products, all 3 products from patient 3 were found in one cluster, but the 3 expanded NK cell products from patient 2 and the 2 from patient 5 did not cluster together.
The expanded NK cell products were also analyzed by multidimensional scaling and this analysis also found that all 3 products from patient 3 clustered together (Figure 2C and D) but the expanded NK cell products from patient 2 and from patient 5 did not. Class comparison analysis of expanded NK cells found no significant differences in gene expression due to patient differences nor due to EBV-TM-LCL lot or duration in culture. Interestingly, the levels of TRAIL expressed by the 3 expanded NK cell products generated from patient 3 tended to be lower than in the other products (Table 1).
Genes Differently Expressed Between Freshly Isolated and Expanded NK Cells
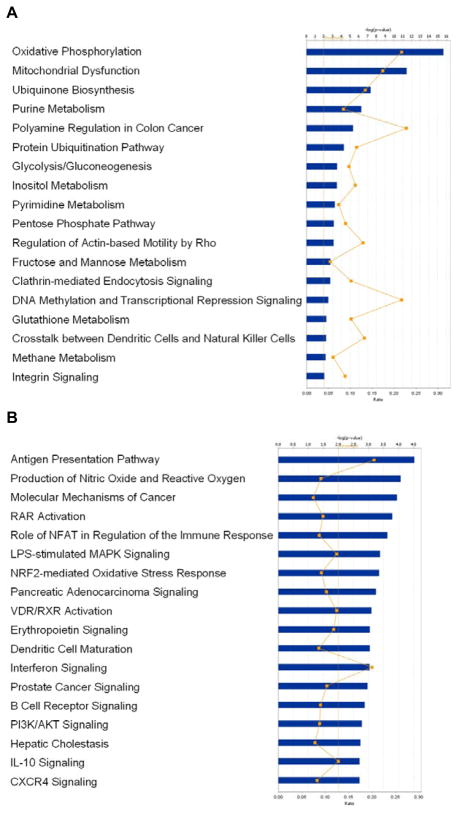
The 1,997 genes that were differently expressed between freshly isolated and expanded NK cells were analyzed by Ingenuity Pathway Analysis (Figure 3). Pathways that included a significant number of genes up-regulated in expanded NK cells include a number of pathways involved in metabolism, ubiquitination, and endocytosis and regulation of actin based motility (Figure 3A). In addition, the genes up-regulated in expanded NK cells belonged to the mitochondrial dysfunction and crosstalk between dendritic cells (DC) pathways. Among the pathways down-regulated in expanded NK cells, there were many related to immune cell function including antigen presentation pathway, LPS-stimulated MAPK signaling, dendritic cell maturation, interferon signaling, B cell receptor signaling, PI3K/AKT signaling, IL-10 signaling, and CXCR4 signaling (Figure 3B).
FIGURE 3. Pathway analysis of differentially expressed genes in expanded NK-cell products compared to freshly isolated NK cells.
(A) Ingenuity Pathway Analysis showed canonical pathways significantly modulated by the genes overexpressed in expanded NK cells (P < 0.01). The p value for each pathway is indicated by the bar and is expressed as -1 times the log of the p value. The line represents the ratio of the number of genes differentially expressed in a given pathway divided by the total number of genes that make up that pathway. (B) Ingenuity Pathway Analysis showed canonical pathways significantly modulated by the genes whose expression was reduced in expanded NK cells (P < 0.01). Only the 18 pathways with the most significant changes are shown.
Gene ontology selection was used to further characterize genes up-regulated in expanded NK cells (Table 2). Genes involved with cell killing, growth and the immune system process were among the up-regulated genes. The 12 cell killing genes up-regulated in expanded NK cells included tubulin β (TUBB) and natural cytotoxicity triggering receptor 3 (NCR3) (Table 3). The 38 up-regulated expanded NK genes associated with cell growth included enolase 1 (ENO1) and isoprenylcysteine carboxyl methyltransferase (ICMT) (Table 3) and the 99 up-regulated immune system process genes included lymphotoxin β (LTB), tubulin β (TUBB), peroxiredoxin 3 (PRDX3), interferon alpha-inducible protein 6 (IFI6), cathepsin C (CTSC), granzyme A (GZMA), and TNF ligand superfamily, member 10 (TNFSF10) (Table 3).
TABLE 2.
Biological Process Associated with Genes Up-Regulated in Expanded NK Cells
| Biological Process | Observed Genes | Total Genes | Expected by Chance | O/E* |
|---|---|---|---|---|
| Cell killing | 12 | 30 | 3.8 | 3.19 |
| Viral reproduction | 16 | 67 | 8.4 | 1.90 |
| Cellular component biogenesis | 126 | 676 | 84.8 | 1.49 |
| Cellular component organization | 284 | 1687 | 211.7 | 1.34 |
| Anatomical structure formation | 122 | 764 | 95.9 | 1.27 |
| Metabolic process | 816 | 5213 | 654.1 | 1.25 |
| Death | 124 | 807 | 101.3 | 1.22 |
| Establishment of localization | 246 | 1692 | 212.3 | 1.16 |
| Cellular process | 1077 | 7440 | 933.5 | 1.15 |
| Localization | 292 | 2022 | 253.7 | 1.15 |
| Growth | 38 | 265 | 33.2 | 1.14 |
| Rhythmic process | 10 | 70 | 8.8 | 1.14 |
| Negative regulation of biological process | 157 | 1107 | 138.9 | 1.13 |
| Multi-organism process | 60 | 438 | 55.0 | 1.09 |
| Immune system process | 99 | 728 | 91.3 | 1.08 |
| Positive regulation of biological process | 161 | 1198 | 150.3 | 1.07 |
| Response to stimulus | 267 | 2017 | 253.1 | 1.06 |
| Biological regulation | 581 | 4497 | 564.2 | 1.03 |
| Regulation of biological process | 543 | 4284 | 537.5 | 1.01 |
| Locomotion | 40 | 319 | 40.0 | 1.00 |
| Developmental process | 227 | 1920 | 240.9 | 0.94 |
| Reproductive process | 44 | 394 | 49.4 | 0.89 |
| Reproduction | 44 | 396 | 49.7 | 0.89 |
| Multicellular organismal process | 219 | 2231 | 279.9 | 0.78 |
| Biological adhesion | 38 | 436 | 54.7 | 0.69 |
| Pigmentation | 1 | 32 | 4.0 | 0.25 |
Ratio of observed number to expected number
TABLE 3.
Genes Up-Regulated in Expanded NK Cells and Associated with Biological Process
| Biological Process | Genes Affected |
|---|---|
| Cell killing | MGEA5, TUBB, PRDX1, NCR3-1, NCR3-2, NCR3-3, GNLY, PTPRC, TUBB2C, IL7R, GIMAP5, CD226 (12 genes) |
| Growth | NDUFS3, ATP6V0E1, EP300, AFG3L2, CDKN2D, CCDC85B, DDX5, PSRC1, CREB1, PARP1, ICMT, C6orf108, ENO3, TSG101, E4F1, CCNB2, CDKN1B, ENO1-1, EI24, NPM1, PDGFB, PRPF19, UBE2E3-1, UBE2E3-2, OGFR, APBB1, RUVBL2, TAF9, IL7R, ENO1-2, BRD8, SOD1, SOCS2, BCAR1, CDKN2A, CHST11, ING1, VPS54 (38 genes) |
| Immune system process | LTB, CCR2-1, LIG1, GPX2, MSH6, NDFIP1, CTSC-1, SKAP1, LTB4R, TNFSF12, CCL25, DHX58, IMPDH2, AP2B1, MIF, CCL4, CD276, XRCC5, MAP4K2, CCND3, TUBB, GZMA, EOMES, CD2, CLPTM1, CORO1A, NUP85, CCNB2, CCL8, IFI6, TRIM22, OAS1-1, PDGFB, IGJ, TNFRSF4, PRDX1, CXCL5, PSMB9, CD8B, DCLRE1C, CTSC-2, TAP1, ITGAL, KIR2DS2, NCR3-1, NCR3-2, FYN, NCR3-3, RELB, GPI, TESC, CKLF, ITGA6, YWHAZ-1, IGHG1, KIR2DS1, CDC42-1, TACC3, PTPRC, KIR3DL1, IGKC, CD79A, CDC42-2, CRIP2, LCK, TUBB2C, DDOST, KIR2DL3, IL7R, YWHAZ-2, C5, CD28, BMI1, TAC1, GIMAP5, NKX2-5, SOD1, PRDX3, C1QBP, TNFSF10, AP2S1, NME1, UBASH3A, C3AR1, FCN1, CCR2-2, MBP-1, BCAR1, JAG2, CDKN2A, CD226, CTSC-3, MBP-2, CD19, FCGR2B, LST1, MYH9, CTSS, OAS1-2 (99 genes) |
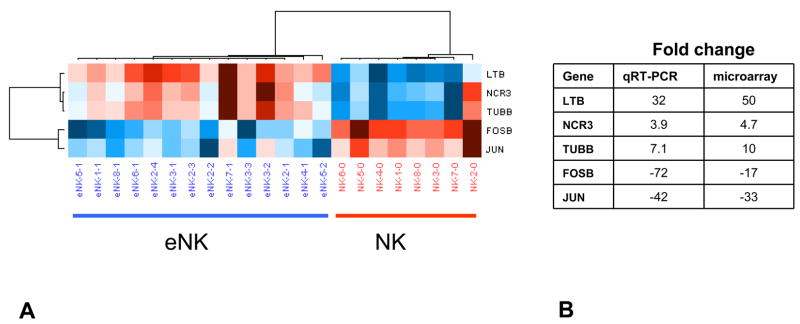
The 100 most up-regulated genes in expanded NK cells are shown in Supplemental Table 1S and the 100 most down-regulated genes in expanded NK cells are shown in Supplemental Table 2S. Those up-regulated the most included matrix metallopeptidase 25 (MMP25), LTB, myxovirus resistance 1 (MX1), BCL2-associated X protein (BAX) and IFI6. Those down-regulated most included jun oncogene (JUN), FBJ murine osteosarcoma viral oncogene homolog B (FOSB), CD83, selectin L (SELL), colony stimulating factor 1 receptor (CSF1R) and interferon-γ receptor 2 (IFNGR2) (Supplemental Table 2S). Quantitative real time PCR confirmed that LTB, NCR3 and TUBB were up-regulated in expanded NK cells while FOSB and JUN were down-regulated (Figure 4).
FIGURE 4. Analysis of selected differently expressed genes by quantitative real time PCR.
The expression of FOSB, JUN, LTB, NCR3 and TUBB were compared among expanded and freshly isolated NKs using quantitative real time PCR. (A) The results were analyzed by hierarchical clustering analysis with up-regulated genes shown in red and down-regulated genes shown in blue. (B) The results were similar to those obtained with the global gene expression microarray.
MicroRNA Expression Profiles of Freshly Isolated and Expanded NK Cells
The 8 isolated NK cells and 14 expanded NK cell products were analyzed by global microRNA analysis. Supervised hierarchical clustering analysis and multidimensional scaling using centered correlation showed obvious discrimination between the fresh NK-cells and the expanded NK-cell products (Figure 5). Within the expanded NK cell group, the 3 products from patient 3 were again within 1 cluster. Comparison of microRNA expressed by expanded NK cells from patient 3 to the other 11 expanded NK cells found that miR-327 (2.99-fold) (P < 0.05) and miR-185 (1.58-fold) (P < 0.05) were up-regulated. However, class comparison analysis found no significant differences among expanded NK cell products due to the EBV-TM-LCL lot, culture duration of EBV-TM-LCL, or patient.
FIGURE 5. MicroRNA expression and multidimensional scaling analysis of NK-cell products.
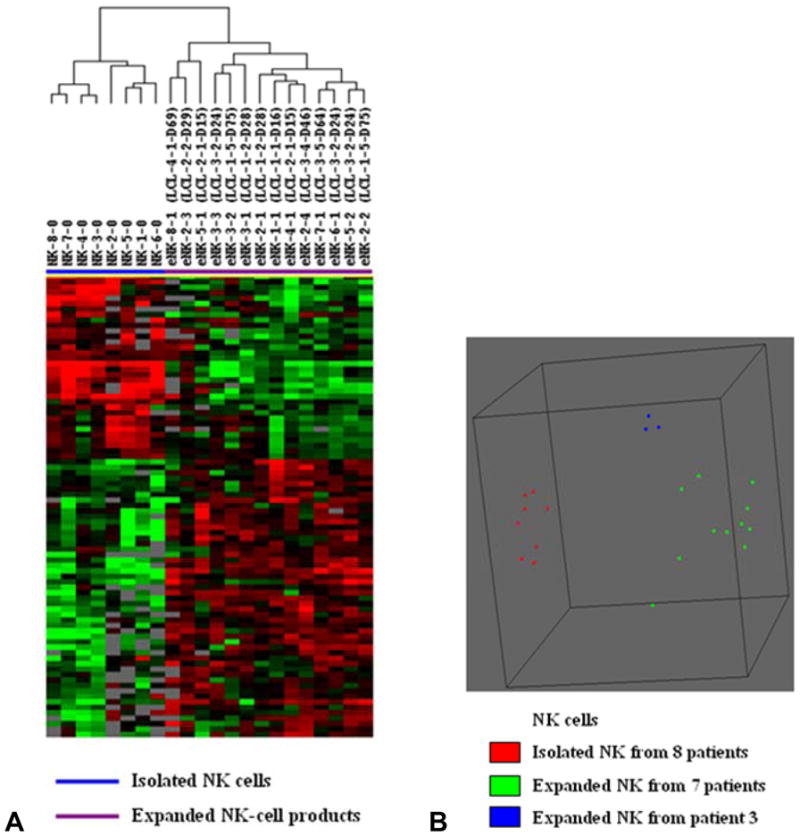
(A) Supervised hierarchical clustering analysis showed obvious discrimination between the freshly isolated NK cells and the expanded NK-cell products. Sample label NK-8-0 indicates the freshly isolated NK cells from patient 8 and eNK-8-1 means the first expanded NK-cell product from patient number 8. (B) Supervised multidimensional scaling using centered correlation separated NK-cell products into the expanded and the freshly isolated cells. Each spot represents one NK cell sample. All three expanded NK-cell products from patient 3 showed closely related microRNA expression profiles, but the 3 expanded NK cell products from patient 2 and the 2 from patient 5 did not cluster together.
MicroRNAs Differently Expressed between Freshly Isolated and Expanded NK Cells
A total of 54 microRNAs were differently expressed between freshly isolated and expanded NK cells (paired t tests, P < 0.05, FDR < 0.10). Among the 28 microRNAs up-regulated in expanded NK cells, 6 microRNAs (miR-1246, miR-583, miR-487b, miR-1207-5p, miR-483-5p, and miR-92b) were overexpressed more than 3-fold (Table 4). Both miR-487b and miR-381, which were up-regulated 3.60-fold and 2.54-fold in expanded NK cells respectively, belong to a microRNA cluster on chromosome 14q32.31 which includes only these two members. The 26 microRNAs down-regulated in expanded NK-cell products are shown by fold-change in Table 5. Among these microRNAs 12 microRNAs (miR-770-5p, let-7g, miR-30e, miR-1323, miR-520c-5p, miR-422a, miR-378, miR-1254, let-7d, miR-30a, miR-575, and miR-18a) were down-regulated more than 3-fold. In addition, the expression of miR-27a and miR-23a, which were down-regulated 2.94-fold and 2.17-fold respectively, belong to a microRNA cluster on chromosome 19p13.12. This cluster is comprised of miR-24-2, miR-27a, and miR-23a. MicroRNA-24-2 was not included in our microarray.
TABLE 4.
MicroRNAs Overexpressed in Expanded NK Cells as Compared to freshly Isolated NK Cells Ranked by Fold-Change
| Symbol | P value | FDR | FC | |
|---|---|---|---|---|
| 1 | hsa-mir-1246 | < 1 × 10−7 | < 1 × 10−7 | 5.69 |
| 2 | hsa-mir-583 | < 1 × 10−7 | < 1 × 10−7 | 3.89 |
| 3 | hsa-mir-487b | 5.00 × 10−7 | 1.71 × 10−5 | 3.60 |
| 4 | hsa-mir-1207-5p | 7.43 × 10−5 | 0.0005245 | 3.18 |
| 5 | hsa-mir-483-5p | 0.0012718 | 0.0053057 | 3.08 |
| 6 | hsa-mir-92b | 3.59 × 10−5 | 0.0003314 | 3.04 |
| 7 | hsa-mir-493 | 1.90 × 10−6 | 5.07 × 10−5 | 2.69 |
| 8 | hsa-mir-765 | 0.0012822 | 0.0053057 | 2.65 |
| 9 | hsa-mir-138-2 | 0.0001875 | 0.0010465 | 2.62 |
| 10 | hsa-mir-1225-5p | 4.35 × 10−5 | 0.0003536 | 2.59 |
| 11 | hsa-mir-17 | 0.0011965 | 0.0051279 | 2.57 |
| 12 | hsa-mir-381 | 0.0001788 | 0.0010465 | 2.54 |
| 13 | hsa-mir-300 | 3.98 × 10−5 | 0.0003411 | 2.50 |
| 14 | hsa-mir-423-5p | 4.40 × 10−6 | 9.60 × 10−5 | 2.44 |
| 15 | hsa-mir-1826 | 9.75 × 10−5 | 0.0006409 | 2.43 |
| 16 | hsa-mir-1268 | 9.69 × 10−5 | 0.0006409 | 2.21 |
| 17 | hsa-mir-320a | 3.15 × 10−5 | 0.0003024 | 2.13 |
| 18 | hsa-mir-370 | 0.0024553 | 0.0087951 | 2.03 |
| 19 | hsa-mir-923 | 0.0004959 | 0.0023803 | 2.01 |
| 20 | hsa-mir-491-5p | 0.0115669 | 0.0319087 | 1.91 |
| 21 | hsa-mir-936 | 0.0001626 | 0.0010006 | 1.88 |
| 22 | hsa-mir-1908 | 0.0189325 | 0.0454380 | 1.81 |
| 23 | hsa-mir-582 | 0.0072223 | 0.0225111 | 1.69 |
| 24 | hsa-mir-140-3p | 0.0175993 | 0.0431003 | 1.67 |
| 25 | hsa-mir-551b | 0.0117146 | 0.0319489 | 1.67 |
| 26 | hsa-mir-25 | 0.0343642 | 0.0743010 | 1.50 |
| 27 | hsa-mir-30c-2 | 0.0270597 | 0.0612672 | 1.48 |
| 28 | hsa-mir-494 | 0.0362466 | 0.0772714 | 1.34 |
FDR indicates false discovery rate; FC, fold-change.
Using t test with P value < 0.05 and false discovery rate < 0.10.
TABLE 5.
MicroRNAs Underexpressed in Expanded NK Cells as Compared to freshly Isolated NK cells Ranked by Fold-Change
| Symbol | P value | FDR | FC | |
|---|---|---|---|---|
| 1 | hsa-mir-770-5p | 1.00 × 10−6 | 3.00 × 10−5 | 9.09 |
| 2 | hsa-let-7g | 1.70 × 10−5 | 0.0001977 | 6.67 |
| 3 | hsa-mir-30e | 6.90 × 10−6 | 0.0001065 | 4.55 |
| 4 | hsa-mir-1323 | 3.50 × 10−6 | 8.40 × 10−5 | 4.35 |
| 5 | hsa-mir-520c-5p | 5.70 × 10−6 | 0.0001052 | 4.35 |
| 6 | hsa-mir-422a | 4.00 × 10−7 | 1.60 × 10−5 | 4.00 |
| 7 | hsa-mir-378 | 3.00 × 10−7 | 1.44 × 10−5 | 4.00 |
| 8 | hsa-mir-1254 | 4.90 × 10−6 | 9.80 × 10−5 | 3.85 |
| 9 | hsa-let-7d | 7.02 × 10−5 | 0.0005105 | 3.33 |
| 10 | hsa-mir-30a | 0.0001548 | 0.0009777 | 3.33 |
| 11 | hsa-mir-575 | 1.73 × 10−5 | 0.0001977 | 3.23 |
| 12 | hsa-mir-18a | 0.0316646 | 0.0703658 | 3.03 |
| 13 | hsa-mir-1182 | 0.0056279 | 0.0182526 | 2.94 |
| 14 | hsa-mir-27a | 0.0023186 | 0.0085610 | 2.94 |
| 15 | hsa-mir-373 | 0.0003116 | 0.0015911 | 2.86 |
| 16 | hsa-mir-30d | 0.0010447 | 0.0045587 | 2.86 |
| 17 | hsa-mir-29c | 0.0206765 | 0.0491323 | 2.86 |
| 18 | hsa-mir-130b | 0.0003756 | 0.0018780 | 2.63 |
| 19 | hsa-let-7i | 0.0447607 | 0.0926083 | 2.63 |
| 20 | hsa-mir-23a | 0.0085896 | 0.0251403 | 2.17 |
| 21 | hsa-mir-143 | 0.0059995 | 0.0190026 | 2.04 |
| 22 | hsa-mir-371-5p | 0.0008938 | 0.0040474 | 2.00 |
| 23 | hsa-mir-1183 | 0.0446072 | 0.0926083 | 1.85 |
| 24 | hsa-mir-150 | 9.40 × 10−6 | 0.0001253 | 1.82 |
| 25 | hsa-mir-202 | 0.0083849 | 0.0248441 | 1.75 |
| 26 | hsa-mir-486-3p | 0.0099476 | 0.0284217 | 1.67 |
FDR indicates false discovery rate; FC, fold-change.
Using t test with P value < 0.05 and false discovery rate < 0.10.
DISCUSSION
We assessed freshly isolated and expanded clinical NK cells using global gene and microRNA expression analysis. As expected, there were marked differences between isolated and expanded NK cells in both gene expression and microRNA expression assays.
Expanded NK cells expressed a number of genes associated with cytotoxicity. NK cell cytotoxicity can be mediated by both death receptors and perforin and granzyme. In addition to expressing high levels of TRAIL, we found that the expression of both granzyme A and K were up-regulated in expanded NK cells. Granzyme A and granzyme K are cell death inducing serine proteases, both of which have trypsine-like protease activity (15;16).
NK cell activating and killing receptors include the Killer-Inhibitory Receptors (KIR) and the triggering receptors: NKG2D and natural cytotoxicity receptors (NCRs). The NCRs include NKp30 (NCR3, CD337), NKp44 (NCR2), and NKp46 (NCR1). NK cell cytotoxocity is reduced in patients with ovarian, breast and cervical cancer (17–19). In patients with cervical cancer and precursor lesions reduced NK cell cytotoxicity is associated with the reduced expression of NKG2D, NKp30 and NKp46 (19). We found that the expression of NCR3 was up-regulated in expanded NK cells. In addition to measuring TRAIL expression, the measurement of NKp30 expression may be a useful expanded NK cell potency biomarker.
An important function of NK cells is their interaction or crosstalk with iDCs and T cells (20;21). The genes up-regulated in expanded NK cells included many in the crosstalk between DCs and NK cells pathway. DCs interact with NK cells by producing IFN-α and IFN-β which increase NK cell cytotoxicity and DC production of IL-12 and IL-18 stimulates the production of IFN-γ by NK cells. NK cells produce IFN-γ and TNF-α which mediates DC differentiation and maturation. Activated NK cells can also kill immature DCs. This DC cytotoxicity is dependent on NK receptors NKp30, NKp46 and DNAM-1. Since expanded cells express high levels of NCR3 which encodes NKp30, they may be effective at editing iDCs. This may be of particular clinical relevance in the context of allogeneic hematopoietic stem cell transplantation, where NK cell killing of dendritic cells has been shown to prevent graft-versus-host disease in animal models of transplantation (22).
LTB and MX1 were among the most up-regulated genes in expanded NKs. This finding is constant with another study that has shown that lymphotoxin B is expressed by activated NK cells (23). Lymphotoxin B is important for the development of secondary lymph nodes and it stimulates the growth of NK and NKT cells (24;25). The up-regulation of LTB may be important for the expansion of cultured NK cells. MX1 is an interferon responsive gene with potent anti-viral activity (26).
The express of the Bcl-2 family protein BAX was up-regulated in expanded NK cells. Bcl-2 proteins play an important role in regulating apoptosis. BAX resides on the outer membrane of mitochondria and can trigger the release of mitochondrial cytochrome c and activate caspase mediated apoptosis (27). The up-regulation of BAX by expanded NK cells may indicate that the NK cells were reaching their limit of cell replication and expansion.
A number of genes up-regulated in expanded NK cells belonged to metabolic, ubiquitination, endocytosis and regulation of actin based motility pathways. The expression of these genes may be secondary to cell proliferation since we have found similar pathways were up-regulated in other expanded cells populations. Pathways associated with up-regulated genes in the expanded NK cell products included oxidative phosphorylation, purine metabolism, pyrimidine metabolism and glycolysis/gluconeogenesis.
Some genes were expressed at higher levels in freshly isolated NK cells than in expanded NK cells. The genes that were down-regulated in expanded NK cells belonged to B cell receptor, interferon signaling, IL-10 signaling and the antigen processing and presentation pathways. The significance of these differences is not certain, however, some T cells and monocyte-derived DCs also express CD56 (28). Although freshly isolated NK cells were obtained from peripheral blood mononuclear cells by CD3+ cell depletion followed by CD56 selection were devoid of T cells, some monocyte-derived DCs may have been present in the isolated NK cell population. The presence of a small proportion of CD56 expressing monocyte-derived DCs in the isolated NK cells that died off during expansion could explain the down-regulation of these pathways in the expanded NK cell population. We cannot, however, exclude the possibility that expanded NK cells down-regulated genes in these pathways.
Clustered microRNAs are generally transcribed into polycistronic primary transcripts (pri-miRNA) and these clusters are indeed single transcriptional units (29;30). We found that the microRNA cluster of miR-487b and miR-381 was overexpressed in expanded NK cell products. On the other hand, expression of the microRNA cluster of miR-27a and miR-23a was down-regulated.
We observed some variability among the expanded NK cell products. Multiple expanded NK cells produced from a single patient’s NK cells tended to cluster together suggesting that inter-patient differences exist, but the number of products studied was too small to detect inter-patient differences that are likely complex and multifactorial. Further studies of larger numbers of patients and products are warranted to determine if significant differences in expanded NK cell products exist due to patient factors and to identify biomarkers associated with these differences. The results of the analysis of expanded NK cell products by flow cytometry, gene expression, and microRNA expression assays should be compared with clinical outcome associated with adoptive NK cell therapy patient to determine if differences among expanded NK cells are clinically important and to identify new biomarkers for predicting the quality and anti-tumor potency of expanded NK cells.
We also analyzed the EBV-TM-LCL to determine if they might contribute to lot-to-lot differences in expanded NK cells. Gene expression analysis showed that there were no significant differences based on EBV-TM-LCL lot and culture duration. In addition class comparison analysis did not find any difference in expanded NK cell products relative to the EBV-TM-LCL lot number used or culture duration.
In summary, many differences in expanded and freshly isolated NK cells were identified. Several genes associated with cytotoxicity and the immune system differed. The expression of up-regulated genes in expanded NK cells must be correlated with patient outcomes following NK cell therapy to determine if any of these genes are clinically important biomarkers. We also found that some expanded NK cell product variability due to patient factors may exist. Further analysis of larger numbers of expanded NK cell products is warranted to determine if real inter-patient differences exist. Product biomarkers found to differ among patients will need to be correlated with patient outcomes following NK cell therapy to determine if the differences are clinically important.
Supplementary Material
Footnotes
Financial disclosure: the authors have declared that there are no financial conflicts of interest in regard to this article.
References
- 1.Miller JS, Soignier Y, Panoskaltsis-Mortari A, McNearney SA, Yun GH, Fautsch SK, et al. Successful adoptive transfer and in vivo expansion of human haploidentical NK cells in patients with cancer. Blood. 2005 Apr 15;105(8):3051–7. doi: 10.1182/blood-2004-07-2974. [DOI] [PubMed] [Google Scholar]
- 2.Srivastava S, Lundqvist A, Childs RW. Natural killer cell immunotherapy for cancer: a new hope. Cytotherapy. 2008;10(8):775–83. doi: 10.1080/14653240802648181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terme M, Ullrich E, Delahaye NF, Chaput N, Zitvogel L. Natural killer cell-directed therapies: moving from unexpected results to successful strategies. Nat Immunol. 2008 May;9(5):486–94. doi: 10.1038/ni1580. [DOI] [PubMed] [Google Scholar]
- 4.Miller JS. Should natural killer cells be expanded in vivo or ex vivo to maximize their therapeutic potential? Cytotherapy. 2009;11(3):259–60. doi: 10.1080/14653240902888000. [DOI] [PubMed] [Google Scholar]
- 5.Dudley ME, Rosenberg SA. Adoptive cell transfer therapy. Semin Oncol. 2007 Dec;34(6):524–31. doi: 10.1053/j.seminoncol.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenberg SA, Dudley ME. Adoptive cell therapy for the treatment of patients with metastatic melanoma. Curr Opin Immunol. 2009 Apr;21(2):233–40. doi: 10.1016/j.coi.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caligiuri MA. Human natural killer cells. Blood. 2008 Aug 1;112(3):461–9. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lundqvist A, Abrams SI, Schrump DS, Alvarez G, Suffredini D, Berg M, et al. Bortezomib and depsipeptide sensitize tumors to tumor necrosis factor-related apoptosis-inducing ligand: a novel method to potentiate natural killer cell tumor cytotoxicity. Cancer Res. 2006 Jul 15;66(14):7317–25. doi: 10.1158/0008-5472.CAN-06-0680. [DOI] [PubMed] [Google Scholar]
- 9.McKenna DH, Jr, Sumstad D, Bostrom N, Kadidlo DM, Fautsch S, McNearney S, et al. Good manufacturing practices production of natural killer cells for immunotherapy: a six-year single-institution experience. Transfusion. 2007 Mar;47(3):520–8. doi: 10.1111/j.1537-2995.2006.01145.x. [DOI] [PubMed] [Google Scholar]
- 10.Berg M, Lundqvist A, McCoy P, Jr, Samsel L, Fan Y, Tawab A, et al. Clinical-grade ex vivo-expanded human natural killer cells up-regulate activating receptors and death receptor ligands and have enhanced cytolytic activity against tumor cells. Cytotherapy. 2009;11(3):341–55. doi: 10.1080/14653240902807034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walter EA, Greenberg PD, Gilbert MJ, Finch RJ, Watanabe KS, Thomas ED, et al. Reconstitution of cellular immunity against cytomegalovirus in recipients of allogeneic bone marrow by transfer of T-cell clones from the donor. N Engl J Med. 1995 Oct 19;333(16):1038–44. doi: 10.1056/NEJM199510193331603. [DOI] [PubMed] [Google Scholar]
- 12.Wang E, Miller LD, Ohnmacht GA, Liu ET, Marincola FM. High-fidelity mRNA amplification for gene profiling. Nat Biotechnol. 2000 Apr;18(4):457–9. doi: 10.1038/74546. [DOI] [PubMed] [Google Scholar]
- 13.Ren J, Jin P, Wang E, Marincola FM, Stroncek DF. MicroRNA and gene expression patterns in the differentiation of human embryonic stem cells. J Transl Med. 2009;7:20. doi: 10.1186/1479-5876-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998 Dec 8;95(25):14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choy JC. Granzymes and perforin in solid organ transplant rejection. Cell Death Differ. 2010 Apr;17(4):567–76. doi: 10.1038/cdd.2009.161. [DOI] [PubMed] [Google Scholar]
- 16.Cullen SP, Brunet M, Martin SJ. Granzymes in cancer and immunity. Cell Death Differ. 2010 Apr;17(4):616–23. doi: 10.1038/cdd.2009.206. [DOI] [PubMed] [Google Scholar]
- 17.Lotzova E, Savary CA, Freedman RS, Bowen JM. Natural killer cell cytotoxic potential of patients with ovarian carcinoma and its modulation with virus-modified tumor cell extract. Cancer Immunol Immunother. 1984;17(2):124–9. doi: 10.1007/BF00200048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lotzova E, Savary CA, Freedman RS, Edwards CL, Wharton JT. Recombinant IL-2-activated NK cells mediate LAK activity against ovarian cancer. Int J Cancer. 1988 Aug 15;42(2):225–31. doi: 10.1002/ijc.2910420214. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Iglesias T, Del Toro-Arreola A, barran-Somoza B, Del Toro-Arreola S, Sanchez-Hernandez PE, Ramirez-Duenas MG, et al. Low NKp30, NKp46 and NKG2D expression and reduced cytotoxic activity on NK cells in cervical cancer and precursor lesions. BMC Cancer. 2009;9:186. doi: 10.1186/1471-2407-9-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferlazzo G, Munz C. Dendritic cell interactions with NK cells from different tissues. J Clin Immunol. 2009 May;29(3):265–73. doi: 10.1007/s10875-009-9283-y. [DOI] [PubMed] [Google Scholar]
- 21.Kalinski P, Mailliard RB, Giermasz A, Zeh HJ, Basse P, Bartlett DL, et al. Natural killer-dendritic cell cross-talk in cancer immunotherapy. Expert Opin Biol Ther. 2005 Oct;5(10):1303–15. doi: 10.1517/14712598.5.10.1303. [DOI] [PubMed] [Google Scholar]
- 22.Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002 Mar 15;295(5562):2097–100. doi: 10.1126/science.1068440. [DOI] [PubMed] [Google Scholar]
- 23.Ware CF, Crowe PD, Grayson MH, Androlewicz MJ, Browning JL. Expression of surface lymphotoxin and tumor necrosis factor on activated T, B, and natural killer cells. J Immunol. 1992 Dec 15;149(12):3881–8. [PubMed] [Google Scholar]
- 24.Drutskaya MS, Efimov GA, Kruglov AA, Kuprash DV, Nedospasov SA. Tumor necrosis factor, lymphotoxin and cancer. IUBMB Life. 2010 Apr;62(4):283–9. doi: 10.1002/iub.309. [DOI] [PubMed] [Google Scholar]
- 25.Franki AS, Van BK, Dewint P, Meeus I, Veys E, Deforce D, et al. Lymphotoxin alpha 1 beta 2: a critical mediator in V alpha 14i NKT cell differentiation. Mol Immunol. 2005 Feb;42(4):413–7. doi: 10.1016/j.molimm.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 26.Haller O, Staeheli P, Kochs G. Interferon-induced Mx proteins in antiviral host defense. Biochimie. 2007 Jun;89(6–7):812–8. doi: 10.1016/j.biochi.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 27.Autret A, Martin SJ. Emerging role for members of the Bcl-2 family in mitochondrial morphogenesis. Mol Cell. 2009 Nov 13;36(3):355–63. doi: 10.1016/j.molcel.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 28.Milush JM, Long BR, Snyder-Cappione JE, Cappione AJ, III, York VA, Ndhlovu LC, et al. Functionally distinct subsets of human NK cells and monocyte/DC-like cells identified by coexpression of CD56, CD7, and CD4. Blood. 2009 Nov 26;114(23):4823–31. doi: 10.1182/blood-2009-04-216374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002 Sep 2;21(17):4663–70. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004 Jun 15;270(2):488–98. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.