Abstract
Kingella kingae is an emerging pathogen causing osteoarticular infections in pediatric patients. Electron microscopy of K. kingae clinical isolates revealed the heterogeneously-sized membranous structures blebbing from the outer membrane that were classified as outer membrane vesicles (OMVs). OMVs purified from the secreted fraction of a septic arthritis K. kingae isolate were characterized. Among several major proteins, K. kingae OMVs contained virulence factors RtxA toxin and PilC2 pilus adhesin. RtxA was also found secreted as a soluble protein in the extracellular environment indicating that the bacterium may utilize different mechanisms for the toxin delivery. OMVs were shown to be hemolytic and possess some leukotoxic activity while high leukotoxicity was detected in the non-hemolytic OMV-free component of the secreted fraction. OMVs were internalized by human osteoblasts and synovial cells. Upon interaction with OMVs, the cells produced increased levels of human granulocyte-macrophage colony-stimulating factor (GM-CSF) and interleuskin 6 (IL-6) suggesting that these cytokines might be involved in the signaling response of infected joint and bone tissues during natural K. kingae infection. This study is the first report of OMV production by K. kingae and demonstrates that OMVs are a complex virulence factor of the organism causing cytolytic and inflammatory effects on host cells.
1. Introduction
The Gram negative bacterium Kingella kingae, a coccobacillus of the Neisseriaceae family, is a component of the human oropharyngeal flora. The highest rate of carriage of K. kingae coincides with the age less than 4 years old [1–3]. Being carried asymptomatically in the respiratory tract, the bacterium may penetrate into the bloodstream and cause invasive infections including septic arthritis, osteomyelitis, infective endocarditis and bacteremia [4, 5]. K. kingae belongs to the HACEK group (Haemophilus spp., Aggregatibacter actinomycetemcomitans, Cardiobacterium hominis, Eikenella corrodens, and Kingella kingae) of oral Gram negative bacteria that share an enhanced capacity to produce infective endocarditis.
While K. kingae infections in adults are still considered to be rare, recent data suggest that the frequency of those infections in children has been underestimated. Improvements in culture techniques and molecular detection methods have increased the recognition of K. kingae as a leading cause of osteoarticular infections in pediatric patients [1, 2, 6–13]. Several reports describe epidemiological cases of invasive K. kingae infections in day care centers showing that the bacterium is able to cause outbreaks of disease in children communities [14–16].
The pathogenesis of K. kingae infection begins with colonization of the upper respiratory tract. It is believed that damage of the respiratory and buccal mucosa allows haematogenous spread of the bacterium to distant organs of the body, especially to the skeletal system [5]. The investigation of K. kingae virulence has been significantly restricted by the lack of genome information and fastidious nature of the organism. Only recently some progress in understanding K. kingae virulence mechanisms has been made [17–20]. The important role in the bacterium virulence has been attributed to the production of a toxin of the RTX-group, RtxA [20]. The toxins of the RTX (repeat in toxin) family are large secreted proteins and found in different Gram negative pathogens: E. coli α-hemolysin [21–23], Aggregatibacter actinomycetemcomitans LtxA [24–26], Bordetella pertussis CyaA [27], Manhemia haemolytica LktA [28], Moraxella bovis MbxA [29], Neisseria meningitides FrpA [30] and others. Consequently, the gene encoding for RtxA was used as a specific molecular marker to diagnose K. kingae infections [6].
Extracellular secretion of metabolic products is the major mechanism by which Gram negative pathogens intoxicate host cells and cause infection. The transport of secreted proteins to the extracellular space may involve high-order complexes with lipid membrane structures, the outer membrane vesicles (OMVs). Bacterial OMVs were shown to serve as secretary delivery vehicles for virulence factors of many bacterial pathogens [31]. OMVs are also studied because of their potential to be used as vaccines [32]. They are enriched with antigenic outer membrane proteins, such as porins, that maintain their native structure being inserted into OMV membrane. The first successful application of this approach was development of a protein-based OMV-vaccine against serogroup B meningococcal disease that is currently in use [33]. In the present study we demonstrated that K. kingae produces toxic OMVs that may play an important role in the organism’s pathogenesis.
2. Results
2. 1. Purification and characterization of OMVs production in clinical isolates
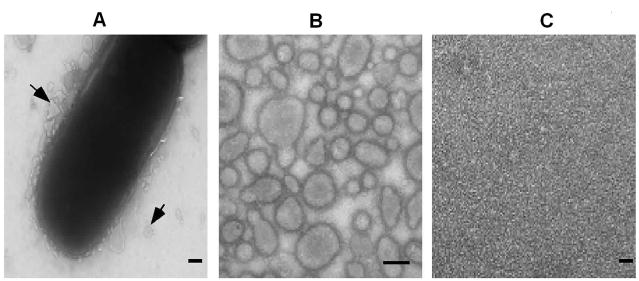
Nine K. kingae clinical isolates from our collection were grown on CA with 5% sheep blood for 40 h and subjected to TEM. Electron micrographs of the strains revealed spherical membranous structures ranging from 50 to 200 nm in diameter around the rod-shared bacteria (Fig. 1A). These structures were blebbing from the outer membrane (OM) of the live bacterial cells and were classified as OMVs.
Fig. 1. TEM micrographs of PYKK081 and its secreted fraction components after negative staining.
A. Secretion of OMVs by the bacterium grown on CA with 5% sheep blood agar for 40 h. The arrow shows OMVs pinching from the OM Direct magnification is 22,000. Bar=100 nm.
B. OMVs isolated from the secreted fraction. Direct magnification is 35,000. Bar=100 nm.
C. OMV-free secreted fraction. Direct magnification is 35,000. Bar=100 nm.
To further study K. kingae OMVs, we designed a procedure for OMVs purification. The bacterium did not express good growth in liquid medium; therefore we isolated its secreted fraction after growing the colonies on solid medium. To do this, the bacteria were cultivated on AAGM agar for 40 h, and the biomass was scraped from plates and subjected to ultracentrifugation, resulting in separation of a compact cell pellet and a supernatant containing concentrated secreted products. The bacterial cells did not lyse during this step as was confirmed by CFU (colony forming unit) count before and after the centrifugation. The liquid fraction was collected and passed through 0.45 μm filter to remove the bacterial cells. The fraction was further subjected to prolonged high speed centrifugation that resulted in the separation of soluble fraction and a pellet. The TEM analysis demonstrated that the obtained pellet, resuspended in OMV-buffer, consisted of concentrated OMVs (Fig. 1B). The schematic diagram of OMVs purification is presented in supplementary data (Fig 1S). Additional purification of OMVs using gel-filtration chromatography resulted in formation of one sharp pick of high molecular weight proteins (data not shown). The carefully separated supernatant was free of OMVs (Fig. 1C) and is referred in the paper as the OMV-free secreted fraction. The yield of purified OMVs from different clinical isolates was compared (Table 1). Using the same growth conditions different K. kingae isolates produced various amounts of OMVs calculated per gram of wet bacterial weight and could be roughly divided into two groups: high- (≥0.03 g) and low-producing (≤0.01 g) OMVs strains. To further study K. kingae OMVs, we selected highly OMV-producing septic arthritis isolate PYKK081. The isolated PYKK081 OMVs sample contained 8 mg/ml protein, 0.5 mg/ml DNA and ≤ 1500 U/ml endotoxin.
Table 1.
Kingella isolates used in the study.
| Strain | Supplier, country of origin | Source | OMVs (mg)/gram of biomass ±SE1 |
|---|---|---|---|
| K. oralis UB-38 | ATCC, USA | Oral cavity | ND2 |
| K. kingae 23330 | ATCC, Norway | Nasal cavity | 0.01±0.003 |
| K. kingae PYP8 | SUMC3, Israel | Oral cavity | 0.01±0.001 |
| K. kingae M200300017 | MDH4, USA | Oral cavity | 0.03±0.007 |
| K. kingae PYKK081 | SUMC, Israel | Joint | 0.10±0.006 |
| K. kingae PYKK135 | SUMC, Israel | Joint | 0.06±0.002 |
| K. kingae PYKK189 | SUMC, Israel | Joint | 0.06±0.008 |
| K. kingae C2003003154 | MDH, USA | Joint | 0.05±0.007 |
| K. kingae PYKK190 | SUMC, Israel | Heart | 0.01±0.002 |
| K. kingae PYKK101 | SUMC, Israel | Bone | 0.01±0.002 |
Standard error, SE was calculated based on results obtained in three independent experiments.
Not determined
Soroka University Medical Center
Minnesota Department of Health
2. 2. Analysis of K. kingae OMV protein content
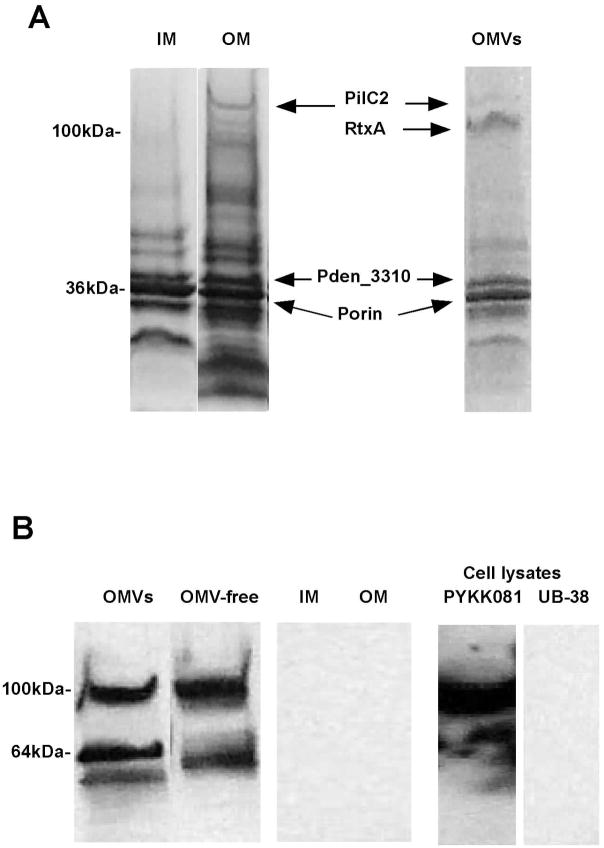
To characterize the protein composition PYKK081 OMV proteins were resolved and visualized by SDS-PAGE. OMV preparation demonstrated several major protein bands that were identified as PilC2 (164 kDa), hypothetical protein Pden_3310 (120 kDa), RtxA (101 kDa), and putative porin (36 kDa) by MALDI-TOF MS. The protein profile of OMVs was compared with protein profiles of PYKK081 OM and inner membrane (IM), prepared by the method of Sarcosyl extraction (Fig. 2). OMVs, IM and OM demonstrated variations in the protein profiles. Three polypeptides (PilC2, Pden_3310 and putative porin) identified in OMVs samples were also present in OM fractions as was confirmed by MALDI-TOF MS analysis. PilC2, known to be an outer membrane protein [34], was identified only in OM and OMVs but not in IM suggesting that our protocol successfully separated the cellular compartments (Fig. 2A). Thus, PYKK081 OMVs appear to contain a subset of OM proteins preparations; however the inclusion of proteins into OMVs is selective.
Fig. 2. Protein analysis of PYKK081 and K. oralis UB-38.
A. One μg of proteins from PYKK081 IM, OM, and OMVs samples was resolved by SDS-PAGE. The proteins were stained with Coomassie blue. The major proteins were identified by MALDI-TOF MS.
B. One μg of proteins from PYKK081 IM, OM, OMVs or PYKK081 and UB-38 cell lysate samples was resolved by SDS-PAGE and subjected to Western blot analysis with 9D4 antibody.
2. 3. Toxicity of K. kingae secreted fraction components on eukaryotic cells
To explore the potential toxicity, we tested the effect of PYKK081 secreted fraction components on different human and mouse cells. Using dye exclusion method, we found that 10 μl (8 μg of protein) of PYKK081 OMV-free secreted fraction caused 20–85% lysis of leukocytes (promyeloblasts, monocytes, T and B lymphoblasts) and about 20% lysis of megakaryoblasts after 24 h; however the maximum toxic effect was reached as early as 3 hours of treatment. (Table 2). The toxicity was not species specific, both human and mouse leukocytes were found to be sensitive. The fraction had insignificant toxic effect on human erythrocytes. The toxicity of OMVs on white blood cells was less pronounced than OMV-free fraction; however OMVs demonstrated some hemolytic activity. About 30% of human or sheep erythrocytes were lysed after 16 h. A cytolytic effect of either OMVs or OMV-free fraction on human epithelial liver, lung cells and osteoblasts was not detected using both dye-exclusion method and ATP-based viability assay performed after after 24 h.
Table 2.
Effect of PYKK081 on eukaryotic cells.
| Designation | Description | Toxicity, % | ||
|---|---|---|---|---|
| Whole cell | OMV-free | OMVs | ||
| HL-60 | Human promyeloblast | +1 | + | −3 |
| K562 | Human lymphoblast | ND4 | + | − |
| CMK | Human megakaryoblast | ND | + | − |
| MOLT-4 | Human T lymphoblast | ND | ++2 | + |
| THP-1 | Human monocyte | ++ | ++ | + |
| Erythrocyte | Human erythrocyte | ++ | − | ++ |
| hFOB 1.19 | Human osteoblast | + | − | − |
| A549 | Human lung epithelial cell | ND | − | − |
| SW 982 | Human synovial fibroblast | + | − | − |
| Hep G2 | Human liver epithelial cell | ND | − | − |
| TIB-192 | Mouse myeloblast | ND | + | − |
| A20 | Mouse B lymphocyte | ND | ++ | + |
-20–30% cells were lysed
30–85% cells were lysed
% of lysed cells ≤4
−Not determined
For toxicity test, 1 × 107 PYKK081 cells, 10 μl PYKK081 OMV-free fraction or OMVs (8 μg of protein) sample were added to 1 × 106 cells and incubated with for 3 h. The cell viability was determined with Trypan blue assay. Hemolytic activity was identified by hemoglobin release after 16 h of incubation. The average result is calculated from three independent measurements, SE ≤ 1.5%.
2. 4. Distribution of RtxA in K. kingae cellular compartments
We detected RtxA as one of the predominant proteins in PYKK081 OMVs. To determine the location of RtxA in the secreted and membrane fractions, we performed Western blot analysis with 9D4 antibody which recognizes the distal portion of the RTX-catalytic domain and that is conserved in many toxins of this group [21]. The immunoreactive bands were detected in equal proportions in both vesicle and OMV-free secreted fraction but not in OM and IM samples (Fig. 2B). Depending on preparation immunoblot resulted in the detection of 100 kDa band (predicted size of RtxA) only or two bands 100 kDa and 60 kDa (Fig. 2B). The 100 kDa protein band was identified as K. kingae RtxA toxin by MALDI-TOF MS. We have failed to identify the smaller band; however we hypothesize it is likely to be a product of RtxA degradation. In support of this claim, we did not find any band detected by 9D4 in cell lysate preparation of K. oralis UB-38, a close relative of K. kingae, that does not have the rtx-operon [20] (Fig. 2B). Using Western blot analysis we investigated OMVs and OMVs-free fractions samples prepared from other K. kingae isolates in the study and found RtxA in both OMVs-associated and as a free form (data not shown). The presence of RtxA in the OMVs samples from different isolates suggests that toxin association with OMVs is highly prevalent phenomenon in K. kingae.
2. 5. OMVs are internalized by osteoblasts and synovial cells
In the human body K. kingae primarily infects joint and bone tissues [5]. Therefore we wished to investigate if OMVs could interact with and cause cytotoxic effects on synovial cells (SW 982) and osteoblasts (hFOB 1.19). Polyclonal anti-OMV antibody was used as an immunological probe to detect OMVs association with host cells. To identify the location of the OMVs, SW 982 and hFOB 1.19 cells were exposed to the purified PYKK081 OMVs overnight. The cells were subsequently reacted with polyclonal anti-OMV antibody and Alexa 488 labeled anti-rabbit IgG and then examined by confocal scanning microscopy. A large number of stainable interacting OMVs was detected in cells exposed to OMVs and permeabilized before the addition of antibodies (Fig. 3A, B). The requirement for permeabilization of the cell membrane in order to stain the OMVs efficiently allowed us to hypothesize their intracellular localization.
Fig. 3. Confocal images of PYKK081 OMVs interaction with human cells.
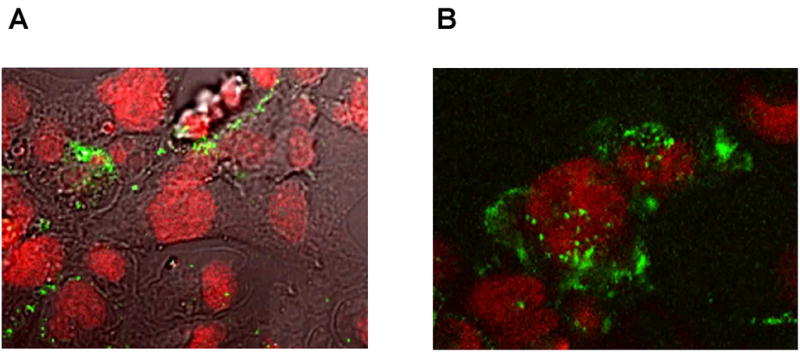
hFOB 1.19 (A) and SW 982 (B) cells were incubated with OMVs overnight followed by treatment with anti-OMVs antibody and were visualized upon interaction with Alexa 488 conjugated anti-rabbit IgG. Nonspecific fluorescence was not observed in the absence of primary antibody. Cell nuclei were labeled with 7-AAD.
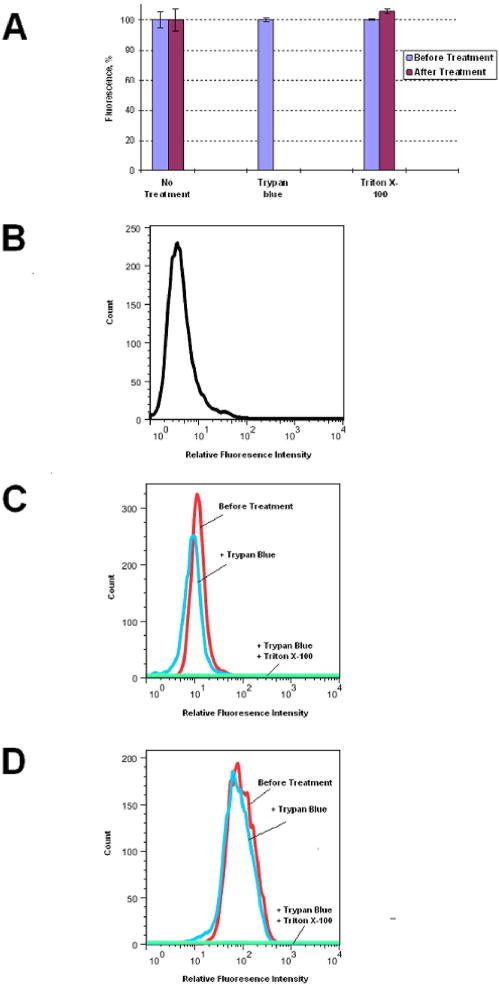
To further study internalization, PYKK081 OMVs were labeled with FITC and assessed by flow cytometry using the impermeable cell dye Trypan blue, which was able to quench FITC-fluorescence (Fig. 4, A). OMV-FITC samples were added to SW 982 or hFOB 1.19 cells and the fluorescence was measured at different time points. The data are shown for SW 982 (Fig. 4B–D, supplementary data Fig. 2S). To quantitatively estimate the amount of internalized fluorescence, Trypan blue was added to OMV-FITC treated cells that led to the quenching of extracellular fluorescence while intracellular fluorescence in the intact cells was not affected. When cells were permeabilized with 0.1% Triton X-100 and Trypan blue was added, no florescence was detected suggesting that internal fluorescence was quenched with Trypan blue, which penetrated into cells. The flow cytometry results based on mean fluorescence intensity (mfi) values demonstrated that some of OMV-FITC fluorescence was associated with cells as early as 15 min and most of it was internalized with the cells (Fig. 4B, C). The maximum increase in total associated fluorescence was observed by two hours of incubation with OMV-FITC and the majority of fluorescence at this time had intracellular location (Fig. 4B, D). Our results indicate that OMVs are rapidly taken up by target cells.
Fig. 4. Flow cytometry analysis of PYKK081 OMVs association with SW 982 cells.
A. Effect of 0.025% Trypan blue and 0.1% Triton X-100 on OMV-FITC fluorescence was estimated after 20 min treatment. The data shown are means and standard deviations of two independent experiments.
B–D. The proportion of OMV-FITC internalized with SW 982 was monitored over time. Controls cells were treated with OMV-buffer (B). In experimental samples the cells were incubated with OMV-FITC for 15 min (C) and 2 h (D). Fluorescence was measured before (total associated, red line), after (intracellular, blue line) the treatment with Trypan blue to quench extracellular fluorescence, and after (light green line) cell permeabilization with 0.1% Triton X-100. Result shown is a representative of two independent experiments.
2. 6. OMVs induce cytokine secretion in osteoblats and synovial cells
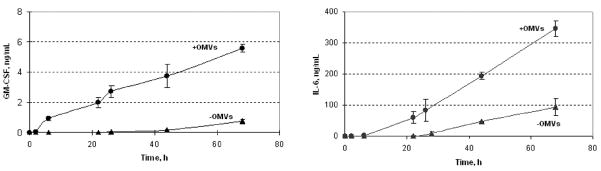
We tested a possible pro-inflammatory effect of OMVs on human synovial cells and osteoblasts through cytokine production. We screened for the production of twenty major inflammatory cytokines by these cells in response to OMVs exposure using an immunoassay. Figure 5 displays results from cytokine production in hFOB 1.19 and SW 982 culture supernatants after 24 h treatment with OMVs. Comparison of control and experimental samples from both cell lines revealed changes in production of human granulocyte-macrophage colony-stimulating factor (GM-CSF) and interleukin-6 (IL-6). In SW 982 cells the production of GM-CSF was induced and IL-6 production was significantly unregulated following PYKK081 OMVs exposure. In hFOB 1.19, expression of these cytokines was constitutive but increased upon the treatment (Fig. 5). To demonstrate the functional response of these cells to OMVs, cytokine secretion was quantified in SW 982 cell culture supernatants collected at different time points using enzyme-linked immunosorbent assays (ELISAs) (Fig. 6). In the experimental cell culture supernatants, GM-CSF concentrations gradually increased over time and reached 5.56 ± 0.26 ng/ml (6.10 fold increase vs. control, P=0.003) while IL-6 concentrations reached 346.82 ± 26.40 ng/ml, (2.67-fold increase vs. control, P=0.002). Thus, our results demonstrated that OMVs could induce a substantial inflammatory response in human joint tissues.
Fig. 5. Qualitative analysis of cytokines in SW 982 and hF0B 1.19 cell culture supernatants.
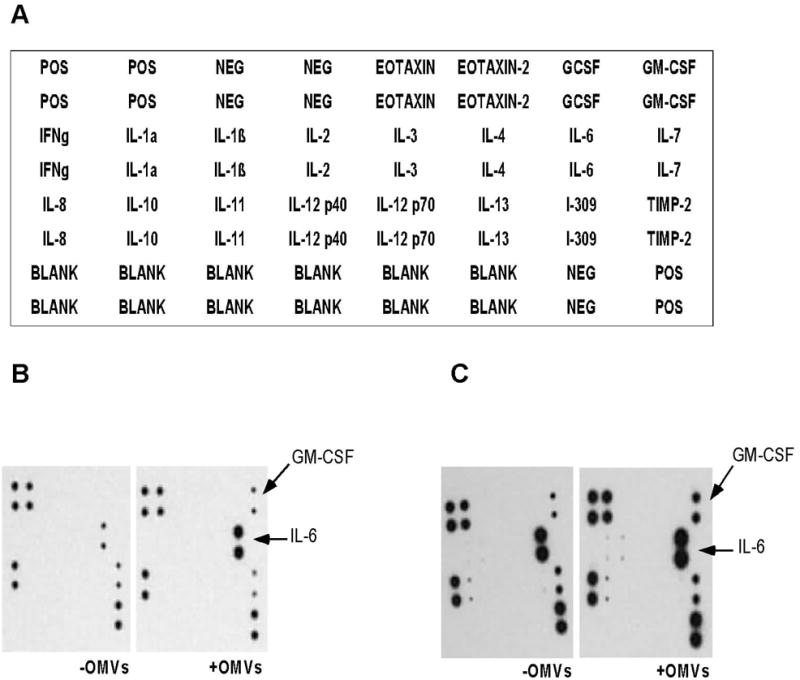
Inflammatory cytokines analysis was performed using RayBio Human Inflammatory Cytokine Antibody Array.
A. The array map represents the location of each antibody on the membrane.
B–C. Increased protein expression levels of GM-CSF and IL-6 (marked by arrows) were detected in the culture supernatants of 24 h PYKK081 OMVs treated SW 982 (B) and hFOB 1.19 (C) in comparison with OMV-buffer (control)-treated samples. The variations in intensity of signals between SW 982 and hFOB 1.19 do not reflect the actual concentration of cytokines in the supernatants. The result is representative of two independent experiments.
Fig. 6. Quantitative analysis of cytokine protein levels in SW 982 cell culture supernatants.
The SW 982 cell culture supernatants of PYKK081 OMVs treated cells were collected at different time-points and used to detect GM-CSF and IL-6 production. Cells treated with OMV-buffer served as a control. Cytokine concentrations in culture supernatants were determined by extrapolation from standard curves. The data shown are means and standard deviations of two independent experiments.
3. Discussion
In this study we demonstrated that K. kingae clinical isolates produce abundant OMVs pinching off the OM during growth. The shape of the heterogeneously sized OMVs resembled those released by other members of Neisseriaceae family [35, 36]. We purified OMVs from eight K. kingae isolates of different origin. The amount of OMVs produced in strains varied and they could be divided into two groups: high- and low- OMVs producing. All joint isolates tested were found to be high- OMV producing strains suggesting that this property might be an advantage in causing septic arthritis. In this study we characterized OMVs from a septic arthritic isolate PYKK081 and investigated their effects on human cells.
It has been shown that native OMVs contain OM and periplasmic proteins [31]. The general protein profiles of PYKK081 OMVs and OM were similar however they had some variations. Although the lack of whole-genome annotation hampered determination of some proteins, we have identified four of the most abundant polypeptides of OMVs. Hypothetical protein Pden_3310, PilC2, and putative porin were shown to also be present in OM of the organism. OM preparations contained some polypeptides that were not detected in OMVs. Thus, the inclusion of proteins into OMVs is selective and may indicate that production of toxic OMVs is intentional and represents a distinct mechanism of K. kingae pathogenesis.
OMVs were found to contain potential protein virulence factors, high endotoxin and DNA concentrations and may contribute to K. kingae pathogenesis by several mechanisms. First, toxic activities of OMVs could be a function of RtxA toxin. Vesicle-mediated toxin delivery is an important virulence mechanism described for several gram negative pathogens [37–43]. Many toxins have been found in both the free soluble form and associated with OMVs [40, 44]. In a previous study, the K. kingae toxic effect on eukaryotic cells was attributed to the production of RtxA [20]. In our experimental conditions we detected leukotoxic and hemolytic activities in the PYKK801 secreted fraction. The toxicity of OMVs on white blood cells was less pronounced than that from the OMV-free fraction, but hemolysis was predominantly associated with the vesicles. Some toxins of the RTX-group have been shown to possess both leukotoxic and hemolytic activities [45–47]. Thus, although we do not exclude the existence of a different hemolysin, it is possible that RtxA possess leukotoxic and hemolytic activities and its cell specificity may be dependent on its location as either a free protein or associated with OMVs. We could not detect lysis of human epithelial liver cells, lung epithelial cells or osteoblasts using OMVs or OMV-free fractions.
Some RTX-toxins have been shown to be both secreted and associated with the bacterial OM [48, 49]. It has been recently demonstrated that another RTX-toxin, EHEC-Hly, secreted extracellularly, has a strong tendency to associate with OMVs after the toxin is secreted [44]. Interestingly, RtxA was not present in OM preparations however it was found to be one of the predominant proteins of OMVs. We presume that RtxA could be also associated with the bacterial OM but might dissociate from it during the OM fraction preparation procedure that includes treatment with detergent (1% Sarkosyl). Indeed, in some our experiments, the treatment of OMVs with denaturing agents, such as 0.8 M Urea, caused toxin degradation and partial dissociation from OMVs (unpublished data).
For pathogenic bacteria, adhesins are particularly important in colonization of host tissues. PYKK081 OMVs were shown to contain PilC2, an adhesin with similarity to PilC2 found in the pathogenic Neisseria species [50, 51]. Previous work demonstrated that type IV pili and the two PilC2 homologs play important roles in mediating K. kingae adherence [17]. Therefore, we hypothesize that K. kingae OMVs may also be involved in biofilm development. Interestingly, the role of OMVs in causing cellular aggregation and the development of dental plaque biofilms was shown for Porphyromonas gingivalis [52, 53].
In the human body, K. kingae causes predominantly osteoarticular infections, such as septic arthritis and osteomyelitis [5], which primarily affect bone and joint tissues. Therefore we focused on investigating the possible toxic effect of OMVs on human osteoblasts and synovial cells. To study the ability of PYKK081 OMVs to internalize within host cells, we tested the interaction between OMVs and human cells using confocal microscopy and flow cytometry. Although the molecular mechanism of K. kingae OMVs uptake by human cells remains to be identified, our results indicate that OMVs are able to be rapidly internalized with human target cells. This is consistent with the reported properties of OMVs from other bacteria [54].
We did not show a direct cytolytic effect of K. kingae OMVs on human osteoblasts and synovial cells. Therefore, we questioned whether interaction of OMVs could induce an inflammatory response in these tissues. Cultured human osteoblasts and synovial cells were exposed to OMVs and subjected to an immunoassay to detect secreted cytokines. We observed changes in GM-CSF and IL-6 production by these cells following interaction with K. kingae OMVs. IL-6, a cytokine with pro-inflammatory characteristics, is known to be an early marker of bacterial infection [55–57]. In some tissues the cytokine secreted in response to lipopolysaccharide produced by gram-negative bacteria [58, 59].
To our knowledge this is the first study to identify GM-CSF expression in synoval cells in response to bacterial cell components. Interestingly, synovial fluids from septic, rheumatoid and osteoarthritis patients contained significant colony-stimulating factors (CSF) activities [60]. GM-CSF was also shown to be elaborated by synovial fibroblasts in rheumatoid arthritis [61]. GM-CSF found in rheumatoid arthritis synovial fluid was shown to sustain viability of neutrophils and activates their functions such as release of reactive oxygen species [62, 63], cytokines [64] and proteases[65] that lead to the inflammation and destruction of the joints. Together, these studies may suggest that similar mechanisms of joint destruction occur in rheumatoid and septic arthritis via the production of GM-CSF.
In addition to functioning as a white blood cell growth factor, GM-CSF was shown to facilitate the early differentiation of myeloid precursors into osteoclast precursors and contribute to the regulation of osteoclastogenesis [66–68]. Thus, K. kingae OMVs can be involved in stimulation of osteoclastogenesis and bone resorption. Significantly, it has been previously reported that Staphylococcus aureus and Salmonella spp. are common causes of bone disease and induced CSF in human osteoblasts [69].
This study is the first report of OMVs production by K. kingae. Having demonstrated the toxic and pro-inflammatory activity of K. kingae OMVs, we hypothesize that they are an important bacterium virulence factor to overcome the barrier of the host immune system defense, can be involved in iron uptake, biofilm formation, horizontal DNA transfer and the development of deep joint and bone infection that are not easily accessible by the bacterium itself.
4. Materials and Methods
4. 1. Bacterial strains and growth conditions
Kingella strains used in the study are listed in Table 1. The bacteria were grown on Colombia agar (CA) (Acumedia Manufactures, Inc., Lansing, MI) with 5% sheep blood (Hemostat laboratories, Dixon, CA) or AAGM agar [70] at 37°C with 10% CO2. The isolates was stored at −80°C in AAGM containing 10% DMSO or lyophilized.
4. 2. Purification of K. kingae secreted fraction and OMVs
K. kingae was grown on AAGM plates for 40 hours, the biomass was scraped from plates, weighed and centrifuged at 150,000 × g for 15 minutes at 4°C. The supernatant was filtered through 0.45 μm and 20% glycerol was added to stabilize the toxic activity in the secreted fraction. The OMVs were further purified by centrifugation of the secreted fraction at 150,000 × g for 120 min at 4°C. The resulted supernatant was carefully removed and the vesicle pellet was weighed, washed and resuspended in 50 mM HEPES pH 7.4 with 20% glycerol and 2 mM CaCl2 (OMV-buffer) to final protein concentration 8 mg/ml. OMVs preparations were stored at −80°C. The sterility of the vesicle preparations was verified by streaking on CA with 5% sheep blood.
4. 3. Negative staining electron microscopy
Transmission electron microscopy (TEM) was performed in UMDNJ Core Imaging lab (Piscataway, NJ). Single K. kingae colonies grown on blood plates were resuspended in 50 mM phosphate buffer saline (PBS) pH 7.4. Purified OMVs samples were diluted twice in 50 mM HEPES pH 7.4. Twenty μl drop of sample was placed on fomavar/carbon coated copper grid and wicked off by a filter paper. The grid was stained with 1% uranyl acetate and viewed with a Philips CM 12 at 80 kV. The images were collected with an AMT XR611 digital camera and analyzed using Engine V600 software.
4. 4. Generation of anti-K. kingae OMV antibody
The OMV material containing 3 mg of protein has been used for stimulation of an antibody response in New Zealand white rabbits (Pacific Immunology Inc., Ramona, CA) The total procedure lasted 13 weeks and included four rabbit injections. The immunoreactivity of antiserum was detected by enzyme-linked immunosorbent assay (ELISA), the antibody titer was estimated as 1:500,000. The antibody were purified from the final rabbit bleed using AminoLink plus kit (Pierce, Rockford, IL) as described previously [71].
4. 5. Toxicity assay
Human and mouse cell lines (Table 2) were obtained from ATCC. A20 mouse lymphoma cell line [72] was kindly provided by Dr. Tsiagbe. The cells were grown according to the supplier’s recommendations in a culture incubator at 37°C with 5% CO2, with the exception of SW 982 that was grown at 37°C without extra CO2 supplied. For toxicity test, 1 × 107 bacterial cells, 10 μl (8 μg of protein) PYKK081 OMVs or OMV-free secreted fraction sample was added to 1 × 106 cells and incubated in the serum-free growth medium for 3 or 24 hours. The cell membrane permeability was determined with Trypan blue assay using Vi-cell machine (Beckman Coulter, Hialeah, FL). ATP production was detected using the Cell Titer-Glo luminescent cell viability assay (Promega, Madison, WI) according to the manufacturer’s instructions. Plates were read in a Synergy HT plate reader in the luminescence mode (Bio-Tek, Winooski, VT). To estimate hemolysis, human blood from a healthy volunteer was collected in a tube containing heparin sulfate. Prior to the assay, both sheep blood and human blood were centrifuged to collect erythrocytes. The erythrocytes were centrifuged at 1000 × g for 5 min and washed in PBS (pH 7.4) until the supernatant was clear. Reactions were performed in 0.5 ml reaction mixtures containing 2% erythrocytes and 5 × 107 of bacterial cells, 50 μl (40 μg of protein) PYKK081 OMVs or OMV-free fractions for 16 h at 37°C. To evaluate hemoglobin release, erythrocytes were removed by centrifugation and the absorbance of the supernatant at 450 nm was measured with a Synergy HT plate reader (BioTek Instruments, Winooski, VT). One hundred percent cell lysis was determined by resuspending erythrocytes in distilled water. Cytotoxicity assays were performed at least three different times.
4. 6. Western blot analysis
Purified proteins (0.5 μg) were resolved by SDS-PAGE and transferred to a nitrocellulose membrane. The membranes were subjected to Western blot analysis with 9D4 antibody (1:1000 dilution) (Santa Cruse, Campbell, CA) or anti-OMV antibody as previously described [71].
4. 7. MALDI-TOF MS
Individual protein bands were excised, digested with trypsin, and peptides were extracted for MALDI-TOF MS analysis using ABI 4700 Proteomics Analyzer (Applied Biosystems, Foster City, CA). A search for the peptide mass fingerprint was carried out with the nrNCBI non-redundant database. All procedures were carried out by Applied Biomics (Hayward, CA).
4. 8. Gel-filtration
The OMV sample was loaded onto a column packed with 40 ml of Sephacryl 300 HR (Sigma, St. Louis, MO). The proteins were eluted in 1 ml fractions with 20 mM Tris pH 7.4, 150 mM NaCl and 2 mM CaCl2 buffer. Sweet potato β-amylase (206 kDa) (Sigma, St. Louis, MO), Aggregatibacter actinomycetemcomitans LtxA (112 kDa) [48], and bovine serum albumin (66 kDa) (Sigma, St. Louis, MO) were used for the column calibration. The protein in the fractions was detected by Bradford reagent (Sigma, St. Louis, MO) or by Western blot analysis using anti-OMV antibody.
4. 9. Fractionation of bacteria
The IM and OM fractions were isolated using Sarkosyl extraction method as previously described [71, 73].
4. 10. Protein assay
Proteins were resolved on a 4–20% SDS-PAGE and visualized by staining with GelCode Blue Stain Reagent (Pierce, Rockford, IL). The protein concentration was measured with a Bicinchoninic Acid Assay Kit (Pierce, Rockford, IL).
4. 11. DNA analysis
DNA associated with OMVs was quantified spectrophotometrically using NanoDrop. Prior the analysis OMVs were lysed by 0.5% (v/v) Sarkosyl and 100 mM EDTA. DNA in OMV samples was also visualized by agarose gel electrophoresis.
4. 12. Endotoxin assay
Endotoxin concentration was measured using E-toxite kit (Sigma, St. Louis, MO). The concentration was determined by extrapolation from standard curve prepared with E.coli 0.55:B5 lipopolysaccharade (LPS) (Sigma, St. Louis, MO).
4.13. Confocal microscopy
Human cells were grown in 60 μ-Dishes (Ibidi, Munich, Germany) and were exposed to 8 μl (6 μg of protein) PYKK081 OMVs for 18 h. All subsequent cell treatment procedures were performed at room temperature, each treatment step followed by wash with PBS pH 7.4. Cells were fixed with freshly prepared 4% paraformaldehyde in PBS for 20 min, permeabilized using 0.1% Triton X-100 for 20 minutes (if required), and blocked for 30 min in PBS pH 7.4 containing 2% bovine serum albumin. OMVs were identified using rabbit polyclonal anti-OMV antibodies for 1 h (1:100 dilution). Primary antibody binding was detected with Alexa 488 goat anti-rabbit IgG (Invitrogen, Carlsbad, CA) (1:200 dilution). Nuclei were stained with 7-aminoactinomycin D (7-AAD) (Invitrogen, Carlsbad, CA) for 30 min. Cells imaging was performed using a scanning confocal microscope (Zeiss LSM 510, Carl Zeiss Inc., Thornwood, NY).
4. 14. Flow cytometry analysis
OMVs were stained with fluorescein isothiocyanate (FITC) using FITC Antibody labeling kit (Pierce, Rockford, IL) as recommended by the manufacturer. Human cells (0.2 × 106 cells/run) were incubated with 10 μl (8 μg of protein) PYKK081 OMV-FITC for 15 min, 2 h and 24 h. The fluorescence measurements were made with a FACS Calibur instrument (BD Biosciences, Franklin Lakes, NJ) and the data were analyzed using Flow Jo software (Ashland, OR). To perform this experiment we adopted the method described previously [54]. 0.025% Trypan blue (Sigma, St. Louis, MO) was used where was used for quenching OMV-FITC fluorescence. Mean fluorescence intensity values (mfi) of cells incubated in the absence of OMV-FITC were subtracted from the values of OMV-FITC pretreated with Trypan blue for 20 min. To quench the intracellular fluorescence cells were permeabilized using 0.1% Triton X-100 (Sigma, St. Louis, MO). To confirm Trypan blue quenched FITC fluorescence, the fluorescence was measured before and after the addition of Trypan blue to OMV-FITC using Synergy HT plate reader in the fluorescence mode (Bio-Tek, Winooski, VT).
4. 15. Cytokine production assay
Human osteoblasts and synovial cells were grown in 6-well plate until confluency, placed in 1 ml of serum-free growth medium and exposed to 10 μl (8 μg of protein) PYKK081 OMVs. The media samples were collected at different time-points (0–68 h) and used to detect inflammatory cytokine production. Cells treated with OMV-buffer served as a control, each condition was repeated twice. Cytokine production analysis and quantitative measurements were performed using a RayBio Human Cytokine Antibody Array kit and RayBio Human GM-CSF and IL-6 ELISA Kits (RayBiotech, Inc., Norcross, GA) according to the manufacturer’s instructions. Cytokine concentrations in culture supernatants were determined by extrapolation from standard curves.
4. 16. Statistical analysis
For statistical analysis, the data were subjected to Student’s t-test, P value of ≤0.05 was considered to be significant.
Supplementary Material
Acknowledgments
We thankful to Dr. Pablo Yagupsky and Dr. Ruth Lynfield for providing K. kingae isolates for this study. We thank Dr. Kabilan Velliyagounder and Raj Patel for the help with TEM, Amy Le and Dr. Erica Salerno for the assistance with flow cytometry experiments, Luke Fritzky and Dr. David Lagunoff for performing confocal microscopy. We are grateful to Dr. Jeffrey Kaplan for the productive suggestions throughout the study. This project was supported by NIH AI080844 and AHA 09SDG2310194 grants to N.V.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yagupsky P, Bar-Ziv Y, Howard CB, Dagan R. Epidemiology, etiology, and clinical features of septic arthritis in children younger than 24 months. Arch Pediatr Adolesc Med. 1995;149:537–40. doi: 10.1001/archpedi.1995.02170180067010. [DOI] [PubMed] [Google Scholar]
- 2.Yagupsky P, Dagan R, Howard CB, Einhorn M, Kassis I, Simu A. Clinical features and epidemiology of invasive Kingella kingae infections in southern Israel. Pediatrics. 1993;92:800–4. [PubMed] [Google Scholar]
- 3.Yagupsky P, Peled N, Katz O. Epidemiological features of invasive Kingella kingae infections and respiratory carriage of the organism. J Clin Microbiol. 2002;40:4180–4. doi: 10.1128/JCM.40.11.4180-4184.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubnov-Raz G, Ephros M, Garty BZ, Schlesinger Y, Maayan-Metzger A, Hasson J, et al. Invasive Pediatric Kingella kingae Infections: A Nationwide Collaborative Study. Pediatr Infect Dis J. doi: 10.1097/INF.0b013e3181d57a6c. [DOI] [PubMed] [Google Scholar]
- 5.Yagupsky P. Kingella kingae: from medical rarity to an emerging paediatric pathogen. Lancet Infect Dis. 2004;4:358–67. doi: 10.1016/S1473-3099(04)01046-1. [DOI] [PubMed] [Google Scholar]
- 6.Ceroni D, Cherkaoui A, Ferey S, Kaelin A, Schrenzel J. Kingella Kingae osteoarticular infections in young children: clinical features and contribution of a new specific real-time PCR assay to the diagnosis. J Pediatr Orthop. 30:301–4. doi: 10.1097/BPO.0b013e3181d4732f. [DOI] [PubMed] [Google Scholar]
- 7.Chometon S, Benito Y, Chaker M, Boisset S, Ploton C, Berard J, et al. Specific real-time polymerase chain reaction places Kingella kingae as the most common cause of osteoarticular infections in young children. Pediatr Infect Dis J. 2007;26:377–81. doi: 10.1097/01.inf.0000259954.88139.f4. [DOI] [PubMed] [Google Scholar]
- 8.Gene A, Garcia-Garcia JJ, Sala P, Sierra M, Huguet R. Enhanced culture detection of Kingella kingae, a pathogen of increasing clinical importance in pediatrics. Pediatr Infect Dis J. 2004;23:886–8. doi: 10.1097/01.inf.0000137591.76624.82. [DOI] [PubMed] [Google Scholar]
- 9.Ilharreborde B, Bidet P, Lorrot M, Even J, Mariani-Kurkdjian P, Liguori S, et al. New real-time PCR-based method for Kingella kingae DNA detection: application to samples collected from 89 children with acute arthritis. J Clin Microbiol. 2009;47:1837–41. doi: 10.1128/JCM.00144-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korach A, Olshtain-Pops K, Schwartz D, Moses A. Kingella kingae prosthetic valve endocarditis complicated by a paravalvular abscess. Isr Med Assoc J. 2009;11:251–3. [PubMed] [Google Scholar]
- 11.Moumile K, Merckx J, Glorion C, Berche P, Ferroni A. Osteoarticular infections caused by Kingella kingae in children: contribution of polymerase chain reaction to the microbiologic diagnosis. Pediatr Infect Dis J. 2003;22:837–9. doi: 10.1097/01.inf.0000083848.93457.e7. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen S, Fayad G, Modine T, Leroy O. Mitral acute bacterial endocarditis caused by HACEK microorganisms. J Heart Valve Dis. 2009;18:353–4. [PubMed] [Google Scholar]
- 13.Verdier I, Gayet-Ageron A, Ploton C, Taylor P, Benito Y, Freydiere AM, et al. Contribution of a broad range polymerase chain reaction to the diagnosis of osteoarticular infections caused by Kingella kingae: description of twenty-four recent pediatric diagnoses. Pediatr Infect Dis J. 2005;24:692–6. doi: 10.1097/01.inf.0000172153.10569.dc. [DOI] [PubMed] [Google Scholar]
- 14.Kraig E, Dailey T, Kolodrubetz D. Nucleotide sequence of the leukotoxin gene from Actinobacillus actinomycetemcomitans: homology to the alpha-hemolysin/leukotoxin gene family. Infect Immun. 1990;58:920–9. doi: 10.1128/iai.58.4.920-929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sena AC, Seed P, Nicholson B, Joyce M, Cunningham CK. Kingella kingae endocarditis and a cluster investigation among daycare attendees. Pediatr Infect Dis J. 29:86–8. doi: 10.1097/INF.0b013e3181b48cc3. [DOI] [PubMed] [Google Scholar]
- 16.Yagupsky P, Erlich Y, Ariela S, Trefler R, Porat N. Outbreak of Kingella kingae skeletal system infections in children in daycare. Pediatr Infect Dis J. 2006;25:526–32. doi: 10.1097/01.inf.0000215243.42501.4f. [DOI] [PubMed] [Google Scholar]
- 17.Kehl-Fie TE, Miller SE, St Geme JW., 3rd Kingella kingae expresses type IV pili that mediate adherence to respiratory epithelial and synovial cells. J Bacteriol. 2008;190:7157–63. doi: 10.1128/JB.00884-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kehl-Fie TE, Porsch EA, Miller SE, St Geme JW., 3rd Expression of Kingella kingae type IV pili is regulated by sigma54, PilS, and PilR. J Bacteriol. 2009;191:4976–86. doi: 10.1128/JB.00123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kehl-Fie TE, Porsch EA, Yagupsky P, Grass EA, Obert C, Benjamin DK, Jr, et al. Examination of type IV pilus expression and pilus-associated phenotypes in Kingella kingae clinical isolates. Infect Immun. 78:1692–9. doi: 10.1128/IAI.00908-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kehl-Fie TE, St Geme JW., 3rd Identification and characterization of an RTX toxin in the emerging pathogen Kingella kingae. J Bacteriol. 2007;189:430–6. doi: 10.1128/JB.01319-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coote JG. Structural and functional relationships among the RTX toxin determinants of gram-negative bacteria. FEMS Microbiol Rev. 1992;8:137–61. doi: 10.1111/j.1574-6968.1992.tb04961.x. [DOI] [PubMed] [Google Scholar]
- 22.Ludwig A, Jarchau T, Benz R, Goebel W. The repeat domain of Escherichia coli haemolysin (HlyA) is responsible for its Ca2+-dependent binding to erythrocytes. Mol Gen Genet. 1988;214:553–61. doi: 10.1007/BF00330494. [DOI] [PubMed] [Google Scholar]
- 23.Ostolaza H, Soloaga A, Goni FM. The binding of divalent cations to Escherichia coli alpha-haemolysin. Eur J Biochem. 1995;228:39–44. [PubMed] [Google Scholar]
- 24.Kachlany SC. Aggregatibacter actinomycetemcomitans Leukotoxin: from Threat to Therapy. J Dent Res. doi: 10.1177/0022034510363682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lally ET, Kieba IR, Sato A, Green CL, Rosenbloom J, Korostoff J, et al. RTX toxins recognize a beta2 integrin on the surface of human target cells. J Biol Chem. 1997;272:30463–9. doi: 10.1074/jbc.272.48.30463. [DOI] [PubMed] [Google Scholar]
- 26.Taichman NS, Dean RT, Sanderson CJ. Biochemical and morphological characterization of the killing of human monocytes by a leukotoxin derived from Actinobacillus actinomycetemcomitans. Infect Immun. 1980;28:258–68. doi: 10.1128/iai.28.1.258-268.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benz R, Maier E, Ladant D, Ullmann A, Sebo P. Adenylate cyclase toxin (CyaA) of Bordetella pertussis. Evidence for the formation of small ion-permeable channels and comparison with HlyA of Escherichia coli. J Biol Chem. 1994;269:27231–9. [PubMed] [Google Scholar]
- 28.Forestier C, Welch RA. Identification of RTX toxin target cell specificity domains by use of hybrid genes. Infect Immun. 1991;59:4212–20. doi: 10.1128/iai.59.11.4212-4220.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hess JF, Angelos JA. The Moraxella bovis RTX toxin locus mbx defines a pathogenicity island. J Med Microbiol. 2006;55:443–9. doi: 10.1099/jmm.0.46366-0. [DOI] [PubMed] [Google Scholar]
- 30.Thompson SA, Wang LL, West A, Sparling PF. Neisseria meningitidis produces iron-regulated proteins related to the RTX family of exoproteins. J Bacteriol. 1993;175:811–8. doi: 10.1128/jb.175.3.811-818.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol. 64:163–84. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Unal CM, Schaar V, Riesbeck K. Bacterial outer membrane vesicles in disease and preventive medicine. Semin Immunopathol. doi: 10.1007/s00281-010-0231-y. [DOI] [PubMed] [Google Scholar]
- 33.Holst J, Martin D, Arnold R, Huergo CC, Oster P, O’Hallahan J, et al. Properties and clinical performance of vaccines containing outer membrane vesicles from Neisseria meningitidis. Vaccine. 2009;27 (Suppl 2):B3–12. doi: 10.1016/j.vaccine.2009.04.071. [DOI] [PubMed] [Google Scholar]
- 34.Jonsson AB, Rahman M, Normark S. Pilus biogenesis gene, pilC, of Neisseria gonorrhoeae: pilC1 and pilC2 are each part of a larger duplication of the gonococcal genome and share upstream and downstream homologous sequences with opa and pil loci. Microbiology. 1995;141 ( Pt 10):2367–77. doi: 10.1099/13500872-141-10-2367. [DOI] [PubMed] [Google Scholar]
- 35.Dewhirst FE, Chen CK, Paster BJ, Zambon JJ. Phylogeny of species in the family Neisseriaceae isolated from human dental plaque and description of Kingella oralis sp. nov [corrected] Int J Syst Bacteriol. 1993;43:490–9. doi: 10.1099/00207713-43-3-490. [DOI] [PubMed] [Google Scholar]
- 36.Perera VY, Penn CW, Smith H. Purification of pili and outer membrane vesicles of Neisseria gonorrhoeae by wheat germ agglutinin affinity chromatography. J Gen Microbiol. 1982;128:1613–22. doi: 10.1099/00221287-128-7-1613. [DOI] [PubMed] [Google Scholar]
- 37.Balsalobre C, Silvan JM, Berglund S, Mizunoe Y, Uhlin BE, Wai SN. Release of the type I secreted alpha-haemolysin via outer membrane vesicles from Escherichia coli. Mol Microbiol. 2006;59:99–112. doi: 10.1111/j.1365-2958.2005.04938.x. [DOI] [PubMed] [Google Scholar]
- 38.Dutta S, Iida K, Takade A, Meno Y, Nair GB, Yoshida S. Release of Shiga toxin by membrane vesicles in Shigella dysenteriae serotype 1 strains and in vitro effects of antimicrobials on toxin production and release. Microbiol Immunol. 2004;48:965–9. doi: 10.1111/j.1348-0421.2004.tb03626.x. [DOI] [PubMed] [Google Scholar]
- 39.Fiocca R, Necchi V, Sommi P, Ricci V, Telford J, Cover TL, et al. Release of Helicobacter pylori vacuolating cytotoxin by both a specific secretion pathway and budding of outer membrane vesicles. Uptake of released toxin and vesicles by gastric epithelium. J Pathol. 1999;188:220–6. doi: 10.1002/(SICI)1096-9896(199906)188:2<220::AID-PATH307>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 40.Kato S, Kowashi Y, Demuth DR. Outer membrane-like vesicles secreted by Actinobacillus actinomycetemcomitans are enriched in leukotoxin. Microb Pathog. 2002;32:1–13. doi: 10.1006/mpat.2001.0474. [DOI] [PubMed] [Google Scholar]
- 41.Lindmark B, Rompikuntal PK, Vaitkevicius K, Song T, Mizunoe Y, Uhlin BE, et al. Outer membrane vesicle-mediated release of cytolethal distending toxin (CDT) from Campylobacter jejuni. BMC Microbiol. 2009;9:220. doi: 10.1186/1471-2180-9-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Negrete-Abascal E, Garcia RM, Reyes ME, Godinez D, de la Garza M. Membrane vesicles released by Actinobacillus pleuropneumoniae contain proteases and Apx toxins. FEMS Microbiol Lett. 2000;191:109–13. doi: 10.1111/j.1574-6968.2000.tb09326.x. [DOI] [PubMed] [Google Scholar]
- 43.Wensink J, Gankema H, Jansen WH, Guinee PA, Witholt B. Isolation of the membranes of an enterotoxigenic strain of Escherichia coli and distribution of enterotoxin activity in different subcellular fractions. Biochim Biophys Acta. 1978;514:128–36. doi: 10.1016/0005-2736(78)90082-2. [DOI] [PubMed] [Google Scholar]
- 44.Aldick T, Bielaszewska M, Uhlin BE, Humpf HU, Wai SN, Karch H. Vesicular stabilization and activity augmentation of enterohaemorrhagic Escherichia coli haemolysin. Mol Microbiol. 2009;71:1496–508. doi: 10.1111/j.1365-2958.2009.06618.x. [DOI] [PubMed] [Google Scholar]
- 45.Balashova NV, Crosby JA, Al Ghofaily L, Kachlany SC. Leukotoxin confers beta-hemolytic activity to Actinobacillus actinomycetemcomitans. Infect Immun. 2006;74:201–21. doi: 10.1128/IAI.74.4.2015-2021.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murphy GL, Whitworth LC, Clinkenbeard KD, Clinkenbeard PA. Hemolytic activity of the Pasteurella haemolytica leukotoxin. Infect Immun. 1995;63:3209–12. doi: 10.1128/iai.63.8.3209-3212.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vanden Bergh PG, Zecchinon LL, Fett T, Desmecht D. Probing of Actinobacillus pleuropneumoniae ApxIIIA toxin-dependent cytotoxicity towards mammalian peripheral blood mononucleated cells. BMC Res Notes. 2008;1:121. doi: 10.1186/1756-0500-1-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diaz R, Ghofaily LA, Patel J, Balashova NV, Freitas AC, Labib I, et al. Characterization of leukotoxin from a clinical strain of Actinobacillus actinomycetemcomitans. Microb Pathog. 2006;40:48–55. doi: 10.1016/j.micpath.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 49.Thompson SA, Sparling PF. The RTX cytotoxin-related FrpA protein of Neisseria meningitidis is secreted extracellularly by meningococci and by HlyBD+ Escherichia coli. Infect Immun. 1993;61:2906–11. doi: 10.1128/iai.61.7.2906-2911.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Backman M, Kallstrom H, Jonsson AB. The phase-variable pilus-associated protein PilC is commonly expressed in clinical isolates of Neisseria gonorrhoeae, and shows sequence variability among strains. Microbiology. 1998;144 ( Pt 1):149–56. doi: 10.1099/00221287-144-1-149. [DOI] [PubMed] [Google Scholar]
- 51.Rahman M, Kallstrom H, Normark S, Jonsson AB. PilC of pathogenic Neisseria is associated with the bacterial cell surface. Mol Microbiol. 1997;25:11–25. doi: 10.1046/j.1365-2958.1997.4601823.x. [DOI] [PubMed] [Google Scholar]
- 52.Grenier D, Mayrand D. Functional characterization of extracellular vesicles produced by Bacteroides gingivalis. Infect Immun. 1987;55:111–7. doi: 10.1128/iai.55.1.111-117.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Inagaki S, Onishi S, Kuramitsu HK, Sharma A. Porphyromonas gingivalis vesicles enhance attachment, and the leucine-rich repeat BspA protein is required for invasion of epithelial cells by “Tannerella forsythia”. Infect Immun. 2006;74:5023–8. doi: 10.1128/IAI.00062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parker H, Chitcholtan K, Hampton MB, Keenan JI. Uptake of Helicobacter pylori outer membrane vesicles by gastric epithelial cells. Infect Immun. 78:5054–61. doi: 10.1128/IAI.00299-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buck C, Bundschu J, Gallati H, Bartmann P, Pohlandt F. Interleukin-6: a sensitive parameter for the early diagnosis of neonatal bacterial infection. Pediatrics. 1994;93:54–8. [PubMed] [Google Scholar]
- 56.Le Moine O, Deviere J, Devaster JM, Crusiaux A, Durand F, Bernuau J, et al. Interleukin-6: an early marker of bacterial infection in decompensated cirrhosis. J Hepatol. 1994;20:819–24. doi: 10.1016/s0168-8278(05)80155-2. [DOI] [PubMed] [Google Scholar]
- 57.Schrum LW, Marriott I, Butler BR, Thomas EK, Hudson MC, Bost KL. Functional CD40 expression induced following bacterial infection of mouse and human osteoblasts. Infect Immun. 2003;71:1209–16. doi: 10.1128/IAI.71.3.1209-1216.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Man P, van Kooten C, Aarden L, Engberg I, Linder H, Svanborg Eden C. Interleukin-6 induced at mucosal surfaces by gram-negative bacterial infection. Infect Immun. 1989;57:3383–8. doi: 10.1128/iai.57.11.3383-3388.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tichomirowa M, Theodoropoulou M, Lohrer P, Schaaf L, Losa M, Uhl E, et al. Bacterial endotoxin (lipopolysaccharide) stimulates interleukin-6 production and inhibits growth of pituitary tumour cells expressing the toll-like receptor 4. J Neuroendocrinol. 2005;17:152–60. doi: 10.1111/j.1365-2826.2005.01286.x. [DOI] [PubMed] [Google Scholar]
- 60.Smith JB, Bocchieri MH, Smith JB, Jr, Sherbin-Allen L, Abruzzo JL. Colony stimulating factor occurs in both inflammatory and noninflammatory synovial fluids. Rheumatol Int. 1990;10:131–4. doi: 10.1007/BF02274828. [DOI] [PubMed] [Google Scholar]
- 61.Katano M, Okamoto K, Arito M, Kawakami Y, Kurokawa MS, Suematsu N, et al. Implication of granulocyte-macrophage colony-stimulating factor induced neutrophil gelatinase-associated lipocalin in pathogenesis of rheumatoid arthritis revealed by proteome analysis. Arthritis Res Ther. 2009;11:R3. doi: 10.1186/ar2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Babior BM. Oxidants from phagocytes: agents of defense and destruction. Blood. 1984;64:959–66. [PubMed] [Google Scholar]
- 63.Smith JA. Neutrophils, host defense, and inflammation: a double-edged sword. J Leukoc Biol. 1994;56:672–86. doi: 10.1002/jlb.56.6.672. [DOI] [PubMed] [Google Scholar]
- 64.Beaulieu AD, McColl SR. Differential expression of two major cytokines produced by neutrophils, interleukin-8 and the interleukin-1 receptor antagonist, in neutrophils isolated from the synovial fluid and peripheral blood of patients with rheumatoid arthritis. Arthritis Rheum. 1994;37:855–9. doi: 10.1002/art.1780370613. [DOI] [PubMed] [Google Scholar]
- 65.Konttinen YT, Lindy O, Kemppinen P, Saari H, Suomalainen K, Vauhkonen M, et al. Collagenase reserves in polymorphonuclear neutrophil leukocytes from synovial fluid and peripheral blood of patients with rheumatoid arthritis. Matrix. 1991;11:296–301. doi: 10.1016/s0934-8832(11)80238-6. [DOI] [PubMed] [Google Scholar]
- 66.Hattersley G, Chambers TJ. Effects of interleukin 3 and of granulocyte-macrophage and macrophage colony stimulating factors on osteoclast differentiation from mouse hemopoietic tissue. J Cell Physiol. 1990;142:201–9. doi: 10.1002/jcp.1041420125. [DOI] [PubMed] [Google Scholar]
- 67.Liggett W, Jr, Shevde N, Anklesaria P, Sohoni S, Greenberger J, Glowacki J. Effects of macrophage colony stimulating factor and granulocyte-macrophage colony stimulating factor on osteoclastic differentiatio of hematopoietic progenitor cells. Stem Cells. 1993;11:398–411. doi: 10.1002/stem.5530110507. [DOI] [PubMed] [Google Scholar]
- 68.Takahashi N, Udagawa N, Akatsu T, Tanaka H, Shionome M, Suda T. Role of colony-stimulating factors in osteoclast development. J Bone Miner Res. 1991;6:977–85. doi: 10.1002/jbmr.5650060912. [DOI] [PubMed] [Google Scholar]
- 69.Bost KL, Bento JL, Ellington JK, Marriott I, Hudson MC. Induction of colony-stimulating factor expression following Staphylococcus or Salmonella interaction with mouse or human osteoblasts. Infect Immun. 2000;68:5075–83. doi: 10.1128/iai.68.9.5075-5083.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fine DH, Furgang D, Kaplan J, Charlesworth J, Figurski DH. Tenacious adhesion of Actinobacillus actinomycetemcomitans strain CU1000 to salivary-coated hydroxyapatite. Arch Oral Biol. 1999;44:1063–76. doi: 10.1016/s0003-9969(99)00089-8. [DOI] [PubMed] [Google Scholar]
- 71.Balashova NV, Park DH, Patel JK, Figurski DH, Kachlany SC. Interaction between leukotoxin and Cu, Zn superoxide dismutase in Aggregatibacter actinomycetemcomitans. Infect Immun. 2007;75:4490–7. doi: 10.1128/IAI.00288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim KJ, Kanellopoulos-Langevin C, Merwin RM, Sachs DH, Asofsky R. Establishment and characterization of BALB/c lymphoma lines with B cell properties. J Immunol. 1979;122:549–54. [PubMed] [Google Scholar]
- 73.Barenkamp SJ, Munson RS, Jr, Granoff DM. Subtyping isolates of Haemophilus influenzae type b by outer-membrane protein profiles. J Infect Dis. 1981;143:668–76. doi: 10.1093/infdis/143.5.668. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.