Abstract
The liver plays a central role in whole-body lipid metabolism by governing the synthesis, oxidization, transport and excretion of lipids. The unfolded protein response (UPR) was identified as a signal transduction system that is activated by ER stress. Recent studies revealed a critical role of the UPR in hepatic lipid metabolism. The IRE1/XBP1 branch of the UPR is activated by high dietary carbohydrates and controls the expression of genes involved in fatty acid and cholesterol biosynthesis. PERK mediated eIF2α phosphorylation is also required for the expression of lipogenic genes and the development of hepatic steatosis, likely by activating C/EBP and PPARγ transcription factors. Further studies to define the molecular pathways that lead to the activation of the UPR by nutritional cues in the liver, and their contribution to human metabolic disorders such as hepatic steatosis, atherosclerosis and type 2 diabetes that are associated with dysregulation of lipid homeostasis, are warranted.
Keywords: Unfolded protein response, Lipogenesis, Gene expression, XBP1, PERK
Introduction
In mammals, the liver plays a crucial role in the control of whole body energy homeostasis. Carbohydrates in the diet are partly consumed for normal bodily functions and metabolized to glycogen or fatty acids in the liver, if in excess [1]. Fatty acids are esterified to triacylglyceride (TG) and then transported to adipose tissue for long-term energy storage. Conversion of carbohydrates to TG involves two steps; glycolysis which generates acetyl CoA from glucose, and lipogenesis which converts acetyl CoA to fatty acids (Fig. 1). Enzymes involved in glycolytic and lipogenic pathways are dynamically regulated at both transcriptional and post-translational levels by various factors such as substrate concentrations and hormones. Pancreatic hormones, insulin and glucagon, play critical roles in the transcriptional regulation of these enzymes. For example, insulin activates the sterol regulatory element binding protein-1c (SREBP-1c) transcription factor which governs the expression of lipogenic genes including fatty acid synthase (FASN) (reviewed in [2, 3]). During fasting, glucagon activates the protein kinase A (PKA), which phosphorylates carbohydrate response element-binding protein (ChREBP), preventing its translocation to the nucleus and the subsequent activation of its target genes involved in glycolysis and lipogenesis (reviewed in [4, 5]).
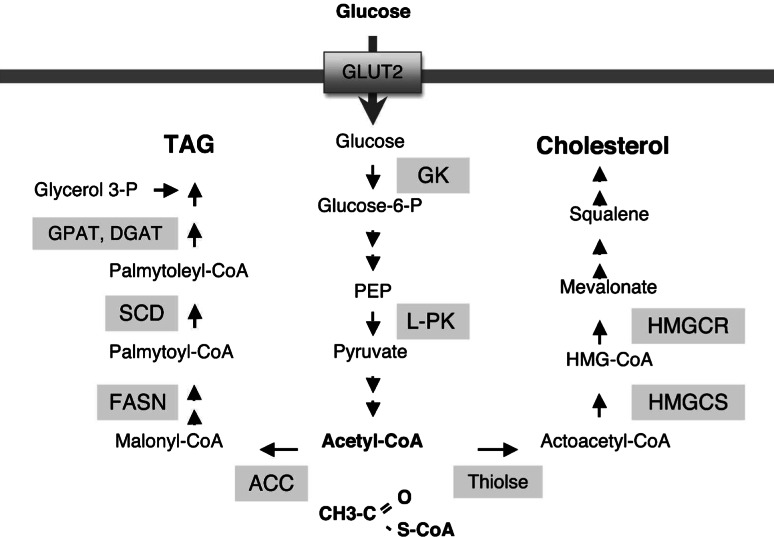
Fig. 1.
Biochemical pathways leading to the synthesis of TAG and cholesterol. Glycolytic pathway generates Acetyl CoA from glucose imported into the cell through GLUT2. TAG and cholesterol are synthesized from Acetyl CoA by sequential enzymatic actions. GLUT2 Glucose transporter 2, GK glucokinase, L-PK liver pyruvate kinase, PEP phosphoenolpyruvate, ACC acetyl-CoA carboxylase, FASN fatty acid synthase, SCD stearoyl-CoA desaturase, GPAT glycerol-3-phosphate acyltransferase, DGAT diacylglycerol acyltransferase, HMG-CoA 3-hydroxy-3-methylglutaryl-coenzyme A, HMGCS HMG-CoA synthase, HMGCR HMG-CoA reductase
The unfolded protein response (UPR) is a signaling system emanating from the endoplasmic reticulum (ER) that is activated when ER protein folding is disturbed [6, 7]. In normal animal physiology, the UPR is critically required for the development and/or secretory function of highly secretory cells in exocrine and endocrine glands. Recent studies revealed novel diverse functions of the mammalian UPR including its role in hepatic lipid metabolism [8–11]. The UPR plays essential roles in hepatic lipogenesis by controlling the expression of genes in the lipogenic pathway. UPR activation has been observed in fatty liver diseases, suggesting the induction of ER stress in these pathological conditions (reviewed in [12]). This review summarizes the physiological functions of the UPR, focusing on studies analyzing the phenotype of mice mutant in each of the critical UPR regulatory genes. It also addresses recent findings regarding the role of the UPR in hepatic lipogenesis and fatty liver diseases (Table 1).
Table 1.
Physiological functions of the UPR: Lessons from genetic alterations of essential UPR genes in humans and mice
| UPR branch | Gene | Alteration | Phenotype | Reference |
|---|---|---|---|---|
| IRE1/XBP1 | Ern1 (IRE1α) | Germline deletion | Embryonic lethal (E10.5) | [181] |
| Ern1−/− Rag2 chimera | Impaired early B cell development | [181] | ||
| Conditional deletion in liver | Altered gene expression upon tunicamycin challenge | [11] | ||
| Ern2 (IRE1β) | Germline deletion | Increased susceptibility to DSS colitis | [182] | |
| Germline deletion | Hyperlipidemia on high cholesterol/fat diet | [11] | ||
| Xbp1 | Germline deletion | Embryonic lethal (E14.5), liver hypoplasia | [87] | |
| Xbp1−/− Rag2 chimera | Absent plasma B cell differentiation | [85] | ||
| Xbp1−/− Rag2 chimera | Impaired dendritic cell differentiation | [88] | ||
| Germline deletion, transgene rescued | Exocrine pancreas malfunction | [84] | ||
| Conditional deletion in GI tract | Inflammatory bowel diseases, Paneth cell loss | [83] | ||
| Conditional deletion in liver | Hypolipidemia, decreased hepatic lipogenesis | [8] | ||
| ATF6 | Atf6 (ATF6α) | Germline deletion | Increased liver toxicity by tunicamycin | [10] |
| Atf6, Atf6b | Germline deletion of ATF6α and β | Embryonic lethal | [81] | |
| PERK/eIF2α | Eif2k3 (PERK) | R587Q mutation in human PERK gene | Wolcott-Rallison syndrome, diabetes, skeletal dysplasias | [107] |
| Germline or conditional deletion | Diabetes, skeletal dysplasias, exocrine pancreas insufficiency | [110–113] | ||
| Conditional deletion in β cells | Diabetes, impaired embryonic pancreatic β cell differentiation | [114] | ||
| Conditional deletion in mammary gland | Mammary gland lipogenesis defect | [136] | ||
| Eif2a (eIF2α) | Homozygous S51A | Perinatal lethality due to defective gluconeogenesis | [129] | |
| Heterozygous S51A | Insulin resistance, defective insulin production on high fat diet | [130] | ||
| Homozygous S51A in liver, transgene rescued | Altered gene expression upon tunicamycin challenge | [11] | ||
| Atf4 | Germline deletion | Fetal anemia, impaired eye lens fiber formation and osteoblast differentiation | [70, 183, 184] |
Transcriptional regulation of hepatic lipogenesis
Ingestion of dietary carbohydrates not only increases the plasma glucose concentration but also promotes the secretion of insulin from pancreatic β cells. These metabolic and hormonal cues activate glycolysis and de novo lipid synthesis in the liver to convert glucose into TG [13], which is carried to adipose tissue in very low-density lipoprotein (VLDL) particles [14]. Several key enzymes catalyzing glycolysis and de novo lipid synthesis such as glucokinase (GK), liver-pyruvate kinase (LPK), acetyl CoA carboxylase (ACC), FAS, and stearyl CoA desaturase-1 (SCD1) are transcriptionally activated in the liver upon ingestion of high carbohydrates [13, 15, 16]. Transcription factors SREBP-1c and ChREBP have been considered as major players in the transcriptional regulation of these glycolytic and lipogenic genes (Fig. 2) [2, 5, 15].
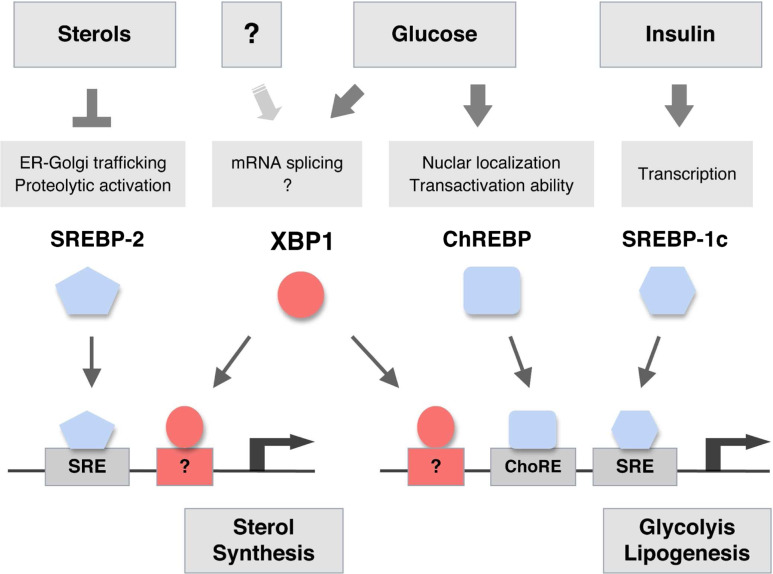
Fig. 2.
Transcriptional control of hepatic lipogenesis. SREBP-2 is activated when cellular sterol levels are low, and controls the expression of genes involved in sterol biosynthesis. SREBP-1c and ChREBP are activated by insulin and glucose, respectively. Ingestion of carbohydrates increases circulating glucose and insulin levels, by inducing insulin production from pancreatic β cells. SREBP-1c and ChREBP bind to specific promoter elements to activate the transcription of genes in glycolytic and lipogenic pathways. XBP1 is required for the de novo synthesis of sterols and fatty acids in the liver and is activated in the presence of high glucose. The mechanism by which XBP1 controls sterol and fatty acid synthesis genes remains to be further investigated
SREBP-1c
SREBP-1c is activated by insulin and plays a major role in the induction of lipogenic genes by insulin [16, 17]. SREBP-1c is a member of a transcription factor family that was originally identified as a mediator of sterol signaling [18]. The SREBP family comprises SREBP-1a, SREBP-1c and SREBP-2 that are encoded by two separate genes [2, 19]. Utilization of alternate promoters in a single gene gives rise to SREBP-1a and SREBP-1c, which are identical in most domains except for the N-terminal region which is derived from an alternate exon 1 [20]. Studies in transgenic and knock-out mice revealed the function of SREBP-1c and SREBP-2 to primarily control the expression of fatty acid and cholesterol biosynthetic enzymes, respectively [2, 15, 16]. SREBPs are produced as precursor proteins containing two transmembrane domains, which anchor the protein in the ER complexed with the SREBP cleavage activating protein (SCAP) and ER retention protein called Insig [21]. Low sterol triggers a conformational change of the sterol sensitive SCAP protein that dissociates the SCAP–SREBP complex from Insig [22]. The SCAP–SREBP complex migrates to the Golgi apparatus, where SREBPs are sequentially cleaved by site 1 (S1P) and site 2 (S2P) proteases to liberate the mature proteins that translocate to the nucleus and act as active transcription factors [23]. While SREBP-2 is primarily controlled at the post-translational level by sterols which regulates its ER-Golgi trafficking, SREBP-1c is controlled at the transcriptional level by insulin [21]. Hepatic SREBP-1c mRNA is downregulated in fasted animals, and dramatically induced upon high carbohydrate ingestion by the action of insulin [20, 24]. The PI(3)-kinase signaling pathway plays an important role in SREBP-1c expression induced by insulin [25, 26]. Liver X receptor (LXR) can also induce SREBP-1c mRNA [27, 28]. Induction of lipogenic genes by a synthetic LXR agonist was blunted in SREBP-1c deficient mice, suggesting that SREBP1-c is essential for the lipogenic activity of LXR [17]. Overexpression of SREBP-1c in the liver by using transgenic mice or adenoviral gene delivery dramatically induced lipogenic genes accompanied by increased hepatic TG content [29–31]. In contrast, mice lacking SREBP-1c in the liver displayed reduced plasma TG levels, and decreased expression of multiple lipogenic genes upon refeeding [17]. It is of interest to note that SREBP-1c deletion only partially suppressed the expression of several key lipogenic genes, such as glucokinase, Fas, and Acc, suggesting the presence of compensatory mechanisms [17].
ChREBP
ChREBP was identified as a transcription factor that binds to a carbohydrate response element (ChRE) in the LPK gene promoter [32]. ChRE is a motif present within several gene promoters responsive to high glucose [13]. As a bZIP transcription factor, ChREBP forms a heterodimeric complex with another bZIP protein Max-like protein X (MLX), and directly binds to ChRE [33]. It was originally proposed that PKA phosphorylates ChREBP under low glucose conditions, preventing its translocation into the nucleus [34]. However, it is controversial if nuclear translocation is sufficient for the activation of ChREBP [35, 36]. Recently, Towle and colleagues demonstrated that the derepression of the transcriptional activity conferred by the N-terminal domain of ChREBP is critical for the activation of ChREBP [37]. ChREBP knock-out mice displayed reduced fat mass, low plasma free fatty acid (FFA), hyperglycemia, high liver glycogen and normal hepatic and plasma TG levels on chow diet [38]. When fed high starch diet, these mice displayed more profound phenotypes, including significant reduction in hepatic TG level, associated with decreased mRNA levels of select lipogenic genes. Fatty acid synthesis rate was reduced by ~65%, indicating the essential role of ChREBP in hepatic lipogenesis [38]. ChREBP knock-out mice exhibited moderate insulin resistance, presumably due to an efficient disposal of glucose [38]. In contrast, downregulation of ChREBP in the liver by using shRNA delivery improved the insulin sensitivity of ob/ob mice [39]. It is conceivable that ChREBP knockdown lowered lipotoxicity by suppressing hepatic lipogenesis.
Unfolded protein response
The ER is a continuous membrane network in the cell that has multiple important functions. First, the ER plays a crucial role in the protein secretory pathway [40–42]. All the nascent polypeptides destined to be secreted are imported into the ER lumen to attain proper three-dimensional protein structures and various post-translational modifications, such as disulfide bond formation and glycosylation, and then sorted for transport to the destined locations. ER resident chaperones, protein disulfide isomerases (PDIs), and various protein modifying enzymes are involved in this process. The ER also serves as a protein quality control check-point, so that misfolded proteins are screened out for retrotranslocation to the cytoplasm followed by proteasomal degradation [43, 44]. Second, the synthesis of neutral and phospholipids occurs in the ER [45–47]. The lipid bilayer expands in the ER and then is mobilized to the membranous organelles as needed. Third, the ER plays a crucial role in drug metabolism with major drug metabolizing enzymes including cytochrome P450 present in the ER [48]. Fourth, the ER is the major storage depot for calcium which is released upon appropriate stimulation to serve as signaling molecules [49].
ER stress can be defined as a condition that compromises the capacity of the ER to handle its client proteins [6, 7, 50]. Increased input of the client proteins into the ER during the development of highly secretory cells can induce ER stress. ER stress can also be induced by treatment of cells with pharmacological reagents that inhibit protein folding in the ER. These include protein glycosylation inhibitors (i.e., tunicamycin), reducing agents (i.e., dithiothreitol) that interfere with protein disulfide bond formation, and calcium channel blockers (thapsigargin) that deplete ER calcium stores. In response to ER stress, eukaryotic cells activate a signaling cascade called the UPR to protect the cell from stress. Mutation of genes in the UPR signaling cascade frequently results in the increased susceptibility of the cell to ER stress-induced cell death.
In yeast, the UPR is initiated by the ER stress sensor IRE1 and executed by the transcription factor HAC1 [51–54]. IRE1 is a type I transmembrane protein, which senses ER stress through its luminal domain [55, 56]. The cytosolic domain of IRE1 possesses dual enzymatic activities of a protein kinase and an endoribonuclease [51, 57–60]. Activated by trans-autophosphorylation, IRE1 specifically cleaves two sites in HAC1 mRNA, which is re-ligated by t-RNA ligase [57, 60, 61]. This unconventional splicing event generates HAC1p protein that activates a variety of genes involved in protein secretory pathways, including ER-resident chaperones and enzymes in phospholipid biosynthesis pathways, as well as those engaged in ER-associated protein degradation (ERAD) [62]. IRE1 or HAC1 mutant yeasts grow normally under standard conditions, but display increased sensitivity to ER stress inducing compounds, such as tunicamycin [53, 63].
Mammals have an IRE1/XBP1 UPR signaling system analogous to yeast IRE1/HAC1 (Fig. 3). IRE1-mediated splicing removes 26 nucleotides in the XBP1 mRNA starting from the 184th codon, resulting in a shift of the translational reading frame [64–66]. XBP1s mRNA encodes a bZIP transcription factor that binds to the CRE-like promoter element to activate a group of UPR target genes [67–69]. While the yeast UPR is controlled solely by HAC1, XBP1 is responsible for the induction of only a subset of UPR target genes, indicating the presence of additional components [68, 70]. Indeed, mammals contain at least two additional UPR branches that are controlled by the PKR-like ER-localized eukaryotic initiation factor 2α (eIF2α) kinase (PERK) and the ER transmembrane transcription factor precursor ATF6 [71, 72]. Phosphorylation of eIF2α by PERK suppresses translational initiation, lowering the input of client proteins into the ER, to mitigate ER stress [73]. In contrast, translation of certain mRNAs (i.e., ATF4 and ATF5) that possess short open reading frames in the 5′ UTR region are facilitated by PERK-dependent eIF2α phosphorylation under ER stress conditions [73, 74]. ATF4 plays a major role in the transcriptional control of PERK-dependent UPR target genes [75]. The ATF6 transcription factor family comprises two transmembrane bZIP transcription factors, ATF6α and ATF6β, that reside in the ER under basal conditions [72, 76–78]. ATF6α and ATF6β display limited amino acid sequence homology only in the bZIP and the ER transmembrane domains, suggesting that they recognize similar cis-acting promoter elements, but may differentially regulate their target genes [76, 79, 80]. Indeed, both ATF6α and ATF6β can bind to the CCAAT-(N9)-CCACG sequences, called the ER stress response element (ERSE) which mediates the transcriptional induction of many ER chaperone genes [67, 69, 81]. It is noteworthy that ATF6 and XBP1 recognize similar DNA target sequences and can form a hetero-dimer, suggesting that they may have similar roles in the ER stress response [67, 82]. ER stress triggers the migration of ATF6 from the ER to the Golgi apparatus, where it is sequentially processed by S1P and S2P proteases to release the N-terminal cytosolic part of the protein, which then enters the nucleus to transactivate gene transcription [77].
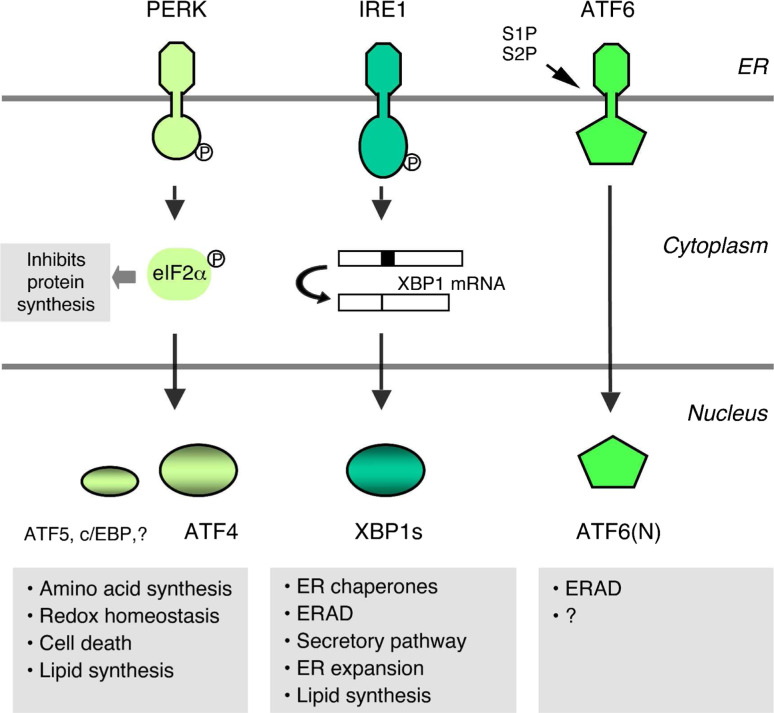
Fig. 3.
Overview of mammalian UPR signaling pathways. IRE1, PERK, and ATF6 are proximal ER stress sensors. IRE1, activated by autophosphorylation, removes 26 nucleotides from the XBP1 mRNA by using its cytosolic endoribonuclease activity. Spliced XBP1s protein is a potent transcription factor that induces diverse target genes. eIF2α phosphorylation by PERK impedes overall protein synthesis, while activating translation of some transcription factors. ATF4 plays a major role in PERK-dependent UPR target gene expression. ATF6 is a member of the family of ER transmembrane transcription factors, and normally resides in the ER associated with the ER chaperone BiP. Upon ER stress, ATF6 moves to the Golgi apparatus where site 1 (S1P) and site 2 proteases (S2P) sequentially cleave ATF6 to release the N-terminal part of the protein that translocates to the nucleus to activate its target genes
Physiological functions of the UPR
IRE1/XBP1
Microarray analyses of XBP1-deficient mouse embryonic fibroblast (MEF) cells and primary B cells stimulated with lipopolysaccharides identified a series of genes involved in ER translocation, protein folding, secretory pathways, and degradation of misfolded proteins as XBP1 target genes [68, 70]. These XBP1 target genes are expressed at basal levels under normal conditions and further induced in an XBP1 dependent manner when cells are ER stressed or differentiate into antibody secreting plasma cells. Consistent with the relationship of its target genes to cellular secretory function, XBP1 was found to be required for the development and protein secretory function of secretory cells [83–85]. XBP1 is highly expressed in exocrine glands and osteoblasts in the skeletal system that produce large amount of secretory proteins [86]. XBP1-deficient mice die during embryonic development due to liver hypoplasia accompanied by defective erythropoiesis [87]. It is not known if embryonic liver development requires a robust UPR or whether XBP1 may have another function in embryonic liver unrelated to the UPR. XBP1 also contributes to the development of dendritic cells [88].
Analysis of recombination activating gene 2 (RAG2) chimeric mice possessing XBP1 deficient lymphoid cells revealed that XBP1 is essential for the terminal differentiation of mature B cells into antibody secreting plasma cells [85]. Similarly, the lack of XBP1 severely impairs the development of other highly secretory cells such as pancreatic acinar cells that secrete zymogens to the small intestine, and intestinal Paneth cells that produce anti-microbial peptides [83, 84]. Normal acinar cells contain tightly packed multilayers of ER in the basal area and zymogen granules at the apical side targeted to the lumen for secretion. Extensive ER is required for the maturation and transportation of zymogens. XBP1-deficient pancreatic acinar cells are completely devoid of mature zymogen granules. Their ER is minimally expanded and contains electron dense particles which appear to be zymogen aggregates that failed to mature into secretory granules [84]. Marked apoptosis of XBP1 deficient pancreatic acinar cells was observed during the embryonic developmental stage when cells dramatically increase zymogen production and expand the ER. Increased expression of a proapoptotic ER stress marker CHOP revealed heightened ER stress in XBP1-deficient pancreatic acinar cells. XBP1 deficiency exerts similar effects on Paneth cells, resulting in complete absence of zymogen granules, impaired ER expansion and apoptotic cell death associated with increased ER stress [83]. These data collectively suggest that XBP1 is required for the biogenesis of cellular secretory machinery and its absence results in apoptotic cell death due to increased ER stress.
XBP1 is absolutely required for only a subset of secretory cells. Moderately secretory cells such as adult hepatocytes, salivary gland acinar cells, pancreatic β cells are only partially dependent on XBP1, suggesting that the requirement of XBP1 correlates with the secretory load of any given cell [8, 84]. XBP1 is required not only for the development of secretory cells, but also to maintain the integrity of the secretory machinery [83]. Inducible deletion of XBP1 in already differentiated Paneth cells or pancreatic acinar cells causes the degeneration of the ER and malformation of the secretory granule, as seen in XBP1 germline deleted mice [83] (Lee and Glimcher, unpublished data).
PERK/eIF2α
In response to various stress signals, mammalian cells control mRNA translation by regulating translational initiation [89]. Phosphorylation of the translational initiation component eIF2α is a central mechanism that regulates mRNA translation. eIF2α can be phosphorylated by different protein kinases, such as protein kinase RNA-activated (PKR), general control nonderepressible-2 (GCN2), PERK, and heme-regulated inhibitor kinase (HRI) under various stressful conditions [71, 89–94]. PKR is activated by double-stranded RNA during viral infection, GCN2 by amino acid starvation, PERK by unfolded proteins in the ER, and HRI by heme deficiency in erythroid cells. Phosphorylation of eIF2α at the Ser51 residue inhibits the translation of most mRNAs, suppressing global protein synthesis to mitigate cellular damage by stress [6]. It is believed that PERK-induced eIF2α phosphorylation lowers the input of cargo proteins into the ER, and thereby reduces ER stress. While global protein translation is suppressed upon eIF2α phosphorylation, the translation of select mRNAs is increased. Short open reading frames in the 5′ UPR of ATF4 mRNA allows translational initiation from the correct start codon, when eIF2α phosphorylation is increased [73]. ATF5 translation is also induced through a similar mechanism [74]. ATF4 is the major transcription factor that executes UPR gene regulation downstream of PERK signaling pathway [75, 95]. In response to ER stress, ATF4 induces genes largely involved in amino acid metabolism and cellular redox control. Therefore, ATF4−/− cells require supplementation with non-essential amino acids and reducing agents for their survival [75]. It has been suggested that oxidative protein folding in the ER generates reactive oxygen species [96], and that ATF4 plays an important role in the removal of reactive oxygen species under ER stress conditions.
The UPR signaling pathway controlled by PERK is important for cell survival under various ER stress conditions. Accumulation of free cholesterol in the ER induces ER stress and apoptosis of macrophages in advanced atherosclerosis [97]. PERK−/− macrophages are protected from free cholesterol induced apoptosis, indicating a cytoprotective role of PERK. Inactivation of the PERK signaling pathway impairs the survival of tumor cells under hypoxic conditions in tumor allograft models [98, 99].
ATF4 is directly upstream of CHOP, an important PERK-regulated UPR target gene [73, 75, 100, 101]. CHOP has long been considered as a proapoptotic transcription factor, as CHOP deficient cells were resistant to ER stress-induced apoptosis [102, 103]. For example, CHOP-deficient macrophages are protected from free cholesterol-induced apoptosis [97]. Similarly, CHOP deletion ameliorates pancreatic β cell loss in Akita mice that express mutant misfolded insulin protein that evokes ER stress [104]. CHOP induces GADD34 transcription, which dephosphorylates eIF2α to restore protein translation during prolonged ER stress [100]. In the absence of CHOP or GADD34, eIF2α phosphorylation is maintained during chronic ER stress, reducing protein synthesis and ER stress. Therefore, mice lacking CHOP or GADD34 were protected from kidney damage in a tunicamycin-induced ER stress model [100].
NF-E2-related factor-2 (Nrf2), a protein that transcriptionally induces enzymes that remove reactive oxygen species in response to oxidative and xenobiotic stress, is also activated by PERK mediated phosphorylation [105, 106]. It is conceivable that PERK maintains the cellular redox status by activating Nrf2 in response to the accumulation of reactive oxygen species that are generated during the unfolded protein response.
PERK is essential for the development of pancreatic β cells and osteoblasts in the skeletal system. Mutation of the PERK gene in humans is responsible for Wolcott-Rallison syndrome (WRS), a syndrome characterized by early onset diabetes due to β cell dysfunction, multiple skeletal dysplasia and growth retardation [107–109]. Germline deletion of the PERK gene in mice results in similar defects of insulin production and skeletal abnormalities [110, 111]. PERK deletion also compromises zymogen production from the exocrine pancreas, accompanied by acinar cell apoptosis [110–113]. These data suggest that PERK is essential for the development of these secretory cells, consistent with its molecular function in transcriptional and translational control in response to ER stress, and that its absence causes apoptotic cell death secondary to increased ER stress. Surprisingly, however, PERK is not required for the postnatal insulin secreting function of β cells [114]. Hence, post-natal deletion of PERK specifically in β cells by using the cre/lox system did not induce β cell death or diabetes, demonstrating a temporal-specific control of β cell development by this branch of the UPR [111]. Similarly, a recent report suggested that the loss of acinar cells in the exocrine pancreas of PERK deficient mice is not related to an impaired ER stress response [113]. Neither the ER structure of the acinar cells, nor the secretagogue-induced exocytosis of zymogens from the pancreatic lobules was compromised in PERK−/− mice. No evidence for increased ER stress in PERK-deficient pancreas was observed. Nonetheless, PERK-deficient mice displayed progressive pancreatic atrophy, losing acinar cells by oncosis, a non-apoptotic form of cell death, starting at ~3 weeks of age. These observations led to the conclusion that PERK is essential for the integrity of acinar cells in a manner which is unrelated to the ER stress response.
ATF6
ATF6 comprises two isoforms, ATF6α and ATF6β, which are encoded by separate genes [76, 79, 80]. ATF6 belongs to a group of transcription factors that are synthesized as precursor proteins with ER transmembrane domains and proteolytically activated by site 1 and site 2 proteases [115, 116]. ATF6 is normally present in the ER forming a stable complex with BiP [117]. ER stress dissociates ATF6 from BiP, which allows the mobilization of ATF6 to the Golgi apparatus for sequential proteolysis by S1P and S2P, releasing the N-terminal cytosolic part of the protein [77–79].
The DNA binding specificity of ATF6 and XBP1 is remarkably similar. Independent studies identified almost identical consensus sequences as binding sites of XBP1 and ATF6 [67, 82]. Clauss et al. demonstrated that recombinant XBP1 protein bound to CRE-like GAT-GACGTG(T/G)NNN(A/T)T sequences [67]. Wang et al. demonstrated that ATF6 bound to TGACGTG(G/A) consensus sequences [82]. The UPRE motif which refers to the ATF6 binding site can be activated by both XBP1 and ATF6 [68]. XBP1 and ATF6 can also form a hetero-dimer, suggesting that they might regulate common target genes [81]. Microarray analysis revealed ATF6α target genes are limited to select ER chaperones and ERAD pathway genes, suggesting that XBP1 has a broader universe of target genes [68, 118]. ATF6α−/− MEF cells and animals show increased susceptibility to tunicamycin, suggesting a protective role of ATF6α in the ER stress response [10, 81]. The physiological functions of ATF6α and ATF6β remain to be determined, however, as mice with targeted disruption of these genes are grossly normal, although ATF6α/β double mutant mice die early during embryonic development [10, 81]. It will be interesting to test if ATF6α has a redundant or supplementary role with XBP1 in the development and/or function of secretory cells.
Role of the UPR in lipid metabolism
Role of XBP1 in hepatic lipogenesis
A crucial function of the liver is to coordinate whole body energy homeostasis by regulating carbohydrate and lipid metabolism. Fasting stimulates glucose production from the liver through glycogenolysis and gluconeogenesis to maintain optimal circulating glucose levels, while suppressing fatty acid synthesis. Hepatic lipogenesis is induced upon ingestion of excess carbohydrates to convert extra carbohydrates to triglyceride (TG) [13]. TG is secreted from the liver as VLDL lipoprotein particles, and then transported to adipose tissue for long-term energy storage. Hepatic lipogenesis is controlled by transcription factors such as SREBP and ChREBP, which are regulated by nutritional and hormonal conditions [2–5] (Fig. 2). SREBP-1c is strongly activated by insulin and increases the expression of genes controlling fatty acid synthesis [119–121]. SREBP-2 activation is controlled by cellular sterol levels, so that low sterol activates SREBP-2 to induce genes in the cholesterol biosynthesis pathway [30, 122]. ChREBP appears to be activated by high glucose to activate genes involved in glycolysis and fatty acid synthesis [34, 38].
Microarray analyses revealed that the majority of XBP1 target genes are involved in protein secretory pathways [68, 70]. XBP1 also activates the phospholipid biosynthesis pathway at least by increasing the level of choline cytidylyltransferase protein, a rate-limiting enzyme in the CDP-choline pathway [123, 124]. The molecular function of XBP1 is highly reminiscent of yeast HAC1 protein, which also controls the expression of genes in protein secretory pathways and lipid synthesis (INO1) [62]. Given that protein secretion requires ER expansion and the trafficking of secretory vesicles, it makes sense that the UPR also activates lipid synthesis to meet the demand of lipid bilayer formation [125]. Consistent with its target gene signature, XBP1 is critically required for the survival and the secretory functions of highly secretory cells [83–85]. As discussed above, however, XBP1 is not essential for the secretory function of hepatocytes [8]. The liver is the major source of plasma proteins but plasma protein levels were only slightly decreased in mice lacking XBP1 in the liver. The ER in XBP1-deficient hepatocytes displayed no morphological abnormality. The secretion and turnover of apolipoprotein B (ApoB) protein was minimally influenced by XBP1 deficiency, collectively suggesting that the basal expression of genes in secretory pathway independent of XBP1 is sufficient to accommodate the secretory load of postnatal hepatocytes.
However, XBP1 deletion in the liver resulted in a dramatic reduction of plasma lipids [8]. Genes encoding key lipogenic enzymes such as Acc2, Dgat2 and Scd1 were dramatically downregulated in XBP1-deficient liver. These lipogenic enzymes were induced upon high carbohydrate diet feeding in WT, but not in XBP1-deficient mouse liver. Consistent with the gene expression profile, de novo synthesis of fatty acids is impaired in XBP1-deficient hepatocytes. Sterol synthesis was also downregulated in XBP1-deficient hepatocytes, although the underlying mechanism is unknown. Given the essential role of XBP1 in the ER stress response, one might ask whether Xbp1 deletion caused ER stress in the liver, and consequently hypodyslipidemia as an indirect effect. Indeed, a recent study demonstrated that acute ER stress induced hepatic steatosis, albeit expression of lipogenic genes was suppressed, suggesting that ER stress in the liver disrupts hepatic lipid metabolism [11]. It has been shown that ER stress inhibits ApoB100 folding and VLDL secretion, which might be responsible for the fat accumulation in the liver in tunicamycin-injected mice [126–128]. Unlike the tunicamycin-injected mice, XBP1-deficient mice did not develop hepatic steatosis, and their hepatocytes did not show any signs of ER stress. In addition to the normal turnover of ApoB protein, XBP1-independent UPR markers were not induced at baseline, and were normally induced upon tunicamycin challenge in the absence of XBP1. These data argue against the possibility that XBP1 deletion disturbed ER homeostasis, which then resulted in defective hepatic lipid metabolism. Instead, chromatin immunoprecipitation assays demonstrated that XBP1 can directly bind to the promoter region of lipogenic genes. Further, enforced expression of XBP1 induced the expression of Acc2 and Dgat2 genes, suggesting that XBP1 is a bona fide transcription factor that controls hepatic lipogenesis (Fig. 2). Active XBP1s protein is induced in mice fed a high fructose diet and in hepatocytes cultured in high glucose media, suggesting that increased glucose availability induces XBP1. The nature of the signals that leads to the activation of IRE1/XBP1 and the role of these proteins in the development of metabolic diseases associated with hyperdislipidemia remain to be further investigated.
PERK/eIF2α
Homozygous eIF2α S51A mutant mice die perinatally due to hypoglycemia caused by defective hepatic gluconeogenesis [129]. In contrast, heterozygous eIF2α S51A mice are prone to weight gain on a high fat diet and develop insulin resistance [130]. To investigate the function of the PERK/eIF2α UPR branch in the liver, Oyadomari et al. generated transgenic mice that overexpressed the c-terminal fragment of GADD34 protein in the liver [9]. GADD34 is a regulatory component of eIF2α phosphatase, induced by PERK signaling upon ER stress [131]. Phosphorylation of eIF2α both at baseline and after ER stress induction was dramatically reduced in GADD34 transgenic mice, which displayed markedly decreased hepatic lipogenesis. Expression of lipogenic enzymes such as Fasn, Acc2 and Scd1 was decreased, especially on high fat diet. Lipid accumulation in the transgenic liver was also reduced, suggesting that PERK/eIF2α signaling contributes to optimal hepatic lipogenesis. Enforced activation of PERK signaling can be achieved by using the chimeric PERK protein fused to a ligand-induced dimerization domain [9, 95]. Ligand-induced PERK activation increased the expression of lipogenic enzymes independent of ER stress, suggesting that PERK activation is sufficient for the activation of lipogenesis in the liver. PERK controls the expression of C/EBPα and C/EBPβ, which play important roles in hepatic lipogenesis [132, 133]. C/EBPα, C/EBPβ, and their downstream transcription factor, PPAR-γ, were significantly downregulated in high fat-diet fed GADD34 transgenic mouse liver. Interestingly, C/EBP gene expression is regulated at both the transcriptional and translational level by XBP1 and eIF2α [9, 134, 135]. Further, the upstream open reading frame (uORF) in C/EBP mRNA directs the production of a truncated form of the C/EBP protein in the absence of eIF2α phosphorylation [135]. eIF2α phosphorylation further increases the translation efficiency of the full-length C/EBP in a manner similar to ATF4 [73]. These data suggest an active role for the PERK-eIF2α signaling pathway in regulation of hepatic lipogenesis (Fig. 4). A recent study demonstrated that PERK is also critical for lipogenesis in the mammary gland. Bobrovnikova-Marjon et al. deleted the Perk gene specifically in mammary glands by using an MMTV-cre [136]. Pups nursed by PERK-deficient female mice showed growth retardation, and mammary glands from PERK-deficient mothers exhibited an immature phenotype with reduced alveolar expansion. Free fatty acid and TG content was significantly reduced in milk produced from these mothers, accounting for the growth retardation of the pups. The reduced lipid content in the milk was the direct consequence of low de novo lipid synthesis as SREBP1 and key lipogenic enzyme genes were significantly downregulated in PERK−/− mammary glands, suggesting that PERK transcriptionally regulates mammary gland lipogenesis via SREBP1. SREBP1 protein level was low in PERK-deficient mammary epithelial cells cultured under ER stress conditions, which activate SREBP processing, although the physiological significance of this observation is unknown [137, 138]. These data suggest that PERK controls mammary gland lipogenesis through controlling activation of SREBP.
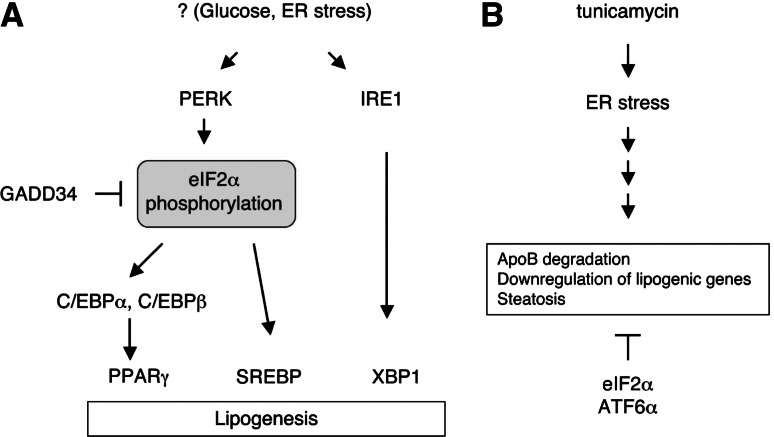
Fig. 4.
Intersection of the UPR and hepatic lipid metabolism. a PERK/eIF2α and IRE1/XBP1 UPR signaling pathways control lipogenesis. eIF2α phosphorylation activates PPARγ and SREBP-1 in liver and mammary gland to increase the lipid synthesis. XBP1 activates the expression of the lipogenic genes, at least partly via direct interaction to target promoters. It is not fully understood what triggers UPR activation leading to increased lipogenesis. ER stress and high glucose may play a role. b Tunicamycin administration induces the ER stress and hepatic steatosis. Ironically, SREBP1 and lipogenic genes are downregulated by tunicamycin. Inhibition of ATF6α and eIF2α signaling pathways further increase fat accumulation, suggesting their protective roles in tunicamycin-induced steatosis. Tunicamycin induces degradation of ApoB protein, which is essential for VLDL secretion from the liver
UPR in Yeast
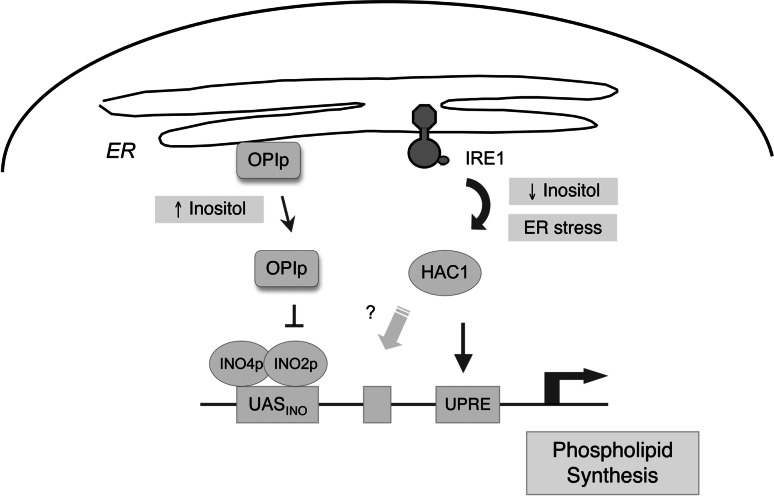
In yeast, phospholipid biosynthesis is dynamically regulated by the availability of inositol in the media [139, 140]. When the inositol level is low, the master negative regulator of the yeast lipogenesis program, OPI1p, is attached to the ER membrane through its interaction with phosphatidic acid and Scs2p (Fig. 5). The addition of inositol consumes phosphatidic acid to produce phosphatidylinositol, resulting in the release of OPI1p from the ER membrane, which translocates to the nucleus and represses the expression of genes involved in phospholipid biosynthesis [141]. This repression occurs through the UASINO promoter element where OPI1p antagonizes the transcriptional activators, Ino2p and Ino4p [142, 143]. Microarray analyses revealed that inositol controls the expression of a group of genes including key lipogenic enzymes, transcription regulators, as well as inositol and choline transporters in an Ino-2p-Ino4p dependent manner [144, 145].
Fig. 5.
Transcriptional regulation of lipid synthesis in yeast. OPI1p is a critical repressor of lipid synthesis in yeast. It is anchored in the ER membrane through its interaction with Scs2p (reviewed in [185]). Inositol starvation activates INO2p–INO4p lipogenic transcription factors by preventing the release of OPI1p from the ER which antagonizes INO2p. Inositol starvation also activates the UPR. INO1 expression is impaired in IRE1 or HAC1 mutant yeasts, which display inositol auxotrophy. The mechanism by which the UPR contributes to the expression of lipogenic genes is not fully understood
Interestingly, inositol deficiency also activates the UPR in yeast, inducing IRE1-mediated HAC1 mRNA splicing, although it is not known whether ER stress is induced under this condition [53]. Induction of UPR target genes including KAR2 by inositol deficiency is dependent on IRE1/HAC1. Notably, HAC1∆ and IRE1∆ strains of yeasts display inositol auxotrophy [146]. INO1 expression is impaired in these mutant yeast strains, and their inositol auxotrophy is rescued by OPI1 deletion or enforced expression of INO1, suggesting that IRE1/HAC1 is required for the induction of INO1 [125]. However, HAC1 is not sufficient for the expression of lipogenic genes that are regulated by OPI1, as constitutively active HAC1 is not sufficient to maintain the expression of these genes after inositol addition [144]. It has been suggested that HAC1 may de-repress OPI1p through heterodimerization to activate lipogenic genes [125]. However, other studies demonstrated the normal expression of INO1 in certain compound mutant yeast strains lacking HAC1, suggesting the minimal role of the UPR in INO1 expression under certain conditions [147, 148]. These data collectively suggest that the IRE1/HAC1 UPR pathway is required for the optimal expression of phospholipid biosynthesis genes in yeast. In addition, considering the role of XBP1 in membrane biogenesis and hepatic lipogenesis in mammals, lipid synthesis may be the evolutionarily conserved molecular function of the IRE1/HAC1/XBP1 pathway.
UPR activation by dysregulation of lipid homeostasis
UPR activation has been reported in the liver, adipose tissue, pancreatic islets, and brain under various genetic and nutritional conditions linked to obesity and insulin resistance [149–155]. Hotamisligil and colleagues first demonstrated the presence of ER stress in the liver of high fat diet-fed or leptin-deficient ob/ob mice [152]. Phosphorylation of PERK and its downstream target eIF2α was increased in the liver of these diet manipulated or genetically modified obese mice. ER stress inhibited insulin signaling by increasing the serine phosphorylation of insulin receptor substrate (IRS)-1, and this phosphorylation occurred via JNK activation by ER stress [156]. Induction of BiP in the liver of diabetic db/db mice was described as a marker of ER stress [151], although this finding was not reproduced in another study [157]. Wang and colleagues examined the effects of various diets on UPR activation in rat liver [150]. High sucrose or high levels of saturated fatty acids in the diet strongly activated the IRE1/XBP1 UPR pathway, as evidenced by increased XBP1 mRNA splicing, although the expression of XBP1 target genes was not explored. In contrast, unsaturated fatty acids did not evoke the UPR, although hepatic TG levels were similarly increased. BiP and CHOP protein levels were ~2-fold increased by dietary sucrose or saturated fatty acids. UPR activation correlated with increased liver damage, suggesting that these diets induce ER stress that causes liver cell death. Several studies demonstrated that saturated fatty acids induced ER stress in cultured liver and pancreatic β cells, while unsaturated fatty acids reduced ER stress [158–161]. One recent study, however, demonstrated that monounsaturated oleic acids also induced ER stress and inhibited VLDL secretion in McA-RH7777 liver cells, suggesting that TG accumulation might induce ER stress in liver cells [128]. In contrast to these reports suggesting the induction of ER stress in fatty liver, others failed to detect the induction of ER stress markers in genetic models of steatosis [157, 162, 163]. Mice lacking microsomal triglyceride transfer protein (MTP) gene are unable to secrete VLDL and accumulate TG in the liver. Despite the TG accumulation, ER stress markers BiP and Grp94 were not induced in the liver of MTP deficient mice [163]. Dgat2 transgenic mice developed hepatic steatosis, but not insulin resistance [162]. JNK and PERK were not activated in the liver of a transgenenic mouse line that developed hepatic steatosis. It is unclear if ER stress is induced only under certain conditions of lipid accumulation in the liver. It is also possible that the limited specificity/sensitivity of the reagents to detect ER stress markers resulted in different outcomes.
ER stress was also reported in the hypothalamus of high fat diet-fed mice [154], although the direct cause of the ER stress was not described. ER stress inhibited leptin signaling in cultured cells, resulting in decreased STAT3 phosphorylation, linking ER stress to leptin resistance in the hypothalamus [154, 155].
Contrary to the induction of ER stress by lipids, recent studies suggested that ER stress can cause hepatic steatosis [10, 11]. Kaufman and colleagues demonstrated that tunicamycin, a potent ER stress inducer, induced hepatic steatosis. Surprisingly, however, tunicamycin markedly decreased the synthesis of lipogenic enzymes as well as SREBP1, suggesting that the tunicamycin-induced steatosis is unrelated to the increased lipogenesis [11]. It is not known why liver cells accumulate lipids under ER stress conditions. Given that ApoB protein folding and lipidation is highly sensitive to ER stress, tunicamycin might induce the accumulation of TG in the liver by inhibiting VLDL secretion, as seen in mice lacking ApoB or microsomal triglyceride transfer protein (MTTP) [126–128]. Indeed, plasma cholesterol and TG levels were markedly decreased in tunicamycin-injected mice, suggesting defective lipid secretion [11]. Interestingly, mice lacking ATF6α or harboring phosphorylation-defective eIF2α A/A mutation displayed further increase in lipid accumulation in the liver upon tuncamycin administration, suggesting a protective role of the UPR in tunicamycin-induced steatosis. Further studies are required to clarify the role of the UPR in this process, and its physiological relevance.
ER stress response in viral hepatitis
Hepatitis B (HBV) and hepatitis C (HCV) viruses are major pathogens causing acute and chronic hepatitis, cirrhosis and liver cancer in humans. Since the synthesis of viral structural proteins and the assembly of viral particles occur in the ER, one might expect that ER homeostasis is important for viral proliferation and disease pathogenesis. Indeed, numerous reports suggested the link between viral infection and the ER stress response. For example, it was reported that the HCV core protein induced ER stress by depleting the ER calcium storage, leading to apoptotic cell death [164]. Another study suggested that ER stress induced by core protein activated protein phosphatase 2A, which in turn inhibited interferon-γ signaling [165]. Activation of the UPR has also been observed in HCV-infected cells [166, 167] and in cells expressing viral proteins [168, 169], but not in another study [170]. Japanese encephalitis virus (JEV) and dengue viruses (DEN), which are flaviviruses like HCV, activated XBP1, which induced ER expansion and protected cells from virus-induced cytotoxicity, suggesting that flaviviruses may take advantage of the UPR for their propagation [171]. On the other hand, Siddiqui and colleagues demonstrated that HCV induced ER stress, but surprisingly, suppressed the transactivation ability of XBP1 and its downstream ERAD pathway, resulting in slow degradation of misfolded ER proteins [169]. Translation of HCV RNA utilizing the internal ribosome entry site was facilitated in IRE1-deficient cells, suggesting that HCV may promote its replication by suppressing the IRE1/XBP1 pathway [169]. Regarding HBV, large HBV surface antigens with deletions at the pre-S1 and pre-S2 regions, which are frequently detected in infected individuals, accumulated in the ER and activated GRP78 and GRP94 expression, suggesting the induction of ER stress [172, 173]. HBx, a regulatory protein encoded by HBV, activated XBP1 and ATF6 branches of the UPR [174]. Further studies are required to clarify the consequences of ER stress in viral infection, and the contribution of the UPR signaling pathways in viral propagation and host responses.
ER stress response in alcoholic liver disease
Alcohol consumption causes various types of liver damage including steatosis, hepatitis and cirrhosis. Interestingly, microarray analysis of liver samples from a murine model of intragastric ethanol feeding, which induced necrotic cell death and steatosis, revealed the induction of genes related to the ER stress response [175]. Ethanol feeding increased the level of GRP78, GRP94, CHOP and caspase-12, as well as SREBP-1c, which may have contributed to the development of fatty liver [176]. It is not completely understood how ethanol consumption induces ER stress in the liver, but the increased level of homocysteine appears to play a role [177]. Plasma homocysteine level is increased upon alcohol consumption both in humans and mice [175, 178]. Homocysteine is known to induce ER stress and activate SREBP-1 processing in liver cells [179]. Betaine, which reduced the plasma homocysteine level in alcohol-fed mice, decreased the fat accumulation and the expression of ER stress response genes, suggesting the crucial role of homocysteine in alcoholic liver disease [175].
Concluding remarks
Recent studies have aroused great interest in the potential role of the UPR in energy metabolism in mammals. XBP1 plays a critical role in the synthesis of TG and cholesterol in the liver by controlling the expression of lipogenic genes. Deletion of XBP1 in the liver drastically lowered circulating TG and cholesterol levels. PERK also plays important role in both hepatic and mammary gland lipogenesis by regulating lipogenic transcription factors such as C/EBP, PPARγ, and SREBPs. It remained to be further investigated how the UPR is activated by nutritional and hormonal cues and interacts with other lipogenesis regulators, such as SREBP and ChREBP. Further studies are also required to define the role of the UPR in metabolic disorders accompanied by the dysregulation of lipid homeostasis.
UPR activation has been reported in the liver, adipose tissue, pancreatic β cells, skeletal muscle and brain under nutritional conditions associated with obesity and type 2 diabetes. Due to the technical difficulty of directly measuring actual ER stress as defined by an increase in unfolded protein species in the ER, or compromise of ER integrity, the UPR itself has been frequently used as a marker of ER stress. Given the active role of the UPR in cell metabolism and the lack of direct evidence of ER stress, one can ask if UPR activation is a causative factor in the development of metabolic dysregulation or is instead the consequence of ER stress. An assay system to measure actual ER stress in the cell [180] will be crucial to a better understanding of the role of the UPR in metabolic disorders. The mammalian UPR is comprised of at least three different signaling pathways, which are not necessarily activated simultaneously. A more thorough investigation of the activation status and function of each branch of the UPR will contribute to a better understanding of the UPR in metabolic disorders.
Acknowledgements
Supported by the National Institutes of Health grants AI32412 and P01 AI56296 (L.H.G.), the Ellison Medical Foundation (L.H.G.), the American Heart Association, AHA0835610P (A.H.L.) and the Harvard University Technology Development Accelerator Fund (L.H.G.).
Contributor Information
Ann-Hwee Lee, Phone: +1-617-4324689, FAX: +1-617-4320084, Email: ahlee@hsph.harvard.edu.
Laurie H. Glimcher, Phone: +1-617-432-0622, FAX: +1-617-432-0084, Email: lglimche@hsph.harvard.edu
References
- 1.Thurman RG, Kauffman FC, Jungermann K. Regulation of hepatic metabolism: intra- and intercellular compartmentation. New York: Plenum; 1986. [Google Scholar]
- 2.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferre P, Foufelle F. SREBP-1c transcription factor and lipid homeostasis: clinical perspective. Horm Res. 2007;68:72–82. doi: 10.1159/000100426. [DOI] [PubMed] [Google Scholar]
- 4.Postic C, Dentin R, Denechaud PD, Girard J. ChREBP, a transcriptional regulator of glucose and lipid metabolism. Annu Rev Nutr. 2007;27:179–192. doi: 10.1146/annurev.nutr.27.061406.093618. [DOI] [PubMed] [Google Scholar]
- 5.Uyeda K, Repa JJ. Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab. 2006;4:107–110. doi: 10.1016/j.cmet.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 7.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 8.Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oyadomari S, Harding HP, Zhang Y, Oyadomari M, Ron D. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 2008;7:520–532. doi: 10.1016/j.cmet.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD, Kaufman RJ. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Rutkowski DT, Wu J, Back SH, Callaghan MU, Ferris SP, Iqbal J, Clark R, Miao H, Hassler JR, Fornek J, Katze MG, Hussain MM, Song B, Swathirajan J, Wang J, Yau GD, Kaufman RJ. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell. 2008;15:829–840. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji C. Dissection of endoplasmic reticulum stress signaling in alcoholic and non-alcoholic liver injury. J Gastroenterol Hepatol. 2008;23(Suppl 1):S16–S24. doi: 10.1111/j.1440-1746.2007.05276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Towle HC, Kaytor EN, Shih HM. Regulation of the expression of lipogenic enzyme genes by carbohydrate. Annu Rev Nutr. 1997;17:405–433. doi: 10.1146/annurev.nutr.17.1.405. [DOI] [PubMed] [Google Scholar]
- 14.Davidson NO, Shelness GS. Apolipoprotein B: mRNA editing, lipoprotein assembly, and presecretory degradation. Annu Rev Nutr. 2000;20:169–193. doi: 10.1146/annurev.nutr.20.1.169. [DOI] [PubMed] [Google Scholar]
- 15.Foufelle F, Ferre P. New perspectives in the regulation of hepatic glycolytic and lipogenic genes by insulin and glucose: a role for the transcription factor sterol regulatory element binding protein-1c. Biochem J. 2002;366:377–391. doi: 10.1042/BJ20020430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang G, Yang J, Horton JD, Hammer RE, Goldstein JL, Brown MS. Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J Biol Chem. 2002;277:9520–9528. doi: 10.1074/jbc.M111421200. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Sato R, Brown MS, Hua X, Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 19.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/S0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 20.Kim JB, Sarraf P, Wright M, Yao KM, Mueller E, Solanes G, Lowell BB, Spiegelman BM. Nutritional and insulin regulation of fatty acid synthetase and leptin gene expression through ADD1/SREBP1. J Clin Invest. 1998;101:1–9. doi: 10.1172/JCI1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldstein JL, Rawson RB, Brown MS. Mutant mammalian cells as tools to delineate the sterol regulatory element-binding protein pathway for feedback regulation of lipid synthesis. Arch Biochem Biophys. 2002;397:139–148. doi: 10.1006/abbi.2001.2615. [DOI] [PubMed] [Google Scholar]
- 22.Radhakrishnan A, Goldstein JL, McDonald JG, Brown MS. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab. 2008;8:512–521. doi: 10.1016/j.cmet.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeBose-Boyd RA. Feedback regulation of cholesterol synthesis: sterol-accelerated ubiquitination and degradation of HMG CoA reductase. Cell Res. 2008;18:609–621. doi: 10.1038/cr.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horton JD, Bashmakov Y, Shimomura I, Shimano H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc Natl Acad Sci USA. 1998;95:5987–5992. doi: 10.1073/pnas.95.11.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fleischmann M, Iynedjian PB. Regulation of sterol regulatory-element binding protein 1 gene expression in liver: role of insulin and protein kinase B/cAkt. Biochem J. 2000;349:13–17. doi: 10.1042/0264-6021:3490013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azzout-Marniche D, Becard D, Guichard C, Foretz M, Ferre P, Foufelle F. Insulin effects on sterol regulatory-element-binding protein-1c (SREBP-1c) transcriptional activity in rat hepatocytes. Biochem J. 2000;350:389–393. doi: 10.1042/0264-6021:3500389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, Li L, Schwendner S, Wang S, Thoolen M, Mangelsdorf DJ, Lustig KD, Shan B. Role of LXRs in control of lipogenesis. Genes Dev. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JM, Shimomura I, Shan B, Brown MS, Goldstein JL, Mangelsdorf DJ. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimano H, Horton JD, Shimomura I, Hammer RE, Brown MS, Goldstein JL. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J Clin Invest. 1997;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horton JD, Shimomura I, Brown MS, Hammer RE, Goldstein JL, Shimano H. Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J Clin Invest. 1998;101:2331–2339. doi: 10.1172/JCI2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Becard D, Hainault I, Azzout-Marniche D, Bertry-Coussot L, Ferre P, Foufelle F. Adenovirus-mediated overexpression of sterol regulatory element binding protein-1c mimics insulin effects on hepatic gene expression and glucose homeostasis in diabetic mice. Diabetes. 2001;50:2425–2430. doi: 10.2337/diabetes.50.11.2425. [DOI] [PubMed] [Google Scholar]
- 32.Yamashita H, Takenoshita M, Sakurai M, Bruick RK, Henzel WJ, Shillinglaw W, Arnot D, Uyeda K. A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc Natl Acad Sci USA. 2001;98:9116–9121. doi: 10.1073/pnas.161284298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma L, Robinson LN, Towle HC. ChREBP*Mlx is the principal mediator of glucose-induced gene expression in the liver. J Biol Chem. 2006;281:28721–28730. doi: 10.1074/jbc.M601576200. [DOI] [PubMed] [Google Scholar]
- 34.Ishii S, Iizuka K, Miller BC, Uyeda K. Carbohydrate response element binding protein directly promotes lipogenic enzyme gene transcription. Proc Natl Acad Sci USA. 2004;101:15597–15602. doi: 10.1073/pnas.0405238101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsatsos NG, Towle HC. Glucose activation of ChREBP in hepatocytes occurs via a two-step mechanism. Biochem Biophys Res Commun. 2006;340:449–456. doi: 10.1016/j.bbrc.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 36.Collier JJ, Zhang P, Pedersen KB, Burke SJ, Haycock JW, Scott DK. c-Myc and ChREBP regulate glucose-mediated expression of the L-type pyruvate kinase gene in INS-1-derived 832/13 cells. Am J Physiol Endocrinol Metab. 2007;293:E48–E56. doi: 10.1152/ajpendo.00357.2006. [DOI] [PubMed] [Google Scholar]
- 37.Davies MN, O’Callaghan BL, Towle HC. Glucose activates ChREBP by increasing its rate of nuclear entry and relieving repression of its transcriptional activity. J Biol Chem. 2008;283:24029–24038. doi: 10.1074/jbc.M801539200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iizuka K, Bruick RK, Liang G, Horton JD, Uyeda K. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc Natl Acad Sci USA. 2004;101:7281–7286. doi: 10.1073/pnas.0401516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dentin R, Benhamed F, Hainault I, Fauveau V, Foufelle F, Dyck JR, Girard J, Postic C. Liver-specific inhibition of ChREBP improves hepatic steatosis and insulin resistance in ob/ob mice. Diabetes. 2006;55:2159–2170. doi: 10.2337/db06-0200. [DOI] [PubMed] [Google Scholar]
- 40.Gaut JR, Hendershot LM. The modification and assembly of proteins in the endoplasmic reticulum. Curr Opin Cell Biol. 1993;5:589–595. doi: 10.1016/0955-0674(93)90127-C. [DOI] [PubMed] [Google Scholar]
- 41.Kleizen B, Braakman I. Protein folding and quality control in the endoplasmic reticulum. Curr Opin Cell Biol. 2004;16:343–349. doi: 10.1016/j.ceb.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 42.Rapoport TA. Transport of proteins across the endoplasmic reticulum membrane. Science. 1992;258:931–936. doi: 10.1126/science.1332192. [DOI] [PubMed] [Google Scholar]
- 43.Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 44.Nakatsukasa K, Brodsky JL. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic. 2008;9:861–870. doi: 10.1111/j.1600-0854.2008.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huijbregts RP, Topalof L, Bankaitis VA. Lipid metabolism and regulation of membrane trafficking. Traffic. 2000;1:195–202. doi: 10.1034/j.1600-0854.2000.010301.x. [DOI] [PubMed] [Google Scholar]
- 46.Voelker DR. Organelle biogenesis and intracellular lipid transport in eukaryotes. Microbiol Rev. 1991;55:543–560. doi: 10.1128/mr.55.4.543-560.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McMaster CR. Lipid metabolism and vesicle trafficking: more than just greasing the transport machinery. Biochem Cell Biol. 2001;79:681–692. doi: 10.1139/bcb-79-6-681. [DOI] [PubMed] [Google Scholar]
- 48.Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW, Gunsalus IC, Nebert DW. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- 49.Michalak M, Robert Parker JM, Opas M. Ca2+ signaling and calcium binding chaperones of the endoplasmic reticulum. Cell Calcium. 2002;32:269–278. doi: 10.1016/S0143416002001884. [DOI] [PubMed] [Google Scholar]
- 50.Todd DJ, Lee AH, Glimcher LH. The endoplasmic reticulum stress response in immunity and autoimmunity. Nat Rev Immunol. 2008;8:663–674. doi: 10.1038/nri2359. [DOI] [PubMed] [Google Scholar]
- 51.Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/S0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 52.Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/S0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 53.Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-A. [DOI] [PubMed] [Google Scholar]
- 54.Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-Q. [DOI] [PubMed] [Google Scholar]
- 55.Zhou J, Liu CY, Back SH, Clark RL, Peisach D, Xu Z, Kaufman RJ. The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc Natl Acad Sci USA. 2006;103:14343–14348. doi: 10.1073/pnas.0606480103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci USA. 2005;102:18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Welihinda AA, Kaufman RJ. The unfolded protein response pathway in Saccharomyces cerevisiae. Oligomerization and trans-phosphorylation of Ire1p (Ern1p) are required for kinase activation. J Biol Chem. 1996;271:18181–18187. doi: 10.1074/jbc.271.30.18181. [DOI] [PubMed] [Google Scholar]
- 58.Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tirasophon W, Lee K, Callaghan B, Welihinda A, Kaufman RJ. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev. 2000;14:2725–2736. doi: 10.1101/gad.839400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shamu CE, Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. Embo J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- 61.Sidrauski C, Cox JS, Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. doi: 10.1016/S0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- 62.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/S0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 63.Mori K, Kawahara T, Yoshida H, Yanagi H, Yura T. Signalling from endoplasmic reticulum to nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells. 1996;1:803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x. [DOI] [PubMed] [Google Scholar]
- 64.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/S0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 65.Shen X, Ellis RE, Lee K, Liu CY, Yang K, Solomon A, Yoshida H, Morimoto R, Kurnit DM, Mori K, Kaufman RJ. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. doi: 10.1016/S0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- 66.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 67.Clauss IM, Chu M, Zhao JL, Glimcher LH. The basic domain/leucine zipper protein hXBP-1 preferentially binds to and transactivates CRE-like sequences containing an ACGT core. Nucleic Acids Res. 1996;24:1855–1864. doi: 10.1093/nar/24.10.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, Kaufman RJ. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee AH, Qian SB, Zhao H, Yu X, Yang L, Tan BK, Rosenwald A, Hurt EM, Petroulakis E, Sonenberg N, Yewdell JW, Calame K, Glimcher LH, Staudt LM. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 71.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 72.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/S1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 74.Zhou D, Palam LR, Jiang L, Narasimhan J, Staschke KA, Wek RC. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J Biol Chem. 2008;283:7064–7073. doi: 10.1074/jbc.M708530200. [DOI] [PubMed] [Google Scholar]
- 75.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/S1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 76.Haze K, Okada T, Yoshida H, Yanagi H, Yura T, Negishi M, Mori K. Identification of the G13 (cAMP-response-element-binding protein-related protein) gene product related to activating transcription factor 6 as a transcriptional activator of the mammalian unfolded protein response. Biochem J. 2001;355:19–28. doi: 10.1042/0264-6021:3550019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–1364. doi: 10.1016/S1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 78.Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/S1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 79.Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol. 2000;20:6755–6767. doi: 10.1128/MCB.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K. Endoplasmic reticulum stress-induced formation of transcription factor complex ERSF including NF-Y (CBF) and activating transcription factors 6alpha and 6beta that activates the mammalian unfolded protein response. Mol Cell Biol. 2001;21:1239–1248. doi: 10.1128/MCB.21.4.1239-1248.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 82.Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J Biol Chem. 2000;275:27013–27020. doi: 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- 83.Kaser A, Lee A-H, Franke A, Glickman JN, Tilg H, Zeissig S, Nieuwenhuis EES, Higgins DE, Schreiber S, Glimcher LH, Blumberg RS. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. Embo J. 2005;24:4368–4380. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 86.Clauss IM, Gravallese EM, Darling JM, Shapiro F, Glimcher MJ, Glimcher LH. In situ hybridization studies suggest a role for the basic region-leucine zipper protein hXBP-1 in exocrine gland and skeletal development during mouse embryogenesis. Dev Dyn. 1993;197:146–156. doi: 10.1002/aja.1001970207. [DOI] [PubMed] [Google Scholar]
- 87.Reimold AM, Etkin A, Clauss I, Perkins A, Friend DS, Zhang J, Horton HF, Scott A, Orkin SH, Byrne MC, Grusby MJ, Glimcher LH. An essential role in liver development for transcription factor XBP-1. Genes Dev. 2000;14:152–157. [PMC free article] [PubMed] [Google Scholar]
- 88.Iwakoshi NN, Pypaert M, Glimcher LH. The transcription factor XBP-1 is essential for the development and survival of dendritic cells. J Exp Med. 2007;204:2267–2275. doi: 10.1084/jem.20070525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wek RC, Cavener DR. Translational control and the unfolded protein response. Antioxid Redox Signal. 2007;9:2357–2371. doi: 10.1089/ars.2007.1764. [DOI] [PubMed] [Google Scholar]
- 90.Shi Y, Vattem KM, Sood R, An J, Liang J, Stramm L, Wek RC. Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mol Cell Biol. 1998;18:7499–7509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Berlanga JJ, Santoyo J, De Haro C. Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2alpha kinase. Eur J Biochem. 1999;265:754–762. doi: 10.1046/j.1432-1327.1999.00780.x. [DOI] [PubMed] [Google Scholar]
- 92.Chen JJ, London IM. Regulation of protein synthesis by heme-regulated eIF-2 alpha kinase. Trends Biochem Sci. 1995;20:105–108. doi: 10.1016/S0968-0004(00)88975-6. [DOI] [PubMed] [Google Scholar]
- 93.Sood R, Porter AC, Olsen DA, Cavener DR, Wek RC. A mammalian homologue of GCN2 protein kinase important for translational control by phosphorylation of eukaryotic initiation factor-2alpha. Genetics. 2000;154:787–801. doi: 10.1093/genetics/154.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Taylor SS, Haste NM, Ghosh G. PKR and eIF2alpha: integration of kinase dimerization, activation, and substrate docking. Cell. 2005;122:823–825. doi: 10.1016/j.cell.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 95.Lu PD, Jousse C, Marciniak SJ, Zhang Y, Novoa I, Scheuner D, Kaufman RJ, Ron D, Harding HP. Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 2004;23:169–179. doi: 10.1038/sj.emboj.7600030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tu BP, Weissman JS. Oxidative protein folding in eukaryotes: mechanisms and consequences. J Cell Biol. 2004;164:341–346. doi: 10.1083/jcb.200311055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Feng B, Yao PM, Li Y, Devlin CM, Zhang D, Harding HP, Sweeney M, Rong JX, Kuriakose G, Fisher EA, Marks AR, Ron D, Tabas I. The endoplasmic reticulum is the site of cholesterol-induced cytotoxicity in macrophages. Nat Cell Biol. 2003;5:781–792. doi: 10.1038/ncb1035. [DOI] [PubMed] [Google Scholar]
- 98.Blais JD, Addison CL, Edge R, Falls T, Zhao H, Wary K, Koumenis C, Harding HP, Ron D, Holcik M, Bell JC. Perk-dependent translational regulation promotes tumor cell adaptation and angiogenesis in response to hypoxic stress. Mol Cell Biol. 2006;26:9517–9532. doi: 10.1128/MCB.01145-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N, Harding H, Novoa I, Varia M, Raleigh J, Scheuner D, Kaufman RJ, Bell J, Ron D, Wouters BG, Koumenis C. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J. 2005;24:3470–3481. doi: 10.1038/sj.emboj.7600777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ma Y, Brewer JW, Diehl JA, Hendershot LM. Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J Mol Biol. 2002;318:1351–1365. doi: 10.1016/S0022-2836(02)00234-6. [DOI] [PubMed] [Google Scholar]
- 102.McCullough KD, Martindale JL, Klotz LO, Aw TY, Holbrook NJ. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol Cell Biol. 2001;21:1249–1259. doi: 10.1128/MCB.21.4.1249-1259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot RT, Remotti H, Stevens JL, Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Oyadomari S, Koizumi A, Takeda K, Gotoh T, Akira S, Araki E, Mori M. Targeted disruption of the Chop gene delays endoplasmic reticulum stress-mediated diabetes. J Clin Invest. 2002;109:525–532. doi: 10.1172/JCI14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cullinan SB, Diehl JA. PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. J Biol Chem. 2004;279:20108–20117. doi: 10.1074/jbc.M314219200. [DOI] [PubMed] [Google Scholar]
- 106.Cullinan SB, Zhang D, Hannink M, Arvisais E, Kaufman RJ, Diehl JA. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol Cell Biol. 2003;23:7198–7209. doi: 10.1128/MCB.23.20.7198-7209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Delepine M, Nicolino M, Barrett T, Golamaully M, Lathrop GM, Julier C. EIF2AK3, encoding translation initiation factor 2-alpha kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat Genet. 2000;25:406–409. doi: 10.1038/78085. [DOI] [PubMed] [Google Scholar]
- 108.Biason-Lauber A, Lang-Muritano M, Vaccaro T, Schoenle EJ. Loss of kinase activity in a patient with Wolcott-Rallison syndrome caused by a novel mutation in the EIF2AK3 gene. Diabetes. 2002;51:2301–2305. doi: 10.2337/diabetes.51.7.2301. [DOI] [PubMed] [Google Scholar]
- 109.Brickwood S, Bonthron DT, Al-Gazali LI, Piper K, Hearn T, Wilson DI, Hanley NA. Wolcott-Rallison syndrome: pathogenic insights into neonatal diabetes from new mutation and expression studies of EIF2AK3. J Med Genet. 2003;40:685–689. doi: 10.1136/jmg.40.9.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Harding HP, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, Sabatini DD, Ron D. Diabetes mellitus and exocrine pancreatic dysfunction in perk−/− mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7:1153–1163. doi: 10.1016/S1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 111.Zhang P, McGrath B, Li S, Frank A, Zambito F, Reinert J, Gannon M, Ma K, McNaughton K, Cavener DR. The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol Cell Biol. 2002;22:3864–3874. doi: 10.1128/MCB.22.11.3864-3874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wei J, Sheng X, Feng D, McGrath B, Cavener DR. PERK is essential for neonatal skeletal development to regulate osteoblast proliferation and differentiation. J Cell Physiol. 2008;217:693–707. doi: 10.1002/jcp.21543. [DOI] [PubMed] [Google Scholar]
- 113.Iida K, Li Y, McGrath BC, Frank A, Cavener DR. PERK eIF2 alpha kinase is required to regulate the viability of the exocrine pancreas in mice. BMC Cell Biol. 2007;8:38. doi: 10.1186/1471-2121-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang W, Feng D, Li Y, Iida K, McGrath B, Cavener DR. PERK EIF2AK3 control of pancreatic beta cell differentiation and proliferation is required for postnatal glucose homeostasis. Cell Metab. 2006;4:491–497. doi: 10.1016/j.cmet.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 115.Bailey D, O’Hare P. Transmembrane bZIP transcription factors in ER stress signaling and the unfolded protein response. Antioxid Redox Signal. 2007;9:2305–2321. doi: 10.1089/ars.2007.1796. [DOI] [PubMed] [Google Scholar]
- 116.Mori K. Frame switch splicing and regulated intramembrane proteolysis: key words to understand the unfolded protein response. Traffic. 2003;4:519–528. doi: 10.1034/j.1600-0854.2003.00112.x. [DOI] [PubMed] [Google Scholar]
- 117.Shen J, Snapp EL, Lippincott-Schwartz J, Prywes R. Stable binding of ATF6 to BiP in the endoplasmic reticulum stress response. Mol Cell Biol. 2005;25:921–932. doi: 10.1128/MCB.25.3.921-932.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Adachi Y, Yamamoto K, Okada T, Yoshida H, Harada A, Mori K. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct. 2008;33:75–89. doi: 10.1247/csf.07044. [DOI] [PubMed] [Google Scholar]
- 119.Foretz M, Guichard C, Ferre P, Foufelle F. Sterol regulatory element binding protein-1c is a major mediator of insulin action on the hepatic expression of glucokinase and lipogenesis-related genes. Proc Natl Acad Sci USA. 1999;96:12737–12742. doi: 10.1073/pnas.96.22.12737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shimomura I, Bashmakov Y, Ikemoto S, Horton JD, Brown MS, Goldstein JL. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc Natl Acad Sci USA. 1999;96:13656–13661. doi: 10.1073/pnas.96.24.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shimano H, Yahagi N, Amemiya-Kudo M, Hasty AH, Osuga J, Tamura Y, Shionoiri F, Iizuka Y, Ohashi K, Harada K, Gotoda T, Ishibashi S, Yamada N. Sterol regulatory element-binding protein-1 as a key transcription factor for nutritional induction of lipogenic enzyme genes. J Biol Chem. 1999;274:35832–35839. doi: 10.1074/jbc.274.50.35832. [DOI] [PubMed] [Google Scholar]
- 122.Shimomura I, Bashmakov Y, Shimano H, Horton JD, Goldstein JL, Brown MS. Cholesterol feeding reduces nuclear forms of sterol regulatory element binding proteins in hamster liver. Proc Natl Acad Sci USA. 1997;94:12354–12359. doi: 10.1073/pnas.94.23.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol. 2004;167:35–41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sriburi R, Bommiasamy H, Buldak GL, Robbins GR, Frank M, Jackowski S, Brewer JW. Coordinate regulation of phospholipid biosynthesis and secretory pathway gene expression in XBP-1(S)-induced endoplasmic reticulum biogenesis. J Biol Chem. 2007;282:7024–7034. doi: 10.1074/jbc.M609490200. [DOI] [PubMed] [Google Scholar]
- 125.Cox JS, Chapman RE, Walter P. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol Biol Cell. 1997;8:1805–1814. doi: 10.1091/mbc.8.9.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Qiu W, Kohen-Avramoglu R, Mhapsekar S, Tsai J, Austin RC, Adeli K. Glucosamine-induced endoplasmic reticulum stress promotes ApoB100 degradation: evidence for Grp78-mediated targeting to proteasomal degradation. Arterioscler Thromb Vasc Biol. 2005;25:571–577. doi: 10.1161/01.ATV.0000154142.61859.94. [DOI] [PubMed] [Google Scholar]
- 127.Liao W, Chan L. Tunicamycin induces ubiquitination and degradation of apolipoprotein B in HepG2 cells. Biochem J. 2001;353:493–501. doi: 10.1042/0264-6021:3530493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ota T, Gayet C, Ginsberg HN. Inhibition of apolipoprotein B100 secretion by lipid-induced hepatic endoplasmic reticulum stress in rodents. J Clin Invest. 2008;118:316–332. doi: 10.1172/JCI32752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell. 2001;7:1165–1176. doi: 10.1016/S1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 130.Scheuner D, Vander Mierde D, Song B, Flamez D, Creemers JW, Tsukamoto K, Ribick M, Schuit FC, Kaufman RJ. Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat Med. 2005;11:757–764. doi: 10.1038/nm1259. [DOI] [PubMed] [Google Scholar]
- 131.Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol. 2001;153:1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schroeder-Gloeckler JM, Rahman SM, Janssen RC, Qiao L, Shao J, Roper M, Fischer SJ, Lowe E, Orlicky DJ, McManaman JL, Palmer C, Gitomer WL, Huang W, O’Doherty RM, Becker TC, Klemm DJ, Jensen DR, Pulawa LK, Eckel RH, Friedman JE. CCAAT/enhancer-binding protein beta deletion reduces adiposity, hepatic steatosis, and diabetes in Lepr(db/db) mice. J Biol Chem. 2007;282:15717–15729. doi: 10.1074/jbc.M701329200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Millward CA, Heaney JD, Sinasac DS, Chu EC, Bederman IR, Gilge DA, Previs SF, Croniger CM. Mice with a deletion in the gene for CCAAT/enhancer-binding protein beta are protected against diet-induced obesity. Diabetes. 2007;56:161–167. doi: 10.2337/db06-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chen C, Dudenhausen EE, Pan YX, Zhong C, Kilberg MS. Human CCAAT/enhancer-binding protein beta gene expression is activated by endoplasmic reticulum stress through an unfolded protein response element downstream of the protein coding sequence. J Biol Chem. 2004;279:27948–27956. doi: 10.1074/jbc.M313920200. [DOI] [PubMed] [Google Scholar]
- 135.Calkhoven CF, Muller C, Leutz A. Translational control of C/EBPalpha and C/EBPbeta isoform expression. Genes Dev. 2000;14:1920–1932. [PMC free article] [PubMed] [Google Scholar]
- 136.Bobrovnikova-Marjon E, Hatzivassiliou G, Grigoriadou C, Romero M, Cavener DR, Thompson CB, Diehl JA. PERK-dependent regulation of lipogenesis during mouse mammary gland development and adipocyte differentiation. Proc Natl Acad Sci USA. 2008;105:16314–16319. doi: 10.1073/pnas.0808517105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Colgan SM, Tang D, Werstuck GH, Austin RC. Endoplasmic reticulum stress causes the activation of sterol regulatory element binding protein-2. Int J Biochem Cell Biol. 2007;39:1843–1851. doi: 10.1016/j.biocel.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 138.Lee JN, Ye J. Proteolytic activation of sterol regulatory element-binding protein induced by cellular stress through depletion of Insig-1. J Biol Chem. 2004;279:45257–45265. doi: 10.1074/jbc.M408235200. [DOI] [PubMed] [Google Scholar]
- 139.Henry SA, Patton-Vogt JL. Genetic regulation of phospholipid metabolism: yeast as a model eukaryote. Prog Nucleic Acid Res Mol Biol. 1998;61:133–179. doi: 10.1016/S0079-6603(08)60826-0. [DOI] [PubMed] [Google Scholar]
- 140.Strahl T, Thorner J. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae . Biochim Biophys Acta. 2007;1771:353–404. doi: 10.1016/j.bbalip.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Loewen CJ, Gaspar ML, Jesch SA, Delon C, Ktistakis NT, Henry SA, Levine TP. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science. 2004;304:1644–1647. doi: 10.1126/science.1096083. [DOI] [PubMed] [Google Scholar]
- 142.Wagner C, Blank M, Strohmann B, Schuller HJ. Overproduction of the Opi1 repressor inhibits transcriptional activation of structural genes required for phospholipid biosynthesis in the yeast Saccharomyces cerevisiae. Yeast. 1999;15:843–854. doi: 10.1002/(SICI)1097-0061(199907)15:10A<843::AID-YEA424>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 143.Bachhawat N, Ouyang Q, Henry SA. Functional characterization of an inositol-sensitive upstream activation sequence in yeast. A cis-regulatory element responsible for inositol-choline mediated regulation of phospholipid biosynthesis. J Biol Chem. 1995;270:25087–25095. doi: 10.1074/jbc.270.42.25087. [DOI] [PubMed] [Google Scholar]
- 144.Jesch SA, Liu P, Zhao X, Wells MT, Henry SA. Multiple endoplasmic reticulum-to-nucleus signaling pathways coordinate phospholipid metabolism with gene expression by distinct mechanisms. J Biol Chem. 2006;281:24070–24083. doi: 10.1074/jbc.M604541200. [DOI] [PubMed] [Google Scholar]
- 145.Jesch SA, Zhao X, Wells MT, Henry SA. Genome-wide analysis reveals inositol, not choline, as the major effector of Ino2p-Ino4p and unfolded protein response target gene expression in yeast. J Biol Chem. 2005;280:9106–9118. doi: 10.1074/jbc.M411770200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Nikawa J, Yamashita S. IRE1 encodes a putative protein kinase containing a membrane-spanning domain and is required for inositol phototrophy in Saccharomyces cerevisiae . Mol Microbiol. 1992;6:1441–1446. doi: 10.1111/j.1365-2958.1992.tb00864.x. [DOI] [PubMed] [Google Scholar]
- 147.Chang HJ, Jones EW, Henry SA. Role of the unfolded protein response pathway in regulation of INO1 and in the sec14 bypass mechanism in Saccharomyces cerevisiae . Genetics. 2002;162:29–43. doi: 10.1093/genetics/162.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chang HJ, Jesch SA, Gaspar ML, Henry SA. Role of the unfolded protein response pathway in secretory stress and regulation of INO1 expression in Saccharomyces cerevisiae . Genetics. 2004;168:1899–1913. doi: 10.1534/genetics.104.032961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gregor MF, Yang L, Fabbrini E, Mohammed BS, Eagon JC, Hotamisligil GS, Klein S. Endoplasmic reticulum stress is reduced in tissues of obese subjects after weight loss. Diabetes. 2009;58:693–700. doi: 10.2337/db08-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang D, Wei Y, Pagliassotti MJ. Saturated fatty acids promote endoplasmic reticulum stress and liver injury in rats with hepatic steatosis. Endocrinology. 2006;147:943–951. doi: 10.1210/en.2005-0570. [DOI] [PubMed] [Google Scholar]
- 151.Nakatani Y, Kaneto H, Kawamori D, Yoshiuchi K, Hatazaki M, Matsuoka TA, Ozawa K, Ogawa S, Hori M, Yamasaki Y, Matsuhisa M. Involvement of endoplasmic reticulum stress in insulin resistance and diabetes. J Biol Chem. 2005;280:847–851. doi: 10.1074/jbc.M411860200. [DOI] [PubMed] [Google Scholar]
- 152.Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Gorgun C, Glimcher LH, Hotamisligil GS. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 153.Laybutt DR, Preston AM, Akerfeldt MC, Kench JG, Busch AK, Biankin AV, Biden TJ. Endoplasmic reticulum stress contributes to beta cell apoptosis in type 2 diabetes. Diabetologia. 2007;50:752–763. doi: 10.1007/s00125-006-0590-z. [DOI] [PubMed] [Google Scholar]
- 154.Zhang X, Zhang G, Zhang H, Karin M, Bai H, Cai D. Hypothalamic IKKbeta/NF-kappaB and ER stress link overnutrition to energy imbalance and obesity. Cell. 2008;135:61–73. doi: 10.1016/j.cell.2008.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hosoi T, Sasaki M, Miyahara T, Hashimoto C, Matsuo S, Yoshii M, Ozawa K. Endoplasmic reticulum stress induces leptin resistance. Mol Pharmacol. 2008;74:1610–1619. doi: 10.1124/mol.108.050070. [DOI] [PubMed] [Google Scholar]
- 156.Hirosumi J, Tuncman G, Chang L, Gorgun CZ, Uysal KT, Maeda K, Karin M, Hotamisligil GS. A central role for JNK in obesity and insulin resistance. Nature. 2002;420:333–336. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- 157.Loffler M, Bilban M, Reimers M, Waldhausl W, Stulnig TM. Blood glucose-lowering nuclear receptor agonists only partially normalize hepatic gene expression in db/db mice. J Pharmacol Exp Ther. 2006;316:797–804. doi: 10.1124/jpet.105.093831. [DOI] [PubMed] [Google Scholar]
- 158.Wei Y, Wang D, Topczewski F, Pagliassotti MJ. Saturated fatty acids induce endoplasmic reticulum stress and apoptosis independently of ceramide in liver cells. Am J Physiol Endocrinol Metab. 2006;291:E275–E281. doi: 10.1152/ajpendo.00644.2005. [DOI] [PubMed] [Google Scholar]
- 159.Kharroubi I, Ladriere L, Cardozo AK, Dogusan Z, Cnop M, Eizirik DL. Free fatty acids and cytokines induce pancreatic beta-cell apoptosis by different mechanisms: role of nuclear factor-kappaB and endoplasmic reticulum stress. Endocrinology. 2004;145:5087–5096. doi: 10.1210/en.2004-0478. [DOI] [PubMed] [Google Scholar]
- 160.Karaskov E, Scott C, Zhang L, Teodoro T, Ravazzola M, Volchuk A. Chronic palmitate but not oleate exposure induces endoplasmic reticulum stress, which may contribute to INS-1 pancreatic beta-cell apoptosis. Endocrinology. 2006;147:3398–3407. doi: 10.1210/en.2005-1494. [DOI] [PubMed] [Google Scholar]
- 161.Cnop M, Ladriere L, Hekerman P, Ortis F, Cardozo AK, Dogusan Z, Flamez D, Boyce M, Yuan J, Eizirik DL. Selective inhibition of eukaryotic translation initiation factor 2 alpha dephosphorylation potentiates fatty acid-induced endoplasmic reticulum stress and causes pancreatic beta-cell dysfunction and apoptosis. J Biol Chem. 2007;282:3989–3997. doi: 10.1074/jbc.M607627200. [DOI] [PubMed] [Google Scholar]
- 162.Monetti M, Levin MC, Watt MJ, Sajan MP, Marmor S, Hubbard BK, Stevens RD, Bain JR, Newgard CB, Farese RV, Sr, Hevener AL, Farese RV., Jr Dissociation of hepatic steatosis and insulin resistance in mice overexpressing DGAT in the liver. Cell Metab. 2007;6:69–78. doi: 10.1016/j.cmet.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 163.Liao W, Hui TY, Young SG, Davis RA. Blocking microsomal triglyceride transfer protein interferes with apoB secretion without causing retention or stress in the ER. J Lipid Res. 2003;44:978–985. doi: 10.1194/jlr.M300020-JLR200. [DOI] [PubMed] [Google Scholar]
- 164.Benali-Furet NL, Chami M, Houel L, De Giorgi F, Vernejoul F, Lagorce D, Buscail L, Bartenschlager R, Ichas F, Rizzuto R, Paterlini-Brechot P. Hepatitis C virus core triggers apoptosis in liver cells by inducing ER stress and ER calcium depletion. Oncogene. 2005;24:4921–4933. doi: 10.1038/sj.onc.1208673. [DOI] [PubMed] [Google Scholar]
- 165.Christen V, Treves S, Duong FH, Heim MH. Activation of endoplasmic reticulum stress response by hepatitis viruses up-regulates protein phosphatase 2A. Hepatology. 2007;46:558–565. doi: 10.1002/hep.21611. [DOI] [PubMed] [Google Scholar]
- 166.Ciccaglione AR, Marcantonio C, Tritarelli E, Equestre M, Vendittelli F, Costantino A, Geraci A, Rapicetta M. Activation of the ER stress gene gadd153 by hepatitis C virus sensitizes cells to oxidant injury. Virus Res. 2007;126:128–138. doi: 10.1016/j.virusres.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 167.Sekine-Osajima Y, Sakamoto N, Mishima K, Nakagawa M, Itsui Y, Tasaka M, Nishimura-Sakurai Y, Chen CH, Kanai T, Tsuchiya K, Wakita T, Enomoto N, Watanabe M. Development of plaque assays for hepatitis C virus-JFH1 strain and isolation of mutants with enhanced cytopathogenicity and replication capacity. Virology. 2008;371:71–85. doi: 10.1016/j.virol.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 168.Vandermeeren AM, Gomez CE, Patino C, Domingo-Gil E, Guerra S, Gonzalez JM, Esteban M. Subcellular forms and biochemical events triggered in human cells by HCV polyprotein expression from a viral vector. Virol J. 2008;5:102. doi: 10.1186/1743-422X-5-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tardif KD, Mori K, Kaufman RJ, Siddiqui A. Hepatitis C virus suppresses the IRE1-XBP1 pathway of the unfolded protein response. J Biol Chem. 2004;279:17158–17164. doi: 10.1074/jbc.M312144200. [DOI] [PubMed] [Google Scholar]
- 170.Deng L, Adachi T, Kitayama K, Bungyoku Y, Kitazawa S, Ishido S, Shoji I, Hotta H. Hepatitis C virus infection induces apoptosis through a Bax-triggered, mitochondrion-mediated, caspase 3-dependent pathway. J Virol. 2008;82:10375–10385. doi: 10.1128/JVI.00395-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yu CY, Hsu YW, Liao CL, Lin YL. Flavivirus infection activates the XBP1 pathway of the unfolded protein response to cope with endoplasmic reticulum stress. J Virol. 2006;80:11868–11880. doi: 10.1128/JVI.00879-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Xu Z, Yen TS. Intracellular retention of surface protein by a hepatitis B virus mutant that releases virion particles. J Virol. 1996;70:133–140. doi: 10.1128/jvi.70.1.133-140.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang HC, Wu HC, Chen CF, Fausto N, Lei HY, Su IJ. Different types of ground glass hepatocytes in chronic hepatitis B virus infection contain specific pre-S mutants that may induce endoplasmic reticulum stress. Am J Pathol. 2003;163:2441–2449. doi: 10.1016/S0002-9440(10)63599-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Li B, Gao B, Ye L, Han X, Wang W, Kong L, Fang X, Zeng Y, Zheng H, Li S, Wu Z. Hepatitis B virus X protein (HBx) activates ATF6 and IRE1-XBP1 pathways of unfolded protein response. Virus Res. 2007;124:44–49. doi: 10.1016/j.virusres.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 175.Ji C, Kaplowitz N. Betaine decreases hyperhomocysteinemia, endoplasmic reticulum stress, and liver injury in alcohol-fed mice. Gastroenterology. 2003;124:1488–1499. doi: 10.1016/S0016-5085(03)00276-2. [DOI] [PubMed] [Google Scholar]
- 176.Ji C, Chan C, Kaplowitz N. Predominant role of sterol response element binding proteins (SREBP) lipogenic pathways in hepatic steatosis in the murine intragastric ethanol feeding model. J Hepatol. 2006;45:717–724. doi: 10.1016/j.jhep.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 177.Kaplowitz N, Ji C. Unfolding new mechanisms of alcoholic liver disease in the endoplasmic reticulum. J Gastroenterol Hepatol. 2006;21(Suppl 3):S7–S9. doi: 10.1111/j.1440-1746.2006.04581.x. [DOI] [PubMed] [Google Scholar]
- 178.Blasco C, Caballeria J, Deulofeu R, Lligona A, Pares A, Lluis JM, Gual A, Rodes J. Prevalence and mechanisms of hyperhomocysteinemia in chronic alcoholics. Alcohol Clin Exp Res. 2005;29:1044–1048. doi: 10.1097/01.ALC.0000169265.36440.EE. [DOI] [PubMed] [Google Scholar]
- 179.Werstuck GH, Lentz SR, Dayal S, Hossain GS, Sood SK, Shi YY, Zhou J, Maeda N, Krisans SK, Malinow MR, Austin RC. Homocysteine-induced endoplasmic reticulum stress causes dysregulation of the cholesterol and triglyceride biosynthetic pathways. J Clin Invest. 2001;107:1263–1273. doi: 10.1172/JCI11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Merksamer PI, Trusina A, Papa FR. Real-time redox measurements during endoplasmic reticulum stress reveal interlinked protein folding functions. Cell. 2008;135:933–947. doi: 10.1016/j.cell.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ. The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest. 2005;115:268–281. doi: 10.1172/JCI21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bertolotti A, Wang X, Novoa I, Jungreis R, Schlessinger K, Cho JH, West AB, Ron D. Increased sensitivity to dextran sodium sulfate colitis in IRE1beta-deficient mice. J Clin Invest. 2001;107:585–593. doi: 10.1172/JCI11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tanaka T, Tsujimura T, Takeda K, Sugihara A, Maekawa A, Terada N, Yoshida N, Akira S. Targeted disruption of ATF4 discloses its essential role in the formation of eye lens fibres. Genes Cells. 1998;3:801–810. doi: 10.1046/j.1365-2443.1998.00230.x. [DOI] [PubMed] [Google Scholar]
- 184.Hettmann T, Barton K, Leiden JM. Microphthalmia due to p53-mediated apoptosis of anterior lens epithelial cells in mice lacking the CREB-2 transcription factor. Dev Biol. 2000;222:110–123. doi: 10.1006/dbio.2000.9699. [DOI] [PubMed] [Google Scholar]
- 185.Gaspar ML, Aregullin MA, Jesch SA, Nunez LR, Villa-Garcia M, Henry SA. The emergence of yeast lipidomics. Biochim Biophys Acta. 2007;1771:241–254. doi: 10.1016/j.bbalip.2006.06.011. [DOI] [PubMed] [Google Scholar]