Abstract
Signal transducer and activator of transcription 5 (STAT5) is activated by numerous cytokines that control blood cell development. STAT5 was also shown to actively participate in leukemogenesis. Among the target genes involved in cell growth, STAT5 had been shown to activate cyclin D1 gene expression. We now show that thrombopoietin-dependent activation of the cyclin D1 promoter depends on the integrity of a new bipartite proximal element that specifically binds STAT5A and -B transcription factors. We demonstrate that the stable recruitment of STAT5 to this element in vitro requires the integrity of an adjacent octamer element that constitutively binds the ubiquitous POU homeodomain protein Oct-1. We observe that cytokine-activated STAT5 and Oct-1 form a unique complex with the cyclin D1 promoter sequence. We find that STAT5 interacts with Oct-1 in vivo, following activation by different cytokines in various cellular contexts. This interaction involves a small motif in the carboxy-terminal region of STAT5 which, remarkably, is similar to an Oct-1 POU-interacting motif present in two well-known partners of Oct-1, namely, OBF-1/Bob and SNAP190. Our data offer new insights into the transcriptional regulation of the key cell cycle regulator cyclin D1 and emphasize the active roles of both STAT5 and Oct-1 in this process.
The signal transducers and activators of transcription (STATs) are latent cytoplasmic transcription factors that were discovered as mediators of cellular response to interferons and cytokines. Following ligand-receptor binding, STATs are rapidly activated by tyrosine phosphorylation, resulting in dimerization via the SH2 domain and translocation to the nucleus. Nuclear STATs regulate the transcription of target genes by binding to a class of palindromic sequences, the cytokine response elements designated gamma interferon activation sequences (GAS) from the prototype sequence found in the promoters of gamma interferon-responsive genes (6, 25). STAT signaling has been implicated in the control of multiple cellular responses to diverse cytokines and growth factors, including cell proliferation, differentiation, and apoptosis. In addition, constitutively activated forms of STAT3 and STAT5 have been observed in a number of tumor-derived cell lines and samples from human cancers and were shown to mediate cell transformation in vivo, consistent with a role of these STATs in oncogenesis (3).
Various cytokines that are responsible for the growth or survival of hematopoietic cells from different lineages activate a particular STAT factor, STAT5. STAT5 activity is associated with two chromosomally colocalized genes that encode proteins that are 95% identical, STAT5A and STAT5B. A potential role of STAT5 in growth regulation has been initially suggested based on the ability of dominant-negative forms to partially reduce cytokine-induced proliferation (32, 36) or on the ability of STAT5 to rescue proliferation-defective mutants of cytokine receptors (27). Mice deficient in both STAT5A and STAT5B genes were first found to exhibit only subtle alterations in peripheral myelopoiesis and erythropoiesis (55). Nevertheless, marked fetal anemia, as well as defects in peripheral T-cell proliferation in vivo, in response to T-cell receptor engagement and to interleukin 2 (IL-2) or IL-4 were subsequently reported. In addition, defects in the growth and survival of bone marrow-derived myeloid precursors and macrophages and in erythropoietin (EPO)-dependent production and survival of fetal liver hematopoietic colonies in vitro were also observed (12, 23, 35, 51). STAT5 was further demonstrated to promote multilineage hematolymphoid development, proliferation, and repopulating potential in vivo through effects on early hematopoietic progenitor cells (4, 50, 55, 61). All these observations indicate that STAT5 promotes cytokine-dependent survival and proliferation of hematopoietic progenitors in situations in which rapid expansion and mobilization of progenitor cells are needed. Studies of primary cells from STAT5 knockout mice and of hematopoietic cell lines identified a limited number of direct STAT5 target genes that regulate cell growth. Among these are G1 cyclins (29, 31, 35), the cell cycle inhibitor p21Waf1 (30), and the antiapoptotic protein bclXL (10, 23, 51).
Thrombopoietin (TPO) is the primary physiological regulator of platelet production and megakaryocytopoiesis. TPO also acts during early hematopoiesis, regulating hematopoietic stem cell production and function (21, 22). TPO exerts its function through binding and activation of the TPO receptor (TPO-R), also called c-mpl, a member of the cytokine receptor superfamily. Activation of TPO-R by TPO leads to the activation of Janus kinases (JAK) and the tyrosine phosphorylation of receptor sites and substrates recruited to the receptor complex, including Shc, MAPK, and STAT1, STAT3, and STAT5 (21). TPO has been shown to favor megakaryocytic development of two human multipotent growth factor-dependent leukemia-derived cell lines, UT7-mpl and F36P-mpl (32, 40). TPO-R expression followed by TPO stimulation sustains the proliferation and survival of these cell lines. In addition, TPO induces morphological differentiation into megakaryocytes. In both models, a prolonged activation of Ras was shown to be required for TPO-induced megakaryocytic differentiation, whereas both the STAT5 and Ras pathways were involved in TPO-induced proliferation (32, 43). However, the identities of the genes that are targets of the TPO-activated signaling pathways are largely unknown. In the present study, we used UT7-mpl cells and searched for genes whose expression was immediately modified in the presence of TPO. We found that TPO induced a strong expression of cyclin D1, and we deciphered the mechanism involved in this expression. We provide evidence that STAT5 participates in cyclin D1 promoter activation through a new regulatory process involving its direct interaction with the POU homeoprotein Oct-1.
MATERIALS AND METHODS
Reagents.
Recombinant human pegylated-megakaryocyte growth and differentiation factor was a kind gift of Kirin (Tokyo, Japan). The TPO mimetic peptide GW395058 (8) was synthesized by Genosys Biotechnologies Ltd. EPO was from Boehringer-Mannheim (Mannheim, Germany). Ovine prolactin (PRL) (NIDDK6-PRL19; 31 IU/mg) was a gift from the National Hormone and Pituitary program (Baltimore, Md.). Specific antibodies to STAT5A or STAT5B were a kind gift from L. Hennighausen (Bethesda, Md.). Anti-cyclin E was kindly provided by V. Baldin (Toulouse, France). Antibodies against STAT5A and -B, Oct-1, cyclin D1, and p21Waf were from Santa Cruz Biotechnology, Inc. Anti-STAT1 antibodies were from Transduction Laboratories. pXM-mSTAT5A was kindly provided by L. Henninghausen. The plasmids pXM-HA-STAT5, encoding hemagglutinin-tagged STAT5A; pGEX, containing the N-terminal region of ovine STAT5A (residues 6 to 131); and pcDNA1, encoding the murine PRL-R, were kindly provided by F. Gouilleux (Paris, France) and were described previously (34). The C-terminal regions of murine STAT5A corresponding to amino acids 545 to 793 (S5-Ct) and amino-acids 545 to 750 (S5-ΔCt) were subcloned in the pGEX 4T3 plasmid (Pharmacia) using the XhoI and NotI sites from the pXM-mSTAT5A plasmid. PGEX STAT5Am767 was prepared by cloning a PCR fragment of STAT5A (545 to 793) containing the appropriate Leu767-to-Pro mutation. A pcDNA3-derived construct encoding human Oct-1 and the pGEX plasmid encoding the Oct-1 POU domain were kindly provided by P. Matthias (Basel, Switzerland) and S. Cereghini (Paris, France), respectively. The −674 D1 luciferase reporter Psp72 D1 luc and the mutated versions Psp72 GAS1m luc (which contained two mutations in GAS1), Psp72 GAS2m (which included three point mutations [underlined] that disrupted both the Oct and the GAS2 sites: 5′-ATTGGCATGTCTATGTA-3′), and Psp72 GAS1-2 (which included mutated GAS1, Oct, and GAS2 elements) were provided by I. Matsumura (Osaka, Japan) and were described previously (31). The mutated cyclin D1 promoter-reporter constructs that included individual mutations in Oct or GAS2 sequences were obtained by exchanging appropriate PCR fragments from the wild-type constructs. The −944 D1-Luc reporter was obtained from R. Muller (Marburg, Germany) (17) and subcloned into the pGL2 basic vector. The −546 D1-Luc construct was derived from the −944 D1-Luc construct by deleting a PfmI-SacI internal fragment.
Cell culture.
Human 293T cells were grown in alpha minimum essential medium supplemented with 10% fetal calf serum. The human UT7-mpl cells were maintained in alpha minimum essential medium supplemented with 10% fetal calf serum and 2 U of EPO/ml as described previously (40). UT7-mpl cells were treated for the appropriate time with 100 ng of recombinant human pegylated-megakaryocyte growth and differentiation factor per ml, or 10 nM TPO mimetic peptide GW395058 (8), a concentration equivalent to 100 ng of the recombinant cytokine per ml, for both proliferation and differentiation of UT7-mpl cells (14) and of megakaryocyte progenitors derived from normal CD34+ human cells (S. Fichelson and E. Cramer, unpublished data).
Northern blotting.
Total RNA was prepared using TRIzol reagent (Gibco). Ten micrograms of RNA was separated in a 1% agarose gel containing formaldehyde and transferred to Hybond-N membranes (Amersham Inc.). The blots were hybridized with 32P-labeled probe corresponding to the human cyclin D1.
Cell transfection.
Transient transfections of UT7-mpl cells were performed in triplicate with 5 × 106 cells per electroporation using 20 μg of luciferase reporter plasmid and 1 μg of pRSVGal as a reference plasmid, as described previously (9). Following electroporation, the cells were resuspended in proliferative medium and split between two flasks, one stimulated with TPO (10 nM) and the other left untreated. The cells were maintained in culture for 15 h. Luciferase assays (Promega) were performed according to the manufacturer's instructions and normalized to β-galactosidase activities (expressed as relative luciferase units).
Transient transfections of 293T cells were performed with pcDNA1-PRL-R (1 μg) and pXM-oSTAT5A (1 μg) using the Fugene method (Roche) as described by the manufacturer. The total amount of plasmid transfected was adjusted to 5 μg by adding empty pcDNA vector. After 48 h, the cells were stimulated with PRL (1 μg/ml) for 30 min or left untreated, and nuclear cell extracts were prepared.
Immunoprecipitation, pull-down assay, and immunoblotting.
Total cell extracts were prepared by lysing the cells for 30 min at 4°C in N250 buffer (0.1% NP-40, 250 mM NaCl, 50 mM HEPES, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, 1 mM EDTA, 1 mM dithiothreitol, 10 μg of leupeptin/ml, 10 μg of aprotinin/ml, 1 μg of pepstatin/ml) and subsequent centrifugation at 20,000 × g for 10 min. Nuclear extracts were prepared (38), diluted in Triton buffer (20 mM Tris, pH 7.4, 137 mM NaCl, 2 mM EDTA, 1% Triton X-100, 10% glycerol supplemented with a cocktail of protease inhibitors and 1 mM vanadate), and incubated overnight at 4°C with 1 μg of antibodies. Immune complexes were pulled down with protein G-Sepharose beads, extensively washed, and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Glutathione S-transferase (GST) fusion proteins were purified from bacteria by using glutathione-Sepharose beads (Pharmacia) as instructed by the manufacturer. Nuclear extracts from 293T cells or UT7-mpl cells (250 to 500 μg of protein) were diluted in Triton buffer and incubated with 25 μg of GST fusion protein precoupled to gluthatione-Sepharose beads. Alternatively, total cell extracts from UT7-mpl cells were preincubated for 30 min at 4°C in the presence or absence of 50 μg of ethidium bromide/ml. The incubation with immobilized GST fusion protein was then allowed to proceed in the presence or absence of ethidium bromide. After incubation for 4 h at 4°C, the beads were extensively washed and analyzed by SDS-PAGE. Western blot analyses were carried out with peroxidase-conjugated anti-mouse or anti-rabbit antibodies (Amersham Pharmacia Biotech) using chemiluminescence, as instructed by the manufacturer.
Oligonucleotide pull-down assays.
Nuclear cell extracts (250 to 500 μg of protein) were incubated with 1 μg of biotin-labeled oligonucleotides in binding buffer (10 mM HEPES, 100 μM EDTA, 50 mM NaCl, 50 mM KCl, 5 mM MgCl2, 4 mM spermidine, 2 mM dithiothreitol, 0.1 mg of bovine serum albumin/ml, 2.5% glycerol, and 4% Ficoll) for 1 h at 4°C. Streptavidin-agarose beads were then added for 2 h at 4°C, washed, and resuspended in SDS-PAGE loading buffer.
EMSA.
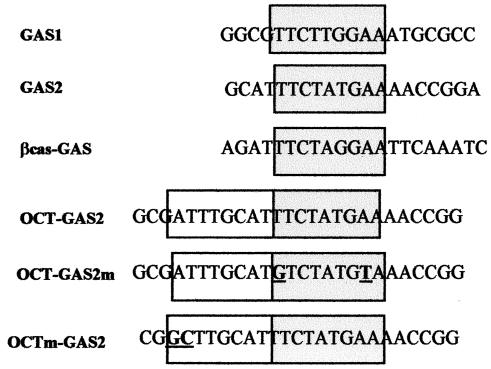
Nuclear extracts (10 μg) were incubated for 30 min with oligonucleotide probes, end labeled with [γ-32P]ATP, and loaded on a 6% nondenaturating polyacrylamide gel as described previously (38). When specified, the reaction proceeded in the presence of 0.2 (limiting amounts) to 2 (excess amounts) μg of the indicated antibodies. The probes used in electrophoresis mobility shift assays (EMSA) are listed in Fig. 1.
FIG. 1.
Sequences of oligonucleotides used in EMSA. Consensus octamer and GAS motifs are boxed, with the GAS motifs shaded.
RESULTS
TPO increases the expression of cyclin D1.
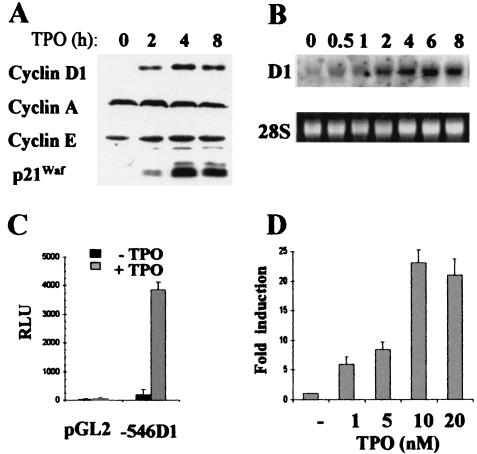
Parental UT-7 cells, as well as UT-7-mpl cells expressing exogenous TPO-R, require the presence of growth factors, such as EPO or granulocyte-macrophage colony-stimulating factor, for their proliferation and survival (24, 40). Upon the addition of TPO, UT-7-mpl cells undergo megakaryocytic differentiation in the absence or presence of the growth factor EPO or granulocyte-macrophage colony-stimulating factor (reference 40 and our unpublished data). The expression patterns of various cell cycle regulators were studied by Western blotting. As shown in Fig. 2A, the expression of cyclin D1 and p21Waf1 strongly increased upon the addition of TPO, while the levels of cyclin E, cyclin A (Fig. 2A), and cdk2, cdk4, and cdk6 (not shown) were unaffected. To delineate the mechanism of cyclin D1 induction, total RNAs were isolated from TPO-treated and untreated cells, and Northern analysis was performed. As shown in Fig. 2B, an early and sustained rise in the levels of cyclin D1 mRNA was observed upon TPO treatment. To study the involvement of the cyclin D1 promoter in this cyclin D1 gene activation, a cyclin D1 promoter fragment spanning positions −546 to +141 was cloned. This construct was transfected in UT7-mpl cells, and promoter activities were assayed in TPO-treated or untreated cells. TPO strongly increased the promoter activity (Fig. 2C) in a time-dependent (not shown) and concentration-dependent (Fig. 2D) manner. A >20-fold increase in cyclin D1 promoter activity was induced at 10 nM TPO. Similar results were obtained with longer promoter fragments spanning positions −944 to +141 (not shown) or positions −674 to +133 (see Fig. 7). These results indicated that TPO enhanced the expression of cyclin D1 by increasing the activity of the cyclin D1 gene promoter.
FIG. 2.
TPO induces the expression of cyclin D1 in UT7-mpl cells. (A) UT7-mpl cells were stimulated with 10 nM TPO. At the indicated times, total cell extracts were prepared and analyzed by SDS-PAGE and Western blotting using the antibodies indicated on the left. (B) RNA was extracted from TPO-treated UT7-mpl cells and analyzed by Northern blotting with a probe from human cyclin D1. 28S, ethidium bromide staining of 28S RNA prior to transfer. (C and D) Transient transfections were performed in UT7-mpl cells with cyclin D1 luciferase reporter construct or the empty pGL2 vector. TPO was added at 10 nM (C) or at the indicated concentrations (D). Fifteen hours after transfection, the cells were harvested for luciferase and β-galactosidase assays. Luciferase activities were expressed relative to β-galactosidase activities (in relative luciferase units [RLU]). The results shown are the means ± standard variations of six experiments. In panel D, the activity of the −546 D1 reporter construct in the presence of TPO was expressed relative to the activity of the construct in the absence of TPO, arbitrarily taken as 1 (i.e., n-fold induction).
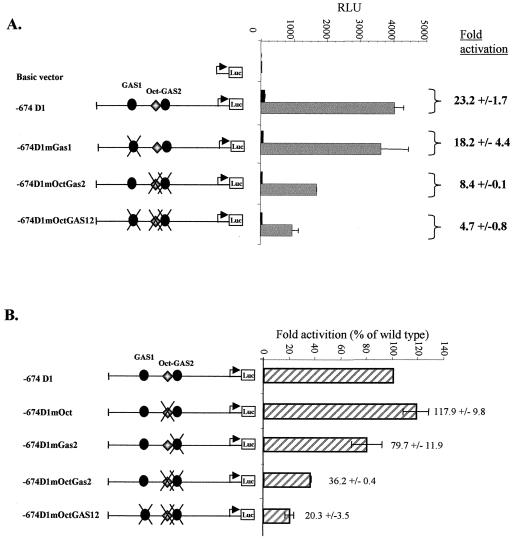
FIG. 7.
Activation of the cyclin D1 promoter by TPO requires the Oct-GAS2 element. Transient transfections were performed in UT7-mpl cells with the −674 D1-luciferase (Luc) reporter or with the same reporter construct containing point mutations in the GAS1 (mGAS1), GAS2 (mGAS2), Oct (mOct), Oct and GAS2 (mOctGAS2), or GAS1, Oct, and GAS2 (mOctGAS12) site. Following transfection, the cells were either treated with TPO (10 nM; shaded bars) or left untreated (solid bars) for 15 h and harvested for luciferase and β-galactosidase assays. (A) The results shown are means ± standard deviations of five experiments and are expressed as relative luciferase units (RLU). Alternatively, activation (n-fold) of the promoter was determined by dividing the relative luciferase activity of cells treated with TPO by the relative luciferase activity of cells transfected with the same construct and left untreated (right). (B) Activation (n-fold) of the various promoter constructs by TPO was expressed relative to the activation of the wild-type −674 promoter.
STAT5 binds to a composite Oct-GAS element.
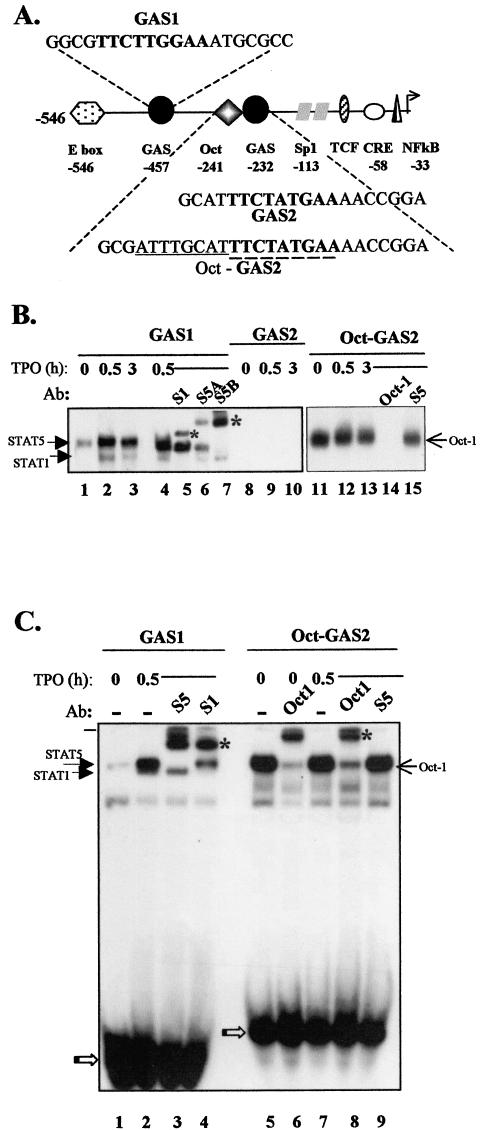
The cyclin D1 promoter contains multiple regulatory elements that may be involved in the transcriptional regulation of the gene (Fig. 3A) (17). Among these are two GAS sites that are potential targets of STAT transcription factors. As TPO can strongly activate STAT1, STAT3, and STAT5 transcription factors in various cellular contexts, including UT7-mpl cells, we tested whether the two cyclin D1-GAS sites could bind TPO-activated STAT factors. Nuclear extracts were prepared from proliferating cells that were incubated in the presence of TPO for various periods. EMSA were performed using the −457 GAS or the −232 GAS as probes, hereafter designated GAS1 and GAS2, respectively (the sequences are shown in Fig. 1). As shown in Fig. 3B, two complexes bound GAS1 in the presence of TPO (lanes 2 and 3). The lower-mobility complex was totally supershifted by anti-STAT5B antibodies, while it was only partially shifted by anti-STAT5A antibodies, suggesting that the complex contained both STAT5B homodimers and STAT5A-STAT5B heterodimers (lanes 6 and 7). The higher-mobility complex was totally supershifted by anti-STAT1 antibodies (lane 5) but not by any of the other anti-STAT antibodies (not shown), suggesting that it contained STAT1 homodimers. A weak complex was also detectable when nuclear extracts from non-TPO-treated cells were used (lane 1). This complex was shifted by anti-STAT5B antibodies and was absent in nuclear extracts prepared from cells made quiescent by growth factor deprivation (not shown). It reflected the continuous activation of STAT5B in proliferating UT7-mpl cells before TPO treatment. EMSA were next performed with the GAS2 probe (Fig. 1). Although the GAS2 probe includes a consensus sequence for STAT binding (45, 52) identical to the critical IL-2-dependent STAT5 element present in the enhancer of the immunoglobulin J chain gene (19), no STAT binding to GAS2 was observed (lanes 8 to 10). Together, these results suggested that TPO-activated STATs bound to GAS1 but not to GAS2.
FIG. 3.
TPO-activated STAT5 and STAT1 bound to cyclin D1 GAS1 but not to GAS2. (A) Schematic view of the transcription factor binding sites present on the human (−546 to +1) cyclin D1 promoter. The positions are numbered relative to the transcription start site. The sequences around the two GAS are shown. Consensus nucleotides for a GAS are shown in boldface, and the canonical octamer motif is underlined. (B and C) Cells were treated with TPO (10 nM) for the indicated times, and nuclear extracts were assayed by EMSA using GAS1, GAS2, or Oct-GAS2 as a probe. Where indicated, antibodies (Ab) against STAT1 (S1), STAT5A (S5A), STAT5B (S5B), STAT5A and -B (S5), or Oct-1 were added to the binding reaction. The asterisks indicate supershifted complexes. The open arrows point to free probes.
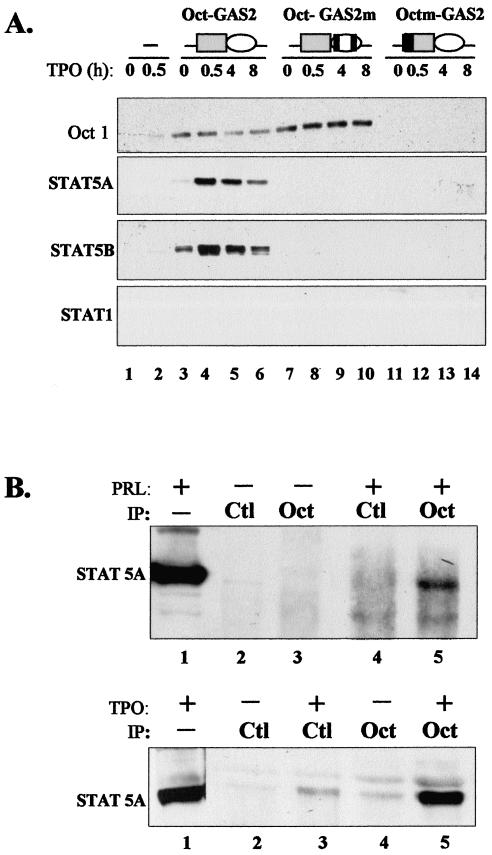
Immediately adjacent (5′) to GAS2 lies a canonical Oct sequence, 5′-ATTTGCAT-3′ (Fig. 3A). We studied possible synergy between the GAS2 and Oct sequences. The octamer motif is recognized by a family of transcription factors containing a conserved DNA binding domain with a unique homeobox-related domain referred to as the POU domain (54). POU domain transcription factors are believed to function as developmental regulators and are known to exert both positive and negative control on transcription (18, 59). Their expression is often tissue specific, but one of the first POU factors described, Oct-1, is ubiquitous and strongly binds the canonical Oct motif. This factor was strongly expressed in UT7-mpl cells, while the highly related Oct-2 factor was undetectable (not shown). We thus extended the GAS2 probe to include the entire Oct motif (referred to as the Oct-GAS2 probe [Fig. 1]) and performed EMSA. As shown in Fig. 3B, extracts from UT7-mpl cells formed a single complex with the Oct-GAS2 probe that was observed irrespective of TPO treatment (lanes 11 to 13). This complex exhibited the same electrophoretic mobility as the STAT5-bound GAS1 complex (Fig. 3C, compare lanes 2 and 5). We therefore performed supershift assays to identify the factors included in this complex. Neither anti-STAT5A and -B antibodies (Fig. 3B, lane 15, and C, lane 9) nor any other anti-STAT antibodies (not shown) affected the Oct-GAS2-bound complex, using either TPO-treated (Fig. 3C lane 9) or untreated (not shown) nuclear extracts in the assays. We also studied the presence of Oct-1 in complex. In the presence of excess anti-Oct-1 antibodies, the previously observed Oct-GAS2 complex totally disappeared (Fig. 3B, lane 14), but no supershifted complex formed (not shown). In the presence of limiting amounts of anti-Oct-1 antibodies, the Oct-GAS2 complexes observed in the absence or presence of TPO treatment were both supershifted (Fig. 3C, lanes 6 and 8). This indicated that Oct-1 was present in the Oct-GAS2 complex, irrespective of TPO treatment.
Oct-1 was highly abundant in UT7-mpl cells; moreover, it exhibited both the same apparent molecular weight and the same isoelectric point as STAT5. The Oct binding site was stuck on GAS2. For these reasons, the binding of Oct-1 to its site may have masked—or overshadowed—STAT binding to the neighboring GAS sequence. In addition, electrophoresis during EMSA may always affect protein conformation and/or impair the binding of some protein complexes to DNA. We thus wished to rule out the possibility that both Oct and STAT factors simultaneously bound to the Oct-GAS2 probe. We incubated biotinylated wild-type Oct-GAS2 oligonucleotides with TPO-treated or untreated cell extracts and then immobilized the biotinylated oligonucleotides on streptavidin-agarose beads. Streptavidin-agarose-bound complexes were then analyzed by conventional denaturating PAGE and subsequent immunoblotting. As shown in Fig. 4A, both Oct-1 and nuclear STAT5A and STAT5B specifically bound to the Oct-GAS2 probe (three upper gels, lanes 3 to 6), while STAT1 did not (bottom gel), indicating that Oct-GAS2 was a functional and restrictive STAT5 binding site. Biotinylated Oct-GAS2 oligonucleotides containing point mutations in Oct or STAT binding sites were next analyzed. Two mutations were generated in the GAS element that changed a single nucleotide within both arms of the palindromic sequence to avoid any remaining functional hemipalindrome. Such mutation totally abolished the binding of STATs to various GAS and, in particular, to cyclin D1-GAS1, as previously reported (31). When such mutations were introduced into the Oct-GAS2 oligonucleotide (referred to as Oct-mGAS [Fig. 4B]), they totally inhibited the binding of STAT5A and STAT5B, while Oct-1 still bound the biotinylated probe (lanes 7 to 10). Interestingly, the amount of Oct-1 bound to the wild-type Oct-GAS2 probe always decreased following TPO treatment, while Oct-mGAS reproducibly bound more Oct-1 from the same TPO-treated extracts. These data suggested that the binding of STAT5 somehow interfered with the conformation of Oct-1 or DNA and weakened Oct-1- DNA interaction. We also analyzed the factors that bound to the biotin-Octm-GAS2 probe that lost a canonical octamer motif. We observed that mutation in the octamer motif indeed abolished the recruitment of Oct-1 (Fig. 4B, upper gel, lanes 11 to 14). However, this mutated probe also lost the capacity to bind STAT5A or STAT5B (lower gels), although Octm-GAS2 contained an intact GAS, and the two mutated nucleotides that disrupted the octamer motif were the most distal nucleotides relative to the GAS motif (i.e., positions −7 and −8 relative to the first nucleotide of the consensus GAS) (Fig. 1). These experiments indicated that binding of STAT5 required both a functional GAS motif in Oct-GAS2 and the presence of DNA-bound Oct-1 in order to be stably recruited to GAS2 by oligonucleotide pull-down assays.
FIG. 4.
STAT5A and -B interacts with Oct-1. (A) Nuclear extracts from TPO-treated or untreated UT7-mpl cells were incubated without (lanes 1 and 2) or with Oct-GAS2 (lanes 3 to 6), Oct-GAS2m (lanes 7 to 10), or Octm-GAS2 (lanes 11 to 14) biotin-labeled oligonucleotides. Complexes bound to the streptavidin beads were analyzed by SDS-PAGE and Western blotting using the antibodies indicated on the left. Schemes showing the different oligonucleotides used are above the lanes, with solid bars representing mutated nucleotides. To avoid possible artifacts with reprobing experiments, all pull-down assays were performed in parallel and immunoblotted once. (B) (Top) 293T cells were transfected with vectors encoding PRL-R and STAT5A and maintained for 48 h in culture. The cells were incubated for 30 min with (+) or without (−) PRL (1 μg/ml), and nuclear extracts were prepared. (Bottom) UT7-mpl cells were treated with (+) or without (−) TPO for 30 min, and nuclear extracts were prepared. Both types of nuclear extracts were immunoprecipitated (IP) using anti-Oct-1 (Oct) or control (Ctl) antibodies, with subsequent Western blotting with anti-STAT5A antibodies. Lanes 1, 1/10 of nuclear extracts from cytokine-treated cells before immunoprecipitation.
STAT5 associates with the POU homeodomain protein Oct-1 in vivo.
As DNA-bound Oct-1 was necessary for STAT5 to stably bind to the Oct-GAS2 sequence, we studied the possible Oct-1/STAT5 protein interaction. We first overexpressed tagged STAT5A in 293T cells, and we cointroduced the PRL receptor to activate STAT5. Nuclear extracts were prepared from PRL-treated or untreated cells, and immunoprecipitations were performed. STAT5 proteins were absent in the nuclear compartment of non-PRL-treated 293T cells, but PRL induced a rapid nuclear translocation of active STAT5 (see Fig. 6). As shown in Fig. 4B, upper gel, anti-Oct-1 antibodies coimmunoprecipitated STAT5A in nuclear extracts from PRL-treated cells (lane 5) while no STAT5A was associated with Oct-1 in the absence of PRL (lane 3). As expected, irrelevant antibodies did not coimmunoprecipitate STAT5A (lanes 2 and 4). These results indicate that PRL-activated STAT5 and Oct-1 can associate in the nucleus.
FIG. 6.
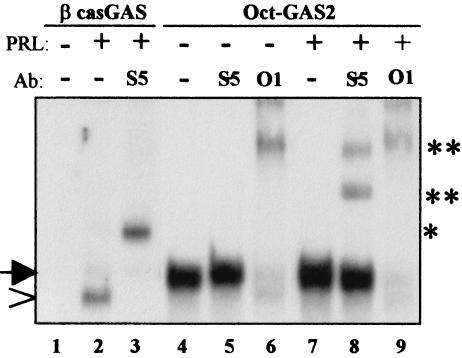
STAT5 forms a unique complex with DNA-bound Oct-1. 293T cells were transfected with vectors encoding PRL-R and STAT5A and maintained for 48 h in culture. The cells were incubated for 30 min with (+) or without (−) PRL (1 μg/ml), and nuclear extracts were prepared. The extracts were assayed for binding activities on βcas-GAS or Oct-GAS2 probes by EMSA. Where indicated, antibodies (Ab) directed against STAT5A and -B (S5) or Oct-1 (O1) were added to the binding reaction. * and **, supershifted complexes; arrow, Oct1-containing complexes; arrowhead, GAS-bound STAT5.
We thus studied whether endogenous STAT5 and Oct-1 factors may interact in TPO-treated UT7-mpl cells. As shown in Fig. 3B, rapid activation and nuclear translocation of STAT1 and STAT5A and -B were observed upon brief TPO treatment of these cells. Nuclear extracts from UT7-mpl cells were prepared and immunoprecipitated with anti-Oct-1 antibodies. We observed that endogenous Oct-1 specifically associated with endogenous nuclear STAT5A (Fig. 4B, lower gel, lane 5) or STAT5B (not shown) following TPO treatment, while no interaction with nuclear STAT1 was observed (not shown). We also did not detect any reproducible association of nuclear STAT5A and -B with irrelevant antibodies (lanes 2 to 3). Overall, these data indicate that two types of cytokines, such as PRL and TPO, can trigger the association of nuclear STAT5A and -B with Oct-1 in vivo in two different cellular contexts (293T and UT7-mpl cells).
STAT5A and -B contain a functional and widespread Oct-1 binding motif.
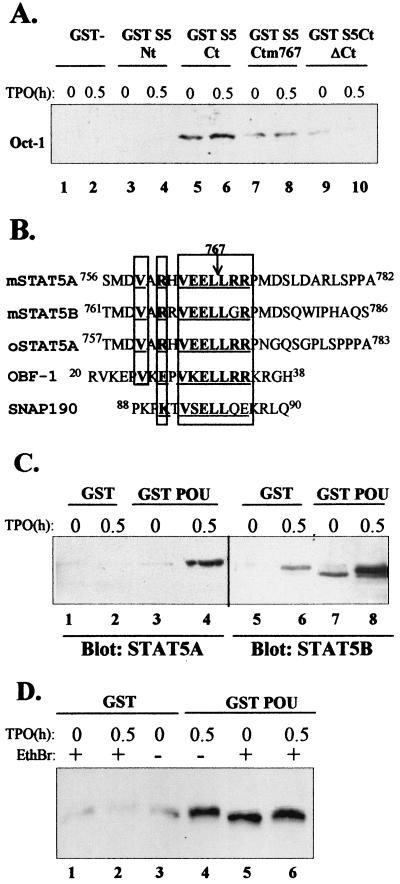
We analyzed the domains of STAT5 required for Oct-1 binding and used GST fusion proteins containing various parts of STAT5A in pull-down assays. As shown in Fig. 5A, the first 131 residues amino terminal of STAT5A linked to the GST moiety did not exhibit any interaction with nuclear Oct-1(lanes 3 and 4, GST S5 Nt), while the last 250 residues carboxy terminal of STAT5A did interact with Oct-1 specifically (lanes 5 and 6, GST S5 Ct). This interaction was not TPO dependent, indicating that Oct-1 does not need any cytokine treatment to interact with this STAT5 domain. This region encompassed SH2 and the transactivation domain of STAT5A. We produced a GST fusion protein with the same carboxy-terminal region of STAT5A with its last 45 residues deleted, thus removing the transactivation domain of the protein (34), and showed that such a deletion greatly impaired the association of Oct-1 with the STAT5 fusion protein (Fig. 5A, lanes 9 and 10).
FIG. 5.
STAT5 associates with Oct-1 through an OBF-like motif. (A) The indicated bacterially expressed GST fusion proteins were bound to glutathione-Sepharose beads and incubated with nuclear extracts of UT7-mpl cells treated for 0 or 0.5 h in the presence of TPO. Proteins bound to the beads were analyzed by SDS-PAGE, followed by immunoblotting with anti-Oct-1 antibodies. (B) Comparison of the sequences of murine STAT5A, murine STAT5B, ovine STAT5A, OBF-1, and SNAP190 in their regions of similarity. Identical amino acids are shown in boldface and underlined, and related amino acids are underlined. (C) Nuclear extracts of TPO-treated or untreated UT7-mpl cells were tested for binding to bacterially expressed GST or GST-POU1 proteins as described above. Bound proteins were analyzed by SDS-PAGE, followed by immunoblotting with antibodiesagainst STAT5A or STAT5B, as indicated below the blots. (D) Total cell extracts of TPO-treated or untreated UT7-mpl cells were prepared and further preincubated for 30 min in the absence (−) or presence (+) of 50 μg of ethidium bromide (EthBr)/ml before being incubated with immobilized GST or GST-POU1 protein as described above. Bound proteins were analyzed by Western blotting with antibodies against STAT5A and -B.
A careful analysis of the 45 residues deleted showed that STAT5A and -B contain a short sequence, 763VEELLRR769 (Fig. 5B), remarkably similar to a sequence present in two well-known Oct-1 binding factors, namely, OBF-1/Bob/OCA-B, a B-cell coactivator, and SNAP190, which is linked to the basal transcription machinery (13, 16). This seven-residue-long motif is absolutely necessary for the direct interaction of either OBF-1 or SNAP190 with Oct-1. To evaluate the importance of this STAT5 motif, we mutated the Leu 767 residue of STAT5A to Pro, since this single mutation was shown to impair OBF-1- Oct-1 interaction on its own (16). As shown in Fig. 5A, lanes 7 and 8, this unique point mutation greatly diminished the binding of Oct-1 to the carboxy-terminal domain of STAT5. This suggests that the OBF-1-like sequence of STAT5 is an Oct-1 binding motif that functions like the similar OBF-1 and SNAP190 motifs.
As all the Oct-1 partners interact with the DNA binding POU domain of Oct-1 (here called POU1) (18, 41), we tested whether STAT5 interacted with POU1. GST fusion proteins containing POU1 were purified and used in pull-down assays with UT7-mpl extracts. Both TPO-activated STAT5A and STAT5B were able to selectively bind the GST-POU1 domain immobilized on glutathione-agarose beads (Fig. 5C). Still, nuclear STAT5B was highly sticky, and a small proportion of the active STAT bound to any type of beads, including the GST-coupled agarose beads (lane 6). GST-POU1 also bound the weakly activated STAT5B present in continuously proliferating cells (Fig. 5C, lane 7) but did not bind any STATs in growth factor-deprived cells (not shown). These data indicate that nuclear STAT5A and STAT5B associate with Oct-1 via its POU domain.
All the coimmunoprecipitation and pull-down experiments described above suggested that Oct-1 and STAT5 can associate in the absence of DNA, but we previously showed (Fig. 4) that binding of STAT5 to GAS2 required the presence of DNA-bound Oct-1 in order to be stably recruited to DNA by oligonucleotide pull-down assays. We therefore wanted to ascertain that Oct-1 and STAT5 did interact in the total absence of contaminating DNA. Total cell extracts were prepared from TPO-treated or untreated cells and were further incubated in the absence or presence of ethidium bromide to disrupt any associated double-stranded DNA, as described previously (15). Lysates were next incubated with GST-POU1 fusion proteins immobilized on glutathione-agarose beads as described above. Figure 5D shows that similar amounts of STAT5 from ethidium bromide-treated (lane 6) and untreated (lane 4) cell extracts bound to GST-POU1. These data demonstrate that Oct-1 and STAT5 can still interact in the total absence of DNA.
STAT5 and Oct-1 form a trimeric complex with the proximal cyclin D1 element.
Although STAT5 and Oct-1 associate in the absence of DNA, we have been unable to observe a DNA-bound Oct-1/STAT5 complex by EMSA. We wished to reconcile these results. We studied nuclear extracts from 293T cells that overexpressed tagged STAT5A and PRL receptors (Fig. 4B) and performed EMSA with PRL-treated or untreated extracts. Gels were run for an extended time to improve the separation between the putative STAT and Oct-1 complexes. As shown in Fig. 6, PRL activated STAT5A that bound to the canonical β-casein (βcas)-GAS probe (lanes 1 to 2). This complex was supershifted by anti-STAT5 (lane 3) but not by anti-Oct-1 (not shown) antibodies, confirming the presence of active STAT5A homodimers in PRL-treated nuclei. When the same extracts were incubated with the Oct-GAS2 probe, a single alternative lower-mobility complex was observed in PRL-stimulated and unstimulated cells (lanes 4 and 7). This complex was weakly increased following PRL treatment (Fig. 6, compare lanes 4 and 7) and was entirely supershifted by anti-Oct-1 antibodies, indicating that it contained Oct-1 (Fig. 6A, lanes 3 and 6). Interestingly, the amount of STAT5 dimer that bound to the high-affinity βcas-GAS probe was rather small compared to that of the Oct-1 complexes bound to Oct-GAS2, indicating that DNA-bound Oct-1 was far more abundant than the overexpressed nuclear STAT5 dimers. We still searched for the presence of STAT5 in Oct-GAS2 complexes. PRL-treated extracts were incubated with the Oct-GAS2 probe in the presence of anti-STAT5 antibodies. Anti-STAT5 antibodies bound to the Oct-1 complexes and led to discrete supershifts (lane 8) that were different from the one formed in the presence of the same antibodies and βcas-bound STAT5 run on the same gel (lane 3). On the other hand, when we incubated untreated extracts with the Oct-GAS2 probe in the presence of the anti-STAT5 antibodies, no supershifts were detected (lane 5). These data indicate the presence of STAT5 in the Oct-1 complexes formed upon PRL stimulation. The partial, albeit significant, supershift observed was concomitant with a small but reproducible decrease in the intensity of the unshifted complex (the gels in Fig. 6 were overexposed to make the supershifted STAT5 complexes easily visible). Altogether, these results suggest that PRL-activated STAT5 associates with Oct-1 and forms a unique DNA-bound complex with Oct-GAS2, which includes both STAT5 and Oct-1. By chance, this complex exhibits an electrophoretic mobility similar to that of DNA-bound Oct-1 on native gels. Interestingly, no classical STAT5A homodimers similar to the ones that strongly bind βcas-GAS were detected (compare lanes 2 and 4), suggesting that the entire active STAT5 present in the nucleus associates with Oct-GAS2 as an Oct-1/STAT5 complex only.
Oct-1/STAT-5 complex functions as a cytokine-dependent transactivator.
Finally, we studied the importance of GAS1 and Oct-GAS2 motifs in the TPO-dependent activation of the cyclin D1 promoter. We therefore introduced point mutations that impair STAT binding to GAS1 or Oct and STAT binding to Oct-GAS2. All of these constructs were introduced into UT7-mpl cells, and their activities were assayed in the presence and absence of TPO. Although GAS1 strongly bound TPO-activated STAT factors, mutations of this site did not significantly affect the TPO responsiveness of the cyclin D1 promoter (Fig. 7A). On the contrary, mutations in the Oct-GAS2 motif resulted in a threefold decrease in TPO responsiveness. Finally, 80% of the TPO-dependent increase in cyclin D1 promoter activity was suppressed when both GAS1 and Oct-GAS2 were mutated. These data indicated the importance of the Oct-GAS2 motif in the cyclin D1 promoter activation by TPO and suggested cooperation between GAS1 and the Oct-GAS2 motif for this activation.
In order to define the respective contributions of Oct-1 and STAT5, we also generated individual mutations of the Oct or STAT binding sites within the composite Oct-GAS2 element. As opposed to the Oct-GAS2 double mutation, we observed that mutating either the Oct or GAS2 sequence individually had no major effect on the activity of the cyclin D1 promoter in the presence or absence of TPO (Fig. 7B and data not shown). These data indicate that the activation of the cyclin D1 promoter following TPO treatment does rely on the formation of the active Oct-1/STAT5 complex per se, but they also suggest that in the context of the whole promoter, the binding of this complex to the Oct-GAS2 element needs only one of the halves of the bipartite Oct-GAS2 in order to be functional.
DISCUSSION
The present study provides evidence that the pleiotropic transcription factor STAT5 activates the transcription of the cyclin D1 promoter by a new, previously unreported mechanism relying on a proximal Oct-GAS bipartite element. It indicates that STAT5 associates with Oct-1 and forms a functional Oct-1/STAT5 complex on the cyclin D1 promoter. We also demonstrate that STAT5 contains a functional Oct-1 binding motif that is similar to the one previously identified in two well-known Oct-1 partners, the ubiquitous SNAP190 and the cell-restricted OBF-1.
Mitogenic and oncogenic signal transduction pathways from diverse classes of receptors converge and strictly require cyclin D-CDK activity (28). In addition to the well-known action of G1 cyclins on cell proliferation, a number of reports suggest that G1 cyclins are also important regulators of cell differentiation, as exemplified by impaired mammary gland development in cyclin D1−/− mice (11) and the obligatory role of cyclin D3 in muscle and megakaryocyte differentiation (7, 57). Cyclin D1 expression has been shown to be regulated by at least three independent but complementary pathways: gene transcription, protein association, and protein stabilization. A wide variety of transcription factors, such as AP1, ATF/CREB complexes, Sp1, E2F, TCF, NF-κB, nuclear receptors, and STATs (17, 19, 26, 31, 44, 58, 60), regulate cyclin D1 promoter activity, suggesting the possibility of multiple cooperative interactions. The present work reveals a new type of cooperativity by showing that STAT5 regulates cyclin D1 expression through the formation of an Oct-1/STAT5 complex. Such data strengthen the suggested function of cyclin D1 to integrate diverse extracellular stimuli (28, 48).
STAT5 binds to a composite Oct-GAS element.
Although TPO activated STAT1, STAT3, and STAT5A and -B, we show that only active STAT1 and STAT5A and -B bind to the distal GAS1 element present in the cyclin D1 promoter, whereas none of the TPO-activated STATs shows discernible binding to the proximal GAS2 sensu stricto. Similar results had been reported in an IL-3-treated human hematopoietic cell line (31). The GAS1 and GAS2 probes we used were of identical sizes, and both exhibited a consensus GAS motif (TTCNNNGAA). Moreover, GAS2 differed by only a single nucleotide change from the core sequence of the canonical high-affinity βcas-GAS (Fig. 1) and was identical to a critical IL-2-dependent STAT5 element present in the enhancer of the immunoglobulin J chain gene (20). These data confirmed that ill-defined nucleotides central to and outside the core GAS are crucial to high-affinity binding of STATs, as previously observed (45, 52). Although different STATs activate distinct sets of genes, the consensus sequences for optimal binding of STAT homodimers were found to be highly similar (45, 52). Nucleotide sequences that favor the binding of a precise STAT are still largely unknown. Recently, DNA binding site selection of dimeric and tetrameric STAT5 proteins revealed a large repertoire of strikingly divergent tetrameric STAT5A binding sites. TTC and GAA half-GAS appeared to be able to function as parts of degenerate GAS motifs anchoring the tetrameric STAT complex to DNA. Moreover, specific mutations in GAS that totally abolished binding of STAT5 dimers were shown to still allow STAT binding as tetramers. This diminished nucleotide stringency is presumably allowed by the increased cooperativity of binding fostered by tetramerization (52). Therefore, enhanced stability of the STAT complex can be favored, or impaired, by protein-DNA, as well as protein-protein, interactions.
Our data indicate that Oct-1 is essential to the detection of strong, stable, and selective recruitment of STAT5 to the cyclin D1 GAS2 element in vitro. Three pieces of evidence support this conclusion. First, none of the TPO-activated STATs could bind to the isolated GAS2 in EMSA, whereas STAT5 bound to Oct-GAS2 and formed a new kind of DNA-bound complex that included Oct-1. On native gels, this complex exhibited an electrophoretic mobility similar to that of DNA-bound Oct-1 but different from those of the conventional STAT5 homodimers. Second, the most distal mutation within the octamer site which impaired Oct-1 binding to Oct-GAS2 simultaneously inhibited STAT5 recruitment to the DNA sequence in oligonucleotide pull-down assays. Third, Oct-1 specifically interacted with nuclear STAT5A and -B through a specific amino acid motif present in the carboxy-terminal region of STAT5A/5B but absent from the other STAT members, and similarly, only the binding of STAT5, but not STAT1 or STAT3, to GAS2 was induced by the presence of an adjacent octamer motif in oligonucleotide pull-down assays. Oct-1 is in huge excess over nuclear STAT5 in activated cells; the relative amount of DNA-bound Oct-1/STAT5 complexes, compared to Oct-1-only complex, was low even in STAT5-overexpressing cells and thus was too small to be seen in supershift assays in untransfected cells. In the presence of overexpressed STAT5, all of the STAT5 that bound to the proximal element was present as an Oct-1/STAT5 complex. However, although Oct-1 and STAT5 interacted in the absence of DNA, the stable recruitment of STAT5 to Oct-GAS2 still required the presence of the core GAS element in vitro. All these observations suggest that Oct-1, by simultaneously binding to the DNA octamer element and STAT5, may enhance the stability of STAT5 and its affinity for neighboring suboptimal GAS sites in the context of a short oligonucleotide element such as Oct-GAS2, much like the stabilizing effect of STAT tetramers previously reported (52). Both Oct and GAS2 would thus be required to allow and/or maintain Oct-1/STAT5 interaction with the oligonucleotide in vitro.
In the context of long DNA sequences, however, such as the cyclin D1 promoter, mutating Oct and GAS2 sequences individually had no major effect on the activation of the promoter, whereas mutating both Oct and GAS2 simultaneously strongly affected cyclin D1 activation. This suggests that additional protein-protein interactions occur within the cyclin D1 promoter that maintain and stabilize the binding of the Oct-1/STAT5 complex to Oct-GAS2 in vivo, so that Oct-1/STAT5 can still bind to Oct-GAS2 even when the Oct or GAS2 sequence, but not both, is impaired.
Overall, our results suggest that the Oct-GAS composite element represents a new repertoire of biologically relevant binding sites for STAT5A and -B. The exact stoichiometry of the DNA-bound Oct-1/STAT5 complex remains to be determined.
The Oct-GAS element is a new cytokine-responsive element.
Despite the inability of STAT dimers to bind the isolated GAS2 in vitro, the integrity of the Oct-GAS2 site, but not GAS1, was shown to be crucial to TPO-dependent activation of the cyclin D1 promoter. Another cytokine, IL-3, was previously reported to trigger the reentry of quiescent serum-deprived hematopoietic cells into the cell cycle through a STAT5-dependent induction of cyclin D1 expression (31). Similarly, PRL was also recently reported to enhance cyclin D1 promoter activity in Chinese hamster ovary (CHO) cells (5). However, in obvious contrast to our results, these two studies showed that only GAS1, not GAS2, was important for the activation of the cyclin D1 promoter. There are two major differences between these data and ours. First, in one of the reports, the search for a STAT5-responsive element(s) in the cyclin D1 promoter was addressed by overexpressing an active mutant of STAT5, namely, STAT5A1*6, in NIH 3T3 cells in the absence of cytokine treatment. The molecular basis for the constitutive activity of STAT5A1*6 is still ill defined. Despite its biological activity, STAT5A1*6 was shown to be unable to strongly bind canonical GAS in the absence of cytokine, and it was reported to need cytokine to maximally activate a STAT5-responsive element, suggesting that STAT5A1*6 activities do not mimic all the activities of a cytokine-activated STAT5 (37). Also, while STAT5A1*6 was constitutively tyrosine phosphorylated with permanent nuclear accumulation and was able to support long-term growth of BAF3 transfectants in the absence of IL-3, it still needed the presence of serum to support cell proliferation, suggesting that additional signals supplied by serum were required to assist STAT51*6 growth potential (37). Studies of the mechanism of activation of cyclin D1 promoter by STAT51*6 in serum-starved NIH 3T3 cells thus may have obscured a complementary role of STAT5 when activated by a cytokine in serum-supplemented media. This would explain the weak activation of the cyclin D1 promoter observed in serum-starved STAT51*6-expressing cells. The second report studied the mechanism underlying a twofold increase in cyclin D1 promoter activity observed in PRL-treated and serum-deprived CHO transfectants that expressed exogenous PRL-R. It showed that a small activation of the cyclin D1 promoter involved the sole GAS1 element. These data fit well with the range of activation of the cyclin D1 promoter obtained in the presence of the STAT51*6 mutant (37), but they also fit with the cooperation of GAS1 we observed in our cellular context. We therefore suggest that GAS1 contributes to a small activation of cyclin D1 promoter activity by active STAT5, while full activation of that promoter involves the additional participation and activation of Oct-GAS2.
Analysis of Oct-1 during progression through the cell cycle revealed a complex temporal program of phosphorylation. Hyperphosphorylation of Oct-1 had been shown to occur as cells complete DNA synthesis and enter mitosis and to be rapidly reversed as cells enter the G1 growth phase of the cell cycle. Mitotic phosphorylation of Oct-1 correlates with a general inhibition of Oct-1 DNA binding activity during mitosis. Moreover, entry into G1 results in removal of most, but not all, of the mitotic phosphates from Oct-1, restoring Oct-1 function (46, 47). Such regulation of Oct-1 DNA binding activities may therefore provide efficient means to restrict STAT5-dependent activation of cyclin D1 to specific phases along the cell cycle. Whether, in addition to regulating its DNA binding properties, some modifications of Oct-1 may be regulated by cytokines or serum signaling pathways and may also influence its association with STAT5 is presently unknown.
Oct-1 has recently been shown to be a potentially important cofactor in regulating basal cyclin D1 expression in a cell line derived from a human breast cancer. Taking into account the frequent overexpression of cyclin D1 in human cancers, the authors suggested that Oct-1 was potentially linked to cell transformation (2). Overexpression of the Oct-1 POU domain in the mouse thymus also led to the development of thymic lymphomas (42). Constitutive activation of STAT5 has been found in T-cell leukemia and lymphoma, in acute lymphoblastic leukemia, and in acute and chronic myeloid leukemia, and STAT5 has been demonstrated to be crucial to BCR/ABL-mediated leukemogenesis (3). Our data further strengthen the idea that both Oct-1 and STAT5 may be oncogenic by cooperating in the aberrant transcription activation of cyclin D1 that underlies the onset or progression of cancer.
Oct-1/STAT5 association involves a well-known Oct-1 binding motif.
Oct-1 is a ubiquitous nuclear factor that is present during all stages of the cell cycle. In agreement with its broad distribution, Oct-1 is a regulator of housekeeping genes, such as those of small nuclear RNA and histone H2B (46, 49). However, octamer recognition sites are widely distributed in the vertebrate genome, and Oct-1 also appears to regulate genes that selectively respond to developmental, cell cycle, or hormonal signals, such as those of immunoglobulins (53). The mechanism by which such a ubiquitous factor can differentially regulate gene transcription is not well understood, but Oct-1 has long been thought to act in a selective manner by adopting distinct conformations on related DNA sites; discrete variations in these conformations are recognized by selective regulators. The specificity of gene expression was therefore proposed to be regulated by the coactivators-corepressors rather than by Oct-1 (18, 33). We observed that the binding of STAT5 on the proximal promoter influences Oct-1 binding to DNA. It is therefore possible that STAT5 affects the conformation of Oct-1 when it binds to it, which then overall leads to promoter activation. The transcriptionally inert POU domain of Oct-1 was recently shown to directly facilitate TATA-binding protein (TBP) recruitment near a core promoter (1). The authors suggested that Oct-1, by locally anchoring a preassembled TBP complex on a promoter, could then be acted upon by distal activators. Oct-1 may thus participate in cyclin D1 gene expression as a scaffold protein that recruits both TBP and STAT5 transactivators in the immediate vicinity.
A number of regulators, such as Sp1, AP1, steroid receptors, POU factor, the recently reported CREB, the ubiquitous SNAP190 subunit of the TBP-containing basal transcription complex called SNAPc, and the B-cell coregulator OBF-1/Bob/OCA-b (18), do interact with Oct-1. Direct interaction with the last two partners was shown to involve similar Oct binding motifs (13, 16). The unexpected finding of the present work was the striking similarity between the core Oct binding motif crucial for OBF or SNAP190/Oct-1 interactions and a stretch of seven residues within the carboxy-terminal tail of STAT5A and -B. This short sequence was highly conserved in STAT5A and STAT5B isolated from mouse, rat, sheep, and humans but was absent from all other STAT factors. Therefore, STAT5 represents a third partner of Oct-1 that exhibits a similar Oct binding motif. Apart from the short Oct-1 POU-interacting motif, OBF-1, SNAP190, and STAT5 do not show any obvious sequence similarity, nor are they known to share any functional role. Our results, therefore, suggest that this stretch of seven residues represents a new protein-protein interaction motif that may be a hallmark of a subclass of Oct-1 partners. Whether binding of each of these three partners to Oct-1 interferes with the binding of the other two remains to be determined.
The Oct-1 binding motif is included in the carboxy-terminal region of STAT5 that was shown to contain both the transactivation domain of STAT proteins and selective information for the rapid deactivation and dephosphorylation of STAT5 only (34, 56). This region was also reported to participate in interaction with other STAT5 partners, such as the transcriptional coactivators CBP/p300 (39). Secondary-structure predictions indicate that 20 amino acids in the central part of this region of STAT5A and -B can adopt an amphipathic α-helical structure that is not present in the other STATs (34, 56). The Oct-1 binding motif is located within this α-helix. Whether interaction with Oct-1 may affect the association of STAT5 with coactivators such as the histone acetyl transferases CBP/p300 or modulate the kinetics of deactivation of STAT5 awaits further investigation.
Overall, our data indicate that cytokine-activated STAT5 can trigger an important expression of cyclin D1 by acting on a new proximal bipartite sequence that binds an Oct-1/STAT5 complex. This action may directly participate in leukemogenesis in cells where STAT5 becomes constitutively activated.
Acknowledgments
Fabrice Gouilleux, Itaru Matsumura, and Lothar Henninghausen are gratefully acknowledged for kind gifts of plasmids and antibodies and for helpful discussions. We thank Claire Francastel, Sylvie Gisselbrecht, Vincent Mignotte, and Paul-Henri Romeo for reading the manuscript and providing valuable comments and Frank Letourneur and Nicolas Lebrun for DNA sequencing.
This work was supported by a grant from the Ligue Nationale Contre le Cancer. S.C. is the recipient of a fellowship from the Ligue Contre le Cancer.
REFERENCES
- 1.Bertolino, E., and H. Singh. 2002. POU/TBP cooperativity: a mechanism for enhancer action from a distance. Mol. Cell 10:397-407. [DOI] [PubMed] [Google Scholar]
- 2.Boulon, S., J. C. Dantonel, V. Binet, A. Vie, J. M. Blanchard, R. A. Hipskind, and A. Philips. 2002. Oct-1 potentiates CREB-driven cyclin D1 promoter activation via a phospho-CREB- and CREB-binding protein-independent mechanism. Mol. Cell. Biol. 22:7769-7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowman, T., R. Garcia, J. Turkson, and R. Jove. 2000. STATs in oncogenesis. Oncogene 19:2474-2488. [DOI] [PubMed] [Google Scholar]
- 4.Bradley, H. L., T. S. Hawley, and K. D. Bunting. 2002. Cell intrinsic defects in cytokine responsiveness of STAT5-deficient hematopoietic stem cells. Blood 100:3983-3989. [DOI] [PubMed] [Google Scholar]
- 5.Brockman, J. L., M. D. Schroeder, and L. A. Schuler. 2002. PRL activates the cyclin D1 promoter via the Jak2/Stat pathway. Mol. Endocrinol. 16:774-784. [DOI] [PubMed] [Google Scholar]
- 6.Bromberg, J., and J. E. Darnell. 2000. The role of STATs in transcriptional control and their impact on cellular function. Oncogene 19:2468-2473. [DOI] [PubMed] [Google Scholar]
- 7.Cenciarelli, C., F. De Santa, P. L. Puri, E. Mattei, L. Ricci, F. Bucci, A. Felsani, and M. Caruso. 1999. Critical role played by cyclin D3 in the MyoD-mediated arrest of cell cycle during myoblast differentiation. Mol. Cell. Biol. 19:5203-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cwirla, S. E., P. Balasubramanian, D. J. Duffin, C. R. Wagstrom, C. M. Gates, S. C. Singer, A. M. Davis, R. L. Tansik, L. C. Mattheakis, C. M. Boytos, P. J. Schatz, D. P. Baccanari, N. C. Wrighton, R. W. Barrett, and W. J. Dower. 1997. Peptide agonist of the thrombopoietin receptor as potent as the natural cytokine. Science 276:1696-1699. [DOI] [PubMed] [Google Scholar]
- 9.Doubeikovski, A., G. Uzan, Z. Doubeikovski, M. H. Prandini, F. Porteu, S. Gisselbrecht, and I. Dusanter-Fourt. 1997. Thrombopoietin-induced expression of the glycoprotein IIb gene involves the transcription factor PU.1/Spi-1 in UT7-Mpl cells. J. Biol. Chem. 272:24300-24307. [DOI] [PubMed] [Google Scholar]
- 10.Dumon, S., S. C. Santos, F. Debierre-Grockiego, V. Gouilleux-Gruart, L. Cocault, C. Boucheron, P. Mollat, S. Gisselbrecht, and F. Gouilleux. 1999. IL-3 dependent regulation of Bcl-xL gene expression by STAT5 in a bone marrow derived cell line. Oncogene 18:4191-4199. [DOI] [PubMed] [Google Scholar]
- 11.Fantl, V., P. A. Edwards, J. H. Steel, B. K. Vonderhaar, and C. Dickson. 1999. Impaired mammary gland development in Cyl-1(−/−) mice during pregnancy and lactation is epithelial cell autonomous. Dev. Biol. 212:1-11. [DOI] [PubMed] [Google Scholar]
- 12.Feldman, G. M., L. A. Rosenthal, X. Liu, M. P. Hayes, A. Wynshaw-Boris, W. J. Leonard, L. Hennighausen, and D. S. Finbloom. 1997. STAT5A-deficient mice demonstrate a defect in granulocyte-macrophage colony-stimulating factor-induced proliferation and gene expression. Blood 90:1768-1776. [PubMed] [Google Scholar]
- 13.Ford, E., M. Strubin, and N. Hernandez. 1998. The Oct-1 POU domain activates snRNA gene transcription by contacting a region in the SNAPc largest subunit that bears sequence similarities to the Oct-1 coactivator OBF-1. Genes Dev. 12:3528-3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia, J., J. de Gunzburg, A. Eychene, S. Gisselbrecht, and F. Porteu. 2001. Thrombopoietin-mediated sustained activation of extracellular signal-regulated kinase in UT7-Mpl cells requires both Ras-Raf-1- and Rap1-B-Raf-dependent pathways. Mol. Cell. Biol. 21:2659-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez, M. I., and D. M. Robins. 2001. Oct-1 preferentially interacts with androgen receptor in a DNA-dependent manner that facilitates recruitment of SRC-1. J. Biol. Chem. 276:6420-6428. [DOI] [PubMed] [Google Scholar]
- 16.Gstaiger, M., O. Georgiev, H. van Leeuwen, P. van der Vliet, and W. Schaffner. 1996. The B cell coactivator Bob1 shows DNA sequence-dependent complex formation with Oct-1/Oct-2 factors, leading to differential promoter activation. EMBO J. 15:2781-2790. [PMC free article] [PubMed] [Google Scholar]
- 17.Herber, B., M. Truss, M. Beato, and R. Muller. 1994. Inducible regulatory elements in the human cyclin D1 promoter. Oncogene 9:1295-1304. [PubMed] [Google Scholar]
- 18.Herr, W., and M. A. Cleary. 1995. The POU domain: versatility in transcriptional regulation by a flexible two-in-one DNA-binding domain. Genes Dev. 9:1679-1693. [DOI] [PubMed] [Google Scholar]
- 19.Hinz, M., D. Krappmann, A. Eichten, A. Heder, C. Scheidereit, and M. Strauss. 1999. NF-κB function in growth control: regulation of cyclin D1 expression and G0/G1-to-S-phase transition. Mol. Cell. Biol. 19:2690-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang, C. J., C. Sheridan, and M. E. Koshland. 1998. A stage-specific enhancer of immunoglobulin J chain gene is induced by interleukin-2 in a presecretor B cell stage. Immunity 8:285-295. [DOI] [PubMed] [Google Scholar]
- 21.Kaushansky, K. 1999. The enigmatic megakaryocyte gradually reveals its secrets. Bioessays 21:353-360. [DOI] [PubMed] [Google Scholar]
- 22.Kaushansky, K. 1999. Thrombopoietin and hematopoietic stem cell development. Ann. N. Y. Acad. Sci. 872:314-319. [DOI] [PubMed] [Google Scholar]
- 23.Kieslinger, M., I. Woldman, R. Moriggl, J. Hofmann, J. C. Marine, J. N. Ihle, H. Beug, and T. Decker. 2000. Antiapoptotic activity of Stat5 required during terminal stages of myeloid differentiation. Genes Dev. 14:232-244. [PMC free article] [PubMed] [Google Scholar]
- 24.Komatsu, N., H. Nakauchi, A. Miwa, T. Ishihara, M. Eguchi, M. Moroi, M. Okada, Y. Sato, H. Wada, Y. Yawata, et al. 1991. Establishment and characterization of a human leukemic cell line with megakaryocytic features: dependency on granulocyte-macrophage colony-stimulating factor, interleukin 3, or erythropoietin for growth and survival. Cancer Res. 51:341-348. [PubMed] [Google Scholar]
- 25.Leonard, W. J., and J. J. O'Shea. 1998. Jaks and STATs: biological implications. Annu. Rev. Immunol. 16:293-322. [DOI] [PubMed] [Google Scholar]
- 26.Lin, S. Y., W. Xia, J. C. Wang, K. Y. Kwong, B. Spohn, Y. Wen, R. G. Pestell, and M. C. Hung. 2000. Beta-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc. Natl. Acad. Sci. USA 97:4262-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lord, J. D., B. C. McIntosh, P. D. Greenberg, and B. H. Nelson. 2000. The IL-2 receptor promotes lymphocyte proliferation and induction of the c-myc, bcl-2, and bcl-x genes through the trans-activation domain of Stat5. J. Immunol. 164:2533-2541. [DOI] [PubMed] [Google Scholar]
- 28.Lukas, J., J. Bartkova, M. Rohde, M. Strauss, and J. Bartek. 1995. Cyclin D1 is dispensable for G1 control in retinoblastoma gene-deficient cells independently of cdk4 activity. Mol. Cell. Biol. 15:2600-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martino, A., J. H. T. Holmes, J. D. Lord, J. J. Moon, and B. H. Nelson. 2001. Stat5 and Sp1 regulate transcription of the cyclin D2 gene in response to IL-2. J. Immunol. 166:1723-1729. [DOI] [PubMed] [Google Scholar]
- 30.Matsumura, I., J. Ishikawa, K. Nakajima, K. Oritani, Y. Tomiyama, J. Miyagawa, T. Kato, H. Miyazaki, Y. Matsuzawa, and Y. Kanakura. 1997. Thrombopoietin-induced differentiation of a human megakaryoblastic leukemia cell line, CMK, involves transcriptional activation of p21(WAF1/Cip1) by STAT5. Mol. Cell. Biol. 17:2933-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsumura, I., T. Kitamura, H. Wakao, H. Tanaka, K. Hashimoto, C. Albanese, J. Downward, R. G. Pestell, and Y. Kanakura. 1999. Transcriptional regulation of the cyclin D1 promoter by STAT5: its involvement in cytokine-dependent growth of hematopoietic cells. EMBO J. 18:1367-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsumura, I., K. Nakajima, H. Wakao, S. Hattori, K. Hashimoto, H. Sugahara, T. Kato, H. Miyazaki, T. Hirano, and Y. Kanakura. 1998. Involvement of prolonged ras activation in thrombopoietin-induced megakaryocytic differentiation of a human factor-dependent hematopoietic cell line. Mol. Cell. Biol. 18:4282-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Misra, V., S. Walter, P. Yang, S. Hayes, and P. O'Hare. 1996. Conformational alteration of Oct-1 upon DNA binding dictates selectivity in differential interactions with related transcriptional coactivators. Mol. Cell. Biol. 16:4404-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moriggl, R., V. Gouilleux-Gruart, R. Jahne, S. Berchtold, C. Gartmann, X. Liu, L. Hennighausen, A. Sotiropoulos, B. Groner, and F. Gouilleux. 1996. Deletion of the carboxyl-terminal transactivation domain of MGF-Stat5 results in sustained DNA binding and a dominant negative phenotype. Mol. Cell. Biol. 16:5691-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moriggl, R., D. J. Topham, S. Teglund, V. Sexl, C. McKay, D. Wang, A. Hoffmeyer, J. van Deursen, M. Y. Sangster, K. D. Bunting, G. C. Grosveld, and J. N. Ihle. 1999. Stat5 is required for IL-2-induced cell cycle progression of peripheral T cells. Immunity 10:249-259. [DOI] [PubMed] [Google Scholar]
- 36.Mui, A. L., H. Wakao, T. Kinoshita, T. Kitamura, and A. Miyajima. 1996. Suppression of interleukin-3-induced gene expression by a C-terminal truncated Stat5: role of Stat5 in proliferation. EMBO J. 15:2425-2433. [PMC free article] [PubMed] [Google Scholar]
- 37.Onishi, M., T. Nosaka, K. Misawa, A. L. Mui, D. Gorman, M. McMahon, A. Miyajima, and T. Kitamura. 1998. Identification and characterization of a constitutively active STAT5 mutant that promotes cell proliferation. Mol. Cell. Biol. 18:3871-3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pallard, C., F. Gouilleux, L. Benit, L. Cocault, M. Souyri, D. Levy, B. Groner, S. Gisselbrecht, and I. Dusanter-Fourt. 1995. Thrombopoietin activates a STAT5-like factor in hematopoietic cells. EMBO J. 14:2847-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfitzner, E., R. Jahne, M. Wissler, E. Stoecklin, and B. Groner. 1998. p300/CREB-binding protein enhances the prolactin-mediated transcriptional induction through direct interaction with the transactivation domain of Stat5, but does not participate in the Stat5-mediated suppression of the glucocorticoid response. Mol. Endocrinol. 12:1582-1593. [DOI] [PubMed] [Google Scholar]
- 40.Porteu, F., M. C. Rouyez, L. Cocault, L. Benit, M. Charon, F. Picard, S. Gisselbrecht, M. Souyri, and I. Dusanter-Fourt. 1996. Functional regions of the mouse thrombopoietin receptor cytoplasmic domain: evidence for a critical region which is involved in differentiation and can be complemented by erythropoietin. Mol. Cell. Biol. 16:2473-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prefontaine, G. G., M. E. Lemieux, W. Giffin, C. Schild-Poulter, L. Pope, E. LaCasse, P. Walker, and R. J. Hache. 1998. Recruitment of octamer transcription factors to DNA by glucocorticoid receptor. Mol. Cell. Biol. 18:3416-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qin, X. F., Y. Luo, H. Suh, J. Wayne, Z. Misulovin, R. G. Roeder, and M. C. Nussenzweig. 1994. Transformation by homeobox genes can be mediated by selective transcriptional repression. EMBO J. 13:5967-5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rouyez, M. C., C. Boucheron, S. Gisselbrecht, I. Dusanter-Fourt, and F. Porteu. 1997. Control of thrombopoietin-induced megakaryocytic differentiation by the mitogen-activated protein kinase pathway. Mol. Cell. Biol. 17:4991-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sabbah, M., D. Courilleau, J. Mester, and G. Redeuilh. 1999. Estrogen induction of the cyclin D1 promoter: involvement of a cAMP response-like element. Proc. Natl. Acad. Sci. USA 96:11217-11222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schindler, U., P. Wu, M. Rothe, M. Brasseur, and S. L. McKnight. 1995. Components of a Stat recognition code: evidence for two layers of molecular selectivity. Immunity 2:689-697. [DOI] [PubMed] [Google Scholar]
- 46.Segil, N., S. B. Roberts, and N. Heintz. 1991. Cell-cycle-regulated phosphorylation of the transcription factor Oct-1. Cold Spring Harbor Symp. Quant. Biol. 56:285-292. [DOI] [PubMed] [Google Scholar]
- 47.Segil, N., S. B. Roberts, and N. Heintz. 1991. Mitotic phosphorylation of the Oct-1 homeodomain and regulation of Oct-1 DNA binding activity. Science 254:1814-1816. [DOI] [PubMed] [Google Scholar]
- 48.Sherr, C. J. 1993. Mammalian G1 cyclins. Cell 73:1059-1065. [DOI] [PubMed] [Google Scholar]
- 49.Sive, H. L., and R. G. Roeder. 1986. Interaction of a common factor with conserved promoter and enhancer sequences in histone H2B, immunoglobulin, and U2 small nuclear RNA (snRNA) genes. Proc. Natl. Acad. Sci. USA 83:6382-6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Snow, J. W., N. Abraham, M. C. Ma, N. W. Abbey, B. Herndier, and M. A. Goldsmith. 2002. STAT5 promotes multilineage hematolymphoid development in vivo through effects on early hematopoietic progenitor cells. Blood 99:95-101. [DOI] [PubMed] [Google Scholar]
- 51.Socolovsky, M., A. E. Fallon, S. Wang, C. Brugnara, and H. F. Lodish. 1999. Fetal anemia and apoptosis of red cell progenitors in Stat5a−/−5b−/− mice: a direct role for Stat5 in Bcl-X(L) induction. Cell 98:181-191. [DOI] [PubMed] [Google Scholar]
- 52.Soldaini, E., S. John, S. Moro, J. Bollenbacher, U. Schindler, and W. J. Leonard. 2000. DNA binding site selection of dimeric and tetrameric Stat5 proteins reveals a large repertoire of divergent tetrameric Stat5a binding sites. Mol. Cell. Biol. 20:389-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Staudt, L. M., and M. J. Lenardo. 1991. Immunoglobulin gene transcription. Annu. Rev. Immunol. 9:373-398. [DOI] [PubMed] [Google Scholar]
- 54.Sturm, R. A., G. Das, and W. Herr. 1988. The ubiquitous octamer-binding protein Oct-1 contains a POU domain with a homeo box subdomain. Genes Dev. 2:1582-1599. [DOI] [PubMed] [Google Scholar]
- 55.Teglund, S., C. McKay, E. Schuetz, J. M. van Deursen, D. Stravopodis, D. Wang, M. Brown, S. Bodner, G. Grosveld, and J. N. Ihle. 1998. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell 93:841-850. [DOI] [PubMed] [Google Scholar]
- 56.Wang, D., R. Moriggl, D. Stravopodis, N. Carpino, J. C. Marine, S. Teglund, J. Feng, and J. N. Ihle. 2000. A small amphipathic alpha-helical region is required for transcriptional activities and proteasome-dependent turnover of the tyrosine-phosphorylated Stat5. EMBO J. 19:392-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, Z., Y. Zhang, D. Kamen, E. Lees, and K. Ravid. 1995. Cyclin D3 is essential for megakaryocytopoiesis. Blood 86:3783-3788. [PubMed] [Google Scholar]
- 58.Watanabe, G., C. Albanese, R. J. Lee, A. Reutens, G. Vairo, B. Henglein, and R. G. Pestell. 1998. Inhibition of cyclin D1 kinase activity is associated with E2F-mediated inhibition of cyclin D1 promoter activity through E2F and Sp1. Mol. Cell. Biol. 18:3212-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wegner, M., D. W. Drolet, and M. G. Rosenfeld. 1993. POU-domain proteins: structure and function of developmental regulators. Curr. Opin. Cell Biol. 5:488-498. [DOI] [PubMed] [Google Scholar]
- 60.Yan, G. Z., and E. B. Ziff. 1997. Nerve growth factor induces transcription of the p21 WAF1/CIP1 and cyclin D1 genes in PC12 cells by activating the Sp1 transcription factor. J. Neurosci. 17:6122-6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang, S., S. Fukuda, Y. Lee, G. Hangoc, S. Cooper, R. Spolski, W. J. Leonard, and H. E. Broxmeyer. 2000. Essential role of signal transducer and activator of transcription (Stat)5a but not Stat5b for Flt3-dependent signaling. J. Exp. Med. 192:719-728. [DOI] [PMC free article] [PubMed] [Google Scholar]