Abstract
RNA interference (RNAi) has been established as an important tool for functional genomics studies and has great promise as a therapeutic intervention for human diseases. In mammalian cells, RNAi is conventionally induced by 19–27-bp RNA duplexes generated by hybridization of two complementary oligonucleotide strands (oligos). Here we describe a novel class of RNAi molecules composed of a single 25–28-nucleotide (nt) oligo. The oligo has a 16-nt mRNA targeting region, followed by an additional 8–10 nt to enable self-dimerization into a partially complementary duplex. Analysis of numerous diverse structures demonstrates that molecules composed of two short helices separated by a loop can efficiently enter and activate the RNA-induced silencing complex (RISC). This finding enables the design of highly effective single-oligo compounds for any mRNA target.
Keywords: RNAi, siRNA, Ago2
INTRODUCTION
Regulation of gene expression by small RNAs is a conserved cellular process known as RNA interference, or RNAi (Castanotto and Rossi 2009). This process has been utilized to study new gene functions and to develop a novel class of therapeutics. The most commonly used molecules to initiate RNAi in mammalian cells are the small interfering RNAs (siRNA) and the small hairpin RNAs (shRNA). The structure of siRNAs is similar to that of cleavage products of long double-stranded RNAs (dsRNA) by the endoribonuclease Dicer (Hutvagner et al. 2001). These molecules possess two complementary 21-nucleotide (nt)-long strands, 19 nt of which are base-paired and the remaining 2 nt protruding as 3′ overhangs on each end. The structure of shRNAs is typically similar to the naturally occurring pre-miRNA hairpins with a double-stranded stem of around 23–25 bp and a loop region of ∼5 nt in length. In contrast to most natural pre-miRNAs, shRNAs are usually designed to have perfect complementarity within the stem region (Paddison et al. 2004; Siolas et al. 2005). One of the main limitations in manufacturing siRNAs is the necessity to synthesize two independent oligonucleotides. Synthetic shRNAs avoid the need to prepare two oligonucleotides; however, there are inherent manufacturing issues with long oligonucleotides. An attempt to use shorter single-oligo RNAi triggers—comprising just the antisense (or “guide”) portion of an siRNA—has been made earlier by Tuschl and co-workers in 2002 (Martinez et al. 2002) but resulted in much lower efficacy of the compounds in cell culture. Further attempts to develop single-oligo RNAi triggers led to the identification of double-stranded self-annealing siRNA compounds containing palindromic sequences (Hossbach et al. 2006). Palindromic motifs are rare, thus restricting the number of possible compounds designed with this approach. Many genes are not amenable for such design altogether.
It is currently perceived that two components are required to activate the RNAi machinery—the double-stranded nucleic acid motif, which is required for recognition by RNAi-associated proteins and the guide strand which serves as the mRNA binding co-factor in the RISC's Argonaute catalytic protein. Taking this into account, we have designed and tested a novel type of RNAi molecule composed of a single short (25–28 nt) oligo capable of self-dimerizing into a partially complementary duplex. The new molecules were demonstrated to efficiently activate the RISC and to produce target mRNA silencing comparable to that obtained with potent conventional RNAi molecules. Importantly, the new configuration enables the design of highly effective single-oligo compounds for any mRNA target. It also allows designing single molecules that can simultaneously target and silence two different genes.
RESULTS AND DISCUSSION
We designed and characterized a novel type of RNAi compound—soloRNA—which is a homodimer originating from a single, short oligonucleotide (Fig. 1A). The soloRNAs have two key functional elements: a minimal duplex structure sufficient for RISC recognition and loading, and a continuous stretch of nucleotides required for target transcript binding and cleavage. It has been previously demonstrated that only 16 nt (positions 2–17) of an RISC-loaded “guide” strand actively participate in target recognition (Chu and Rana 2008; Patel et al. 2006; Wang et al. 2008). In the soloRNA design, the targeting segment is extended by an additional 9–11 nt complementary to the 5′ sequence of the oligo itself. This results in a single, 26–28-nt oligo that is capable of self-annealing to form a partially complementary duplex that can trigger RISC-mediated gene silencing.
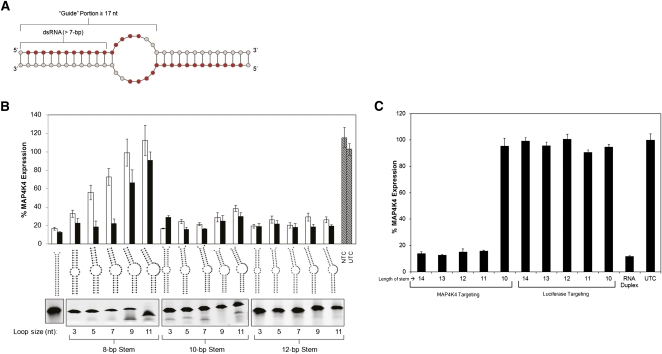
FIGURE 1.
Structural requirements for self-dimerizing single-oligo RISC substrates. (A) 27-nt oligonucleotides were designed to contain a 5′ portion complementary (at least 16 nt, shown in red) to the target mRNA transcript, and a 3′ portion (8–10 nt) enabling oligo self-dimerization into a partially complementary duplex. (B) RNA duplexes composed of two different oligonucleotides (heterodimers that mimic the soloRNA design) were designed and tested in HEK293 cells to investigate the capability of the RNAi machinery to accommodate structurally diverse compounds. The size of the guide strand of the heteroduplex was fixed at 19, 23, and 27 nt for the 8-, 10-, and 12-bp stems, respectively. The loop size was varied through the sequence of the passenger (nontargeting) strand of the duplex. Duplexes were transfected at either 0.5 nM (white bar) or 5 nM (black bar). The panel below the structures depicts a native polyacrylamide gel analyzing the efficiency of the duplex formation. The gel image has been inverted for visual clarity. (C) A panel of soloRNAs (homodimers with stem lengths varying between 10 and 14 bp) targeting MAP4K4 were transfected into HEK293 cells at 1 nM, and gene expression (normalized to PPIB) was evaluated at 48 h. Matching stem length luciferase-targeting soloRNAs were used as negative controls. Untreated cells and a MAP4K4 targeting conventional RNAi duplex (duplex 11546) (Supplemental Table 1) were used as positive controls. Error bars represent the standard deviation of biologic triplicates.
To explore the structural requirements for the bulged dsRNAs to enter and activate RISC, we synthesized a panel of heterodimers where the helix and loop sizes were altered systematically. Heterodimers were used in this case to minimize the potential contribution of multiple conformations on RNAi efficacy (see also Fig. 2 and below). MAP4K4 mRNA silencing in HEK293 cells was used as a functional readout (Fig. 1B). Bulges as large as 14 nt (in total) were well-tolerated when flanked by 10-bp duplexes. Naturally occurring RISC substrates (pre-miRNAs) are known to have bulges of up to 4 nt (Lagos-Quintana et al. 2001), but the effect of larger bulges on RISC recognition has not been described. Constructs with duplex regions of 8 bp or shorter tolerated small bulges, while introduction of larger bulges resulted in decreased functionality. Such relatively large bulges flanked with 10- or 12-bp duplex regions supported RNAi activity well. Based on these results, various homodimer constructs were designed with duplex regions of 10–14 bp (noted as stem lengths) and containing a 19-nt continuous stretch complementary to the target starting from the 5′ end of the oligo (Fig. 1C). All molecules (with the exception of 10-bp-containing structures) were fully functional, demonstrating that single-oligo, bulge-containing homodimer duplexes can be used for effective gene silencing.
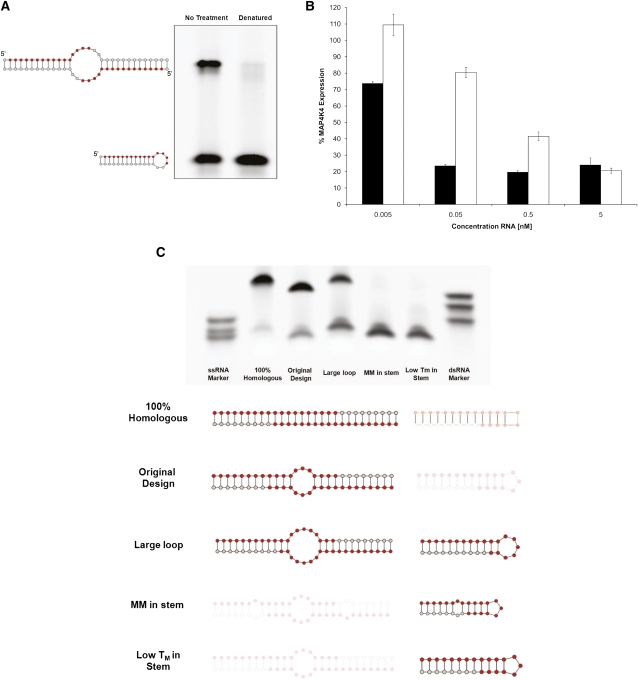
FIGURE 2.
Single oligo compound conformation (monomer versus homodimer) affects RNAi activity. (A) Native polyacrylamide gel electrophoresis demonstrates the ability of the soloRNAs to fold into two conformations—the homodimer (upper band) and the self-annealing mini-hairpin monomer (lower band). The monomer formation is favored upon heat-denaturation followed by snap cooling at low concentration (right lane). The gel image has been inverted for visual clarity. (B) Analysis of the efficacy of original (black bars) and monomer-enriched conformations (white bars) in tissue culture experiments reveals substantial differences in efficacy. MAP4K4-targeting constructs were transfected at various concentrations in HEK293 cells, and gene expression was measured after 48 h using the QuantiGene bDNA assay. Error bars represent the standard deviation of biological triplicates. (C) Native gel analysis of a panel of RNA oligonucleotides with various stem and loop structures. Lack of shading (bottom panel) indicates a preferred conformation based on native polyacrylamide gel (top panel). Oligonucleotides were resuspended at 10 mM, and a fraction of the sample (10 pmol) was run on a native 20% polyacrylamide gel. The gel was run alongside ssRNA markers (17, 19, and 25 nt; left-most lane; bottom to top) and dsRNA markers (17, 19, and 25 bp; right-most lane; bottom to top). The gel was stained with SYBR Green, and the image was inverted to increase visual clarity.
As expected and demonstrated by native polyacrylamide gel electrophoresis (PAGE) analysis, soloRNA can exist in two conformations: a homodimeric duplex and a monomeric small hairpin (Fig. 2A, left lane). To demonstrate that the homodimer is the active conformation, we used conditions to preferentially form the soloRNA monomeric hairpin (heat denaturation and snap cooling at relatively low [100 uM] concentration) (Fig. 2A, right lane). Transfection with such samples resulted in an approximately >10-fold reduction in efficacy when compared to the original samples, suggesting that the monomeric hairpin is a less active or inactive conformation (Fig. 2B; see Supplemental Fig. 1 for a detailed dose response analysis and EC50 calculations).
To investigate the conditions required for preferential homodimer formation, multiple factors were varied—salt concentration, annealing temperature, oligonucleotide concentration, sequence composition, algorithm-predicted conformations—and the resultant soloRNAs were analyzed by native PAGE (Fig. 2C; Supplemental Fig. 2). The results indicated that stem destabilization by introducing mismatches and increasing the loop size both had a negative impact on homodimer formation. An increase in stem stability and minimization of loop size are, therefore, predicted to be favorable for generation of potent soloRNAs. The annealing conditions were further optimized to minimize the kinetic advantage of monomeric self-annealing. Consistent with expectations, denaturation of the samples and re-annealing at a very high concentration (>5 mM) favors second-order chemical reaction resulting in an increase in dimer formation (Fig. 2C, original design; Supplemental Fig. 2B). Following homodimer formation, the molecule remained stable as a duplex for an extended period of time and after multiple serial dilutions (data not shown). Another attribute of the soloRNA structure may make it a substrate for Dicer cleavage. We evaluated the susceptibility of unmodified soloRNAs and soloRNA-like structures to be Dicer substrates (Supplemental Fig. 3). The sequence of the soloRNA has a large influence on its folding, just as the structure has an influence on Dicer processing. The central bulge (loop) size and the stem size contribute an affinity or ability for the Dicer enzyme to cleave the RNA target. In addition to structure, application of chemical modifications to RNA structures will prevent and inhibit Dicer cleavage in vitro and in vivo, allowing for the 5′ end and seed sequence of the RNA to remain constant, thus giving a target cleavage site across from the 10th nucleotide (Salomon et al. 2010).
Numerous studies have been conducted to establish the rules and algorithms for selecting conventional siRNA molecules with silencing activity on a given target (Li and Cha 2007). We demonstrated that a significant fraction of functional siRNA sequences can be simply converted into the soloRNA configuration. Conversion was done by assigning the sequence of the 5′ end of the siRNA's guide strand to the 5′ segment of the soloRNA, extended by additional nucleotides to allow the self-dimerization. A panel of such converted soloRNAs has been tested, and at least 50% of them produced silencing comparable to the corresponding siRNAs (Fig. 3). Consistent results were obtained for another six gene targets (data not shown). As is the case with conventional siRNAs, de novo screening for functional soloRNAs identified both active and inactive compounds (Supplemental Fig. 4). Titration curves obtained with the most potent soloRNAs demonstrated EC50s in the range of 10 pM, comparable to the best-known conventional siRNAs (Supplemental Fig. 5). Development and refinement of bioinformatic algorithms is expected to further improve the target hit rate and overall efficacy. Thus, soloRNAs can be an alternative for use in both functional genomics and therapeutics.
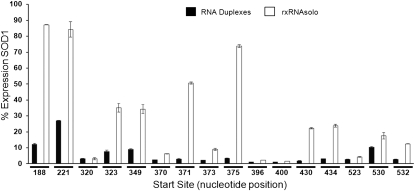
FIGURE 3.
Identification of highly potent soloRNA compounds. A panel of soloRNAs corresponding (identical seed region) to previously identified functional 25-bp RNAi duplexes against the SOD1 gene were transfected at 1 nM into HEK293 cells. SOD1 expression (normalized to PPIB) was evaluated at 48 h using a bDNA assay. Error bars represent the standard deviation of biological triplicates.
All commercially available synthetic RNAi compounds require synthesis and equimolar annealing of two independent 21–27-nt RNA oligonucleotides. In contrast, the soloRNA requires synthesis of just one 25–28-nt oligo which does not require quantification before the annealing step. This may make high-throughput production of RNAi reagents more efficient and less expensive. The de facto absence of the “passenger” strand in soloRNAs may also improve specificity by eliminating the potential miRNA-associated off-target effects through only having a single seed sequence in the molecule, whereas conventional RNAi molecules have two potential seed sequences. Furthermore, the soloRNA configuration offers an added advantage for therapeutic RNAi development by reducing the cost of large-scale synthesis and significantly simplifying the QA/QC process. The soloRNA is amenable to chemical modification to introduce favorable therapeutic properties. For example, modified soloRNAs have an increased half-life in serum and a reduced capacity to induce the interferon response when tested in vitro (Supplemental Figs. 6, 7).
The systematic analysis of the structural requirements for RISC substrate recognition described above (Fig. 1) demonstrated that molecules with a bulge flanked by two relatively small duplexes are able to induce gene silencing. This concept was applied to the design of a dual-targeting RNAi compound consisting of a bulge-containing duplex formed by two oligos, each targeting a different gene. Each strand in this dual-targeting design contains a 16–19-nt stretch complementary to a corresponding target and an extended 3′ end to allow dimerization and formation of the “stem–loop–stem” structure similar to the soloRNA (Fig. 4A). A dual-targeting construct was tested for ability to silence both the SOD1 (blue squares) and PPIB (red circles) genes (Fig. 4B). The dual-targeting compound was highly efficacious in silencing both genes with EC50 values of ∼100 pM. This configuration can be used to design RNAi compounds that can silence two genes as long as the seed pairs are selected to have similar RISC loading activity. Dual-targeting RNAi compounds could be particularly useful for therapeutic applications such as cancer, where targeting multiple genes at once may be essential for an effective treatment.
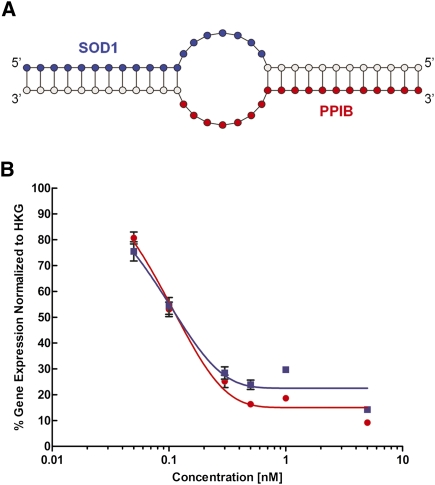
FIGURE 4.
Design and testing of dual-targeting RNAi compounds. (A) Dual targeting RNAi compound structure: A bulge-containing duplex is formed by two oligos, each containing 19 nt targeting for two different genes. The 3′ portions serve as adaptors to allow formation of the imperfect (containing internal bulge) duplex reminiscent of the homodimer soloRNA shown in Figure 1A. The guide portions for SOD1 (blue) and PPIB (red) are highlighted. (B) Dose response of dual targeting RNAi compounds: SOD1 (blue) or PPIB (red). HEK293 cells were transfected with varying concentrations of the RNAi construct, and gene silencing was measured 24 h post-transfection in reference to the housekeeping gene (HKG; MAP4K4, in this case).
The results described in this paper demonstrate the flexibility of the RNAi machinery and its capability to accommodate novel, unconventional compounds that have advantageous properties for both research and therapeutic use.
MATERIALS AND METHODS
Synthesis of single-stranded and double-stranded RNAs
All single-stranded RNAs were synthesized and desalted by Integrated DNA technologies. All double-stranded RNAs were synthesized by Dharmacon RNAi Technologies (ThermoScientific). Sequences of single-stranded and double-stranded RNAs are listed in Supplemental Tables 1 and 2.
Cell culture, transfection, and gene expression analysis
HEK293 cells (ATCC) were cultured in Dulbecco's Modified Eagle Media (DMEM) with 10% Fetal Bovine Serum and 1% penicillin/streptomycin. Cells were incubated at 37°C with 10% CO2 as recommended by ATCC. Cultured cells were reverse-transfected in 96-well plates using antibiotic-free media and Lipofectamine RNAiMAX (Invitrogen) transfection reagent as described by the manufacturer. Transfection complexes were prepared with a concentration of active duplex ranging from 0.005 nM to 10 nM, and the final concentration of RNA was adjusted to 25 nM using the RISC-free filler RNA control (Dharmacon). Cells were lysed 24 h or 48 h after transfection, and then mRNA levels were measured using the QuantiGene bDNA hybridization assay (Affymetrix) as described by manufacturer instructions. Gene specific probes (Affymetrix) were used to detect target and housekeeping gene mRNA expression levels. Target gene expression was measured by normalizing to a housekeeping gene (PPIB in all cases, except the experiments described in Fig. 4) that is unaffected by the specific target gene silencing. The percent silencing and EC50 values were based on comparing the normalized values of the samples transfected to that of the normalized expression for the RISC-free filler alone (referred as nontargeting control, NTC) or untreated cells (UTC). Error bars represent the standard deviation of biological triplicates.
Analysis and visualization of RNA structure formation
Lyophilized oligonucleotides were resuspended to 10 mM using 1× siRNA buffer (60 mM KCl, 6 mM HEPES pH 7.5, 0.2 mM MgCl2), unless otherwise noted. The monomer structure of the soloRNA was created by incubation at 95°C for 5 min then immediately placing on ice. The structures were confirmed on a native 20% polyacrylamide TBE-buffered gel. For folding optimization experiments, 10 pmol of RNA was loaded on a 20% polyacrylamide TBE-buffered gel. Single-stranded and double-stranded RNA markers (New England Biolabs) were run alongside samples for size comparison. The single-stranded marker sizes were 17, 19, and 25 nt, bottom to top, and double-stranded RNA markers (17, 19, and 25 bp, bottom to top). Gels were stained with SYBR Green II (Invitrogen) for 20 min and then visualized using a UV transillumintor in the Biochemi imaging system (UVP). Images were captured using the LabWorks software (UVP), and the images were inverted to increase visual clarity. RNA structures were generated using RNA Structure, version 5.02 (Mathews Lab, University of Rochester Medical Center) (Mathews et al. 2004).
Serum stability assay
The soloRNAs were combined with either human serum (Bioreclamaton) or RNase-free water and incubated at 37°C for 0, 0.5, 6, 12, 24, or 48 h. After incubation the reaction was stopped by addition of 9 volumes 2× Proteinase K buffer (200 mM Tris-HCl pH 7.5, 25 mM EDTA, 300 mM NaCl, 2% [w/v] SDS with Proteinase K concentration at 1.2 mg/mL) and incubation at 65°C for 15 min. RNA was isolated by a phenol-chloroform extraction, heat-denatured, and then loaded onto a 20% nondenaturing polyacrylamide gel. The gel was stained with SYBR Gold II (Invitrogen) and visualized under a UV transilluminator. All images were inverted to increase visual clarity. The RNA stability half-life was measured by densitometry of the individual time points respective of the untreated RNA (time 0) using LabWorks software (UVP). The densitometry values were graphed and fit to an exponential decay curve with the half-life calculated using Graphpad Prism 5 (GraphPad Software).
HeLa IFN induction assays
HeLa S3 cells were reverse-transfected with 50 nM soloRNAs or RNA duplexes as described above, with the exception of the Poly I:C (Sigma) positive control, which was transfected at a final concentration of 5 ng/mL. Following incubation for 24 h, total cell lysate was generated and collected for QuantiGene bDNA analysis (Affymetrix/Panomics) as described by the manufacturer. The bDNA assay was carried out according to manufacturer recommendations using a species-specific probe set to IFIT-1 and the PPIB housekeeping gene control for normalization. Percentage IFIT-1 gene induction was calculated. All transfections were carried out as biological triplicates. Fold immune induction was calculated with respect to the nonstimulatory RNA duplex (11546, 2′OMe RNA duplex). Poly I:C (Sigma) was used at 5 ng/ul as a positive control for stimulating IFIT gene expression.
PBMC IFN induction assay
Human PBMCs (Astarte Biologics) were thawed according to manufacturer recommendations, then seeded in triplicate in a 96-well plate at 2 × 105 cells/well in RPMI 1640 medium with 10% FBS, 2 mM L-glutamine, and 1% penicillin/streptomycin. Cells were incubated at 37°C and 5% CO2 for 24 h, then transfected with soloRNAs or RNA duplexes using Lipofectamine 2000 (Invitrogen) according to manufacturer recommendations. Following incubation for 24 h, total cell lysate was generated and collected for QuantiGene bDNA analysis (Affymetrix/Panomics) as described by the manufacturer. The bDNA was carried out according to manufacturer recommendations using human probe sets to IFIT-1 or TNF and the PPIB housekeeping gene control for normalization. Fold immune induction was calculated with respect to untreated cells.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
The authors thank Craig Mello (University of Massachusetts Medical School), Greg Hannon (Cold Spring Harbor Laboratory), and Victor Ambros (University of Massachusetts Medical School) for critical review of these data and helpful discussion, and Olke Uhlenbeck (Northwestern University) for his advice on homodimer folding conditions. We thank the staff of RXi Pharmaceuticals for support and helpful discussion while carrying out these studies and in preparation of the manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2399411.
REFERENCES
- Castanotto D, Rossi JJ 2009. The promises and pitfalls of RNA-interference-based therapeutics. Nature 457: 426–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CY, Rana TM 2008. Potent RNAi by short RNA triggers. RNA 14: 1714–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossbach M, Gruber J, Osborn M, Weber K, Tuschl T 2006. Gene silencing with siRNA duplexes composed of target-mRNA-complementary and partially palindromic or partially complementary single-stranded siRNAs. RNA Biol 3: 82–89 [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838 [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853–858 [DOI] [PubMed] [Google Scholar]
- Li W, Cha L 2007. Predicting siRNA efficiency. Cell Mol Life Sci 64: 1785–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563–574 [DOI] [PubMed] [Google Scholar]
- Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH 2004. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci 101: 7287–7292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison PJ, Cleary M, Silva JM, Chang K, Sheth N, Sachidanandam R, Hannon GJ 2004. Cloning of short hairpin RNAs for gene knockdown in mammalian cells. Nat Methods 1: 163–167 [DOI] [PubMed] [Google Scholar]
- Patel DJ, Ma JB, Yuan YR, Ye K, Pei Y, Kuryavyi V, Malinina L, Meister G, Tuschl T 2006. Structural biology of RNA silencing and its functional implications. Cold Spring Harb Symp Quant Biol 71: 81–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon W, Bulock K, Lapierre J, Pavco P, Woolf T, Kamens J 2010. Modified dsRNAs that are not processed by Dicer maintain potency and are incorporated into the RISC. Nucleic Acids Res 38: 3771–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siolas D, Lerner C, Burchard J, Ge W, Linsley PS, Paddison PJ, Hannon GJ, Cleary MA 2005. Synthetic shRNAs as potent RNAi triggers. Nat Biotechnol 23: 227–231 [DOI] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ 2008. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 456: 921–926 [DOI] [PMC free article] [PubMed] [Google Scholar]