Abstract
The TREX (transcription/export) complex has been conserved throughout evolution from yeast to man and is required for coupled transcription elongation and nuclear export of mRNAs. The TREX complex in mammals and Drosophila is composed of the THO subcomplex (THOC1, THOC2, THOC5, THOC6, and THOC7), THOC3, UAP56, and Aly/THOC4. In human and Drosophila, various studies have shown that THO is required for the export of heat shock mRNAs, but nothing is known about other mRNAs. Our previous study using conditional THOC5 (or FMIP) knockout mice revealed that the presence of THOC5 is critical in hematopoietic cells but not for terminally differentiated cells. In this study, we describe the establishment of a mouse embryo fibroblast cell line (MEF), THOC5 flox/flox. Four days after infection of MEF THOC5 flox/flox with adenovirus carrying Cre-recombinase gene (Ad-GFP-Cre), THOC5 is down-regulated >95% at the protein level, and cell growth is strongly suppressed. Transcriptome analysis using cytoplasmic RNA isolated from cells lacking functional THOC5 reveals that only 2.9% of all genes were down-regulated more than twofold. Although we examined these genes in fibroblasts, one-fifth of all down-regulated genes (including HoxB3 and polycomb CBX2) are known to play a key role in hematopoietic development. We further identified 10 genes that are spliced but not exported to the cytoplasm in the absence of THOC5. These mRNAs were copurified with THOC5. Furthermore, Hsp70 mRNA was exported in the absence of THOC5 at 37°C, but not under heat shock condition (42°C), suggesting that THOC5 may be required for mRNA export under stress and/or upon signaling-induced conditions.
Keywords: mRNA export complex THOC, Fms interacting protein (FMIP), transcriptome analysis, conditional knockout mouse fibroblasts, mRNP complex
INTRODUCTION
During expression of protein-coding genes, pre-mRNAs are transcribed in the nucleus and undergo several RNA-processing steps. The mature mRNA is then exported from the nucleus to the cytoplasm for translation. The TREX (transcription/export) complex has been shown to contribute to cotranscriptional mRNP assembly and mRNA export from the nucleus (Reed and Hurt 2002; Aguilera 2005; Reed and Cheng 2005; Kohler and Hurt 2007). The subcomplex of TREX, the THO complex, was first identified in Saccharomyces cerevisiae as a five-protein complex (THO2/THOC2, Hpr1/THOC1, Mft1, Thp2, and Tex1/THOC3) that plays a role in transcriptional elongations, nuclear RNA export, and genome stability (Chavez and Aguilera 1997; Piruat and Aguilera 1998; Chavez et al. 2001; Jimeno et al. 2002; Strasser et al. 2002). In higher eukaryotes such as Drosophila melanogaster or human, three proteins (THOC1, THOC2, and THOC3) and three additional unique proteins were identified, namely, THOC5/FMIP/fSAP79, THOC6/fSAP35, and THOC7/fSAP24 (Rehwinkel et al. 2004; Masuda et al. 2005). In the yeast system, it was previously shown that yeast strains lacking THO complex components showed nuclear accumulation of poly(A)+ RNA at the restrictive temperature (Gallardo et al. 2003). However, in the Drosophila system, only 20% of all genes were suppressed by depletion of THOC1 and THOC2 (Rehwinkel et al. 2004). In addition, using the HeLa human cell line, it has been shown that depletion of THOC5 did not cause an accumulation of poly(A)+ RNAs in the nucleus, but depletion of Tap-p15 or Aly causes a dramatic accumulation of poly(A)+ RNAs in the nucleus (Katahira et al. 2009), suggesting that Aly and Tap-p15, but not THOC5, represent a crucial function in the nuclear export of a wide range of poly(A)+ RNA. It remains uncertain which genes are THO function-dependent. THOC5/Fms-interacting protein (FMIP), a member of the THO complex, was originally identified as a substrate for the Macrophage Colony Stimulating Factor (M-CSF) receptor tyrosine kinase, Fms (Tamura et al. 1999). THOC5 is phosphorylated not only by several tyrosine kinases (Pierce et al. 2008), but also by protein kinase C (Mancini et al. 2004), by the downstream kinase from insulin stimulus (Gridley et al. 2005) or ATM kinase (Matsuoka et al. 2007), suggesting that extracellular stimulation regulates the function of THOC5. We have previously shown that depletion of THOC5 by siRNA or ectopic expression causes abnormal hematopoiesis and abnormal muscle differentiation in myeloid progenitor or mesenchymal progenitor cell lines, indicating that the THO complex is essential for the differentiation process in mammals (Tamura et al. 1999; Mancini et al. 2007; Carney et al. 2009). Furthermore, using interferon inducible THOC5 knockout mice, we have shown that THOC5 is essential at an early stage of mouse development and that this gene is essential for survival in adult mice. In these knockout mice, bone marrow cells become apoptotic, hematopoietic progenitor cell numbers collapse, and the animals become anemic. Although the THOC5 gene was deleted in liver, kidney, and heart, pathological alterations to these organs were not observed (Mancini et al. 2010). These data suggest that the THO complex plays a key role in early embryogenesis and in hematopoietic differentiation but does not play a role in differentiated cells. To obtain further insight into the role of THOC5 at a molecular level, we performed a transcriptome analysis using mouse embryo fibroblasts (MEF THOC5flox/flox) in the presence or absence of THOC5. The depletion of THOC5 expression suppressed the cell growth of MEF THOC5flox/flox cells. Surprisingly, only 2.9% of the genes are affected by the depletion of THOC5. We further identified mRNAs that were detected in the nucleus as spliced forms but were not exported into the cytoplasm in the absence of THOC5. Furthermore, these mRNAs were copurified with THOC5, suggesting that these are THO complex-dependent.
RESULTS
Depletion of the THOC5 gene in fibroblasts down-regulates cell growth
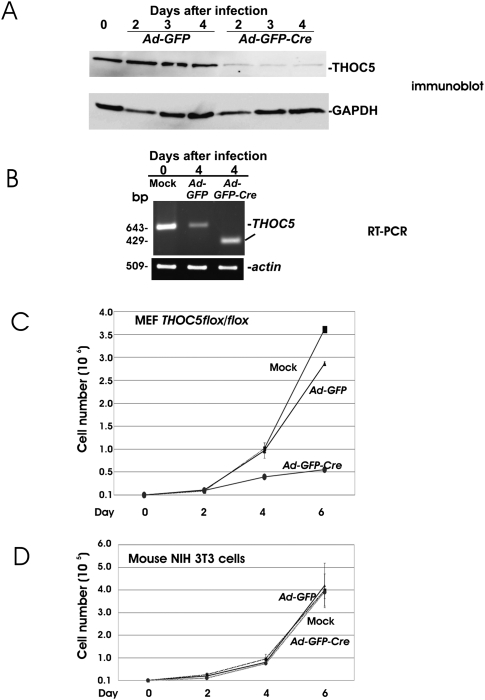
We have reported previously on the THOC5 flox/flox mouse that is generated by flanking exons IV and V of the THOC5 gene with loxP sites (Mancini et al. 2010). To examine gene regulation by THOC5, we first established a mouse embryo fibroblast (MEF) THOC5 flox/flox cell line. Upon infection of MEF THOC5 flox/flox with adenovirus carrying the Cre-recombinase gene (Ad-GFP-Cre), THOC5 is down-regulated >95% within 4 d at the protein level (Fig. 1A). As control, we infected the same cells with adenovirus carrying only the GFP gene (Ad-GFP). The level of THOC5 did not change after control virus infection. By RT-PCR, a PCR product lacking exons IV and V (429 bp) was detected after infection with Ad-GFP-Cre virus (Fig. 1B), indicating that delta exons IV/V transcript was synthesized in the cells. We have previously shown that the depletion of the THOC5 gene causes apoptosis of hematopoietic cells (Mancini et al. 2010). To examine whether the depletion of THOC5 alters the phenotype of fibroblasts, we examined the growth rate of THOC5-deficient fibroblasts. As shown in Figure 1C, THOC5-deficient fibroblasts slow down growth drastically 2 d after Ad-GFP-Cre virus infection. In agreement with these data, yeast THO-null mutants are slow growers (Chavez et al. 2001; Garcia-Rubio et al. 2008). As control, mouse fibroblast cell line NIH3T3 cells were infected with Ad-GFP or Ad-GFP-Cre virus. NIH 3T3 cells grew equally well before and after infection with Ad-GFP or Ad-GFP-Cre virus (Fig. 1D). Furthermore, the deletion of THOC5 in hematopoietic cells causes apoptosis (Mancini et al. 2010); however, no apoptotic cells were detected by poly (ADP-ribose) polymerase cleavage assay (data not shown), suggesting that the THO complex may play different roles in the differentiation/proliferation of distinct cell types.
FIGURE 1.
Depletion of the THOC5 gene in fibroblasts down-regulates cell growth. (A) MEF THOC5 flox/flox were infected with adenovirus carrying the GFP gene (Ad-GFP) or adenovirus carrying the GFP and Cre-recombinase genes (Ad-GFP-Cre) (Vector Biolab). Cell extracts were then examined by THOC5- and GAPDH-specific immunoblot. (B) RNAs were isolated from cells treated as above and were then analyzed by RT-PCR using actin- and THOC5-specific primers: forward primer: 5′-tctgccttttcacctggaag-3′; reverse primer: 5′-ctcggtacttttctgccagc-3′ (product size = 643 bp [wild type] or 429 bp [without exon IV/V]). (C,D) 104 MEF THOC5 flox/flox or mouse NIH3T3 cells were infected with Ad-GFP or Ad-GFP-Cre virus, and cells were counted each day. Results represent the average values ±SD from three independent experiments.
The relevance of the THO function in mammals is limited to a subset of genes, but key genes for mouse development and hematopoiesis
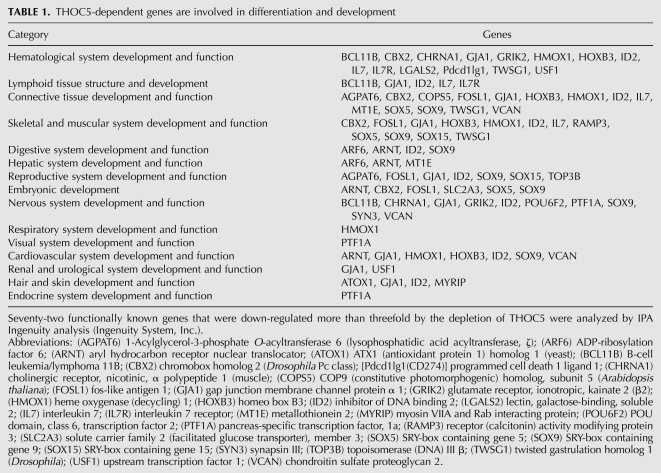
It has been previously suggested that the metazoan THO complex does not play a genome-wide role (Rehwinkel et al. 2004; Farny et al. 2008; Katahira et al. 2009). To examine whether the significance of the THOC5 function in mammals is a consequence of a general genome-wide role or whether its role is limited to a subset of genes, we applied a microarray technique using THOC5-depleted fibroblasts. The cytoplasmic RNA was isolated from cells infected with Ade-GFP-Cre, Ade-GFP virus, or mock-infected on days 2 and 4 and then applied for Affymetrix transcriptome analyses. We selected genes that were down-regulated more than twofold by Ad-GFP-Cre virus infection but were down-regulated <1.5-fold by Ad-GFP control virus. By selecting genes that were down-regulated more than twofold, 559 genes (2.9%) were found. Similarly, in the Drosophila system, ∼20% of genes were down-regulated by depletion of THOC1 and 2 (Rehwinkel et al. 2004), indicating that the THO complex plays a role in expression of a subset of genes. Furthermore, only 19 genes were up-regulated more than twofold by the depletion of THOC5. Among 198 genes that were down-regulated more than threefold by depletion of THOC5, 143 genes were functionally known genes (Supplemental Table 1). We uploaded these lists to the Ingenuity Pathway Analysis (IPA, Ingenuity System, Inc.) application for biological function analysis. Seventy-one out of 143 genes mapped to the IPA knowledge database. Forty-five percent of the genes are involved in development and cell differentiation (Table 1). This is in agreement with the previous data that in differentiating cells THO depletion has a severe effect. It has been shown that THOC1 and THOC5 are required for viability of the early mouse embryo (Wang et al. 2006, 2009; Mancini et al. 2010). As shown in Table 1, six genes are involved in embryo development. The expression of one of these genes, solute carrier family 2 (facilitated glucose transporter), member 3 (Slc2a3 or Glut3) was reduced 7.4-fold (Supplemental Table 1). Mutation of this gene causes early pregnancy loss and fetal growth restriction (Ganguly et al. 2007), suggesting that this gene may participate in early embryonic lethality of THOC5−/− mice (Mancini et al. 2010). In addition, we have shown previously that overexpression of THOC5 enhances muscle differentiation of C2C12 cells and down-regulation of THOC5 suppressed muscle differentiation (Mancini et al. 2007). In agreement with these data, 12 down-regulated genes are involved in skeletal and muscle development. Our previous study also showed that hematopoietic differentiation was impaired severely by THOC5 depletion in adult mice (Mancini et al. 2010). Although we analyzed the down-regulated genes in fibroblasts, one-fifth of all down-regulated genes, including homeobox B3 (HoxB3) and polycomb CBX2 are known to play a key role in hematopoietic development. These data indicate that THOC5 plays a role in the export of only a subset of genes, but it seems to play an important role in mouse development and the differentiation processes. Since the deletion of THOC5 strongly affected fibroblast proliferation, we examined the genes that are involved in cell proliferation. Indeed, one-fifth of down-regulated genes are involved in cell proliferation. In addition, among 559 genes, 50 genes were known to be transcriptional regulators, indicating that not all 559 genes are regulated directly by THOC5.
TABLE 1.
THOC5-dependent genes are involved in differentiation and development
Identification of THOC5-dependent mRNA
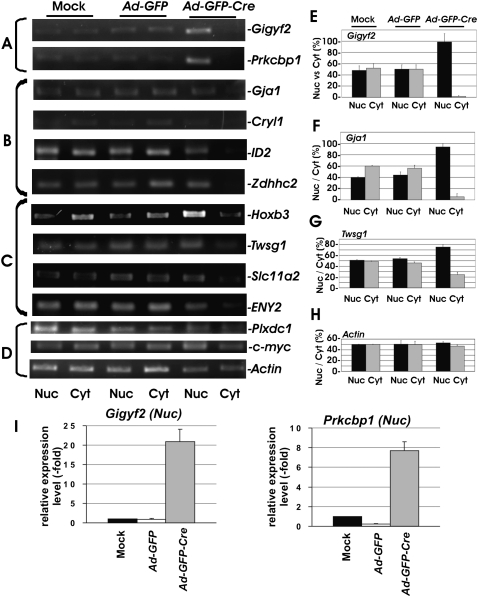
Since yeast TREX is important for both transcription and mRNA export (Aguilera 2005), we further addressed the mechanism through which THOC5 regulates expression of the identified genes. To identify the genes that are inhibited to export by the depletion of THOC5, we next examined the nuclear export of mRNAs that are down-regulated more than threefold. We isolated nuclear and cytoplasmic RNA from THOC5-depleted MEFs, and samples were standardized by equal levels of actin mRNA in both fractions (Fig. 2D,H, Actin). Then, we analyzed 10 candidate genes, such as Grb10 interacting GYF protein 2 (Gigyf2 or Tnrc15), protein kinase C binding protein 1 (Prkcbp1), gap junction membrane channel protein alpha 1 (Gja1), twisted gastrulation homolog 1 (Twsg1), zinc finger, enhancer of yellow 2 homolog (ENY2), crystalline, lamda 1 (Cryl1), solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 (Slc11a2), zinc finger, DHHC domain containing 2 (Zdhhc2), inhibitor of DNA binding 2 (ID2), or HoxB3 genes by RT-PCR. In this experiment, only spliced forms of mRNA can be detected since we have chosen the primer pair for RT-PCR that is located at two different exons. As shown in Figure 2, nuclear accumulation of the spliced form of Gigyf2 and Prkcbp1 mRNA but almost no cytoplasmic mRNAs was observed after the depletion of THOC5 (Fig. 2A,E, Gigyf2). Additionally, four genes—Gja1, Cryl1, ID2, and Zdhhc2 mRNAs—were hardly detected in the cytoplasm; however, no nuclear accumulation of these genes was observed (Fig. 2B,F, Gja1). The export of HoxB3, Twsg1, Slc11a2, and ENY2 mRNAs was only reduced after the depletion of THOC5 (Fig. 2C,G, Twsg1). As control, genes that were not down-regulated in our transcriptome data, such as c-Myc, plexin domain containing 1 (plxdc1), and actin genes, were examined. As expected, these genes were detected equally in all fractions (Fig. 2D,H, actin). Interestingly, we observed the spliced form of all mRNAs that were examined here. In agreement with these data, it has been reported that human THO associates with proteins of the spliceosome and with spliced RNAs, this latter interaction being independent of transcription (Masuda et al. 2005; Cheng et al. 2006). In addition, to quantify the nuclear accumulation of Gigyf2 and Prkcbp1 mRNA in the nuclear fraction of Mock MEF THOC5 flox/flox, and MEF THOC5 flox/flox infected with Ad-GFP or Ad-GFP-Cre virus, we performed TaqMan-RT-PCR. Relative expression levels compared to hypoxanthine guanine phosphoribosyl transferase 1 (HPRT1) were normalized to gene expression in mock-infected MEF THOC5 flox/flox. In agreement with RT-PCR data (Fig. 2A,E), in the absence of THOC5, Gigyf2 or Prkcbp1 mRNA was accumulated 21-fold or 7.8-fold, respectively, compared to the level in the presence of THOC5 (Fig. 2I). To validate our assay, we treated MEF THOC5 flox/flox cells with Aly-specific siRNA, which has been shown to cause an accumulation of poly(A)+ mRNA in the nucleus in human cells (Katahira et al. 2009) and to be a crucial export factor for a wide range of poly(A)+ mRNAs in the Drosophila system (Gatfield and Izaurralde 2002). We then examined the expression of nuclear export of THOC5-dependent genes (Gigyf2 and Prkcbp1) and THOC5-independent genes (c-Myc and Plxdc1).
FIGURE 2.
Down-regulation of mRNA export by the depletion of THOC5 in fibroblasts. MEF THOC5 flox/flox cells were infected with adenovirus carrying the GFP gene (Ad-GFP) or adenovirus carrying the GFP and Cre-recombinase genes (Ad-GFP-Cre). Four days after infection, nuclear (Nuc) and cytoplasmic (Cyt) RNAs were isolated and applied for RT-PCR using primers as described in Materials and Methods. cDNA from all samples were standardized by an equal level of actin mRNA in both fractions. We have performed three to five independent experiments, and we show one example of representative data (A–D). (A) mRNAs were accumulated in the nucleus and were not exported. (B) mRNAs were not accumulated in the nucleus and also were not exported. (C) mRNAs were not accumulated in the nucleus, and mRNA export was reduced. (D) There was no significant alteration by the depletion of THOC5. (E–H) Signal intensity from Gigyf2- (E), Gja1- (F), Twsg1- (G), and actin-specific (H) RT-PCR products was quantified using TINA 2.0 software. The percent signal intensity from the nuclear (Nuc) or the cytoplasmic (Cyt) fraction of total intensity (Nuc + Cyt) in Mock MEF THOC5 flox/flox (Mock) or MEF THOC5 flox/flox infected with Ad-GFP or Ad-GFP-Cre. Average values ±SD from three independent experiments are shown. (I) Aliquots of cDNA samples from nuclear fractions of A were applied for quantitative RT-PCR analysis of Gigyf2 and Prkcbp1 mRNA. Relative expression levels compared to HPRT1 were normalized to gene expression in mock-infected MEF THOC5 flox/flox. Average values from four independent PCR reactions ±SEM are shown.
In agreement with these data, Aly knockdown caused the suppression of mRNA export of all four genes that were examined (data not shown). Since two genes, Gigyf2 and Prkcbp1, were clearly accumulated in the nuclear fraction after the depletion of THOC5 (Fig. 2E), we further examined the complex formation of THOC5 with these two genes.
Gigyf2 and Prkcbp1 are associated with a complex containing THOC5
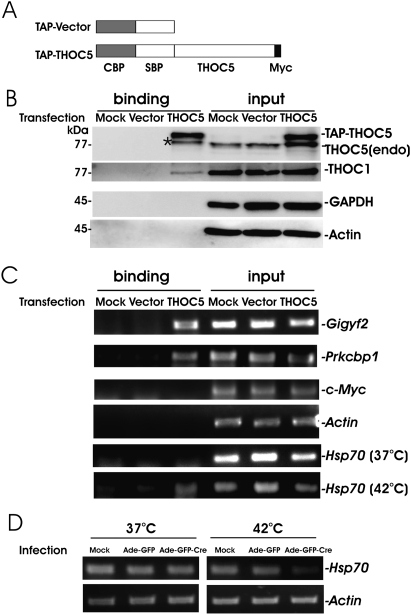
To further examine whether the identified mRNAs are regulated directly by THOC5, we examined whether these mRNAs form a complex with THOC5. For this experiment, the streptavidin-binding peptide (SBP) was fused to Myc-tagged THOC5 (Fig. 3A) and expressed in MEF THOC5 flox/flox cells, then THOC5-containing mRNP was purified with streptavidin conjugated Sepharose from cell extracts (Fig. 3B). Empty vector was transfected into MEF THOC5 flox/flox cells as negative control. As shown in Figure 3B, mRNP with THOC5 contains only a slight amount of THOC1. It may be due to the fact that endogenous THOC1 forms a tight complex with endogenous THOC5 and most of the exogenous TAP-THOC5 is not able to form a new complex. Two bands of 89- and 79-kDa proteins were detected from TAP-THOC5 transfectants by THOC5-specific immunoblot, indicating that the upper band represents exogenous TAP-THOC5 and the lower band represents endogenous THOC5 (Fig. 3B, input). After the purification with streptavidin conjugated Sepharose, two bands were also detected (Fig. 3B, binding). The lower band (Fig. 3B, asterisk [*]) may be the result of the degradation product of TAP-THOC5 or THOC5 forming a homodimer with endogenous THOC5. In the same mRNP complex, bound mRNAs were purified and applied for Gigyf2-, Prkcbp1-, c-Myc-, and actin-specific RT-PCR (Fig. 3C). Gigyf2 and Prkcbp1 genes, but not c-Myc and actin genes, were detected from the THOC5 RNP complex, and no mRNA was detected in the RNP complex obtained from mock and vector control cells (Fig. 3C).
FIGURE 3.
Isolation of mRNP complex by THOC5 as bait. pNTAP (TAP-Vector, [CBP] calmodulin-binding peptide; [SBP] streptavidin-binding peptide) and pNTAP carrying myc-tagged THOC5 (TAP-THOC5) (A) were transfected into MEF THOC5 flox/flox. After 24 h, RNP complex was isolated using streptavidin Sepharose. (B) Cell extract (input) and binding fractions with streptavidin Sepharose (binding) were analyzed by immunoblotting using THOC5-, Actin-, THOC1-, and GAPDH-specific antibodies. (C) The same samples were analyzed: Gigyf2-, Prkcbp1-, c-Myc-, actin-, or (37°C) Hsp70-specific RT-PCR. In addition, MEF THOC5 flox/flox cells were transfected as in B and were then incubated for 2 h at 42°C. After isolation of RNP, samples were applied for Hsp70 (42°C) RT-PCR. (D) The depletion of THOC5 causes down-regulation of the Hsp70 gene export under heat shock conditions. MEF THOC5 flox/flox cells were infected with Ad-GFP or Ad-GFP-Cre virus for 4 d. Cells were incubated for 2 h at 37°C or 42°C, and RNAs were then isolated from cytoplasmic fraction followed by Hsp70- or actin-specific RT-PCR.
Hsp70 mRNA is associated with a THOC5-containing complex exclusively under heat shock condition
In human and Drosophila, various studies have shown that THO is required for the export of heat shock protein (Hsp) 70 mRNAs (Rehwinkel et al. 2004; Katahira et al. 2009). Our array data in non-heat-shock condition, however, revealed that the Hsp70 gene did not significantly change the expression level after depletion of THOC5. In addition, Hsp70 mRNA was not detected in THOC5-containing mRNP at 37°C (Fig. 3C). Interestingly, THOC5–RNP complex was isolated from cells incubated for 2 h at 42°C, and Hsp70 mRNA was isolated from this complex (Fig. 3C), suggesting that THOC5 plays a role in Hsp70 mRNA export under heat shock condition. We, therefore, then examined the Hsp70 mRNA export in MEF THOC5 flox/flox cells in the presence or absence of THOC5 at 37°C or at 42°C. In agreement with previous data (Rehwinkel et al. 2004; Katahira et al. 2009), in heat shock condition, the levels of Hsp70 mRNA (Fig. 3D) as well as Hsp70 protein (data not shown) were drastically reduced in the cytoplasm in the absence of THOC5, suggesting that the Hsp70 mRNA transport mechanism may be different under the non-heat-shock condition (THOC-independent) than under the heat shock condition (THOC-dependent).
DISCUSSION
Cell differentiation is paralleled by a timely ordered expression of a set of genes. Most genes are regulated at transcriptional and epigenetic levels. However, recent data reveal that regulatory RNAs and RNA binding proteins have emerged as developmental and differentiation regulators. THOC5, a member of the mRNA export complex, is modified post-translationally under differentiation and DNA damage signaling. Firstly, M-CSF stimulation of myeloid cells causes phosphorylation of tyrosine residue 225 of THOC5 (Tamura et al. 1999; Pierce et al. 2008). Secondly, insulin signaling for adipocyte differentiation leads to threonine 328 phosphorylation (Gridley et al. 2005). Thirdly, the phosphorylation of THOC5 by protein kinase C impairs the THOC5 nuclear/cytoplasm shuttling (Mancini et al. 2004). Fourthly, DNA damage leads to phosphorylation of THOC5 by ATM kinase (Matsuoka et al. 2007), followed by inhibition of the THO complex formation (S Ramachandran and T Tamura, unpubl.). These facts suggest that the THO complex may play a role in fine-tuning differentiation processes dependent on the extracellular conditions. Our data obtained from THOC5 knockout mice reveal that hematopoietic cells not only stopped growing, but also became apoptotic (Mancini et al. 2010). On the other hand, we show here that THOC5/FMIP-deficient fibroblasts just cease growing. Similarly, yeast THO-null mutants are slow growers (Chavez et al. 2001; Garcia-Rubio et al. 2008), and THOC1 and/or THOC2 depletion has a negative effect on growth rate of the Drosophila cell line (Rehwinkel et al. 2004). Interestingly, the deletion of THOC1, a major conserved component of the THO complex, causes apoptosis in transformed cells but not in normal fibroblasts (Li et al. 2007). The bulk of these data shows that the THO complex plays different roles in distinct cell types.
Our data revealed that THOC5 is only involved in the expression of 2.9% of all genes. In agreement with our data, it has recently been shown that depletion of THOC5 does not affect bulk poly(A)+ RNA export but does affect Hsp70 mRNA export in HeLa cells under the heat shock condition (Katahira et al. 2009). It has also been shown that in the Drosophila system, 20% of all genes were down-regulated by knockdown of THOC1 and THOC2 (Rehwinkel et al. 2004). In the same system, ∼80% of all genes were down-regulated by depletion of NXF1 (TAP in mammals), UAP56, or p15, which are required for bulk poly(A)+ RNA export (Herold et al. 2001, 2003), indicating that the relevance of the THOC function is limited to a subset of genes also in the Drosophila system. These data suggest that various nuclear mRNA export pathways, which may be dictated by different adaptor RNA binding proteins, exist in higher eukaryotes. It is not yet clear, however, whether knockout of THOC5 suppresses the function of the entire THO complex or THOC5 is associated with a specific role in particular mRNA export. Although it has been shown that members of THOC form a tight complex in the nucleus (Cheng et al. 2006), the THO complex may not function as a single subunit. By affinity purification using mRNPs under physiological conditions, all members of the THO complex were copurified with Adenovirus pre-mRNA, but only THOC2 was copurified with beta globin pre-mRNA. In addition, one mutant Adenovirus pre-mRNA associated only with THOC1 (Merz et al. 2007). These data suggest that a certain population of the THO complex may be loosely associated with other THOC members, or an individual member of the THO complex binds to only a certain population of mRNAs. On the other hand, we have previously shown that knockdown of THOC5 depleted THOC1 at the protein level in mice and in MEF THOC5 flox/flox cells (Mancini et al. 2010), suggesting that the inhibition of mRNA export that we observed in our experiment might be the result of both THOC1 and THOC5 depletion.
How does the THO complex select a subset of mRNAs? The selection is not based on a common feature, such as homological domain (or motif) among the 10 mRNAs that have been identified here as THOC5-dependent mRNAs. In the yeast system, several motifs such as “GC-rich” or “internal repeats” were suggested as criteria for the THO complex dependency (Chavez et al. 2001; Voynov et al. 2006). In those reports, the THO complex is required for efficient transcription of genes with long tandem repeats upon induction or under some conditions, such as under stress and/or upon signaling-induced conditions. The genes described as THOC-dependent in this report are all spliced, but not exported efficiently and not especially GC-rich. It should be noted, however, that two genes, Gigyf2 and Prkcbp1, contain common motif CAG repeats, suggesting that “internal repeats” may be one of the factors involved in THOC dependency. Since other genes do not exhibit such a motif, it is presently not clear which structure is required for the THOC dependency.
We show here that Hsp70 mRNA is THOC5-dependent under only the heat shock condition. In agreement with these data, THOC1 depletion does not influence the expression of Hsp70 under the non-heat-shock condition in several human cancer cell lines (Li et al. 2007). Similarly, it has been discussed in the yeast system that some genes, such as TIR1 (Voynov et al. 2006), may require the THO complex upon induction or some environmental stress condition that requires enhanced transcription. Furthermore, since Hsp70 mRNA was isolated from the THOC5 complex at 42°C but not at 37°C, genes whose transcription creates aberrant structures under stress conditions could also require the THO complex.
MATERIALS AND METHODS
Cell culture and virus infection
Mouse embryo fibroblast (MEF) THOC5 flox/flox cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) FCS. Adenovirus carrying GFP and Cre-recombinase (Ad-Cre-GFP) or GFP (Ad-GFP) genes were obtained from Vector Biolab. Cells were infected with Ad-Cre-GFP or Ad-GFP at M.O.I. 200.
Western blot procedures
Cells were extracted with lysis buffer containing 10 mM Tris HCl (pH 7.6), 50 mM NaF, 1 mM PMSF, 10 mM EDTA, 1% (w/v) Triton-X 100, 0.1% SDS, and 1% Trasylol (Bayer Vital). Details of immunoblotting have been described previously (Koch et al. 2008). The monoclonal antibody against GAPDH was purchased from Santa Cruz, and the monoclonal antibody against THOC5 was generated as described previously (Mancini et al. 2007). Corresponding proteins were visualized by incubation with peroxidase conjugated anti-mouse immunoglobulin followed by incubation with SuperSignal West FemtoMaximum Sensitivity Substrate (Pierce). Results were documented on an LAS4000 imaging system (GE Healthcare Bio-Sciences). The signal intensity of chemoluminescence was quantified using TINA 2.0 software (Raytest Isotopenmessgeraete GmbH).
Transcriptome analysis
Cytoplasmic RNA was isolated from MEF THOC5 flox/flox infected with Ad-Cre-GFP or Ad-GFP (control virus) virus, using the QIAGEN RNeasy kit (QIAGEN) according to the manufacturer's recommendations. For transcriptome analysis, the quality and integrity of the total RNA were confirmed using an Agilent Technology 2100 Bioanalyzer (Agilent Technology). Biotin-labeled target synthesis was performed by Affymetrix. About 12.5 μg of each biotinylated cRNA preparation was fragmented and placed in a hybridization cocktail that contained four biotinylated hybridization controls (BioB, BioC, BioD, and Cre) as recommended by the manufacturer. All samples were hybridized to the same lot of Affymetrix MGU74A for 16 h. The GeneChips were washed, stained with streptavidin-phycoerythrin, and read by using an Affymetrix GeneChip fluidic station and scanner. Analysis of microarray data was performed using the Affymetrix Microarray Suite 5.0, Affymetrix Micro DB 3.0, and Affymetrix Data Mining Tool 3.0. For normalization, all array experiments were scaled to a target intensity of 150 (Kroger et al. 2007).
RT-PCR analysis
Cytoplasmic RNA was isolated as described above. Reverse transcription was carried using oligo(dT) primers and the Omniscript reverse transcriptase kit (QIAGEN) following the instructions provided. The primer pairs for each PCR were c-myc: forward primer, 5′-GCTGGATTTCCTTTGGGCGT-3′; reverse primer, 5′-CGCAACATAGGATGGAGAGCA (274 bp); Cryl1: forward primer, 5′-AGGAGTGTGTTCCAGAGAACC-3′; reverse primer, 5′-TGGTGGATTGACAGGATGAGC-3′ (176 bp); Eny2: forward primer, 5′-ATGGTGGTTAGCAAGATGAACAA-3′; reverse primer, 5′-TGCCTTTAGCTGATCCTTCCA-3′ (144 bp), Gigyf2: forward primer, 5′-CGCCGACTGGAAGAGAACC-3′; reverse primer, 5′-TTGCTGTGTTAGACTGCTGAC-3′ (235 bp); Gja1: forward primer, 5′-CAGCCTCCAAGGAGTTCCAC-3′; reverse primer, 5′-GAGAGATGGGGAAGGACTTGT-3′ (292 bp); Hoxb3: forward primer, 5′-AGCGATGCAGAAAGCCACCTACTA-3′; 5′-TGTACTTCATGCGACGGTTCTGGA-3′ (743 bp); Hsp70: forward primer, 5′-TGGTGCAGTCCGACATGAAG-3′; reverse primer, 5′-GCTGAGAGTCGTTGAAGTAGGC-3′ (219 bp); Id2: forward primer, 5′-CAGGCGTCCAGGACGCCGCT-3′; reverse primer, 5′-CCGGAGAACGACACCTGGGC-3′ (310 bp); Plxdc1: forward primer, 5′-AAGACCCAGCTAAGCCAGGA-3′; reverse primer, 5′-GGATCTTCACATGACTCCGGT (208 bp); Prkcbp1: forward primer, 5′-AAAACCCGACAAGACTTCCAC-3′; reverse primer, 5′-CGGCTGTATAAACATCCTCTGAG-3′ (245 bp); Slc11a2: forward primer, 5′-CAATGT CTTTGTCGTGTCCGT-3′; reverse primer, 5′-GCGACCATTTTAGGTTCAGGAAT-3′ (293 bp); β-actin: forward primer, 5′-AACACCCCAGCCATGTACGTAG-3′; reverse primer, 5′-GTGTTGGCATAGAGGTCTTTACGG-3′ (509 bp); Twsg1: forward primer, 5′-ACTGTGTCGGTATGTGCAACC-3′; reverse primer, 5′-GGAGACGATGTTCCAGTTCAG-3′ (146 bp); Zdhhc2: forward primer, 5′-CTACTACGCCTACGCCATCC-3′; reverse primer, 5′-TCCAGCAATTCTTTCTCTGCAT-3′ (189 bp expected product size). PCRs were set up according to the following profile: an initial denaturation step of 94°C for 3 min, 35 cycles of 94°C for 30 sec, 60° or 56°C for 30 sec, and 72°C for 30 sec followed by a final extension step at 72°C for 10 min. Separation of the DNA fragments was carried out on 2.0% (w/v) agarose gels stained with ethidium bromide (2 μg/mL) and photographed under UV light. Signal intensity was quantified using TINA 2.0 software (Raytest Isotopenmessgeraete GmbH). Quantitative (TaqMan) RT-PCR was performed as previously described (Dhamija et al. 2010). TaqMan probes (Applied Biosystems), assay-IDs were as follows: Mm00525091_m1 (Gigyf2); Mm00835499_m1 (Prkcbp1); Mm00446968_m1 (HPRT1); and were used with TaqMan Fast Universal PCR Master Mix (2×) (Applied Biosystems).
THOC5-mRNA complex isolation using TAP-purification
MEF cells were transfected with the pNTAP vector (Stratagene) and the same vector carrying Myc-tagged THOC5 using the Polyfect reagent as described by the manufacturer (QIAGEN), and the cells were allowed to grow for 48 h. After washing three times, cells were lysed with lysis buffer (10 mM Tris, 150 mM NaCl, 1 mM PMSF, 0.4% NP-40, protease inhibitor cocktail [Sigma-Aldrich]) and RNase inhibitor, and were then frozen and thawed three times. After centrifugation, supernatants were incubated with streptavidin Sepharose (GE Healthcare) in streptavidin binding buffer (lysis buffer with 10 mM β-mercaptoethanol and 0.5 mM EDTA) and then washed three times. All steps were carried out in the cold. Bound RNAs were supplied for RT-PCR analysis.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Sabine Klebba-Faerber for technical assistance, Anette Melk for helping with qRT-PCR, and C. Bruce Boschek for critically reading the manuscript. This research was supported by Sonderforschungsbereich 566 (B2), the MD/PhD program, and Leistungsorientierte Mittelvergabe with Frauenfaktor from MHH. A.K. was supported by Wiedeking-Stiftung, Habilitationsstipendium (MHH), and the HiLF program (MHH).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2607011.
REFERENCES
- Aguilera A 2005. Cotranscriptional mRNP assembly: from the DNA to the nuclear pore. Curr Opin Cell Biol 17: 242–250 [DOI] [PubMed] [Google Scholar]
- Carney L, Pierce A, Rijnen M, Gonzalez Sanchez MB, Hamzah HG, Zhang L, Tamura T, Whetton AD 2009. THOC5 couples M-CSF receptor signaling to transcription factor expression. Cell Signal 21: 309–316 [DOI] [PubMed] [Google Scholar]
- Chavez S, Aguilera A 1997. The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev 11: 3459–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez S, Garcia-Rubio M, Prado F, Aguilera A 2001. Hpr1 is preferentially required for transcription of either long or G+C-rich DNA sequences in Saccharomyces cerevisiae. Mol Cell Biol 21: 7054–7064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R 2006. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell 127: 1389–1400 [DOI] [PubMed] [Google Scholar]
- Dhamija S, Doerrie A, Winzen R, Dittrich-Breiholz O, Taghipour A, Kuehne N, Kracht M, Holtmann H 2010. IL-1-induced post-transcriptional mechanisms target overlapping translational silencing and destabilizing elements in IκBζ mRNA. J Biol Chem 285: 29165–29178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farny NG, Hurt JA, Silver PA 2008. Definition of global and transcript-specific mRNA export pathways in metazoans. Genes Dev 22: 66–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo M, Luna R, Erdjument-Bromage H, Tempst P, Aguilera A 2003. Nab2p and the Thp1p–Sac3p complex functionally interact at the interface between transcription and mRNA metabolism. J Biol Chem 278: 24225–24232 [DOI] [PubMed] [Google Scholar]
- Ganguly A, McKnight RA, Raychaudhuri S, Shin BC, Ma Z, Moley K, Devaskar SU 2007. Glucose transporter isoform-3 mutations cause early pregnancy loss and fetal growth restriction. Am J Physiol 292: E1241–E1255 [DOI] [PubMed] [Google Scholar]
- Garcia-Rubio M, Chavez S, Huertas P, Tous C, Jimeno S, Luna R, Aguilera A 2008. Different physiological relevance of yeast THO/TREX subunits in gene expression and genome integrity. Mol Genet Genomics 279: 123–132 [DOI] [PubMed] [Google Scholar]
- Gatfield D, Izaurralde E 2002. REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J Cell Biol 159: 579–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gridley S, Lane WS, Garner CW, Lienhard GE 2005. Novel insulin-elicited phosphoproteins in adipocytes. Cell Signal 17: 59–66 [DOI] [PubMed] [Google Scholar]
- Herold A, Klymenko T, Izaurralde E 2001. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA 7: 1768–1780 [PMC free article] [PubMed] [Google Scholar]
- Herold A, Teixeira L, Izaurralde E 2003. Genome-wide analysis of nuclear mRNA export pathways in Drosophila. EMBO J 22: 2472–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno S, Rondon AG, Luna R, Aguilera A 2002. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J 21: 3526–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J, Inoue H, Hurt E, Yoneda Y 2009. Adaptor Aly and co-adaptor Thoc5 function in the Tap-p15-mediated nuclear export of HSP70 mRNA. EMBO J 28: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch A, Scherr M, Breyer B, Mancini A, Kardinal C, Battmer K, Eder M, Tamura T 2008. Inhibition of Abl tyrosine kinase enhances nerve growth factor-mediated signaling in Bcr-Abl transformed cells via the alteration of signaling complex and the receptor turnover. Oncogene 27: 4678–4689 [DOI] [PubMed] [Google Scholar]
- Köhler A, Hurt E 2007. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 8: 761–773 [DOI] [PubMed] [Google Scholar]
- Kroger A, Stirnweiss A, Pulverer JE, Klages K, Grashoff M, Reimann J, Hauser H 2007. Tumor suppression by IFN regulatory factor-1 is mediated by transcriptional down-regulation of cyclin D1. Cancer Res 67: 2972–2981 [DOI] [PubMed] [Google Scholar]
- Li Y, Lin AW, Zhang X, Wang Y, Wang X, Goodrich DW 2007. Cancer cells and normal cells differ in their requirements for Thoc1. Cancer Res 67: 6657–6664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini A, Koch A, Whetton AD, Tamura T 2004. The M-CSF receptor substrate and interacting protein FMIP is governed in its subcellular localization by protein kinase C-mediated phosphorylation, and thereby potentiates M-CSF-mediated differentiation. Oncogene 23: 6581–6589 [DOI] [PubMed] [Google Scholar]
- Mancini A, El Bounkari O, Norrenbrock AF, Scherr M, Schaefer D, Eder M, Banham AH, Pulford K, Lyne L, Whetton AD, et al. 2007. FMIP controls the adipocyte lineage commitment of C2C12 cells by downmodulation of C/EBP alpha. Oncogene 26: 1020–1027 [DOI] [PubMed] [Google Scholar]
- Mancini A, Niemann-Seyde SC, Pankow R, El Bounkari O, Klebba-Farber S, Koch A, Jaworska E, Spooncer E, Gruber AD, Whetton AD, et al. 2010. THOC5/FMIP, an mRNA export TREX complex protein, is essential for hematopoietic primitive cell survival in vivo. BMC Biol 8: 1 doi: 10.1186/1741-7007-8-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R 2005. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev 19: 1512–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166 [DOI] [PubMed] [Google Scholar]
- Merz C, Urlaub H, Will CL, Luhrmann R 2007. Protein composition of human mRNPs spliced in vitro and differential requirements for mRNP protein recruitment. RNA 13: 116–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce A, Carney L, Hamza HG, Griffiths JR, Zhang L, Whetton BA, Gonzalez Sanchez MB, Tamura T, Sternberg D, Whetton AD 2008. THOC5 spliceosome protein: a target for leukaemogenic tyrosine kinases that affects inositol lipid turnover. Br J Haematol 141: 641–650 [DOI] [PubMed] [Google Scholar]
- Piruat JI, Aguilera A 1998. A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J 17: 4859–4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R, Cheng H 2005. TREX, SR proteins and export of mRNA. Curr Opin Cell Biol 17: 269–273 [DOI] [PubMed] [Google Scholar]
- Reed R, Hurt E 2002. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell 108: 523–531 [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Herold A, Gari K, Kocher T, Rode M, Ciccarelli FL, Wilm M, Izaurralde E 2004. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat Struct Mol Biol 11: 558–566 [DOI] [PubMed] [Google Scholar]
- Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–308 [DOI] [PubMed] [Google Scholar]
- Tamura T, Mancini A, Joos H, Koch A, Hakim C, Dumanski J, Weidner KM, Niemann H 1999. FMIP, a novel Fms-interacting protein, affects granulocyte/macrophage differentiation. Oncogene 18: 6488–6495 [DOI] [PubMed] [Google Scholar]
- Voynov V, Verstrepen KJ, Jansen A, Runner VM, Buratowski S, Fink GR 2006. Genes with internal repeats require the THO complex for transcription. Proc Natl Acad Sci 103: 14423–14428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chang Y, Li Y, Zhang X, Goodrich DW 2006. Thoc1/Hpr1/p84 is essential for early embryonic development in the mouse. Mol Cell Biol 26: 4362–4367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chinnam M, Wang J, Wang Y, Zhang X, Marcon E, Moens P, Goodrich DW 2009. Thoc1 deficiency compromises gene expression necessary for normal testis development in the mouse. Mol Cell Biol 29: 2794–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]