Abstract
Systemic RNAi in Caenorhabditis elegans requires the widely conserved transmembrane protein SID-1 to transport RNAi silencing signals between cells. When expressed in Drosophila S2 cells, C. elegans SID-1 enables passive dsRNA uptake from the culture medium, suggesting that SID-1 functions as a channel for the transport of double-stranded RNA (dsRNA). Here we show that nucleic acid transport by SID-1 is specific for dsRNA and that addition of dsRNA to SID-1 expressing cells results in changes in membrane conductance, which indicate that SID-1 is a dsRNA gated channel protein. Consistent with passive bidirectional transport, we find that the RNA induced silencing complex (RISC) is required to prevent the export of imported dsRNA and that retention of dsRNA by RISC does not seem to involve processing of retained dsRNA into siRNAs. Finally, we show that mimics of natural molecules that contain both single- and double-stranded dsRNA, such as hairpin RNA and pre-microRNA, can be transported by SID-1. These findings provide insight into the nature of potential endogenous RNA signaling molecules in animals.
Keywords: RISC, RNAi, SID-1, dsRNA channel
INTRODUCTION
Systemic RNAi is the spreading of RNAi silencing to cells that do not initially encounter a dsRNA trigger. Systemic RNAi occurs in many organisms (Jose and Hunter 2007), but has been best described in plants, where it serves primarily as an anti-viral mechanism (Baulcombe 2004), and in Caenorhabditis elegans. In C. elegans, silencing triggered by injected, ingested, or locally expressed dsRNA can move throughout the organism to reduce endogenous gene expression in all non-neuronal cells, including the germline, thus transmitting silencing to the next generation (Fire et al. 1998; Tabara et al. 1998; Timmons and Fire 1998; Winston et al. 2002; Jose and Hunter 2007). A key component required for systemic RNAi in C. elegans is the broadly expressed transmembrane protein SID-1, which we identified in a screen for systemic RNAi defective mutants (Winston et al. 2002; Feinberg and Hunter 2003). The process of systemic RNAi requires both export and import of silencing signals between cells, and analysis of sid-1 genetic mosaic animals shows that SID-1 is required for the import of RNAi silencing signals (Winston et al. 2002). SID-1 homologs are present in a variety of other invertebrates and in all sequenced vertebrate genomes, indicating an ancient origin and likely conserved function.
To characterize SID-1 activity we have expressed wild-type and mutant forms of C. elegans SID-1 in Drosophila S2 cells (Feinberg and Hunter 2003; Shih et al. 2009). Drosophila S2 cells were initially selected for this analysis because Drosophila lacks a sid-1 homolog and systemic RNAi (Winston et al. 2002; Roignant et al. 2003), thus, SID-1 activity could be analyzed in the absence of a competing endogenous activity. This has proven to be a powerful system to investigate the activity and mechanism of SID-1 dsRNA transport as SID-1 expressing S2 cells show rapid and sensitive silencing in response to low dsRNA concentrations (Feinberg and Hunter 2003; Shih et al. 2009). Consistent with observed dsRNA-induced silencing, Drosophila S2 cells expressing SID-1 rapidly internalize radiolabeled dsRNA (Feinberg and Hunter 2003; Shih et al. 2009). Treating these cells with oligomycin to deplete ATP levels or performing the dsRNA uptake assay at 4°C has little effect on the extent or rate of dsRNA uptake, indicating that rather than acting as a pump or receptor, SID-1 functions as a dsRNA channel (Feinberg and Hunter 2003; Shih et al. 2009). Similarly, overexpression of SidT1, a mammalian SID-1 homolog, enhances siRNA uptake and silencing in human cells (Duxbury et al. 2005), and SidT1 knockdown disrupts the internalization of cholesterol-modified siRNA in human hepatic cells (Wolfrum et al. 2007). These results are consistent with a conserved dsRNA transport function for SID-1.
SID-1 was discovered because of its requirement for systemic RNAi in C. elegans. This led to the demonstration that cells expressing SID-1 passively import dsRNA. However, whether dsRNA transport is a selected function for SID-1 or whether SID-1 is a general transporter that can also transport dsRNA is unknown. Here we show that adding dsRNA to SID-1 expressing cells results in an increase in membrane conductance, consistent with the opening of dsRNA conducting channels. Using this assay and a modified version of a nucleic acid transport assay we demonstrate that SID-1 transport is selective and specific for dsRNA-containing molecules. Consistent with the passive nature of dsRNA transport by SID-1, we demonstrate bidirectional dsRNA transport and show that the RISC is required for the retention of a portion of the internalized dsRNA. Further, the RISC-retained dsRNA remains largely unprocessed, suggesting a dsRNA retention role for RISC that may function to extend silencing beyond the time of exposure and/or that may buffer systemic RNAi responses to small doses of dsRNA. Finally, we show that SID-1 dsRNA transport can accommodate single-stranded regions to transport microRNAs precursors and larger hairpin RNAs. These results, although constraining SID-1 substrates to dsRNA molecules, dramatically expand the possible role of SID-1 and noncoding RNAs in intercellular signaling in C. elegans.
RESULTS
SID-1 is activated by dsRNA
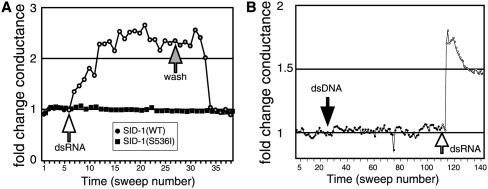
The structure and activity of SID-1 indicate that it may function as a dsRNA channel (Feinberg and Hunter 2003). A channel with a pore large enough to transport dsRNA must be gated, and when open may produce a measurable current across the cell membrane. To test this idea we applied whole-cell electrophysiology methods to determine whether addition of dsRNA to SID-1 expressing Drosophila S2 cells would result in a measurable change in membrane conductance. We first established and verified a whole-cell patch connection by measuring the activity of an endogenous Cl− channel in Drosophila S2 cells (Asmild and Willumsen 2000). We then measured the change in current and membrane conductance of voltage clamped cells following the addition of 500-base pair (bp) dsRNA to the bath solution (for voltage ramp and current response, see Supplemental Fig. S1). In nearly all cases, the current and membrane conductance increased, consistent with the opening of dsRNA conducting channels (Fig. 1A). To verify that dsRNA was causing this conductance change, we washed away the dsRNA-containing bath solution with fresh bath solution and noted that the conductance returned to the baseline level (Fig. 1A). The dsRNA-induced increase in membrane conductance is SID-1 dependent as cells expressing the strong loss-of-function sid-1(qt2) mutant, which encodes an amino acid substituted SID-1(S536I) (Winston et al. 2002), did not show a conductance change (Fig. 1A).
FIGURE 1.
Double-stranded RNA-induced conductance increases in Drosophila S2 cells expressing SID-1. (A) Changes in whole-cell conductance of a SID-1(WT) (circles) or a SID-1(S536I) (squares) expressing cell in response to dsRNA addition to (white-filled circles) and subsequent replacement of the bath solution (gray-filled circles). (B) The addition of dsDNA (black-filled circles) and then dsRNA (white-filled circles) to the bath solution of a patched, SID-1(WT) expressing S2 cell at the indicated sweeps.
The charge carrier in these experiments is unknown. The charge equivalent of naked dsRNA is 2e− per base pair, or 103 e− per 500 bp dsRNA. In these experiments the SID-1-dependent portion of the current averaged 3.4 pA, which corresponds to ∼2 × 107e− equivalent charges/second or 2 × 104 500 bp dsRNA molecules per second. Since under simple diffusion conditions up to ∼3 × 103 500 bp dsRNA molecules per cell could be detected intracellularly (see the following), it is not unreasonable that dsRNA could be the sole charge carrier in these experiments. However, during the experiment the cells are maintained in a simple salt solution, thus the ions in the buffer will both reduce the effective charge on dsRNA and may also contribute to all or a portion of the measured SID-1-dependent current. Thus, these experiments provide information only about SID-1 gating properties. In summary, SID-1 dsRNA transport is associated with changes in membrane conductance consistent with the activity of a dsRNA-activated channel.
RISC is important for retention of internalized dsRNA
We have previously shown that Drosophila S2 cells transfected with C. elegans SID-1 can import dsRNA in an energy-independent manner (Feinberg and Hunter 2003). This indicates that SID-1 can function as a passive dsRNA transporter. Consequently the net direction of dsRNA transport should depend on the concentration of dsRNA; flowing into cells when the extracellular dsRNA concentration is high and flowing out of cells during wash steps. Our observation that SID-1 expressing cells retain dsRNA likely reflects dsRNA-binding activity in the S2 cells; consequently only molecules that are retained would be detected as transported. To test this idea we developed a transport assay that is independent of retention and used RNAi to identify the major dsRNA retention activity in S2 cells.
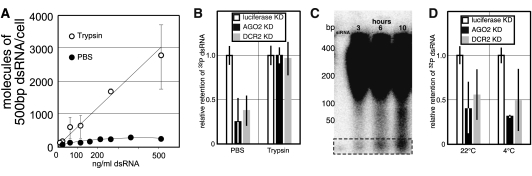
Incubating control S2 cells with labeled dsRNA, followed by extensive washing results in significant SID-1-independent retention of dsRNA, reflecting either surface bound or internalized dsRNA. In developing our previously described dsRNA uptake assay (Feinberg and Hunter 2003), we determined that treating these extensively washed cells with trypsin significantly reduced the retention of dsRNA, therefore we inferred that most of the dsRNA was binding nonspecifically to cell surface proteins. Similarly, trypsin treatment of SID-1 expressing cells should cleave the SID-1 extracellular domain and inactivate SID-1-dependent dsRNA transport. To test this idea, we first incubated the SID-1 expressing cells in dsRNA. By passive diffusion the net flow of dsRNA is into the cell, until the intracellular dsRNA concentration is similar to the dsRNA concentration in the medium. We then either washed the cells first with PBS followed by trypsin proteolysis (PBS-first protocol) or treated the cells first with trypsin followed by PBS washes (trypsin-first protocol). In the PBS-first protocol the dsRNA concentration in the wash solution is very low and by passive diffusion the net flow of dsRNA is out of the cell. The expectation is that only the dsRNA that is not free to diffuse out (retained) will remain associated with the cells. In the trypsin-first protocol the N-terminal half of SID-1 is presumably digested, inactivating SID-1-dependent dsRNA transport (passive diffusion into or out of cells); the dsRNA that was passively transported in during the initial incubation is now “trapped” inside the cell. Consistent with these expectations, we find that the amount of dsRNA trapped inside the cell is linearly proportional to the amount originally in the medium (Fig. 2A). In contrast, the amount of dsRNA retained in cells following the PBS-first protocol plateaued at the mass equivalence of 300 molecules of 500-bp dsRNA per cell (Fig. 2A). We conclude that the nonretained dsRNA escaped during the wash steps. These results provide direct experimental support for bidirectional SID-1-dependent transport in S2 cells.
FIGURE 2.
Transport, retention, and processing of internalized dsRNA. (A) Trypsin protease treatment traps internalized dsRNA that would otherwise escape during PBS washes. SID-1 expressing S2 cells soaked in increasing concentrations of radiolabeled 500-bp dsRNA were washed extensively in PBS buffer before (PBS) or after (trypsin) trypsin protease treatment (see text). For concentrations higher than 10 ng/mL, increasing amounts of unlabeled 500 bp dsRNA was added and molecules per cell extrapolated. (B) RISC components are required for retention not transport. Five hundred-bp radiolabeled dsRNA was added to Drosophila S2 cells that had been cotransfected with SID-1 expressing plasmids and dsRNA targeting RISC components DCR2 or AGO2 (Supplemental Fig. S1), or luciferase (control). Assays were performed for 30 min and then were washed extensively in PBS buffer before (PBS) or after (trypsin) trypsin protease treatment. The amount of internalized dsRNA was normalized to the control cells cotransfected with luciferase dsRNA. (C) Retained dsRNA is largely unprocessed. 6% polyacrylamide gel electrophoresis of retained radiolabeled 500-bp dsRNA extracted from SID-1 expressing Drosophila S2 cells washed with PBS prior to trypsin treatment, then incubated in fresh media for the indicated hours. siRNA size marker (21 bp) is in the left-most column, siRNA is highlighted in the box. Other dsRNA sizes are indicated on the far left. (D) RISC-dependent retention is energy independent. Five hundred-bp radiolabeled dsRNA was added to Drosophila S2 cells that had been cotransfected with SID-1 expressing plasmids and dsRNA targeting RISC components DCR2 or AGO2 (Supplemental Fig. S1) or luciferase (control). Retention assays with PBS wash prior to trypsin protease treatment were performed at 22°C or 4°C for 30 min and the amount of internalized dsRNA was normalized to the control cells cotransfected with luciferase dsRNA. Error bars = 1 standard deviation; P < 0.005, Student's t-test.
The amount of dsRNA retained after extensive washing plateaued (Fig. 2A), suggesting the saturation of available dsRNA binding protein activity in S2 cells. Because SID-1 transported dsRNA causes dose-dependent RNAi, we hypothesized that the RNA-induced silencing complex (RISC) may function to retain some or all of the imported dsRNA (Feinberg and Hunter 2003).To determine whether RISC is important for dsRNA retention or whether other unknown dsRNA binding proteins sequester imported dsRNA in the cytoplasm, we used RNAi to knockdown select RISC components and then assayed radiolabeled dsRNA retention in Drosophila S2 cells expressing SID-1. We targeted two proteins, Argonaute2 (AGO2) and Dicer-2 (DCR2). DCR2 efficiently processes long dsRNA into siRNA, which then associate with AGO2 in RISC (Hammond et al. 2001; Lee et al. 2004). Additionally, knockdown of these components can affect the levels of other RISC components, thereby affecting the overall level of functional RISC (Liu et al. 2003, 2006; Rehwinkel et al. 2006; Kim et al. 2007). We found that knockdown of AGO2 or DCR2 (Supplemental Fig. S2), relative to RNAi directed at luciferase as a control, reduced the amount of dsRNA retained in cells by 50%–80% (Fig. 2B, PBS) indicating that either RISC has a role in dsRNA retention or that it plays an active role in SID-1-mediated import; for example, binding to and ratcheting dsRNA into the cell through a SID-1 pore. To discriminate between these two possibilities, we determined whether knockdown of AGO2 and DCR2 would affect the amount of dsRNA trapped in S2 cells using the retention-independent, trypsin protease-first protocol. We found that reducing functional RISC had no effect on dsRNA transport (Fig. 2B, trypsin), thus, RISC is not involved in regulating or modulating SID-1 import of dsRNA; rather RISC is required for retention of imported dsRNA.
The RISC components AGO2 and DCR2 actively metabolize RNA. In vitro studies in Drosophila cell extracts demonstrate that DCR2 processing of long dsRNA into siRNA requires ATP, whereas AGO2 in active RISC requires ATP for efficient release of cleaved mRNA (Zamore et al. 2000; Bernstein et al. 2001; Haley and Zamore 2004). Thus, the apparent requirement for intact RISC for dsRNA retention may simply reflect an indirect effect of RNAi associated dsRNA processing. To determine whether RISC processing of imported dsRNA is important for dsRNA retention, we analyzed internalized radiolabeled dsRNA by polyacrylamide gel electrophoresis (Fig. 2C). This analysis showed that some of the retained dsRNA appears to be 500 bp in size, but the majority of the retained RNA migrates with mobility between 100 and 500 bp. However, only a small fraction of retained dsRNA was apparently processed to siRNA, the vast majority remaining >100 bp even after 10 h. Because of this remarkable stability, we assume the intracellular RNA remains double stranded. In addition, ATP-dependent processing of RNA is likely to be limited by performing the retention assays at 4°C. However, at both 4°C and 22°C knockdown of AGO2 and DCR2 reduced the amount of radiolabeled dsRNA retained in cells (Fig. 2D). These results suggest the possibility that AGO2 and DCR2 have a dsRNA retention role that is independent of dsRNA cleavage or target mRNA slicing.
SID-1 nucleic acid transport is selective for dsRNA
The above-mentioned results indicate that our previous inability to detect uptake of other nucleic acids may have been compromised by the requirement for retention of transported molecules. Therefore, we used both patch-clamp electrophysiology and the transport assay described previously to broadly investigate the specificity of nucleic acid transport by SID-1. For each potential substrate, the membrane conductance of S2 cells expressing wild-type SID-1 was compared to cells expressing SID-1(S536I) and for the transport assay we compared addition of trypsin before and after the PBS washes. In all cases, we found that nucleic acids that induced SID-1-dependent currents were also transported into cells, and those that failed to activate the channel were not transported.
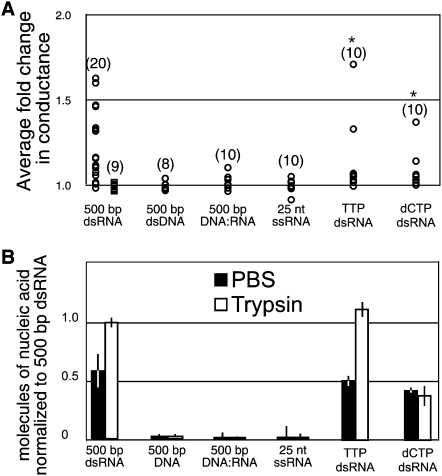
We first asked whether DNA could either activate SID-1 or be transported into SID-1 expressing cells. We found that addition of 500-bp dsDNA failed to activate the channel (Figs. 1B, 3A). To confirm that the cells were properly patched, we added dsRNA toward the end of each recording to confirm the expected change in conductance. Such confirmation was standard for all substrates that failed to cause activation. Consistent with the electrophysiology data, we failed to detect transport of radiolabled 500-bp dsDNA in the trypsin-first transport assay (Fig. 3B). To investigate the apparent selection for dsRNA we next determined that a 500-bp DNA:RNA heteroduplex failed to activate SID-1 or be transported into SID-1 expressing S2 cells, indicating selection for dsRNA or against DNA (Fig. 3). Consistent with a selection for dsRNA, we found that a 25-nucleotide single-stranded RNA with no predicted secondary structure or self-complementarity failed to either activate SID-1 or be transported into SID-1 expressing S2 cells (Fig. 3).
FIGURE 3.
Specificity of SID-1 activation, transport, and retention. (A) Summary of normalized conductance change of SID-1(WT) (circles) or SID-1(S536I) (squares) expressing cells in response to the addition of the indicated nucleic acids. The normalized fold change in membrane conductance was calculated from the average conductance of 10 consecutive time points after dsRNA addition divided by the average conductance of 10 consecutive time points before dsRNA addition. The magnitude of the conductance change following addition of dsRNA was variable from cell to cell, likely reflecting relative SID-1 expression levels in these transiently transfected cells. The measured conductance for S2 cells ranged from 2.5 to 17 μS (microSiemens) in the absence of dsRNA and 3.5–25 μS in the presence of dsRNA, likely reflecting the variability in cell size. *P < 0.001 (Kologonov-Smirnoff test) for all results compared to SID-1(S536I) cells exposed to 500-bp dsRNA. (n) Number of patched cells. (B) Summary of transport and retention properties for the indicated nucleic acids. SID-1 expressing S2 cells soaked in radiolabeled dsRNA were washed extensively in PBS buffer before (PBS) or after (trypsin) trypsin protease treatment. In order to increase assay sensitivity 5 ng/mL of nucleic acid was used instead of the standard 10 ng/mL. Error bars = 1 standard deviation; P < 0.005, Student's t-test.
RNA is distinguished from DNA by the presence of a 2′ hydroxyl on the ribose sugar and the base uridine rather than thymidine, either of which may be used to select for dsRNA. We first synthesized 500-bp dsRNA using TTP rather than UTP (TTP dsRNA) and found that 500-bp TTP dsRNA was indistinguishable from 500-bp dsRNA in our assays (Fig. 3). We then used a modified RNA polymerase to synthesize a mixed 500-bp dsRNA, substituting dCTP for CTP (dCTP dsRNA). We found that although 500-bp dCTP dsRNA activated the SID-1 channel, its transport into cells was reduced compared to both 500-bp dsRNA and 500-bp TTP dsRNA (Fig. 3). This indicates that the presence of deoxyribose interferes with transport. Further, the amount of retained dCTP dsRNA was independent of whether the cells were treated with trypsin before the wash steps. This result is consistent with several explanations: (1) because fewer molecules were transported the available RISC was not saturated, (2) the incorporated dCTP may bind to non-RISC intercellular binding proteins, (3) export of dCTP dsRNA is more sensitive to the structural requirements for dsRNA than is import, thus less of it is exported after import. In summary, the electrophysiology and transport assay results indicate that SID-1 specificity for dsRNA structure is mediated by the ribose 2′ hydroxyl in dsRNA.
SID-1 can transport dsRNA with single-stranded regions
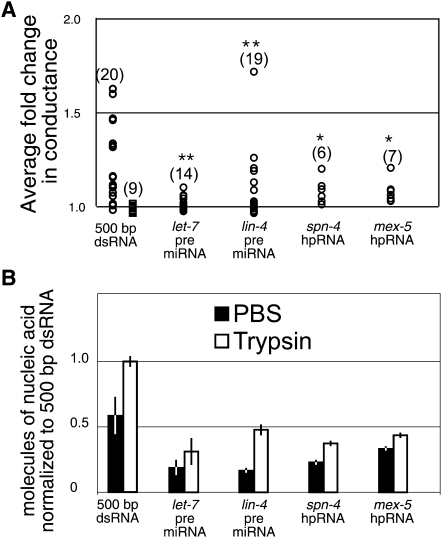
Many endogenous dsRNA-containing molecules, such as microRNA precursors (pre-miRNA) and hairpin RNAs, include single-stranded RNA regions. To determine whether these kinds of molecules should be considered candidate substrates for SID-1 activity we tested two artificially constructed long hairpin RNA molecules (spn-4 and mex-5) that are in vivo triggers for systemic RNAi and two different double-stranded pre-miRNA hairpin molecules (let-7 and lin-4). We found that all of the above-mentioned RNA molecules can activate SID-1 (Fig. 4A) indicating that modified and partially double-stranded RNA molecules can stimulate SID-1 activity. Further, all four were transported into and retained in Drosophila S2 cells, though not at the same efficiency as the 500 bp dsRNA (Fig. 4B). This suggests that single-stranded domains may impede SID-1 transport and intracellular retention. In summary, dsRNAs that contain some single-stranded regions can activate SID-1 and be transported by SID-1.
FIGURE 4.
Transport of natural-like RNAs that contain single-stranded RNA regions. (A) Summary of normalized conductance change of SID-1(WT) (circles) or SID-1(S536I) (squares) expressing cells in response to the addition of the indicated nucleic acids (see Fig. 3 legend). *P < 0.001; **P < 0.05 (Kologonov-Smirnoff test) for all results compared to SID-1(S536I) cells exposed to 500-bp dsRNA. (B) Summary of transport and retention properties for the indicated nucleic acids (see Fig. 3 legend). Error bars = 1 standard deviation; P < 0.005, Student's t-test.
DISCUSSION
We have applied whole-cell electrophysiology methods and developed a retention-independent nucleic-acid transport assay to gain significant insights into the selectivity of the SID-1 dsRNA channel. Most significantly, the specificity of nucleic acid transport for dsRNA-molecules indicates selection for dsRNA transport. Thus, systemic RNAi is not a fortuitous phenomenon that occurs only in the presence of nonphysiological levels of dsRNA. This is consistent with the observation that sid-1 is important for efficient transgene silencing in C. elegans (Jose et al. 2009). Further, although our analysis is limited to Drosophila S2 cells, RISC is identified as an important intercellular dsRNA retention activity. Retention of unprocessed dsRNA may serve to extend the length of the silencing response. Finally, we show that SID-1 has the capacity to transport partially double-stranded RNAs analogous to endogenous noncoding RNAs, expanding the possibilities for RNA mediated cell signaling in animals.
SID-1 discrimination between dsRNA and dsDNA
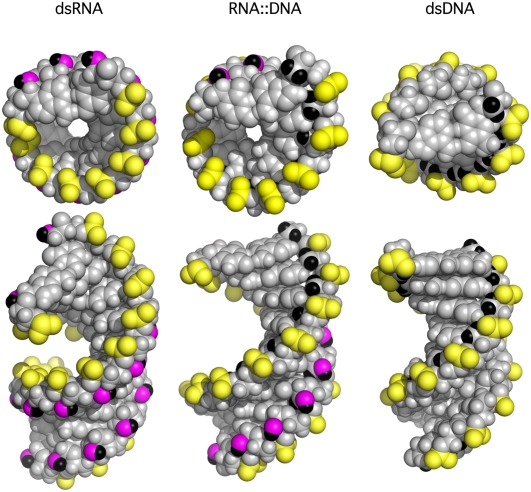
The chemical difference that allows SID-1 to discriminate between RNA and DNA is a hydroxyl versus a hydrogen on the ribose 2′ carbon. This chemical difference results in profoundly different structures (Fig. 5). dsRNA forms an A-form helix in which the bases stack more compactly and the phosphate groups orient inward toward the deep major groove. In contrast, hydrated dsDNA forms a B-form helix in which the phosphate groups orient outward and are exposed on the surface of the helix. However, the structural differences between dsRNA and A-form helix-like RNA:DNA hybrid molecules are likely the most instructive. Although NMR solution structures of hybrid molecules more strongly resemble a B-form helix (Conn et al. 1999), even the X-ray crystal data show that the phosphate groups are surface exposed on one half of the helix.
FIGURE 5.
Models of double-stranded nucleic acids. Top row, end-on view of the indicated double-stranded nucleic acid. Bottom row, view oriented to highlight the major and minor grooves. Phosphate groups (PO3) = yellow, Ribose 2′ O = magenta, 2′ H = black. Note that in dsRNA the yellow phosphate groups are oriented into the major groove, whereas in DNA and RNA:DNA hybrid molecules they are oriented toward the surface. Conversely, the magenta and black ribose 2′ hydroxyl are oriented toward the surface in dsRNA such that they are accessible along the length of the helix (end on view), whereas in RNA:DNA hybrid molecules they are displaced into the minor groove. Nucleic acid models generated using PDB files 1ZF0 for dsDNA, 280D for dsRNA, and 1FIX for the RNA:DNA hybrid from RSCB Protein Data Bank (http://www.pdb.org).
Considering how these differences might function in SID-1 substrate recognition, we note two distinctions. First, in dsRNA the 2′ hydroxyl groups in each RNA chain lie across from each other in the minor groove. Further, end-on views show that these hydroxyls are surface accessible throughout the length of the helix. Thus, SID-1 may select for these paired hydroxyls in the minor groove. Second, the orientation of the phosphate groups in A-form dsRNA shifts their strong negative charge from the surface to the inside of the major groove. As a result the surface of the A-form helix is composed primarily of polar hydroxyl groups and nonpolar CH2 groups. This surface is significantly less electronegative than that of DNA. As described previously, although the RNA:DNA heteroduplex resembles dsRNA in the stacking of nucleotide bases, it resembles dsDNA in the placement of phosphate groups, and the 2′ hydroxyl groups on the RNA strand are recessed in the minor groove and no longer surface accessible. The sum effect is that in A-form dsRNA the C4′H and C5′H2 hydrocarbon groups are maximally surface exposed. This more neutral surface charge may underlie the selectivity for dsRNA and the diffusion-mediated transport of dsRNA through a protein pore. In summary, the minor chemical difference between RNA and DNA results in significant structural differences and it is these large structural differences that likely guide the selectivity of nucleic acid transport by SID-1.
SID-1-dependent bidirectional dsRNA transport in Drosophila S2 cells
In Drosophila S2 cells, SID-1 enables both rapid and passive dsRNA import and export. The most likely explanation is that SID-1 is acting as a bidirectional, dsRNA activated, dsRNA-conducting channel, but our experiments cannot rule out the possibility that SID-1 could activate endogenous Drosophila proteins that are transporting the dsRNA into or out of cells. If so, these Drosophila proteins must passively and, therefore, bidirectionally, transport dsRNA, but only in the presence of SID-1. Although remote, it is possible that Drosophila S2 cells maintain an active dsRNA export system that is sensitive to trypsin digestion. Further, it is important to emphasize that we have not yet directly demonstrated that nucleic acids pass through a SID-1 pore. To show directly that SID-1 is transporting nucleic acids would require reconstituting purified SID-1 in synthetic membranes or isolating SID-1 mutants with altered substrate specificity or transport rates.
Passive retention of SID-1 internalized dsRNA by RISC
The export of a portion of internalized dsRNA is inhibited by RISC, most likely by direct binding. We demonstrated that partial knockdown of AGO2 or DCR2, and, by proxy, knockdown of RISC levels in general, dramatically reduced dsRNA retention (Fig. 2B), suggesting that RISC is the primary “available” dsRNA binding activity in the cytosol. In principle, any dsRNA binding protein could contribute to retention. If any dsRNA binding protein were generally available to bind cytoplasmic dsRNA, it would likely compete with RISC for access to dsRNA triggers. Such genes have been identified in C. elegans as enhancers of RNAi and several of them are putative dsRNA binding proteins (Kennedy et al. 2004; Duchaine et al. 2006; Lee et al. 2006; Fischer et al. 2008).
Unexpectedly, the RISC-dependent intracellular pool of dsRNA remains largely unprocessed (Fig. 2C). Although our observation is limited to Drosophila S2 cells, an unprocessed pool of dsRNA is also apparent in C. elegans following exposure to dsRNA (Tabara et al. 2002). A pool of dsRNA may function as a reserve, to extend the period of silencing beyond the time of the initial exposure. Alternatively, when the abundance of initiating dsRNA is low, the sequestering of long dsRNA may serve to limit the spread of systemic silencing. Dose-dependent RNAi effects are apparent in many organisms, and have been reported for systemic RNAi in C. elegans (Timmons et al. 2003; Tijsterman et al. 2004).
Our characterization of SID-1 in Drosophila S2 cells has important implications for practical applications, such as large-scale RNAi screens using insect and vertebrate cell lines that do not have efficient uptake of dsRNA. For example, heterologous expression of SID-1 in Wnt pathway sensitive Drosophila Kc167 cells has enabled use of existing dsRNA resources to screen these cells to identify Wnt pathway genes (Bartscherer et al. 2006). Similarly, expression of C. elegans SID-1 in embryonic stem cells enables initiation of RNAi by addition of siRNA to the growth media (Tsang et al. 2007). Our findings show that at high dsRNA concentrations the available pool of RISC-dependent dsRNA-binding activity is rapidly saturated and that the off-rate is negligible (Shih et al. 2009). These results indicate little concern for the half-life of dsRNA in the extracellular environment. However, these results do suggest that serial dsRNA additions are unlikely to be effective.
SID-1-dependent transport of RNA structurally analogous to endogenous noncoding RNAs
We detected transport in SID-1 expressing S2 cells of dsRNA-containing molecules structurally identical to or similar to endogenously expressed RNAs (Fig. 4). So far, no evidence of nonautonomous effects has been described for these molecules, but very few have been tested (Zhang and Fire 2010). Consistent with this possibility, sid-1 mutant developmental and physiological phenotypes have recently been observed (J Brooks and CP Hunter, unpubl.) and the extent of experimentally induced systemic RNAi can be regulated by starvation and exposure to nonspecific RNA (Winston et al. 2002; Timmons et al. 2003). Further, small RNAs and their double-stranded precursors are modified (Chen 2007), which may regulate their transport by SID-1. Indeed, we show that transport of dsRNA by SID-1 is sensitive to slight dsRNA modifications (Fig. 3). Similarly, synthetic siRNAs, particularly those designed for therapeutic applications (Manoharan 2004), are often synthesized with modified sugar-phosphate backbones and nonstandard terminal nucleotides, modifications often designed to stabilize siRNA in sera. Such modifications may restrict uptake in SID-1 expressing mammalian cells, thus an in-depth characterization of SID-1 substrates may have importance for the design and delivery of RNAi-based therapeutics to target tissues expressing the SID-1 protein. Such studies may also provide insight into the nature of endogenous RNAs that mediate general systemic RNA signaling in the worm.
MATERIALS AND METHODS
Cell culture
Drosophila S2 were maintained at 27°C in Schneider's Drosophila medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) and 50 U/mg penicillin and streptomycin (Invitrogen). Transient transfections of S2 cells with pHC235 (Act-5::sid-1-FLAG) and pHC247 (Act-5:sid-1(qt2)-FLAG) were performed as described (Feinberg and Hunter 2003) using an Effectene transfection kit (Qiagen). To knockdown Drosophila RISC components, transfection was performed as described in the manufacturer's protocol, except gene specific dsRNA was cotransfected with pHC235 or pHC247 at a 3:2 dsRNA:plasmid ratio.
Nucleic acid strand synthesis
Primer oligonucleotide sequences are listed (Supplemental Table S1). Five hundred-bp luciferase DNA was PCR amplified (plasmid pGL3-basic [Promega], primers Luc forward, and Luc 500 reverse). GFP dsRNA, spn-4 hairpin RNA, and mex-5 hairpin RNA were all assembled from plasmid DNA templates (pPD128.110[T7::GFP::7T] [Timmons et al. 2001] pHC203[T7::spn-4::GST::4-nps], and pHC209[T7::mex-5::GST::5-xem]). RNA/DNA hybrid was assembled from the PCR product of 500-bp luciferase sense DNA (plasmid pGL3-basic [Promega], primers Luc forward, and Luc 500 reverse) and 500-bp antisense RNA (plasmid pGL3-basic, primers Luc forward, and Fluc 500 reverse T7). The RNA/DNA hybrid was treated with DNaseIII (NEB) to remove any double stranded DNA.
Dual T7 PCR DNA templates for dsRNA: 500-bp dsRNA (plasmid pGL3-Basic, primers Fluc forward T7 and Fluc 500 reverse T7), let-7 pre-miRNA (C. elegans gDNA template, primers pre let-7 forward T7, pre let-7 reverse), lin-4 pre-miRNA (C. elegans gDNA template, primers pre lin-4 forward T7, pre lin-4 reverse), Argonaute2 dsRNA (plasmid RE04347 (DGRC), primers AGO2 forward T7 and AGO2 reverse T7), Dicer-2 dsRNA (plasmid SD11113 (DGRC), primers Dicer-2 forward T7 and Dicer-2 reverse T7). NTP's were provided by Epicentre Technologies. Thymidine triphosphate on a ribose backbone (TTP) was provided by TriLink Biotechnologies. dCTP was incorporated into 500-bp dsRNA using T7 D&RNA Polymerase (Sousa and Padilla 1995) (Epicentre Technologies). Twenty-five-bp ssRNA was verified for the absence of a secondary structure by FastPCR 4.0 and RNAfold 1.5.
RNA was synthesized using Ampliscribe T7 kit (Epicentre). siRNA was prepared by treating 500-bp dsRNA with recombinant Dicer (Genlantis). Single strands were annealed by heating to 90°C followed by cooling 1°C/8 sec until reaching a temperature of 25°C. dsRNA was purified using Micro Bio-Spin 6 Chromatography Columns (Bio-Rad). Concentration and purity were verified by agarose gel electrophoresis and spectroscopy.
Blunt and overhanging termini for the 500-bp dsRNA and 500-bp dsDNA were tested in activation and transport assays. There was no difference in results from these assays, indicating that there are minimal differential effects from different termini. The precursor miRNA naturally have overhanging termini and were tested that way. The long hairpin RNA, other dsRNA constructs, and the DNA:RNA hybrid were created and tested with overhanging termini.
Radiolabeled RNA retention assay
All retention assays were performed in duplicate. In all radiolabeled dsRNA assays the amount of radiolabeled nucleic acid in S2 cells expressing the sid-1(qt2) mutant, SID-1(S536I), was subtracted from the radiolabeled nucleic acid in S2 cells expressing the SID-1(WT) to eliminate background non-SID-1-dependent transport. In the original protocol (Feinberg and Hunter 2003) cells were first incubated in Schnieder's media for 30 min. Then 10 ng/mL of 500-bp radiolabeled dsRNA was added at room temperature for 30 min. Media containing the dsRNA was removed and cells were then washed with ice-cold PBS to remove unbound dsRNA. Three-tenths mL of 0.25% trypsin without EDTA (Invitrogen) was added for 15 min. Cold 0.7 mL of Schnieder's media supplemented with 10% FBS and pen/strep was then added and cells were transferred to microcentrifuge tubes. Cells were pelleted, washed with PBS, and then solubilized with 0.1% SDS in water. Scintillation measurements were conducted in Ecoscint-A (National Diagnostics) on a LS1801 scintillation counter (Beckman). For the trypsin protease first uptake assay, after the initial incubation in dsRNA and removal of the media, 0.3ml of trypsin protease solution was added to the cells for 15 min. To impede export of internalized radiolabeled dsRNA into the trypsin solution, 500 ng/mL of appropriate length unlabeled dsRNA was added with the trypsin. These cells were then washed twice in PBS and prepared for dsRNA quantification as described previously. Phenol/chloroform extracted RNA from 0.1% SDS solubilized cells was run on 6% polyacrylamide gel, dried and exposed on a Fuji Type BAS-III imaging plate, which was read on a GE (Amerohan) Typhoon Trio Imager.
Immunofluorescence
Cells were allowed to adhere to poly-L-lysine coated slides, fixed with 4% paraformaldehyde, and permeabilized with 0.2% Triton X-100. Fixed cells were costained with anti-FLAG M2 monoclonal antibody (Sigma) and either rabbit anti-AGO2 polyclonal antibody (Förstemann et al. 2007) or rabbit anti-DCR2 antiserum (Liu et al. 2003) as the primary antibodies. Secondary antibodies used were rhodamine (TRITC)-conjugated donkey anti-rabbit and fluorscein (FITC)-conjugated goat anti-mouse antibodies (Jackson Immuoresearch). Fluorescent images were collected on a Zeiss Axiovert 200M microscope using Axiovision 4.6 software.
Whole-cell recording
Drosophila S2 cells were cotransfected with pHC235 or pHC247 and pEGFP-1 (Act-5:EGFP) at a 10:1 ratio. Currents were recorded 48–96 h after transfection. Cells were allowed to settle on 12 mm round glass coverslips (Ted Pella, Inc.) and positioned on a Diaphot 300 inverted microscope (Nikon) placed on an anti-vibration table surrounded by a Faraday cage. Pipettes were pulled from borosilicate glass capillaries (World Precision Instruments) and fire polished to a resistance of 5–10 MΩ on a MF-900 fire polisher (Narishiga). Cells with GFP expression were selected for patching. The tip of the pipette was brought into membrane contact with a MP-225 micromanipulator (Sutter Instrument Co.) and a slight negative pressure was applied to the pipette until a GΩseal was achieved. Whole-cell configuration was set up by additional suction after seal formation. Whole-cell currents were recorded on an Axopatch 200B amplifier controlled by a Digidata 1322A and pClamp 9.2 software (Molecular Devices), with data sampling at 5–10 kHz. Voltage ramps 2000 msec long from −100 mV to +100 mV were applied every 5 sec from a holding potential of 0 mV. Nucleic acids were added directly to cells plated in bath solution. 0.3–8 nM of nucleic acid products were added to each experiment. Data were analyzed and displayed with Clampfit 9.2 (Axon Instruments). The bath solution was a modified version of standard SF9 Ringer: 17 mM NaCl, 110 mM NaGluconate, 5 mM KCl, 5 mM MgCl2, 4 mM CaCl2, 10 mM TES, 10 mM glucose, 50 mM sucrose, adjusted to pH 7.2 with NaOH with an osmolarity of 361 mOsm. The pipette solution contained 130 mM NaMethanesulfonate, 25 mM NaCl, 10 mM EGTA, 10 mM HEPES, adjusted to pH 7.2 with NaOH with an osmolarity of 330 mOsm.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Antony Jose, Jennifer Whangbo, Jacqueline Brooks, Daniel Schott, Andrea Hinas, and Deborah de Jong for critical reading of this manuscript, all the members of the Hunter laboratory for helpful comments and suggestions, Rachelle Gaudet for use of her electrophysiology equipment, Victoria D'Souza for discussions on nucleic acid structures, and Christopher Phelps and Venkatesh Murthy for technical advice. Antibodies and antisera were provided by Silas Maniatis and Sam Kunes. Drosophila cDNAs used in this work were provided by the Drosophila Genomics Resource Center (DGCR). This work was supported by NIH grant no. GM069891.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2596511.
REFERENCES
- Asmild M, Willumsen NJ 2000. Chloride channels in the plasma membrane of a foetal Drosophila cell line, S2. Pflugers Arch 439: 759–764 [DOI] [PubMed] [Google Scholar]
- Bartscherer K, Pelte N, Ingelfinger D, Boutros M 2006. Secretion of Wnt ligands requires Evi, a conserved transmembrane protein. Cell 125: 523–533 [DOI] [PubMed] [Google Scholar]
- Baulcombe D 2004. RNA silencing in plants. Nature 431: 356–363 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Chen X 2007. A marked end. Nat Struct Mol Biol 14: 259–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn GL, Brown T, Leonard GA 1999. The crystal structure of the RNA/DNA hybrid r(GAAGAGAAGC). d(GCTTCTCTTC) shows significant differences to that found in solution. Nucleic Acids Res 27: 555–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D Jr, Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, et al. 2006. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124: 343–354 [DOI] [PubMed] [Google Scholar]
- Duxbury MS, Ashley SW, Whang EE 2005. RNA interference: a mammalian SID-1 homologue enhances siRNA uptake and gene silencing efficacy in human cells. Biochem Biophys Res Commun 331: 459–463 [DOI] [PubMed] [Google Scholar]
- Feinberg EH, Hunter CP 2003. Transport of dsRNA into cells by the transmembrane protein SID-1. Science 301: 1545–1547 [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811 [DOI] [PubMed] [Google Scholar]
- Fischer SE, Butler MD, Pan Q, Ruvkun G 2008. Trans-splicing in C. elegans generates the negative RNAi regulator ERI-6/7. Nature 455: 491–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förstemann K, Horwich MD, Wee LM, Tomari Y, Zamore PD 2007. Drosophila microRNAs are sorted into functionally distinct Argonaute complexes after production by Dicer-1. Cell 130: 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B, Zamore PD 2004. Kinetic analysis of the RNAi enzyme complex. Nat Struct Mol Biol 11: 599–606 [DOI] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150 [DOI] [PubMed] [Google Scholar]
- Jose AM, Hunter CP 2007. Transport of sequence-specific RNA interference information between cells. Annu Rev Genet 41: 305–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose AM, Smith JJ, Hunter CP 2009. Export of RNA silencing from C. elegans tissues does not require the RNA channel SID-1. Proc Natl Acad Sci 106: 2283–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S, Wang D, Ruvkin G 2004. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427: 645–649 [DOI] [PubMed] [Google Scholar]
- Kim K, Lee YS, Carthew RW 2007. Conversion of pre-RISC to holo-RISC by Ago2 during assembly of RNAi complexes. RNA 13: 22–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81 [DOI] [PubMed] [Google Scholar]
- Lee RC, Hammell CM, Ambros V 2006. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA 12: 589–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X 2003. R2D2, a bridge between the initiation and effector steps of the Drosophia RNAi pathway. Science 301: 1921–1925 [DOI] [PubMed] [Google Scholar]
- Liu X, Jiang F, Kalidas S, Smith D, Liu Q 2006. Dicer-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. RNA 12: 1514–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoharan M 2004. RNA interference and chemically modified small interfering RNAs. Curr Opin Chem Biol 8: 570–579 [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Natalin P, Stark A, Brennecke J, Cohen SM, Izaurralde E 2006. Genome-wide analysis of mRNAs regulated by Drosha and Argonaute proteins in Drosophila melanogaster. Mol Cell Biol 26: 2965–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignant JY, Carré C, Mugat B, Szymczak D, Lepesant JA, Antoniewski C 2003. Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila. RNA 9: 299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih JD, Fitzgerald MC, Sutherlin M, Hunter CP 2009. The SID-1 double-stranded RNA transporter is not selective for dsRNA length. RNA 15: 384–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa R, Padilla R 1995. A mutant T7 RNA polymerase as a DNA polymerase. EMBO J 14: 4609–4621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara H, Grishok A, Mello CC 1998. RNAi in C. elegans: soaking in the genome sequence. Science 282: 430–431 [DOI] [PubMed] [Google Scholar]
- Tabara H, Yigit E, Siomi H, Mello CC 2002. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell 109: 861–871 [DOI] [PubMed] [Google Scholar]
- Tijsterman M, May RC, Simmer F, Okihara KL, Plasterk RH 2004. Genes required for systemic RNA interference in Caenorhabditis elegans. Curr Biol 14: 111–116 [DOI] [PubMed] [Google Scholar]
- Timmons L, Fire A 1998. Specific interference by ingested dsRNA. Nature 395: 854 doi: 10.1038/27579 [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263: 103–112 [DOI] [PubMed] [Google Scholar]
- Timmons L, Tabara H, Mello CC, Fire AZ 2003. Inducible systemic RNA silencing in Caenorhabditis elegans. Mol Biol Cell 14: 2972–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang SY, Moore JC, Huizen RV, Chan CW, Li RA 2007. Ectopic expression of systemic RNA interference defective protein in embryonic stem cells. Biochem Biophys Res Commun 357: 480–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston WM, Molodowitch C, Hunter CP 2002. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295: 2456–2459 [DOI] [PubMed] [Google Scholar]
- Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK, Rajeev KG, Nakayama T, Charrise K, Ndungo EM, et al. 2007. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol 25: 1149–1157 [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP 2000. RNAi double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33 [DOI] [PubMed] [Google Scholar]
- Zhang H, Fire AZ 2010. Cell autonomous specification of temporal identity by Caenorhabditis elegans microRNA lin-4. Dev Biol 344: 603–610 [DOI] [PMC free article] [PubMed] [Google Scholar]