Abstract
Polypyrimidine tract binding protein (PTB) is an RNA-binding protein with four RNA-binding domains (RBDs). It is a major regulator of alternative splicing and also stimulates translation initiation at picornavirus IRESs (internal ribosome entry sites). The sites of interaction of each RBD with two picornaviral IRESs have previously been mapped. To establish which RBD–IRES interactions are essential for IRES activation, point mutations were introduced into the RNA-binding surface of each RBD. Three such mutations were sufficient to inactivate RNA-binding by any one RBD, but the sites of the other three RBD–IRES interactions remained unperturbed. Poliovirus IRES activation was abrogated by inactivation of RBD1, 2, or 4, but the RBD3-IRES interaction was superfluous. Stimulation of the encephalomyocarditis virus IRES was reduced by inactivation of RBD1, 3, or 4, and abrogated by mutation of RBD2, or both RBDs 3 and 4. Surprisingly, therefore, the binding of PTB in its normal orientation does not guarantee IRES activation; three native RBDs are sufficient for correct binding but not for activation if the missing RBD–IRES interaction is critical.
Keywords: tethered hydroxyl radical probing, poliovirus, encephalomyocarditis virus, IRES, RRM (RNA recognition motif), RBD (RNA-binding domain)
INTRODUCTION
Polypyrimidine tract binding protein (PTB) has four RNA-binding domains (RBDs), also known as RRMs (RNA recognition motif), of the RNP1/RNP2 class (Oberstrass et al. 2005; Auweter and Allain 2008), although these elements are rather atypical in their lack of aromatic residues. PTB is a monomer in solution, but it has an extended conformation (Simpson et al. 2004; Monie et al. 2005), with flexible linkers between RBDs 1 and 2 and between RBDs 2 and 3. In contrast, RBDs 3 and 4 are separated by a short linker and seem to act as a pair or didomain (Oberstrass et al. 2005). In addition to the prototypic species (PTB-1), there are two isoforms resulting from alternative splicing (Wollerton et al. 2001) which differ from PTB-1 by the insertion of 19 (PTB-2) or 26 amino acids (PTB-4) in the linker between RBDs2 and 3.
PTB is predominantly nuclear, where its main function is as a regulator of alternative splicing of many pre-mRNAs (Spellman et al. 2005; Auweter and Allain 2008). However, PTB (which is also known as hnRNP I) is a shuttling protein (Ghetti et al. 1992), and there is a significant amount of it in the cytoplasm. An influence of PTB on several different cytoplasmic events has been reported (for review, see Sawicka et al. 2008), and the best studied case is the stimulation of translation initiation dependent on picornavirus internal ribosome entry sites/segments (IRESs) (Jackson and Kaminski 1995).
The PTB requirement for picornavirus IRESs varies quite widely according to the species. The majority of picornavirus IRESs are generally classified into two main groups (Alexander et al. 1994; Jackson and Kaminski 1995): type I IRESs, exemplified by enteroviruses (e.g., polioviruses) and human rhinoviruses, and type II, exemplified by encephalomyocarditis virus (EMCV) and foot-and-mouth-disease virus (FMDV). There is quite strong conservation of primary sequence and even stronger conservation of secondary structure within each group but very little similarity between the two groups. PTB is absolutely required for internal initiation on all the type I IRESs that have been tested so far, namely polioviruses (PV) and human rhinovirus-2 (Hunt and Jackson 1999), but type II picornavirus IRESs show more variability in their response to PTB. The FMDV IRES shows strong dependency on PTB (Niepmann 1996; Pilipenko et al. 2000), but for the encephalomyocarditis virus (EMCV) IRES, the PTB requirement is conditional on the nature of the reporter and the IRES variant used (Kaminski and Jackson 1998).
The binding sites of PTB on its pre-mRNA targets and on IRESs have been mapped by a combination of gel-shift and UV-cross-linking assays with mutagenesis of the putative interaction sites. However, these approaches provide no information on the orientation of the PTB binding to its target RNAs—the question of which RBD binds to which site on the RNA. We have recently generated a battery of PTB mutants, each with a single cysteine positioned at sites flanking the actual RNA-binding surface of each RBD, and we have used these derivatives in tethered hydroxyl radical probing to map the orientation of PTB binding to the EMCV and poliovirus IRESs (Kafasla et al. 2009, 2010). The results showed a very different pattern of PTB interaction with these two IRESs. The core EMCV IRES bound a single PTB, with RBDs1 and 2 binding near the 3′-end of the IRES and RBDs3 and 4 interacting with the 5′-end of the core IRES (Kafasla et al. 2009). This suggested that PTB binding would constrain the flexibility of the three-dimensional structure of the IRES and probably help to stabilize the optimum structure for internal initiation. In contrast, PTB binding to the poliovirus IRES was much more localized and was confined to the base of just one (Domain V) of the five irregular stem–loop structures present in the core IRES (Kafasla et al. 2010). In this case, the PTB binding site partially overlapped with the binding site of initiation factor eIF4G on the IRES, and the presence of PTB caused the position and orientation of eIF4G binding to be subtly modified.
While this tethered hydroxyl radical probing has provided considerable insight into how PTB interacts with these target RNAs, it does not answer the question of whether the interactions of all four RBDs with the target are essential for the biological activity of PTB. To address this issue, we have made several point mutations in the RNA-binding surface of each RBD. We found that three amino acid substitutions in the β2- or β4-strand suffice to disrupt the interaction of the mutated RBD with the EMCV (strain R) and poliovirus type 1 Mahoney (PV-1) IRESs, without altering the sites of interaction of the other three RBDs with these IRESs. These mutants were then tested in translation assays to assess the relative importance of each RBD for the PTB-dependent activation of the two viral IRESs.
RESULTS
Mutants generated
This investigation had four distinct steps: (1) generating the point mutations in the RNA-binding surface of each RBD; (2) verifying that the RNA-binding potential of the mutated RBDs has, indeed, been destroyed and determining which mutations are the most effective in this aim; (3) verifying that the interaction of the other three (non-mutated) RBDs with the target RNA has not been significantly perturbed; and (4) assaying the biological activity of the mutants in each RBD to assess which interactions are the most critical for this activity.
All the mutants were made in the background of a PTB-1 construct with a deletion of the N-terminal domain (NTD) (amino acids 1–54), which has nuclear import and export signals but is not involved in the interaction of PTB with RNA (Auweter and Allain 2008). This parent ΔNTD–PTB construct, hereafter designated PTB′, also has an N-terminal His-tag, and when the two remaining cysteine residues (C250 and C251 in RBD2) were replaced by serines (Fig. 1C), it resulted in the Cys-less PTB′ construct which is used as the control in tethered hydroxyl radical probing assays of the interaction of each RBD with the target RNA, as in our previous work (Kafasla et al. 2009, 2010).
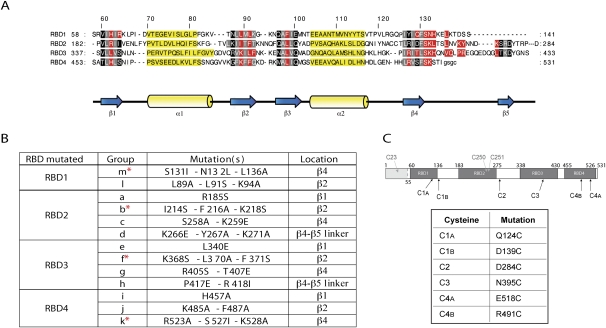
FIGURE 1.
Generation of PTB′ mutants to disrupt the RNA-binding potential of individual RBDs. (A) Sequence alignment of the four RBDs of PTB, adapted from Oberstrass et al. (2005), with the same color coding: amino acid residues interacting with the RNA in red; residues in black and gray are located in the β-sheets, with those in gray pointing toward the hydrophobic core of the RBD; and residues in yellow are in the α-helices. (B) The mutation groups introduced into each RBD to disrupt RNA-binding. Their location in each RBD is given, and the asterisks identify the most effective inactivating mutation group in each RBD. (C) Schematic representation of PTB1 showing the positions of the four RBDs, the first (N-terminal) 54 amino acids that were deleted in PTB′ (gray shading) and the native cysteines of PTB (in gray) which were substituted with serines in PTB′. The positions of the single cysteines introduced into each RBD and used as reference standards are also shown, and the actual amino acid replacements are listed.
The choice of sites for mutation was based on the NMR structures of each RBD complexed with an oligonucleotide ligand (Oberstrass et al. 2005), which identified the amino acid residues (colored red in Fig. 1A) in intimate contact with the RNA. The 29 mutated sites are listed in Figure 1B, with the most effective combinations for disrupting RNA-binding highlighted by an asterisk; all except two of these mutations were at positions colored red in Figure 1A. Single mutations were made in the β1-strand of each RBD (apart from RBD1), but no mutations were made in the β3-strands because Oberstrass et al. (2005) reported that this strand does not contribute to the RNA-binding of any of the four RBDs. Groups of 2 or 3 mutations were introduced by multiple site-directed mutagenesis into the β2- and β4-strands in each RBD and in the β4–β5 linker of RBDs2 and 3.
For the first RBD examined (RBD2), we constructed several combinations of the four mutation groups (a–d), in the expectation that complete inactivation of RNA-binding would require mutations in more than one β strand. However, this proved unnecessary, because the group b mutations were as effective as any combination, as shown in the next section. Moreover, combining mutation groups had the disadvantage that the increased number of mutations generally resulted in more recombinant protein appearing in the bacterial inclusion bodies (although there was always sufficient soluble protein to work with). For the other three RBDs, we, therefore, made a preliminary screen to identify which single mutation group was the most effective at disrupting the RBD–IRES interaction and then tested whether pairwise combinations of this mutation group with others in the same RBD were even more effective. We also made a mutant (designated Δ1) in which the whole RBD1 of PTB′ (amino acids 55–139 of PTB-1) was replaced with a 7-residue flexible segment (GSGSGSG).
Tethered hydroxyl radical probing was used to verify whether the RNA-binding potential of the mutated RBD had, indeed, been disrupted. To this end, the point mutations in the RNA-binding surface of a particular RBD were initially put into the background of a single cysteine derivative where the cysteine is located in the same RBD. From our library of 16 single cysteine PTB′ derivatives which all reacted efficiently with Fe(II)-BABE (Kafasla et al. 2009), we selected six for these assays— one in each of RBDs2 and 3 and two in each of RBDs1 and 4 (Fig. 1C). These six derivatives were chosen because they gave strong cleavages with both viral IRESs (Kafasla et al. 2009, 2010). The designation of such mutants takes the form C2-ab (for example) signifying that the a and b groups of point mutations in RBD2 were combined with a single cysteine derivative (D284C) with the Cys in RBD2 (hence, the designation C2).
Tethered hydroxyl radical probing was likewise used to test whether disruption of the RNA-binding potential of a particular RBD perturbed the interactions of the other three RBDs with the IRESs. For this purpose, up to two additional cysteines (Fig. 1C) were introduced into other (non-mutated) RBDs (one cysteine per RBD). The designation of these derivatives takes the form C2C3C4A-b (for example), indicating that the group b mutations and the cysteine substitution (D284C) in RBD2 were combined with a cysteine (N395C) in RBD3 and another (E518C) in RBD4.
Verifying the disruption of the RNA-binding potential of the mutated RBDs
Following conjugation of the mutant proteins with Fe(II)-BABE, tethered hydroxyl radical probing was used to determine whether the mutations in each RBD had disrupted its interactions with the two viral IRESs. For the EMCV IRES, we used the whole 5′ UTR downstream from the poly(C) tract as the target (Duke et al. 1992), because this binds two PTB molecules, whereas the minimal core IRES, which has Domain H at its 5′-end (Fig. 2B; Jang and Wimmer 1990), binds only one (Kafasla et al. 2009). All the analyses included assays with the single cysteine derivatives (C1–C4) (see Fig. 1C) in the background of wild-type (unmutated) RNA-binding surfaces of all four RBDs. These serve as reference standards, showing the sites of cleavage generated by Fe(II)-BABE conjugated at these positions. In addition, an assay with C-less PTB′ (which had been taken through the conjugation step) was always included as a background control. A band in the lanes with the single cysteine reference standards was considered to be a genuine cleavage product due to hydroxyl radical action only if it was distinctly more intense than the corresponding band in the C-less control lane (Kafasla et al. 2009). Mutations in the RNA-binding surface of a particular RBD were considered to have disrupted its RNA-binding potential if all the expected cleavages generated by the reference Cys derivative in this RBD disappeared at all three PTB binding sites (one in the PV-1 IRES and two in the EMCV probe).
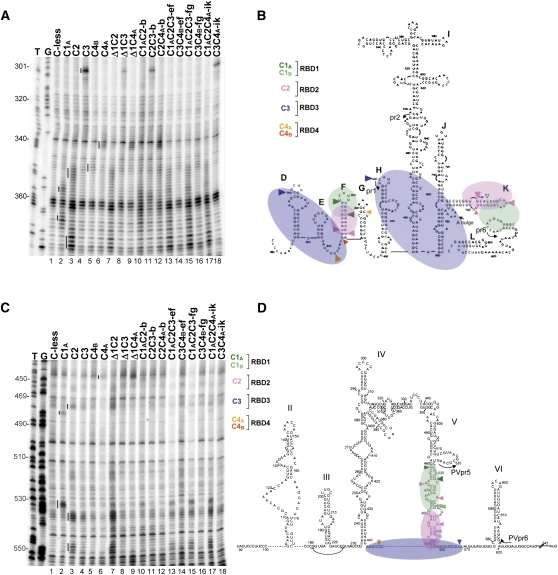
FIGURE 2.
Representative primer extension analyses of directed hydroxyl radical probing of the EMCV and PV IRESs with Fe(II)-BABE PTB′ mutants. (A) Directed hydroxyl radical probing was carried out under standard conditions with the designated Fe(II)-PTB′ derivatives, and the cleavage products were analyzed by primer extension using EMCVpr1 to examine nt 300–375 of the EMCV IRES. Sites of hydroxyl radical cleavages are indicated by vertical lines to the left of each reference standard (control) lane. Lanes T,G are from sequencing ladders using the same primer. (B,D) Schematic representation of the orientation of PTB binding to the EMCV IRES (panel B), adapted from Kafasla et al. (2009), and to the PV-1 IRES (panel D), adapted from Kafasla et al. (2010). RBD1 is in transparent green, pink denotes RBD2, and the RBD3 + 4 didomain is in transparent blue. The same color coding is used to show the sites of cleavage by the designated Fe(II)-PTB′ reference standard derivatives. Cleavages are classified as strong (large filled arrowhead), medium (medium filled arrowhead), or weak (smallest open arrowhead). For clarity, only the cleavages used in the present study as reference standards are shown; for complete cleavage site maps, see Kafasla et al. (2009, 2010). The annealing positions of the primers used for the primer extension analyses in the present study are also shown. (C) Tethered hydroxyl radical probing of PV IRES RNA with the specified Fe(II)-PTB′ mutants, using primer PVpr6 for primer extension analysis of nt 445–555. Cleavage sites are highlighted by vertical lines, as in panel A.
Cleavage sites were detected by reverse transcription using primers that bind to the sites shown in Figure 2B (EMCV IRES) and Figure 2D (PV-1 IRES). Representative autoradiographs of full-length gels showing the primer extension analyses of cleavage sites in the EMCV IRES and PV-1 IRES, using one particular primer in each case, are presented in Figure 2, A and C, respectively. Figure 3 shows the parts of gels (with the same and other primers) where the main cleavage products generated by Fe(II)–BABE conjugated to different RBDs are found.
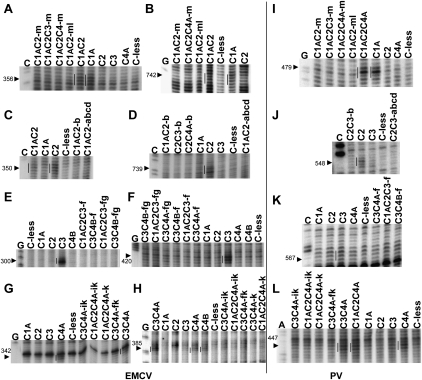
FIGURE 3.
Primer extension analyses of directed hydroxyl radical probing experiments to test for disruption of the interaction of mutated RBDs with the EMCV and PV IRESs. Assays of the designated PTB′ mutants with the EMCV IRES are shown in panels A–H, with panels A,C,E,G examining interactions with Domains D–F, and panels B,D,F,H, the interactions with Domains G–K. Assays with the PV-1 IRES are in panels I–L. The cleavage products arising from Fe(II) conjugated to the relevant non-mutated RBD are indicated by vertical lines to the left of the reference standard lanes. The inactivating mutations analyzed in each subpanel are as follows: A,B,I—RBD1 mutations; C,D,J—RBD2 mutations; E,F,K—RBD3 mutations; and G,H,L—RBD4 mutations. Lanes A,C,G are from sequencing ladders with the same primers as the primer extension lanes. The arrowheads to the left of each panel indicate the position of the reference standard cleavage site in the IRES.
These data show that the mutations in group m in the RBD1 β4-strand are sufficient to disrupt its binding to Domains F (nt 356) and K (nt 742) of the EMCV IRES and Domain V (nt 479) of the PV-1 IRES (Fig. 3A,B,I); the ml mutation combination was not significantly different from m alone. Similarly the group b mutations in the β2-strand inactivate interaction of RBD2 with Domains F (nt 350) and K (nt 739) of the EMCV IRES (Fig. 2A, cf. lanes 10–12 and 3; Fig. 3C,D), in addition to Domain V of the PV IRES (Fig. 2C, cf. lanes 10–12 and 3; Fig. 3J), and were as effective as the abcd combination (Fig. 3C,D,J).
For RBD3, the mutation group f in the β2-strand clearly disrupted the strong interaction of RBD3 with Domain D (nt 300) and Domain H (nt 420) of the EMCV IRES, and we saw no evidence that the ef and fg combinations were more effective (Fig. 2A, cf. lanes 13–16 and 4; Fig. 3E,F). Mutation group f in RBD3 also appeared to disrupt the interaction with the PV-1 IRES (Fig. 3K), although this is harder to judge because Fe(II)-N395C PTB′ generates no strong cleavages in the PV IRES but only one somewhat variable medium-weak cut and a few other extremely weak cleavages (Kafasla et al. 2010). Finally, the mutation group k in the β4-strand disrupted the interaction of RBD4 with the Domain E–F linker (nt 342) and Domain G (nt 385) of the EMCV IRES and the Domain IV–V linker in the PV-1 IRES; at all three sites, it appeared to be no less effective than the ik combination (Fig. 2A, cf. lanes 16–18 and 6; Fig. 3G,H,L) or the jk combination (data not shown).
Importantly, there was no target site specificity with respect to which mutations were the most effective in abrogating the interactions of each RBD with these IRESs. This suggests that the m, b, f, and k mutations will successfully disrupt the interactions of PTB RBDs1, 2, 3, or 4, respectively, with any target RNA.
Testing whether the binding of the other three (non-mutated) RBDs is maintained
In the next step, the most effective inactivating mutations in the RNA-binding surface of each RBD were combined with mutants having single cysteines in the other RBDs to enable the use of tethered hydroxyl radical probing for evaluating whether the interactions of these non-mutated RBDs with the IRESs had been maintained. For these purposes, we examined the interactions with both IRESs, and a representative set of results is shown in Figure 4, which includes a tabulated semi-quantitative summary of the consensus outcome of these and numerous other similar experiments. Both PTB binding sites in the EMCV IRES were examined, but in order to keep the figure to a manageable size, Figure 4 shows only the autoradiographs for the 5′-proximal binding site.
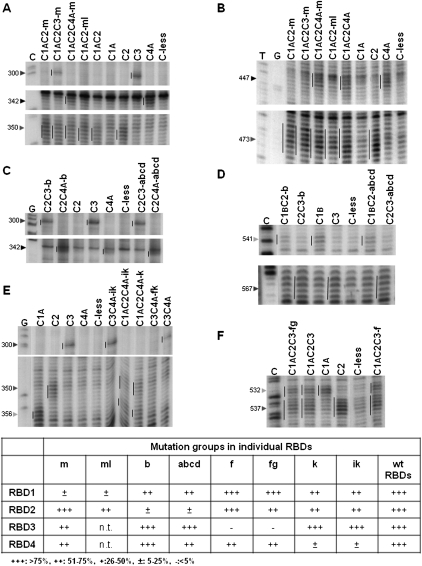
FIGURE 4.
Representative primer extension analyses of directed hydroxyl radical probing experiments to test whether inactivation of one RBD affects the interaction of the other three RBDs with the EMCV and PV IRESs. (A,C,E) Assays of interactions with the 5′-proximal part of the EMCV IRES, using mutants inactivated in RBD1 (panel A), RBD2 (panel C), and RBD4 (panel E). (B,D,F) Assays of interactions with the PV-1 IRES, using mutants inactivated in RBD1 (panel B), RBD2 (panel D), and RBD3 (panel F). In each panel, the vertical line to the left of the reference standard lane highlights the cleavage products generated (in the absence of any inactivating mutations) by Fe(II) conjugated to the particular RBD under examination, and they are repeated in all the other lanes assaying PTB′ derivatives with Fe(II) conjugated to the same RBD. The arrowheads to the left of each panel indicate the reference standard cleavage site position in the IRESs. Lanes C,G,T are from sequencing ladders using the same primer. The cleavage band intensities in these and many other similar experiments were quantitated as described in Materials and Methods, and the cleavage band intensity determined for PTB′ derivatives with inactivating mutations in one RBD and Fe(II)-BABE conjugated to one or more non-mutated RBDs was compared with that found for the corresponding reference standard (WT) with no inactivating mutations (taken as 100%). The consensus of the relative intensity values obtained from many such experiments, covering all three PTB binding sites (two in the EMCV IRES and one in the PV IRES), is tabulated in a semi-quantitative form at the bottom of the figure.
The general conclusion from the results shown in Figures 2A,C and 4, plus numerous other similar experiments, is that inactivating the RNA-binding activity of any one RBD had no effect on the actual positions where the IRESs were cleaved by Fe(II)-BABE conjugated to the other (non-mutated) RBDs. No cleavage bands were seen at new sites where there were no cuts seen with control Fe(II)-PTB′ derivatives in which all four RBDs were active in RNA-binding. This leads to the important conclusion that inactivating one RBD does not perturb the orientation of binding of PTB to the IRESs.
However, in some cases inactivating one RBD resulted in a clear reduction in the intensity of the cleavage bands arising from Fe(II) conjugated to some of the non-mutated RBDs. Although there appear to be enhancements of the Fe(II)-RBD4A cleavage at EMCV nt 342 in Figure 2A (cf. lanes 12 and 6), the Fe(II)-RBD4A cleavage at PV nt 447 in Figure 2C (cf. lanes 9 and 6), and the several Fe(II)-RBD2 cleavages of the PV IRES in Figure 2C (cf. lanes 7 and 3), a visual inspection (supported by quantitation using the SAFA software as described in Materials and Methods) of the background bands in these lanes shows that these apparent enhancements can be explained by a slight underloading of the reference standard lanes (Fig. 2A, lane 6; Fig. 2C, lanes 3,6) and overloading of lane 12 in Figure 2A and lanes 7 and 9 in Figure 2C. When these considerations were taken into account, we found no evidence for any genuine enhancements of cleavage band intensity.
The reduction in the intensity of some of the cleavage bands arising from Fe(II)-BABE conjugated to a non-mutated RBD implies that over the 15-min duration of the probing assay, the average position of that particular non-mutated RBD was more distant from the RNA than when all four RBDs were active. One possible but unlikely explanation is that the non-mutated RBD was permanently located for the 15-min period at a position somewhat more distant from the RNA than in the control in which all four RBDs were active in RNA-binding. However, the more likely explanation is that the particular non-mutated RBD was fully engaged with the RNA (i.e., as tightly bound as in the controls) for relatively less time and fully disengaged or dissociated for longer. This raises the question of whether the temporary dissociation of one non-mutated RBD in this way will lead to complete dissociation of the PTB from the RNA, and this obviously depends on whether there is simultaneous dissociation of both of the other two non-mutated RBDs. Nevertheless, complete dissociation in these circumstances is a real possibility warranting consideration, given the indications that all four RBDs contribute to the binding of PTB to its target RNAs (Simpson et al. 2004; Oberstrass et al. 2005), and, in particular, the finding that deletion mutants which retain just two RBDs have significantly lower binding affinity than full-length PTB for the EMCV IRES (and also for a pre-mRNA target of PTB), with the affinity of the RBD3 + 4 combination distinctly lower than that of the RBD1 + 2 pair (Simpson et al. 2004).
If the reduced intensities in the cleavage bands arising from Fe(II) conjugated to the non-mutated RBDs are entirely due to complete dissociation of the PTB from the IRES, we would expect to see equivalent reductions in intensity for all three non-mutated RBDs. By this criterion, the results suggest that inactivating one RBD by introducing point mutations causes very little complete dissociation of the PTB from the RNA, because, for each inactivated RBD, there was at least one non-mutated RBD that showed significantly less reduction in cleavage band intensity than one (or both) of the other two (as summarized in Fig. 4). In addition, with the Δ1 replacement of RBD1, there was no reduction in cleavage band intensity arising from Fe(II)-RBD4A at EMCV nt 342 (Fig. 2A, lanes 6,9) or PV nt 447 (Fig. 2C, lanes 6,9), and no reduction of the several Fe(II)-RBD2 cleavages on the PV IRES (Fig. 2C, lanes 3,7), although reductions in intensity were seen in the case of the Fe(II)-RBD2 cleavages on the EMCV IRES (Fig. 2A, lanes 3,7) and, more especially, the Fe(II)-RBD3 cleavage at EMCV nt. 300 (Fig. 2A, lanes 4,8).
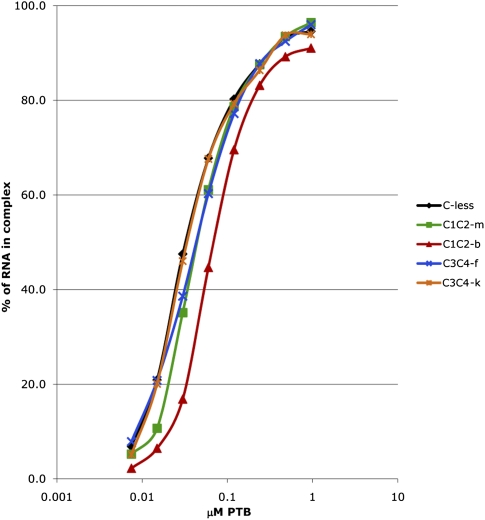
We conclude that even though the inactivation of one particular RBD may increase the susceptibility of one or two specific non-mutated RBDs to dissociate from their usual binding site(s) on the IRESs, it does not provoke a large increase in the frequency of complete dissociation of the PTB. Nevertheless, in order to examine this question more directly and more quantitatively, we have done filter-binding assays, using the full-length EMCV probe with Cys-less PTB′ as the control and the four mutants with the group m, b, f, and k mutations (Fig. 5). These assays gave the following average Kd values (with standard deviations in parentheses): C-less PTB′, 33 nM (±9 nM); RBD1 m mutant, 46 nM (±16 nM); RBD2 b mutant, 65 nM (±13 nM); RBD3 f mutant, 43 nM (±14 nM); and RBD4 k mutant, 34 nM (±7 nM). This value for the Cys-less PTB′ positive control is similar to those reported previously for wild-type PTB (Kafasla et al. 2009). In addition, the hierarchy of affinity of the inactivated mutants, namely RBD4 k > RBD3 f > RBD1 m > RBD2 b, is entirely consistent with the observation that a deletion mutant which retains just RBDs3 and 4 (and therefore lacks RBDs1 and 2) binds with significantly lower affinity than the reciprocal deletion which retains just RBDs1 and 2 (Simpson et al. 2004).
FIGURE 5.
Filter-binding assays of the interaction of various PTB′ derivatives with the full-length EMCV probe. The assay was performed in triplicate as described in Materials and Methods. The average values for the fraction of RNA bound to PTB′ are plotted against the concentration of PTB′ derivative, which is plotted on a log10 scale. For clarity, error bars have been omitted, and instead the standard deviations for the calculated Kd values are given in the text.
Assay of the mutants in translation assays of EMCV and PV IRESs
Finally, we assessed the effect of disrupting the RNA-binding potential of each RBD on the activity of PTB in stimulating translation driven by the EMCV and PV-1 IRESs. The most effective group of inactivating mutations in each RBD was used for these assays, with wild-type full-length PTB as the positive control. The maximum stimulation observed with the PTB′ mutant was compared to the maximum observed with the PTB-1 positive control and the background observed with no added protein. The PTB′ mutants (and the wild-type PTB-1 positive control) were added at 24, 47, 94, or 188 nM final concentration. Our previous work has shown that, despite the modest reduction in binding affinity of some of the inactivated mutants noted in the previous section, a concentration of 188 nM should be sufficient to achieve effective saturation with both IRESs (Kafasla et al. 2009, 2010), without the complication of the downturn in stimulation seen at higher protein concentrations, especially with the PV IRES (Kafasla et al. 2010), but also with the EMCV IRES in some experiments (Fig. 7G, see below).
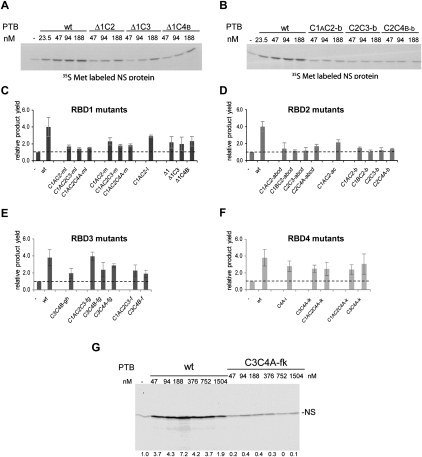
FIGURE 7.
Translation assays of the stimulation of EMCV IRES activity by PTB′ derivatives with mutations in the RNA-binding surface of each RBD. (A,B) Representative autoradiographs showing the translation products synthesized in the presence of various concentrations of the wild-type full-length PTB-1 control (WT) and the designated PTB′ mutants. (C–F) Summary of translation assay results for the designated mutants of RBD1 (panel C), RBD2 (panel D), RBD3 (panel E), and RBD4 (panel F). The maximum translation product yield obtained with each mutant was compared to the yield observed when no PTB was added to the translation reaction (−PTB; set at 1.0 and shown by the dashed horizontal line). The results shown are the mean (with standard deviation) of the values obtained for each mutant in three independent experiments. (G) Assay of wild-type full-length PTB (WT) and the C3C4A-fk PTB′ derivative with inactivating mutations in both RBDs3 and 4. Translation products were analyzed by SDS-PAGE, and the resulting autoradiograph is shown. The concentration of added wild-type PTB or C3C4A-fk PTB′ in the assays is given above each lane, and the numbers below each lane show the actual stimulation (i.e., the increment) in translation product yield expressed relative to the yield (assigned a value of 1.0) in the control assay lacking any added PTB or PTB′. The PTB-depleted lysate used for this experiment was a different batch from that used for panels A–F, and the depletion was likely to have been more efficient, resulting in a greater stimulation.
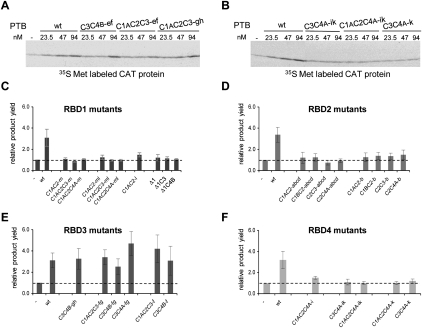
For the PV IRES, standard nuclease-treated reticulocyte lysate was used, which contributes ∼40 nM endogenous PTB to the assay (Kafasla et al. 2010), with a monocistronic mRNA consisting of the complete PV 5′ UTR fused to a CAT reporter. The results showed that inactivation of either RBD1, RBD2, or RBD4 completely abrogated all stimulation (Fig. 6C,D,F, respectively). By contrast, inactivation of RBD3 had a much smaller effect (Fig. 6E), which is entirely consistent with our previous results of tethered hydroxyl radical probing with Fe(II)-N395C (Kafasla et al. 2010) (see Discussion).
FIGURE 6.
Translation assays of the stimulation of PV-1 IRES activity by PTB′ derivatives with mutations in the RNA-binding surface of each RBD. (A,B) Representative autoradiographs showing the translation products synthesized in the presence of various concentrations of the wild-type full-length PTB-1 control (WT) and the designated PTB′ mutants. (C–F) Summary of translation assay results for the designated mutants of RBD1 (panel C), RBD2 (panel D), RBD3 (panel E), and RBD4 (panel F). The maximum translation product yield obtained with each mutant was compared to the yield observed when no PTB was added to the translation reaction (−PTB; set at 1.0 and shown by the dashed horizontal line). The results shown are the mean (with error bars showing standard deviations) of the values obtained for each mutant in three independent experiments.
For the EMCV IRES, we used a PTB-depleted lysate (Kaminski et al. 1995) because the affinity of PTB for the EMCV IRES is higher than for the PV-1 IRES, and yet the stringency of the PTB-requirement is lower (Kaminski and Jackson 1998). A monocistronic mRNA was used consisting of the entire EMCV 5′ UTR downstream from the poly(C) tract (Duke et al. 1992) (i.e., from nt 260) fused to an influenza virus NS reporter ORF, and the IRES sequence was a mutant with an enlarged 7A-bulge at the three-way junction between Domains J and K (Fig. 2B), because this shows considerably higher PTB-dependency than the wild-type IRES which has a 6A-bulge in this position (Kaminski and Jackson 1998).
The results of these assays showed that inactivation of RBD2 completely eliminated all stimulation by PTB, whereas RBD1 inactivation had a considerably smaller negative effect (Fig. 7C,D). Inactivation of either RBD3 or RBD4 caused a ∼50% reduction (Fig. 7E,F), rather than complete abrogation of the PTB-dependent stimulation. In view of the finding that RBDs3 and 4 can act as a coordinated pair, or didomain (Oberstrass et al. 2005), and that Fe(II)-N395C in RBD3 generates a strong cleavage in the apical loop of IRES Domain H, while Fe(II)-E518C in RBD4 cuts in the neighboring Domains G and the base of I (Kafasla et al. 2009), it seemed possible that when either RBD3 or RBD 4 had been inactivated, binding of the other (intact) RBD to this region of the IRES could still result in some partial stabilization of the three-dimensional structure of the IRES, which is thought to be the underlying explanation for the stimulatory effect of PTB (Kafasla et al. 2009). We, therefore, tested the effect of inactivating both RBDs3 and 4, which had the outcome that stimulation of the 7A-bulge IRES was almost completely abolished and could not be rescued by raising the concentration of this PTB′ mutant to extremely high levels (Fig. 7G).
DISCUSSION
The important general conclusion is that it is possible to selectively inactivate the RNA-binding potential of any of the RBDs in PTB by introducing no more than three point mutations at strategically chosen positions. Moreover, inactivating the RNA-binding of any one RBD had no effect on the binding positions of the other (non-mutated) RBDs on the EMCV and PV-1 IRESs.
Although complete inactivation of RNA-binding required three point mutations, it should be noted that there were other derivatives with triple mutations that still retained significant binding, notably the group l mutations in the β2-strand of RBD1 and the group d mutations in the β4–β5 linker of RBD2 (data not shown). Thus, it is not just the number of mutations that is the important determinant but also their position, as well as perhaps the nature of the amino acid substitutions. The most effective inactivating triple mutations were either in the β2-strand (RBDs2 and 3) or β4-strand (RBDs1 and 4). The NMR structures of each wild-type RBD complexed with a CUCUCU hexanucleotide ligand (Oberstrass et al. 2005) show that the amino acids substituted in these triple mutants primarily contact the two nucleotides at the core of the hexameric oligonucleotide. Moreover, inspection of the ensemble of the 20 lowest energy structures for each RBD (Oberstrass et al. 2005) suggests that these contacts hold the core nucleotides in place in the complex (by contrast, the outer nucleotides are not fixed in position) and, therefore, probably contribute significantly to RNA-binding.
The translation assay results show that, apart from the RBD3 mutant and the PV IRES, inactivation of any one RBD generally reduces the stimulation of IRES activity, though to different extents according to which IRES is tested and which RBD has been mutated. The decreased stimulation cannot be explained simply by reduced affinity of the PTB′–IRES interaction. Even in the case of the RBD2 b mutations, which showed the greatest decrease in binding affinity (Fig. 5), we calculate that at 188 nM added PTB′ derivative, 73% of the input mRNA with the EMCV IRES will be associated with this mutant PTB′, compared to 84% with the Cys-less PTB′ or wild-type PTB controls. Thus, reduced affinity of the PTB–IRES interaction in the case of this particular mutant could only account for a ∼13% reduction in IRES stimulation, whereas stimulation of both IRESs was actually abrogated (Figs. 6D, 7D). It follows that the decrease in IRES stimulation seen with all the inactivated mutants is due to loss of specific contacts between the mutated RBD and the IRES and not to a general decrease in PTB binding to the IRES. This further implies that PTB binding per se is not sufficient to stimulate IRES functionality if the missing RBD–IRES interaction plays a critical role.
Our previous investigation of the PV-1 IRES showed that, under conditions in which stimulation by PTB is observed, the IRES binds a single PTB, with RBDs1 and 2 binding to closely neighboring sites in the basal half of IRES Domain V and RBD4 in close proximity to the short Domain IV–V linker. Initiation factor eIF4G (or eIF4F) also binds to the basal half of Domain V (de Breyne et al. 2009), and we found that PTB caused a subtle change in the orientation of eIF4G binding, either as a result of competition between PTB and eIF4G for overlapping binding sites or through PTB binding remodeling the RNA structure of this region (Kafasla et al. 2010). This reorientation of eIF4G is likely to play a pivotal role in the activation of the IRES by PTB, and we would therefore expect at least one of the two interactions (of RBD1 and RBD2) with Domain V to be critical. In fact, binding of both RBDs1 and 2 was clearly essential for IRES activation, as the mutation of either completely abrogated stimulation (Fig. 6C,D). This strong negative effect is unlikely to be due to a secondary effect in which the inactivation of one of these RBDs destabilizes the binding of the other, because neither the group m mutations nor the complete Δ1 replacement reduced the binding of RBD2 to Domain V (Fig. 2C, cf. lanes 3 and 7; Fig. 4B), and the group b mutations in RBD2 caused only a rather modest reduction in RBD1 binding to Domain V (Fig. 4D). RBD4 binding was also found to be important (Fig. 6F), perhaps because its interaction with the Domain IV–V linker places constraints on the relative spatial orientation of these two large domains. On the other hand, the RBD3–IRES interaction seemed superfluous (Fig. 6E). This is consistent with the tethered hydroxyl radical probing results (Kafasla et al. 2010), which showed that the strongest cleavage generated by the Fe(II)-N395C derivative in RBD3 was only a rather variable medium-weak cleavage just downstream from Domain V, whereas Fe(II)-E518C (in RBD4) gave a medium strength cut in the Domain IV–V linker, and the RBD1 and RBD2 derivatives generated strong cleavages in Domain V. From these data, we deduced that RBD3 is not in close contact with the PV IRES (Kafasla et al. 2010), and thus, it is hardly surprising that disrupting its RNA-binding potential has such a minor effect on stimulation of IRES activity.
In the only previous publication relevant to this issue, where poliovirus IRES activity in transfection assays showed significant stimulation when cotransfected with a wild-type PTB expression construct, a PTB-1 derivative lacking RBD1 not only failed to stimulate but in fact quite strongly inhibited the basal IRES activity sustained by the endogenous PTB (Back et al. 2002). This implies that the lack of RBD1 does not perturb PTB binding to its usual site on the PV-1 IRES but does inhibit its functionality, which is entirely consistent with our results. Overexpression of derivatives comprising just RBDs1 and 2, or just RBDs3 and 4, also failed to stimulate, but in both these cases, the inhibitory effect was much weaker, suggesting that binding affinity was considerably reduced (Back et al. 2002).
In our previous study of PTB binding to the EMCV IRES, we found that the complete 5′ UTR downstream from the poly(C) tract (as shown in Fig. 2B) bound two PTBs, but the core IRES, which has Domain H at its 5′-end, bound a single PTB, with RBDs1 and 2 binding to closely neighboring sites in the distal part of Domain K, RBD3 binding strongly to the apex of Domain H, and RBD4 interacting with the base of Domain I, as well as with Domain G when the target RNA was the complete 5′ UTR (Kafasla et al. 2009). It was suggested that this would place limits on the distance separating the apex of Domain K from Domains G, H, and the base of Domain I, thus constraining the three-dimensional structure of the IRES to one more favorable for internal initiation. We would, therefore, expect that the interaction of at least two RBDs with the RNA would be essential, one at the Domain K site and the other in the 5′-proximal region. In fact, we found that mutation of RBD2 had the greatest negative effect (Fig. 7D), and it appears that the interaction of PTB with Domain K is more critically dependent on RBD2 than RBD1 (Fig. 7C). The importance of the other two RBDs is more complicated. Although maximum stimulation required both RBD3 and RBD4, inactivation of either one of them only reduced the stimulation by ∼50%, suggesting the possibility of some partial redundancy between them (Fig. 7E,F). This suggestion is supported by the fact that almost no stimulation was seen in the presence of the double mutation, not even when its concentration was raised to extremely high levels (Fig. 7G). A possible explanation is that the binding of just one RBD (either RBD3 or RBD4) to the 5′-end of the core IRES does not constrain the three-dimensional flexibility of the IRES as effectively as when both interactions are occurring.
A previous report on the stimulation of the FMDV IRES (which is structurally similar to the EMCV core IRES) by PTB deletion mutants found that deletion of RBD1 had no discernible effect (Song et al. 2005), and deletion of RBD2, or both RBDs1 and 2, reduced stimulation by <50%, far less than observed here with point mutations inactivating RBD2. On the other hand, deletion of RBD3 or RBD4 almost eliminated all stimulation, particularly the RBD4 deletion. The authors concluded that RBDs3 and 4 were sufficient for stimulation (Song et al. 2005), in contrast to our conclusions for the EMCV IRES.
It is quite possible that this apparent discrepancy reflects a genuine difference between the two IRESs, because our previous study (Kaminski et al. 1995) of similar PTB deletion mutants (tested as GST–PTB fusion proteins) gave results remarkably consistent with those we have reported here. In sharp contrast to the FMDV results (Song et al. 2005), a deletion mutant which retained just RBDs3 and 4 failed to stimulate the EMCV IRES (with a 7A-bulge at the Domain J–K three-way junction) but did bind to this IRES, according to the results of UV-cross-linking assays, and this was confirmed by the fact that it inhibited stimulation by full-length wild-type GST-PTB. Precisely the same results were found for a mutant that retained just RBDs1 and 2. The deletion mutant which showed by far the highest stimulation of the EMCV IRES (∼60% with respect to wild-type GST-PTB) was one with just RBD1 deleted, consistent with our conclusion that RBD1 binding is of relatively marginal importance. Very weak stimulation was seen with a PTB mutant that had intact RBDs1, 2, and 3, with a deletion of the C-terminal 27 amino acids which removes part of RBD4 (including the whole β4-strand), and this mutant also inhibited stimulation by wild-type GST-PTB. The negative effect of this mutation on stimulation was, therefore, greater than we found with our RBD4 point mutants (Fig. 7F), but this difference could well be due to a type of dominant negative effect exerted by the truncated RBD4. Apart from this disagreement, the results we obtained previously with these deletion mutants (Kaminski and Jackson 1998) and with the point mutations described here lead to very similar conclusions: RBD2 binding is essential for stimulation of the EMCV IRES, RBD1 is of relatively minor significance, and maximum stimulation needs both partners of the RBD3/RBD4 didomain.
While this agreement is highly satisfactory, the potential caveats and artifacts associated with extensive deletions make it preferable that point mutants of the type developed here should be exploited for further investigations into the significance of each RBD for biological activity. Together with our single cysteine PTB derivatives for use in directed hydroxyl radical probing (Kafasla et al. 2009), a complete set of reagents is now available for detailed investigation of the interaction of PTB with its other RNA targets, such as cellular mRNA IRESs or pre-mRNAs which are subject to PTB-regulated alternative splicing. Moreover, the approaches we have used should be applicable to the analysis of any other protein with multiple RBDs (for example, proteins of either the RNP1/RNP2 or KH-domain families), particularly those cases where structural data on the RBD complexed with an RNA ligand are available.
MATERIALS AND METHODS
Construction of PTB′ mutants
The construction of expression plasmids encoding PTB′ (a cysteine-less PTB-1 derivative lacking the N-terminal 54 amino acids) and single cysteine derivatives of PTB′, the isolation of these N-terminally hexahistidine-tagged recombinant PTB′ mutant proteins, and their conjugation with Fe(II)-BABE have been described previously (Kafasla et al. 2009). Groups of up to three mutations in the RNA-binding surface of each RBD of PTB′ were introduced, usually with a single mutagenic oligonucleotide, using a standard (DpnI-dependent) site-directed mutagenesis protocol. To combine different mutation groups into the same RBD, a multichange mutagenesis protocol was used, either the Stratagene Multi Site Mutagenesis kit, or an in-house adaptation of this method. The appropriate reference cysteine (Fig. 1C) was then introduced into the same RBD by site-directed mutagenesis. These mutations were subsequently combined with single cysteines in other RBDs by making use of the unique restriction enzyme sites in the inter-RBD linkers (Kafasla et al. 2009). In every case, the mutations were verified by sequencing the whole ORF.
Tethered hydroxyl radical probing assays
The preparation of the EMCV IRES RNA (nt 260–848, and with a wild-type 6A-bulge at the Domain J–K three-way junction) was as described previously (Kafasla et al. 2009), and the PV IRES RNA (nt 94–630) was prepared as described in Kafasla et al. (2010). Standard conditions for tethered hydroxyl radical probing were 50 nM EMCV IRES RNA and 100 nM Fe(II)-PTB′ derivative, or 100 nM PV-1 IRES RNA with 700 nM Fe(II)-PTB′. The procedure was as described previously (Kafasla et al. 2010), except that the binding buffer for the EMCV IRES assays was 100 mM in KCl, rather than the 70 mM as used for the PV IRES (Kafasla et al. 2010). Following initiation of the Fenton reaction by adding ascorbic acid and H2O2, incubation was for 15 min on ice with both RNA probes before re-isolation of the RNA by phenol extraction. Primer extension analysis using AMV-RT was carried out as described previously (Kafasla et al. 2010), using primers PVpr5 and PVpr6 (described therein) in the case of the PV IRES and the following primers for the EMCV IRES:
EMCVpr1: 5′-GACCCCTAGGAATGCTCGTC-3′;
EMCVpr2: 5′-GTCTTCAAGAAGCTTCCAGAG-3′;
EMCVpr6: 5′-GTTGTGGCCATATTATC-3′.
For quantitative analysis of cleavage bands, SAFA (Semi-Automated Footprinting Analysis) software (Das et al. 2005; Laederach et al. 2008) was used as described previously (Kafasla et al. 2010). To compensate for possible variations in loading, the band intensity values generated by the SAFA software were first normalized with respect to background bands, whose intensity was largely invariant in all lanes (including the Cys-less control lane) but sometimes fluctuated in a few lanes in parallel with each other (and with the full-length primer extension product when it was visible). This normalized intensity for each individual band in a given experimental lane was then compared, as a ratio, to the intensity of the corresponding normalized band in the Cys-less control lane.
Filter-binding assays
The affinity of the various PTB mutants for 32P-labeled EMCV IRES RNA (nt 260–848) was assessed using filter-binding assays. PTB–RNA complexes were assembled by incubation for 20 min at room temperature in 50 μL reactions in 10 mM HEPES pH 7.4, 100 mM KCl, 2.5 mM MgCl2, 5% glycerol, 1 mM DTT, 50 μg/mL yeast tRNA, 50 μg/mL bovine serum albumin, and 8 nM probe RNA. Assays were performed using the protein-binding Protran NBA-085B nitrocellulose membrane (Whatmann) to immobilize the PTB–RNA complexes and a lower layer of Hybond-N membrane (Amersham Life Sciences) to immobilize the unbound free RNA. The membranes were washed extensively in 10 mM HEPES pH 7.4, 2.5 mM MgCl2, 5% glycerol, 1 mM DTT and mounted on a 96-well dot-blotter. Before and after application of 45 μL of the binding reaction, the membrane was washed with 180 μL of wash buffer. Following the experiment, the membrane was dried, and the fraction of input RNA bound to PTB was determined by densitometric analysis of the autoradiograph using TotaLab TL120 software (Nonlinear Dynamics).
In vitro translation assays
Uncapped monocistronic mRNA with the PV-1 IRES (nt 1–747) fused in frame with a CAT ORF was prepared by transcription with T7 RNA polymerase as described previously (Kafasla et al. 2010). The translation assay reactions contained 60% (by volume) standard nuclease-treated rabbit reticulocyte lysate, and the following additional components at the final concentration stated: 10 μM haemin, 50 μg/mL creatine kinase, 10 mM creatine phosphate, 0.1 mM each amino acid (except methionine), 70 mM KCl, 0.5 mM MgCl2, 4 mM 2-aminopurine and [35S]methionine. The standard mRNA concentration was 12 nM mRNA, and PTB′ derivatives (or wild-type PTB-1 positive control) were added at 23.5, 47, 94, or 188 nM. Incubation was at 30°C for 60 min, then the products were separated by SDS-PAGE, and the yield of the full-length product band was quantified. The maximum stimulation observed with the PTB′ derivatives was compared with the maximum seen with the PTB-1 control.
Uncapped monocistronic mRNA with the EMCV IRES (nt 260–846 and with an enlarged 7A-bulge at the Domain J–K three-way junction) fused to the influenza virus NS1 ORF was prepared by transcription with T7 polymerase of plasmid pEMCV-NS (Kaminski and Jackson 1998) that had been linearized with Eco RI. It was translated in nuclease-treated reticulocyte lysate that had been depleted of PTB by the procedure described previously (Kafasla et al. 2009). Translation assay conditions were otherwise as stated above except that the final concentration of added KCl was 100 mM. The standard mRNA concentration was 20 nM, with PTB′ derivatives or wild-type PTB-1 at the same concentrations as for the mRNA with the PV IRES.
ACKNOWLEDGMENTS
We thank Jenny Reed for infrastructure and technical assistance. This work was supported by BBSRC Project Grants BB/E004857/1 (to R.J.J.) and BB/H004203/1 (to Prof. C.W.J. Smith and R.J.J.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2549411.
REFERENCES
- Alexander L, Lu HH, Wimmer E 1994. Polioviruses containing picornavirus type 1 and/or type 2 internal ribosomal entry site elements: Genetic hybrids and the expression of a foreign gene. Proc Natl Acad Sci 91: 1406–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auweter SD, Allain FHT 2008. Structure-function relationships of the polypyrimidine tract binding protein. Cell Mol Life Sci 65: 516–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back SH, Kim YK, Kim WJ, Cho S, Oh HR, Kim JE, Jang SK 2002. Translation of polioviral mRNA is inhibited by cleavage of polypyrimidine tract-binding proteins executed by polioviral 3C(pro). J Virol 76: 2529–2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB 2005. SAFA: Semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA 11: 344–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Breyne S, Yu Y, Unbehaun A, Pestova TV, Hellen CU 2009. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc Natl Acad Sci 106: 9197–9202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke GM, Hoffman MA, Palmenberg AC 1992. Sequence and structural elements that contribute to efficient encephalomyocarditis virus RNA translation. J Virol 66: 1602–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghetti A, Pinol-Roma S, Michael WM, Morandi C, Dreyfuss G 1992. hnRNP-I, the polypyrimidine tract-binding protein: Distinct nuclear localization and association with hnRNAs. Nucleic Acids Res 20: 3671–3678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt SL, Jackson RJ 1999. Polypyrimidine-tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA 5: 344–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Kaminski A 1995. Internal initiation of translation in eukaryotes: The picornavirus paradigm and beyond. RNA 1: 985–1000 [PMC free article] [PubMed] [Google Scholar]
- Jang SK, Wimmer E 1990. Cap-independent translation of encephalomyocarditis virus RNA: Structural elements of the internal ribosome entry site and involvement of a cellular 57-kDa RNA-binding protein. Genes Dev 4: 1560–1572 [DOI] [PubMed] [Google Scholar]
- Kafasla P, Morgner N, Pöyry TA, Curry S, Robinson CV, Jackson RJ 2009. Polypyrimidine tract binding protein stabilizes the encephalomyocarditis virus IRES structure via binding multiple sites in a unique orientation. Mol Cell 34: 556–568 [DOI] [PubMed] [Google Scholar]
- Kafasla P, Morgner N, Robinson CV, Jackson RJ 2010. Polypyrimidine tract binding protein stimulates the poliovirus IRES by modulating eIF4G binding. EMBO J 29: 3710–3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski A, Jackson RJ 1998. The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA 4: 626–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski A, Hunt SL, Patton JG, Jackson RJ 1995. Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA 1: 924–938 [PMC free article] [PubMed] [Google Scholar]
- Laederach A, Das R, Vicens Q, Pearlman SM, Brenowitz M, Herschlag D, Altman RB 2008. Semi-automated and rapid quantification of nucleic acid footprinting and structure mapping experiments. Nat Protoc 3: 1395–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monie TP, Hernandez H, Robinson CV, Simpson P, Matthews S, Curry S 2005. The polypyrimidine tract binding protein is a monomer. RNA 11: 1803–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepmann M 1996. Porcine polypyrimidine tract-binding protein stimulates translation initiation at the internal ribosome entry site of foot-and-mouth-disease virus. FEBS Lett 388: 39–42 [DOI] [PubMed] [Google Scholar]
- Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, et al. 2005. Structure of PTB bound to RNA: Specific binding and implications for splicing regulation. Science 309: 2054–2057 [DOI] [PubMed] [Google Scholar]
- Pilipenko EV, Pestova TV, Kolupaeva VG, Khitrina EV, Poperechnaya AN, Agol VI, Hellen CU 2000. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev 14: 2028–2045 [PMC free article] [PubMed] [Google Scholar]
- Sawicka K, Bushell M, Spriggs KA, Willis AE 2008. Polypyrimidine-tract-binding protein: A multifunctional RNA-binding protein. Biochem Soc Trans 36: 641–647 [DOI] [PubMed] [Google Scholar]
- Simpson PJ, Monie TP, Szendroi A, Davydova N, Tyzack JK, Conte MR, Read CM, Cary PD, Svergun DI, Konarev PV, et al. 2004. Structure and RNA interactions of the N-Terminal RRM domains of PTB. Structure 12: 1631–1643 [DOI] [PubMed] [Google Scholar]
- Song Y, Tzima E, Ochs K, Bassili G, Trusheim H, Linder M, Preissner KT, Niepmann M 2005. Evidence for an RNA chaperone function of polypyrimidine tract-binding protein in picornavirus translation. RNA 11: 1809–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman R, Rideau A, Matlin A, Gooding C, Robinson F, McGlincy N, Grellscheid SN, Southby J, Wollerton M, Smith CW 2005. Regulation of alternative splicing by PTB and associated factors. Biochem Soc Trans 33: 457–460 [DOI] [PubMed] [Google Scholar]
- Wollerton MC, Gooding C, Robinson F, Brown EC, Jackson RJ, Smith CW 2001. Differential alternative splicing activity of isoforms of polypyrimidine tract binding protein (PTB). RNA 7: 819–832 [DOI] [PMC free article] [PubMed] [Google Scholar]