Abstract
Influenza A is a negative sense RNA virus of significant public health concern. While much is understood about the life cycle of the virus, knowledge of RNA secondary structure in influenza A virus is sparse. Predictions of RNA secondary structure can focus experimental efforts. The present study analyzes coding regions of the eight viral genome segments in both the (+) and (−) sense RNA for conserved secondary structure. The predictions are based on identifying regions of unusual thermodynamic stabilities and are correlated with studies of suppression of synonymous codon usage (SSCU). The results indicate that secondary structure is favored in the (+) sense influenza RNA. Twenty regions with putative conserved RNA structure have been identified, including two previously described structured regions. Of these predictions, eight have high thermodynamic stability and SSCU, with five of these corresponding to current annotations (e.g., splice sites), while the remaining 12 are predicted by the thermodynamics alone. Secondary structures with high conservation of base-pairing are proposed within the five regions having known function. A combination of thermodynamics, amino acid and nucleotide sequence comparisons along with SSCU was essential for revealing potential secondary structures.
Keywords: influenza, RNA, secondary structure, structure prediction, codon suppression
INTRODUCTION
Influenza A virus is a significant public health threat causing more than an estimated 200,000 severe infections and 41,400 deaths each year in the United States (Dushoff et al. 2006). The influenza A virus is a negative (−) sense RNA virus composed of eight discrete genomic segments. Each segment is packaged as a ribonucleoprotein (RNP) complex that contains multiple structural NP proteins and the heterotrimeric polymerase consisting of the PB1, PB2, and PA protein subunits (Compans 1972; Noda et al. 2006; Ye et al. 2006). Each genome segment serves as a template for the synthesis of two distinct positive (+) sense RNA molecules within the nucleus of infected cells. The (+)RNAs serve protein coding (mRNA) and genomic replication (cRNA) functions and are generated by distinct mechanisms in vivo (Bouloy et al. 1978; Plotch et al. 1981; Shapiro and Krug 1988).
In general, RNA secondary structure is important for viral viability. For example, internal ribosome entry sites (IRES), which allow mRNA to internally initiate translation and bypass canonical ribosomal scanning, are heavily structured and found in many viruses (Kieft 2008). Other examples are the Hepatitis Delta Virus (HDV) ribozyme that is used for maturation of the viral RNA (Kuo et al. 1988), the tRNA-like structures found in the 3′ untranslated region (UTR) of many plant viruses (Weiner and Maizels 1987; Dreher 2009), the frameshifting signals that allow some viruses to encode overlapping open reading frames (ORFs) (Jacks et al. 1988; Dam et al. 1990), viral packaging signals (Clever et al. 1995), and many more. Recently, widespread secondary structure of an HIV-1 genome was deduced using free energy minimization coupled with chemical probing (Watts et al. 2009).
The de novo discovery of structured regions in long RNA strands, such as viral RNAs, has been approached with a variety of techniques (Washietl et al. 2005a; Schroeder 2009; Mathews et al. 2010). One method is based on searching for regions predicted to be unusually stable thermodynamically (Washietl et al. 2005b; Uzilov et al. 2006). When the search occurs in coding regions, it is possible to also consider the effect of RNA structure on codon evolution (Pedersen et al. 2004). In particular, RNA structural constraints lead to suppression of variation in the third (wobble) position of amino acid codons. Suppression of synonymous codon usage (SSCU) has been used to identify structured RNA elements in viral genomic RNAs (Simmonds and Smith 1999; Tuplin et al. 2004). Here, we use a combination of thermodynamics, SSCU, amino acid and RNA sequence comparison to reveal potential secondary structures in influenza.
The influenza virus is an interesting target for structural analysis because it uses RNA exclusively throughout its life cycle; no DNA intermediate is involved. Each (−)RNA carries conserved 5′ and 3′ sequences that can base-pair to circularize the molecule (Hsu et al. 1987). This pairing provides a binding site for the heterotrimeric polymerase that carries out the synthesis of both mRNA and cRNA molecules (Hagen et al. 1994). Additionally, two studies have described structures in the segment 8 (+)RNA that encode the nonstructural (NS1) and nuclear export protein (NEP, formerly NS2) (Gultyaev et al. 2007; Ilyinskii et al. 2009). Segment 8 (+)RNA has been the most extensively studied with regard to secondary structure. RNA secondary and tertiary structures have also been proposed to be important in the splicing of segment 8 mRNA (Plotch and Krug 1986; Nemeroff et al. 1992) and in viral packaging of (−)RNA (Muramoto et al. 2006; Marsh et al. 2007, 2008; Hutchinson et al. 2008; Liang et al. 2008). Outside of segment 8, very little is known about RNA secondary structure.
Improved knowledge of secondary structure in influenza may shed light on important aspects of influenza biology and lead to new therapeutic targets. RNA structural motifs may be targeted with oligonucleotides (Childs et al. 2002, 2003; Disney et al. 2004) or small molecules (Mei et al. 1998; Sucheck and Wong 2000; Wilson and Li 2000; Gallego and Varani 2001; Childs-Disney et al. 2007; Disney et al. 2008; Lee et al. 2009; Pushechnikov et al. 2009) to disrupt viral function.
MATERIALS AND METHODS
Sequences
Sequence data were obtained from the National Center for Biotechnology Information (NCBI) Influenza Virus Resource page (Bao et al. 2008). For prediction of conserved structural RNA, six full influenza A genome sets were used from human, avian, and swine strains, H5N1 and H1N1 (Taxonomy IDs: 755298 [Human H5N1], 279728 [Avian H5N1], 287864 [Swine H5N1], 865618 [Human H1N1], 768723 [Avian H1N1], 762299 [Swine H1N1]). For analysis of synonymous codon suppression, sets of sequences were obtained for each separate segment by downloading all nonredundant influenza A sequences.
Prediction of conserved structural regions
The six influenza A genome sets were divided by segment. Coding regions were translated in silico with BioEdit (Hall 2001), and protein sequences were aligned with ClustalW (Larkin et al. 2007) using the default protein parameters. The aligned sequences were converted back into nucleotides, now aligned based on the protein sequence, and submitted to RNAz 2.0 (Gruber et al. 2010) for prediction of potentially conserved structures. It was important to use protein-based alignments, as the quality was much improved over nucleotide alignments. The amino acid alignments also allowed analysis of how RNA structure may influence codon evolution.
RNAz predictions were run in both strand orientations (+/−) using a 120-nt window size, 10-nt step size, and with the program's default filtering parameters. RNAz uses a support vector machine (SVM) to make predictions based on the following five criteria: minimum predicted free energy (MFE) from single sequence structure calculations, Z-score, structure conservation index (SCI), average pairwise sequence identity (APSI), and number of sequences in the alignment. The Z-score measures the “excess” predicted minimum free energy of folding for a native sequence versus random sequence. A dinucleotide shuffling model for calculating Z-scores was used that reduces background over the default mononucleotide model setting. The SCI is the consensus structure free energy divided by the average of the individual sequence free energies in the alignment and is a measure of how well represented the consensus structure is in individual sequence folds. Based on the above criteria, RNAz assigns a classification value, herein referred to as p-class, to indicate the probability that a given region contains structure. For this study, RNAz predictions with a p-class of >0.5 are considered structured. RNAz was trained on representative sequences including rRNAs, spliceosomal RNAs, tRNAs, miRNAs, small nucleolar RNAs, nuclear RNase P, and SRP RNA (Gruber et al. 2010).
Structural predictions in both the (+)RNA and (−)RNA strand orientation can arise for the same window because base-pairing is also possible in the reverse complement sequence of the strand that contains the true or functionally conserved structure (Reiche and Stadler 2007). To check predictions for these structural “echoes,” fragments corresponding to overlapping RNAz hits were concatenated and submitted to the program RNAstrand (Reiche and Stadler 2007), which uses an SVM to predict strand bias. RNAstrand uses four criteria for identifying asymmetries between structure in the (+)RNA and (−)RNA strand orientations: average folding free energy of individual sequences in the alignment, folding free energy of the consensus secondary structure as calculated by RNAalifold (Bernhart et al. 2008), mean free energy Z-score of the individual sequences in the alignment, and SCI. The resulting “P-value” ranges from 0 to 1, where 1 implies high likelihood for structure in the given strand orientation. The RNAstrand SVM was trained on a similar set of structures as RNAz (Reiche and Stadler 2007).
Analysis of suppression of synonymous codon usage
All nonredundant sequences for each segment were aligned using MAFFT (Katoh et al. 2002, 2005) with the FFT-NS-1 strategy optimized for very large alignments. A Perl script was written to randomly select sequences from the alignment while simultaneously restricting the APSI to <95% to avoid selecting very similar sequences. For segments 4 and 6, the two segments encoding the antigenic proteins, 300 sequences were selected, while 100 sequences were used for all other segments. In each case, the larger sequence sets were appended to the alignments used in the RNAz analysis. Gaps were removed from the sequences, which were then translated in silico and submitted to ClustalW as above.
The resulting amino acid alignments were converted back into RNA sequences (now aligned with respect to encoded amino acids) and submitted to the Simmonics package (Simmonds and Smith 1999) for analysis of the suppression of synonymous codon usage (SSCU). Sequence scans were run analyzing the synonymous sites using mean pairwise distance measurements. The SSCU was calculated for windows of 15 nt, with a 3-nt step size. In segments 7 and 8, the calculation of SSCU was done separately for each alternatively spliced product. This analysis was repeated with resampled alignments to check for sampling bias, and results were not significantly different.
Secondary structure modeling
Preliminary structural models were built when RNAz or strong SSCU predictions correlated with functional annotations. Initial models were built with RNAalifold (Bernhart et al. 2008), the same algorithm used to calculate the free energies in RNAz and RNAstrand. RNAalifold was run with the default program parameters. The alignments submitted to RNAalifold contained the six genome sequences used for the RNAz analysis or the alignments used for the SSCU analysis. The RNAalifold secondary structure predictions were compared to predictions from Dynalign (Mathews and Turner 2002; Mathews 2004; Harmanci et al. 2007), which simultaneously optimizes sequence alignment and consensus structure for two sequences. The Dynalign calculation input consisted of the two most distant sequences (by APSI) in the alignment. Sequence comparison for each secondary structure model was carried out with all available unique sequences. Free energies for nonpseudoknot structures were predicted using nearest-neighbor free energy parameters (Xia et al. 1998; Mathews et al. 2004).
To scan for potential pseudoknots, which are forbidden in the RNAz and Dynalign prediction algorithms, the program DotKnot was used (Sperschneider and Datta 2010). DotKnot is a heuristic algorithm that searches for stems in long RNAs from a secondary structure prediction dotplot and then assembles candidate pseudoknots (Sperschneider and Datta 2010). When RNAs were >1000 nt in length, the maximum length allowed in DotKnot, they were cut into overlapping windows (∼100 nt overlap) and submitted to the program. Free energies for pseudoknots were predicted using published parameters optimized either by comparison of predictions to known structures (Dirks and Pierce 2003) or to results from a diamond lattice model (Cao and Chen 2006, 2009).
In vitro folding of RNA
Representative cluster a (5′-GGGUGAUGCCCCAUUCCUUGAUCGGCUUCGCCGAGA UCAGAAGUCCCUAAGAGGAAGAGGCAGCACUC-3′) and cluster b (5′-GGGUGAUGCUC CCUUUGAUGACAGACUCAGAAGAGAUCAAAAGGCAUUAAAGGGAAGAGGCAGCACUC-3′) sequences for segment 8 region 81–148 were synthesized from deoxyoligonucleotide templates with an Ambion MEGAscript T7 transcription kit. Purified RNA was 5′-end-labeled with [γ-32P]ATP and purified with an Ambion NucAway spin column. Labeled RNA was renatured in 10 mM Tris-HCl (pH 7.0) and 100 mM KCl by heating for 2 min to 90°C and then slow-cooling to 37°C. MgCl2 was added to a final concentration of 5, 10, or 15 mM, and the RNA was incubated for 20 min at 37°C. Folded RNAs were fractionated on a nondenaturing 8% polyacrylamide gel. The dried gel was exposed to a phosphorscreen, and bands were detected with a Bio-Rad Personal Molecular Imager.
RESULTS
Evidence for conserved RNA secondary structure in influenza virus genome segments
Segment 8 (NS1/NEP)
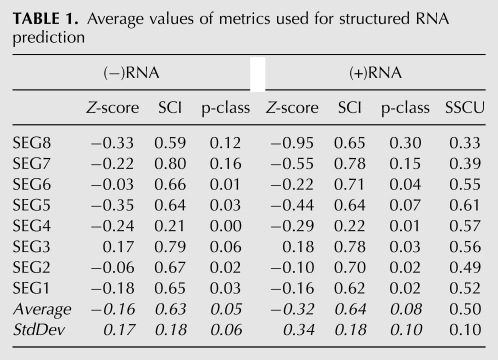
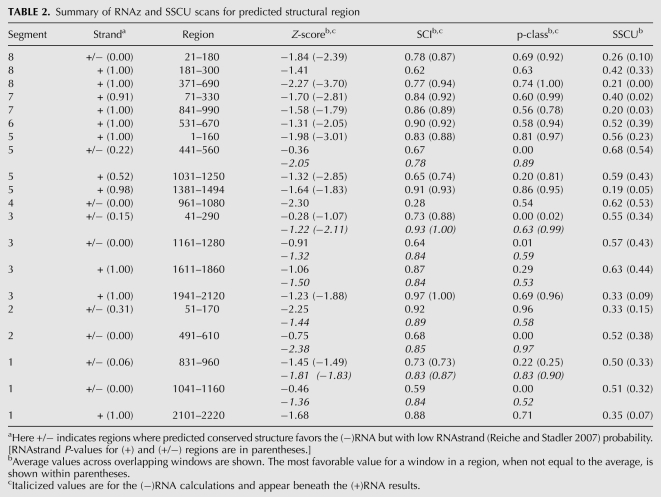
Segment 8 is the smallest influenza RNA and one of the two (+)RNA segments that undergoes splicing. The alignment of six sequences used in the RNAz analysis has a length of 838 nt and APSI of 86.4%. Of all the influenza segments, segment 8 has the most widespread distribution of RNAz predictions of conserved secondary structure, which is consistent with its having the most negative average Z-score across the alignment [−0.95 in the (+)RNA sense] (Table 1). It also has the highest average RNAz p-class of any segment [0.30 in the (+)RNA sense]. Thus, segment 8 has the strongest bias toward predicted structure in the (+)RNA (Tables 1, 2).
TABLE 1.
Average values of metrics used for structured RNA prediction
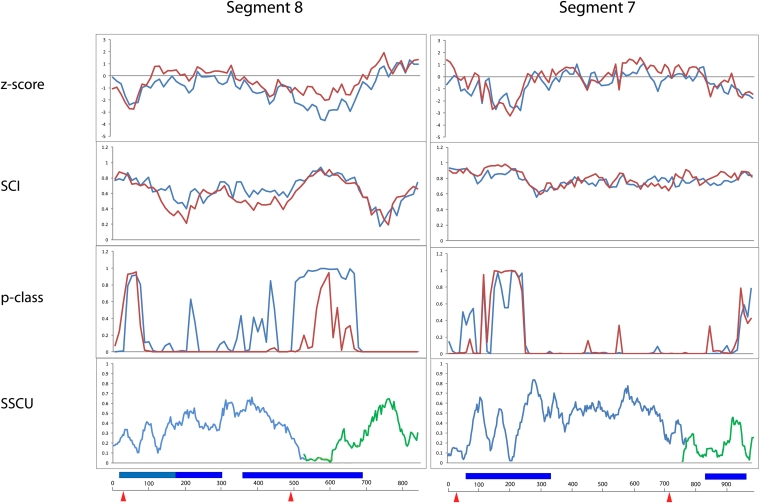
Three regions are predicted to contain conserved secondary structure in segment 8 (+)RNA (Fig. 1; Table 2). Region 371–690 has the most favorable single window Z-score, p-class, and SSCU of any predicted region in any segment (−3.70, 1.00, and 0.00, respectively). This region also contains the 3′ splice site at position 487. After 487, the codon use in the NS1 ORF is severely suppressed because of overlap with the frameshifted NS2 ORF. Codon suppression in the NS2 ORF starts to abate toward the end of the structural region at 371–690 (Fig. 1).
FIGURE 1.
Results of RNAz calculations and suppression of synonymous codon usage (SSCU) studies for coding regions of segments 8 and 7. (Red lines) (−)RNA; (blue lines) (+)RNA. The first plot gives the calculated Z-score, which is a measure of the “excess” free energy of folding a native RNA sequence vs. random sequence. The second plot gives the structure conservation index (SCI), which indicates how well represented the consensus structure is in predictions of individual sequence secondary structures (y-axis indicates the fraction of conservation). The third plot shows the RNAz probability (p-class) of the presence of conserved structural RNA. The bottom panel shows the results for the SSCU calculations, which measure the variation at the third codon position (y-axis gives the distance at the third codon position). Here, low distance/variation indicates strong SSCU. Results for the larger ORF (blue) and the smaller (green). (Below the bottom panel) The common x-axis indicates the input alignment position in nucleotides. (Dark blue bars) Overlapping RNAz predictions clearly in the (+) sense; (light blue bars) RNAz predictions with ambiguous strand bias; (red arrows) the splice sites.
TABLE 2.
Summary of RNAz and SSCU scans for predicted structural region
Regions 21–180 and 181–300 of segment 8 have moderate values for Z-score, SCI, p-class, and SSCU (Table 2). Region 21–180, however, has individual windows with values that strongly predict structure. The fifth most favorable local Z-score occurred in 21–180. Strong SSCU occurs near positions 60, 150, and 550. The 5′ splice site of segment 8 occurs at position 30.
Segment 7 (M1/M2)
Segment 7 is the second smallest viral segment and is also spliced to produce two protein products. The alignment used in the RNAz analysis has a length of 982 nt and an APSI of 92.5%. As shown for (+)RNA in Table 1, segment 7 has the second most favorable average Z-score (−0.55) and the second strongest average SSCU and RNAz p-class (0.39 and 0.15), indicating a high overall probability of structure in the (+)RNA (Table 1).
RNAz predicts a high probability of structure in two regions of segment 7 (Fig. 1; Table 2). Regions 71–330 and 841–990 have a high RNAstrand probability for structure in the (+)RNA. Region 841–990 is notable for having, respectively, the second and third strongest average and single window SSCU with respect to all other segments. The SSCU that overlaps with this region continues upstream and also overlaps with the nearby 3′ splice site at position 715 (Fig. 1).
Region 71–330 has moderate values for Z-score, SCI, p-class, and SSCU, but single window values are very strong (Table 2). This region has the second strongest single window p-class and SSCU value of any region. This predicted structural region occurs just downstream from the 5′ RNA splice site at position 26, which falls within the region of suppressed synonymous codon usage (Fig. 1).
Segment 6 (NA)
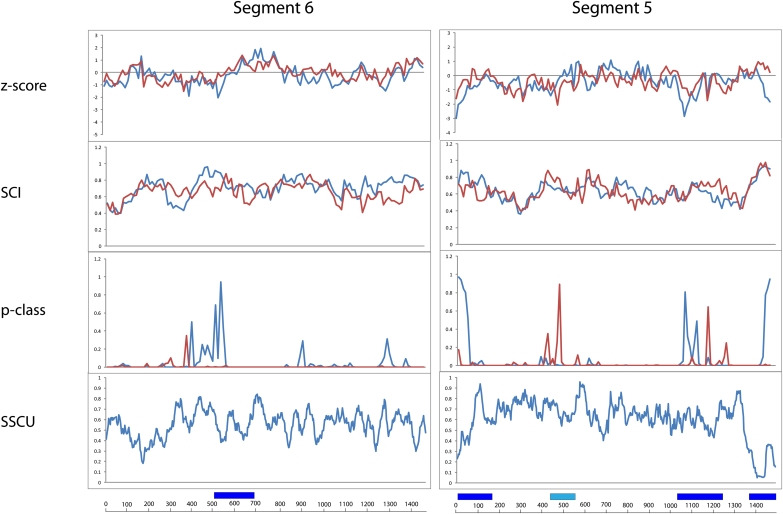
The alignment for segment 6 is 1460 nt in length and has an APSI of 86.7%. As shown in Table 1, segment 6 has the third highest average SCI for the (+)RNA, but the Z-score of −0.22 averaged over the entire coding region predicts a roughly average amount of structure for segment 6 when compared to the other segments.
RNAz predicts a moderate probability of structure in region 531–670, but the single window 551–670 has favorable values for Z-score, SCI, and p-class (Fig. 2; Table 2). The RNAz predictions are for structure in the (+)RNA, consistent with RNAstrand predictions (Fig. 2; Table 2). Synonymous codon usage appears to be highly suppressed only at positions 100–250, which does not overlap with the RNAz predicted conserved structural regions (Fig. 2).
FIGURE 2.
Results of RNAz calculations and suppression of synonymous codon usage (SSCU) studies for segments 6 and 5. Figure annotations are as in Figure 1.
Segment 5 (NP)
The alignment for segment 5 is 1494 nt in length and has an APSI of 88.1%. Average Z-score across the alignment is −0.35 and −0.44 for the (−)RNAs and (+)RNAs, respectively, suggesting an above average amount of structure in segment 5 compared to the other segments (Fig. 2; Table 1).
In segment 5, RNAz predicts a high probability of structure solely in (+)RNA at regions 1–160, 1031–1250, and 1381–1494, and of ambiguous RNAstrand predicted strand bias at position 441–560 (Fig. 2; Table 2). Region 1–160 has the fourth most favorable average Z-score of any (+)RNA region (Table 2).
Toward the ends of segment 5, there are two regions with strong SSCU that match up with the two high-probability RNAz predicted regions of structure in the (+)RNA. The region of SSCU that overlaps with region 1381–1494 is, on average, the strongest for any region (Table 2).
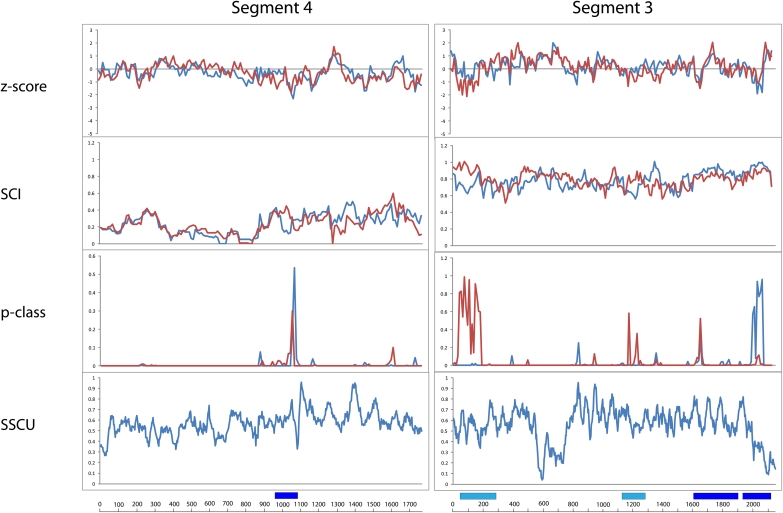
Segment 4 (HA)
The alignment for segment 4 is 1770 nt in length and has an APSI of 73.4%. On average, segment 4 has the lowest SCI of any of the influenza segments in both strands (0.21 and 0.22) (Fig. 3; Table 1). Average Z-scores of −0.24 and −0.29 in the (−)RNA and (+)RNA, respectively, suggest a slightly above average amount of structure in segment 4 compared to the other segments (Table 1).
FIGURE 3.
Results of RNAz calculations and suppression of synonymous codon usage (SSCU) studies for segments 4 and 3. Figure annotations are as in Figure 1.
In segment 4, RNAz predicts a single conserved structural region at 961–1080 in the (+)RNA (Fig. 3), but RNAstrand favors structure in the (−)RNA with very low confidence (Table 2).
There is below average SSCU corresponding to the RNAz prediction at 961–1080 (Fig. 3). There is also moderate SSCU toward the 5′ end of the RNA, which does not correspond to any RNAz predictions (Fig. 3).
Segment 3 (PA)
The alignment for segment 3 is 2151 nt in length and has an APSI of 91.9%. The positive Z-scores of 0.17 and 0.18 for the (−)RNA and (+)RNA, respectively, suggest a lack of overall conserved structure in both strands (Table 1).
In segment 3, however, RNAz predicts four regions with potentially conserved structure (Fig. 3). Two regions are predicted by RNAstrand to have bias for structure in the (+)RNA, 1611–1860, and 1941–2120, while regions 41–290 and 1161–1280 are predicted to have ambiguous strand bias (Table 2).
Region 1941–2120 has the highest average SCI of any (+)RNA region, a high average and single window p-class, and strong SSCU (Table 2). Another region, ∼500–800, has strong SSCU but does not occur near any RNAz predictions of structure or any known features of the viral RNA.
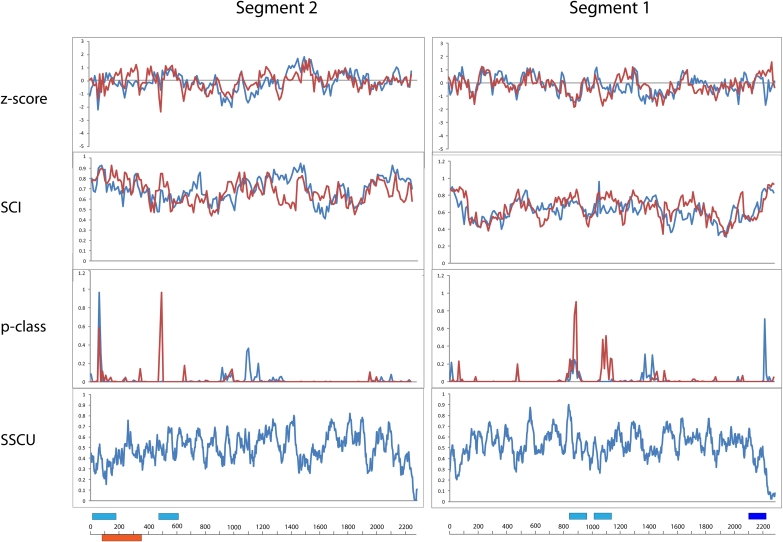
Segment 2 (PB1/PB1-F2)
The alignment for segment 2 is 2151 nt in length and has an APSI of 89.3%. The average Z-score for segment 2 in the (+)RNA is −0.10 (Table 1), suggesting a below average amount of conserved structure compared to the other segments. Nevertheless, RNAz predicts structure in two regions, 51–170 and 491–610: RNAstrand predictions are ambiguous in both regions (Fig. 4; Table 2).
FIGURE 4.
Results of RNAz calculations and suppression of synonymous codon usage (SSCU) studies for segments 2 and 1. Figure annotations are as in Figure 1; (orange bar) the internal ORF for the PB1-F2 product.
Region 51–170 has strong RNAz scores in the (+)RNA, while region 491–610 has strong RNAz scores in the (−)RNA. Segment 2 has two regions with strong SSCU (Fig. 4). One region spans positions 50–150, which corresponds to both the RNAz-predicted region of structure at 51–170 and the start of the internal ORF for PB1-F2 at position 91 (Fig. 4). The second region with SSCU is at the 3′ end of the RNA and does not overlap with known or predicted features of the RNA.
Segment 1 (PB2)
The alignment for segment 1 is 2280 nt in length and has an APSI of 88.8%. The average Z-scores of −0.18 and −0.16 for the (−)RNA and (+)RNA, respectively, suggest the presence of an average amount of conserved structure in segment 1 compared to the other segments (Table 1).
In segment 1, RNAz predicts three regions with potentially conserved secondary structure: 831–960, 1041–1160, and 2101–2220 (Fig. 4). Only region 2101–2220 has unambiguous strand bias, as predicted by RNAstrand (Table 2). This region is also the only one to overlap with an area of strong SSCU (Fig. 4; Table 2). Toward the 5′ end of the RNA is another region that shows SSCU (Fig. 4).
Comparisons to previous predictions of RNA structures
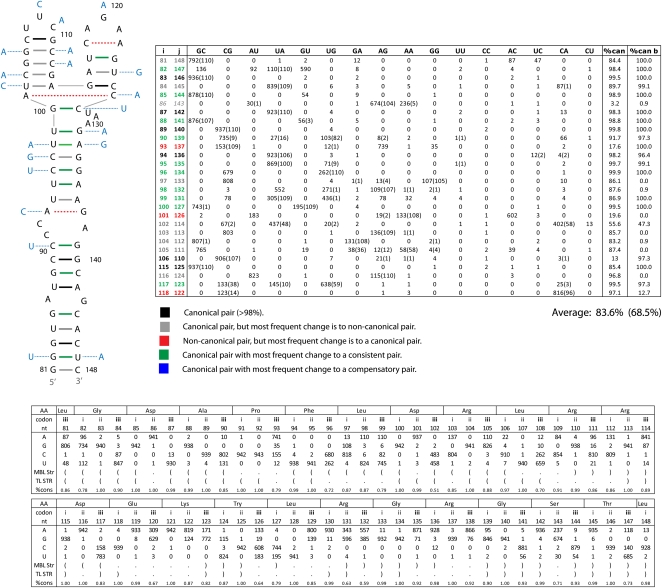
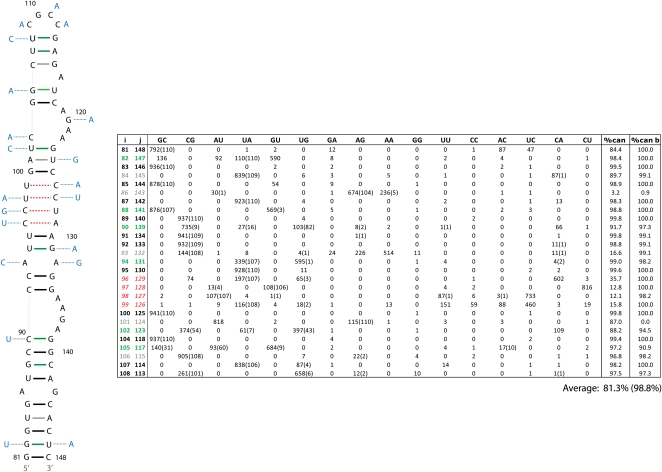
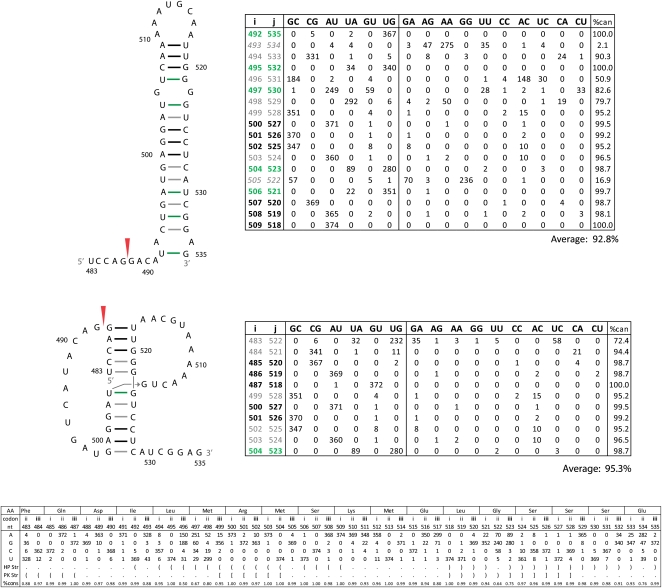
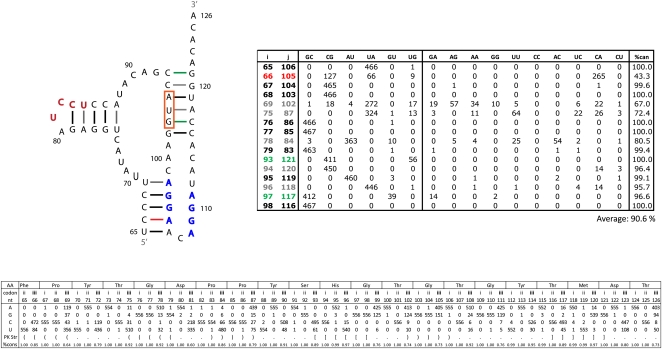
Two of the RNAz-predicted regions of conserved structure in segment 8 contain previously predicted secondary structures (Gultyaev et al. 2007; Ilyinskii et al. 2009). Region 21–180 contains a fragment that has been proposed to fold into a structure that influences NS protein expression (Ilyinskii et al. 2009). Using the RNAz alignment, RNAalifold predicts a consensus structure for this region similar to the published one (Fig. 5). Using the SSCU alignment for RNAalifold, however, results in a different predicted secondary structure (Fig. 6). The two models are identical at the basal stem, but the nucleotides between positions 90 and 139 are folded differently (Figs. 5, 6). In the alternative model of Figure 6, the multi-branch loop from nucleotides 100–130 (Fig. 5) is folded into a tetraloop hairpin. The tetraloop model has less mutational support, a slightly lower (81.3% vs. 83.6%) conservation of canonical base-pairing, and much less favorable predicted free energy (−9.6 vs. −19.7 kcal/mol at 37°C) than the multi-branch loop in Figure 5. A similar tetraloop structure has been proposed for clade B influenza strains (Gultyaev et al. 2010).
FIGURE 5.
RNAalifold predicted secondary structure from the RNAz alignment for fragment of 5′ predicted secondary structure region from segment 8 (+)RNA. This structure was also predicted by Ilyinskii et al. (2009). Base pairs are color annotated with information from base pair counts (tabulated to the right of the structure) from an alignment of all available unique sequences. The color annotation key is given below the table. Pairing type is given at the top of the table; canonical pairs to the left, and noncanonical to the right. The “%can” column gives the percentage of canonical pairs found in those aligned positions. Italicized alignment positions (i–j) are for symmetric internal loop bases. The average percent conservation of the whole structure is given below the table. Base pair counts for all unique sequences and cluster b sequences are given without and with parenthesis, respectively. Cluster b consensus sequence is indicated by light blue nucleotides. The predicted free energies of folding, ΔG37° (Mathews et al. 2004), for the consensus sequence of all unique sequences is −19.7 kcal/mol and for cluster b sequences is −8.6 kcal/mol. Nucleotide composition by alignment position is summarized at the bottom of the figure. The structure is notated in bracket notion, codon position is indicated by roman numerals, and consensus amino acid sequence is notated at the top of the table. The percent conservation for each position is also given.
FIGURE 6.
Alternative secondary structure for the region shown in Figure 5. This was predicted by RNAalifold using the SSCU alignment of segment 8 sequences. Figure annotations and base pair counts are as described in Figure 5. Base pair counts for all unique sequences and cluster b sequences are given without and with parentheses, respectively. Cluster b consensus sequence is indicated by light blue nucleotides. The predicted free energies of folding, ΔG37° (Mathews et al. 2004) for the consensus sequence of all unique sequences is −9.6 kcal/mol and for cluster b sequences is −23.8 kcal/mol.
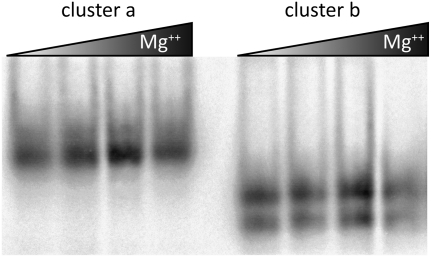
To test whether a subset of sequences prefers the tetraloop structure, sequences were clustered using the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) (Sokal and Michener 1958). All unique sequences clustered into two broad groups consistent with the previously described phylogeny for segment 8 (Kawaoka et al. 1998; Basler et al. 2001). As shown in the table in Figure 6, a cluster of 110 sequences (cluster b) increases the average canonical pairing of the tetraloop structure to 98.8%, much higher than the 68.5% for cluster b in the multi-branch loop structure (see table in Fig. 5). Specifically, cluster b has canonical base pairs at positions 93–132, 96–129, 97–128, 98–127, and 99–126. For cluster b, the predicted free energies for the multi-branch loop versus the tetraloop structure are −8.6 and −23.8 kcal/mol at 37°C, respectively. The results provide strong support for the tetraloop structure in cluster b sequences. The limited understanding of all factors affecting in vivo folding of RNA, however, leaves both the multi-branch and tetraloop structures as viable alternatives for all influenza sequences. To test whether cluster a and b sequences adopt different conformations, in vitro–synthesized RNA was folded under different Mg++ concentrations at 37°C. As shown in Figure 7, cluster b sequences migrate distinctly from cluster a and appear to adopt two different folds.
FIGURE 7.
Nondenaturing 8% polyacrylamide gel of in vitro folded cluster a and cluster b sequences from Figures 5 and 6 (see Materials and Methods). Final Mg++ concentrations are 0, 5, 10, and 15 mM. Two bands are apparent in the clade b samples: slower and faster migrating products that account for 57% and 43% of the integrated band intensity, respectively.
The other previously described structure in segment 8 occurs in the 440–690 region, which encompasses the 3′ splice site. The RNAalifold predicted consensus structure for this region contains a hairpin identical to previous reports (Fig. 8, top; Gultyaev et al. 2007). The predicted secondary structure of this fragment did not change with the SSCU alignment and is also predicted by Dynalign. The overall conservation of canonical base-pairing in this hairpin is 92.8%, and the predicted free energy at 37°C is −18.9 kcal/mol. An alternative pseudoknot conformation has been proposed for this region with average canonical pairing of 95.3% (Fig. 8, bottom; Gultyaev et al. 2007). The predicted free energy for the pseudoknot is −9 kcal/mol at 37°C, using parameters from Dirks and Pierce (2003) or Cao and Chen (2009). Current knowledge, however, does not allow inclusion of tertiary interactions in these predictions. Such interaction can be very favorable (Theimer et al. 1998; Liu et al. 2009). Gultyaev et al. (2007) provided evidence for equilibrium between hairpin and pseudoknot structures in some strains of influenza.
FIGURE 8.
Secondary structure models for fragment of 3′ predicted secondary structure region from segment 8 (+)RNA. The top structure is for the hairpin predicted by RNAalifold on the SSCU alignment, while the alternative pseudoknot conformation is shown below. These structures were also predicted by Gultyaev et al. (2007). Figure annotations and base pair counts are as described in Figure 5. The predicted free energy of folding, ΔG37° (Mathews et al. 2004), for the consensus hairpin is −18.9 kcal/mol. Predicted free energies for the pseudoknot were calculated as −9 kcal/mol (Dirks and Pierce 2003; Cao and Chen 2009).
Structure prediction in regions of known function
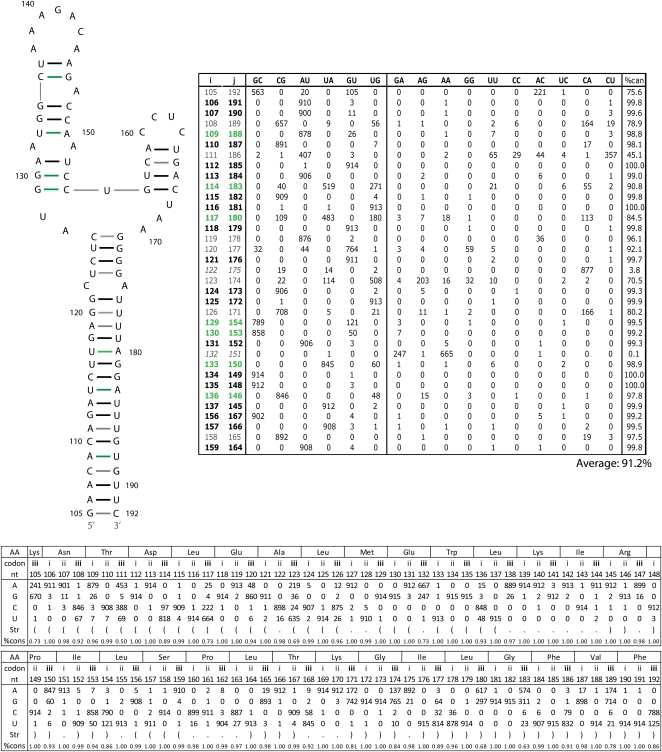
RNAalifold calculations of the 5′ region of segment 7 using the six sequence RNAz alignment and the SSCU alignment generate a structure with a stem topped by a multi-branch loop and having 91.2% overall base-pair conservation (Fig. 9). Dynalign calculations of the two most distant sequences predict the same structure with a predicted free energy of −30.0 kcal/mol at 37°C (Fig. 9). The size of this structure is 88 nt compared to the 68-nt structure in segment 8 (Fig. 5). The segment 7 and 8 putative structures are, respectively, 79 and 51 nt downstream from 5′ splice sites.
FIGURE 9.
RNAalifold predicted secondary structure for fragment of 5′ predicted secondary structure region from the RNAz and the SSCU alignments of segment 7 (+)RNA. Figure annotations and base pair counts are as described in Figure 5. The predicted free energy of folding, ΔG37° (Mathews et al. 2004), for the consensus sequence is −30.0 kcal/mol.
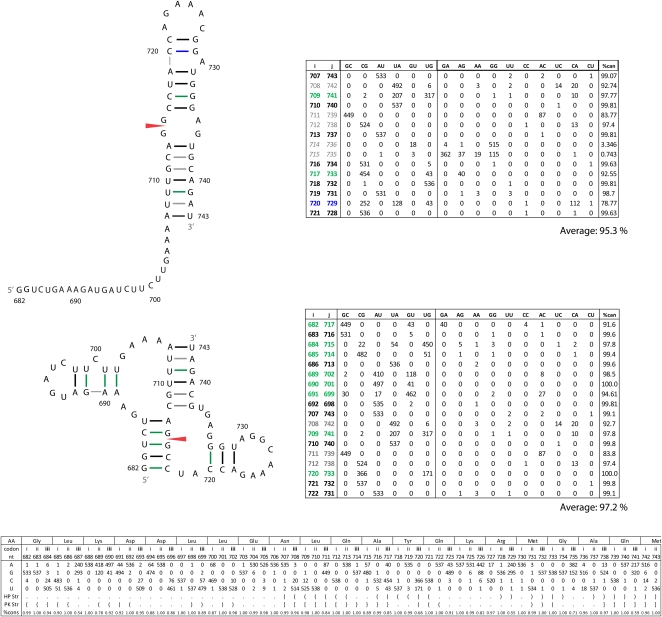
To assess whether the region surrounding the 3′ splice site of segment 7 could form a pseudoknot structure similar to the one proposed by Gultyaev et al. (2007) for segment 8 (Fig. 8), the program DotKnot (Sperschneider and Datta 2010) was used to scan the entire segment 7 (+)RNA coding sequence from genome set 755298. A potential pseudoknot structure was identified that incorporates the 3′ splice site (Fig. 10, bottom). Additional base pairs were manually added in the loop regions between nucleotides 690 and 701. As an alternative folding, RNAalifold calculations on alignments from this region generated a consensus hairpin where two additional conserved base pairs could be manually added at positions 720–729 and 721–728 (Fig. 10, top). Dynalign (Mathews and Turner 2002) calculations of the two most distant sequences also predict the hairpin with a predicted free energy of −14.3 kcal/mol at 37°C. The pseudoknot has slightly higher pair conservation than the hairpin (97.2% vs. 95.3%, respectively) and has more consistent mutations (Fig. 10). Predicted free energies for the pseudoknot range from −7 to −4 kcal/mol at 37°C, using parameters from Dirks and Pierce (2003) and Cao and Chen (2006), respectively. If the bulged C700 is slipped to pair with G691 and an A692–U699 pair forms, then the predicted free energies range from −12 to −9 kcal/mol at 37°C using parameters from Dirks and Pierce (2003) and Cao and Chen (2006), respectively. This pairing scheme, however, reduces the pair conservation to 79.4% and 89.2%, respectively, for these two base pairs. Co-transcriptional folding would be expected to generate preferentially the pseudoknot.
FIGURE 10.
Secondary structures predicted for a fragment of the 3′ region of segment 7 (+)RNA. The top structure is for the hairpin predicted by RNAalifold on the SSCU alignment, while the alternative pseudoknot conformation is shown below. Figure annotations and base pair counts are as described in Figure 5. The predicted free energy of folding, ΔG37° (Mathews et al. 2004), for the consensus hairpin is −14.3 kcal/mol. Predicted free energies for the pseudoknot were calculated as −7 (DP) or −4 (CC) kcal/mol, depending on the parameters used (Dirks and Pierce 2003; Cao and Chen 2006). A slipped helix with G691–C700 and A692–U699 pairs results in a more favorable predicted free energy range of −12 (DP) to −9 (CC), but less favorable percentage of canonical pairing of 79.4% and 89.2%, respectively, for these base pairs.
DotKnot (Sperschneider and Datta 2010) was also used to scan the whole segment 2 (+)RNA sequence from genome set 755298. Three potential pseudoknot folds are predicted for the region encompassing positions 65–121. All three models incorporate the start codon of the PB1-F2 ORF into a helix with base-pair conservation of 98.0% (Fig. 11, nucleotides 93–98 and 116–121). The other helix of the pseudoknot is formed by bases upstream of and downstream from the PB1-F2 start codon. The most conserved helix predicted is shown in Figure 11. The other possibilities are represented by the blue-shaded nucleotides, which can base-pair with the red-shaded nucleotides to form two alternative helices. Predicted free energies for the most conserved pseudoknot range from −14 kcal/mol (Dirks and Pierce 2003) to −8 kcal/mol (Cao and Chen 2009) at 37°C. Interestingly, the nucleotides at positions 66 and 105 always form canonical or CA pairs (Fig. 11).
FIGURE 11.
DotKnot (Sperschneider and Datta 2010) secondary structure model for 5′ region 65–126 from segment 2 of genome set 755298. Other predictions by DotKnot for this region are represented by blue shaded nucleotides, which can base-pair with the red shaded nucleotides to form two alternate 5′ helices leaving nucleotides 65–80 unpaired. Base pair counts from an alignment of all unique segment 2 sequences are shown to the right. The nucleotides boxed in orange in the 3′ pseudoknot helix are the start codon for the internal ORF for the PB1-F2 product. This ORF is shifted +1 compared to the PB1 coding region. Figure annotations and base pair counts are as described in Figure 5. Predicted free energies for the pseudoknot were calculated as −14 (DP) or −8 (CC) kcal/mol, depending on the parameters used (Dirks and Pierce 2003; Cao and Chen 2009).
To determine if structurally distinct subsets of sequences exist for Figures 8–11, an UPGMA analysis was conducted as described for segment 8. No obvious pattern for these structures was detected between the different sequence clusters.
Base-pair and codon conservation in secondary structure models
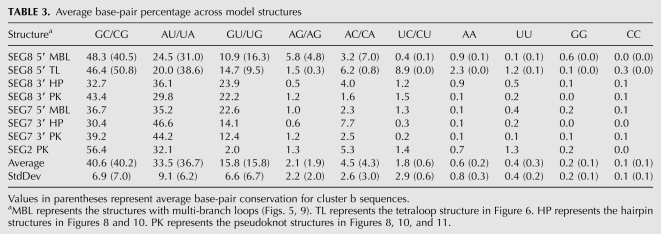
Table 3 summarizes the average base-pair composition of the structures in Figures 5–6 and 8–11. When noncanonical pairing is observed, it is predominantly AC followed by AG. Occurrence of AA, UU, GG, and CC is almost always <1%. The one apparent exception is the 5′ tetraloop model for segment 8 (Fig. 6), but virtually all of the noncanonical pairs are confined to the cluster a sequences, which better support the multi-branch loop structure rather than the tetraloop structure. If cluster a sequences are eliminated from the tetraloop percentages, then the pattern for noncanonical pairing follows the average pattern.
TABLE 3.
Average base-pair percentage across model structures
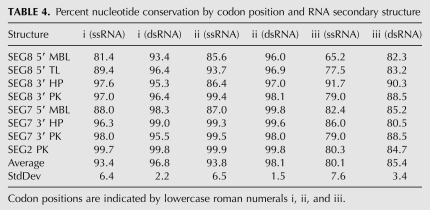
In Table 4, nucleotide conservation is analyzed in the context of codon position between predicted double-stranded and single-stranded regions. On average, the predicted double-stranded regions are more constrained at all three codon positions. This is statistically significant for the second and third codon positions (statistical P-values of 0.05). The only cases in which predicted single-stranded regions are significantly more constrained than predicted double-stranded regions are for the 3′ HP/PK model structures in segments 7 and 8. In segment 8 this occurs mainly where a nucleotide is predicted to be single-stranded in one structure and double-stranded in the other. Many positions where consistent and compensatory mutations are observed show lower conservation than average. When compared to the amino acid sequence, almost all of these changes maintain the coding potential of protein sequence and predicted RNA secondary structure. For example, there are several third codon positions that have fourfold degeneracy, yet observed mutations are usually restricted to two nucleotides that preserve predicted base-pairing (Figs. 5, 6, 8–11).
TABLE 4.
Percent nucleotide conservation by codon position and RNA secondary structure
DISCUSSION
The bioinformatics analysis of influenza coding regions provides evidence for many areas with potentially conserved RNA secondary structure and supports previous SSCU studies of influenza A (Gog et al. 2007). With the exception of segment 3, Z-scores are negative in both strand orientations across the eight coding regions. Results for the coding regions predict more conservation of RNA secondary structure in the (+)RNA over the (−)RNA. No predicted structured region strongly favors the (−)RNA (Table 2). This is a reasonable result as the influenza (−)RNA is tightly associated with multiple copies of the NP protein to form viral ribonucleoproteins (vRNPs) that are packaged into viral particles. NP protein binding melts RNA secondary structure (Baudin et al. 1994). Except for interactions between the (−)RNA UTRs, which are important for associating with viral polymerase (Fodor et al. 1994), it is unlikely that most of the (−)RNA possesses extensive conserved secondary structure in the areas analogous to the (+)RNA coding region.
Of the 20 structured regions predicted in influenza RNA, 11 are clearly predicted to be in the (+)RNA, while RNAstrand predictions for the remaining nine are ambiguous (Table 2). These ambiguous regions occur mainly where RNAz predicts structure solely in the (−)RNA, but RNAstrand predicts strand bias toward the (−)RNA with extremely low probability.
Additional evidence for the predicted structured regions comes from the observed overlap of RNAz predictions with areas of strong SSCU and with biologically significant sections of influenza RNA. In all but segments 2 and 3 (Figs. 3, 4), areas with high SSCU overlapped regions with predicted conserved RNA secondary structure. The constraint on synonymous codon site variation may be a manifestation of the preservation of RNA secondary structure (Simmonds and Smith 1999; Pedersen et al. 2004; Tuplin et al. 2004). This is supported by the observed constraint on double-stranded versus single-stranded RNA at third codon positions (Table 4).
Especially compelling are the instances in which predictions of conserved RNA secondary structures overlap with both SSCU and with known biologically important regions. In segment 8, there are two regions where the predicted conserved secondary structure overlaps with sites of SSCU and with viral splice sites (Fig. 1). Others have also described structured RNA elements within these regions of predicted conserved secondary structure (Gultyaev et al. 2007; Ilyinskii et al. 2009). Using the RNAz alignment, RNAalifold predicted a multi-branch loop for the 5′ region of segment 8 (Fig. 5) that is similar to the one proposed previously (Ilyinskii et al. 2009). Additionally, a tetraloop structure was predicted using the SSCU alignment. This structure is not as well supported by all unique sequences for this area. However, the cluster b sequences support this structure quite well, raising the possibility that some strains may adopt a tetraloop structure in this region. Perhaps the RNAalifold calculation revealed the tetraloop structure because the SSCU alignment was biased toward cluster b strains. A recent review article also suggests a tetraloop structure as an alternative fold for clade B influenza strains (Gultyaev et al. 2010). It is likely that the cluster b described in this study corresponds to clade B influenza phylogeny (Kawaoka et al. 1998; Basler et al. 2001). In vitro folding of representative cluster a and b sequences have distinct migration on native gels, thus supporting the prediction that these groups adopt different conformations in nature (Fig. 7).
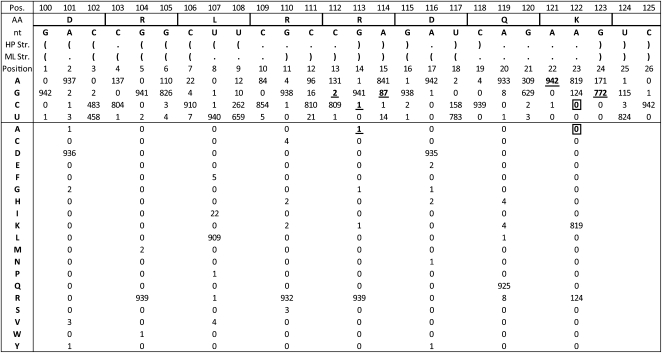
Interestingly, a reported mutant (NS1mut3841) to the 5′ region of segment 8 was shown to inhibit NS1 protein expression (Ilyinskii et al. 2009). It was proposed that this inhibition occurred due to disruption of RNA secondary structure. The expanded sequence data set, however, provides an alternative possibility. A nucleotide count on an alignment of all unique sequences for this region shows that four of the five substitutions in NS1mut3841 occur naturally (Fig. 12). Alignment position 122, however, is never observed to mutate naturally to the C residue in NS1mut3841. This apparently critical position is predicted to be single-stranded in both structural models (Figs. 5, 6). Therefore, it is less likely that RNA secondary structure is responsible for the inhibition of protein production.
FIGURE 12.
Segment 8 sequence from nucleotides 100–125 compared to amino acid coding for region with mutations reported by Ilyinskii et al. (2009). The top row has the alignment positions. Below are given the consensus amino acid sequence and primary nucleotide sequence. Position 102 is a C to represent the consensus sequence for this region, but a U is used at this position in Figure 5 to more accurately represent the canonical pairing in the structural model. Natural occurrences of the NS1mut3841 mutations made by Ilyinskii et al. (2009) are underlined, while the mutations never observed naturally are boxed.
An alternative explanation for the observed inhibition of protein expression is in the mutant amino acid sequence. The mutations in NS1mut3841 lead to a protein product with two new alanine residues (only one of which is observed in the alignment of all unique wild-type sequences) (Fig. 12). These alanines are separated by two intervening residues (AXXA). Previous studies have found that AXXA is able to substitute for RXXL in the destruction box motif (Yamano et al. 1996) and signal the cell's proteolytic machinery to degrade the protein product. The reported effect of the NS1mut3841 mutation (Ilyinskii et al. 2009) may be due to AXXA marked protein destruction in the eukaryotic 293 human embryo kidney cell (HEK) expression system used.
Near the 3′ splice site of segment 8, Gultyaev et al. (2007) described an RNA secondary structure in equilibrium between a hairpin and pseudoknot. RNAalifold calculations on this fragment also predict the hairpin (Fig. 8). As verified by native gel electrophoresis, Gultyaev et al. (2007) demonstrated that more evolutionarily recent sequences of H5N1 influenza A favor the hairpin over the pseudoknot. This is consistent with free energy predictions for the hairpin (−18.9 kcal/mol) versus the pseudoknot (−9 kcal/mol). However, nucleotides 493 and 496–498 at the bottom of the hairpin model (Fig. 8, top) exhibit lower conservation than the rest of the structure and are predicted to be single-stranded in the pseudoknot (Fig. 8, bottom). Thus, it appears that some mutations in these sequences favor the pseudoknot. However, the UPGMA clustering in this analysis did not reveal any clear pattern for sequences that prefer one structure over the other.
The proximity of the predicted structured region shown in Figures 5 and 6 to the 5′ splice site, coupled with the structured region at the 3′ splice site in Figure 8, suggests that these structures may be involved in segment 8 splicing. Others have noted that RNA secondary structure may play an important role in splicing of influenza virus segment 8 (Plotch and Krug 1986; Nemeroff et al. 1992). Interestingly, segment 7 RNA displays a similar pattern of predicted secondary structure surrounding the 5′ and 3′ splice sites (Figs. 9, 10). Both structures have similar sizes and spacing in relation to the 5′ and 3′ splice sites as their segment 8 counterparts. The 3′ splice sites contain a putative hairpin with a possible alternative pseudoknot conformation in both segments 7 and 8. The putative structures at the 5′ and 3′ splice sites in segments 7 and 8 suggest experiments to test the proposed structural models.
Segment 2 provides another correspondence between a predicted structured region and a known feature of influenza RNA. The predicted structured region at 51–170 (Fig. 4) has moderate SSCU and includes the start site of the internal ORF for PB1-F2, a small protein product thought to increase virulence through a pro-apoptotic mechanism (Chen et al. 2001; Gibbs et al. 2003; Chanturiya et al. 2004; Zamarin et al. 2005, 2006). DotKnot predicts a pseudoknot in this region that encompasses the PB1-F2 start codon in a 3′ helix that displays 98.0% base-pair conservation and consistent mutations. Other possibilities predicted by DotKnot for the 5′ helix are described in the Figure 11 caption. AC is the most common noncanonical pair at position 66–105, which is intriguing. In fact, AC is the most common noncanonical pair for almost all structural models (Table 3).
There are many examples where noncanonical base pairs play important roles in RNA secondary structure (Nagaswamy et al. 2002). In particular, AC pairs can induce little perturbation to the A-form helix and can play a role in protein recognition (Jang et al. 1998; Lima et al. 2002). AC is the only noncanonical base pair that can preserve a canonical C1′–C1′ distance in the helix. This has been observed primarily when the N1 position of the adenosine is protonated (Leontis et al. 2002), but recent NMR spectra reveal an almost identical C1′–C1′ distance even in the absence of protonation (Y Lerman, SD Kennedy, N Shankar, M Parisien, F Major, and DH Turner, unpubl.). In segment 2, nucleotides at positions 99–101 and 113–115 (Fig. 11) offer the possibility of two additional AC base pairs capped with a sheared purine–purine base pair, similar to the motif found in helix 8 of the signal recognition particle RNA (Ataide et al. 2011), which would maintain helicity between the predicted pseudoknot helixes.
Another interesting possibility for this pseudoknot (Fig. 11) is suggested by the sequences at positions 89–92 and 122–126, which are identical to the Tetrahymena thermophila group I ribozyme sequences that facilitate a 180° turn in the 3D structure (Cate et al. 1996; Guo et al. 2004). Thus, it is possible that a similar turn orients the helix between nucleotides 75 and 87. The mechanism for expression of PB1-F2 product is not definitively known but is presumed to occur via ribosomal scanning (Chen et al. 2001). RNA structure in this region may facilitate the initiation of translation at the PB1-F2 ORF as seen in other viruses with expressed internal ORFs (Ryabova and Hohn 2000; Pooggin et al. 2006).
There are three instances in which predicted structured regions overlap with strong SSCU but do not correspond to known annotations: at the 5′ and 3′ ends of segments 5 (Fig. 2) and in the 3′ end of segments 3 and 1 (Figs. 3 and 4, respectively). These are intriguing regions of the influenza (+)RNA. There is apparent conserved structure that may be constraining codon variation within regions with no current annotation. These predictions suggest RNA structures with novel function in the influenza virus.
The remaining regions with predicted conserved structure do not overlap with strong SSCU or current annotations. These may be false positives, or the RNA structure conservation may not be constraining enough to have noticeable SSCU.
There are also two regions where strong SSCU does not overlap with predicted structural regions: positions 500–800 in segment 3 and toward the 3′ end of segment 2 (Figs. 3, 4). These constrained sites may be due to other factors acting on the evolution of the sequence (e.g., RNA–protein interactions), or they could be due to RNA structure not predicted by RNAz. Long-range base pairs would be missed in the structure prediction, which divides sequences into windows.
Although only a subset of sequences was used for SSCU studies and secondary structure predictions, structural models are supported by sequence data from all unique sequences for each region. Examining the conservation of each codon position in the context of the proposed secondary structure models adds additional support. Due to the constraint to maintain amino acid coding potential, few compensatory mutations were observed. However, Table 4 shows that when all unique sequences are considered for each structured region, predicted double-stranded nucleotides are more constrained than predicted single-stranded nucleotides. When nucleotides did vary more than average, it was at tolerant codon positions that preserve the amino acid identity and often resulted in consistent and compensatory mutations in the structural model (Figs. 5, 6, 8–11). This conservation recapitulates the global SSCU findings and favors RNA secondary structure as the source of the suppression.
This analysis paves the way for a focused approach to experimental secondary structure determination in influenza A virus. Most importantly, the secondary structures predicted here surround previously defined functional annotations, therefore maximizing the possibility that they are functionally important.
ACKNOWLEDGMENTS
S.F.P. is a trainee in the Medical Scientist Training Program funded by NIH T32 GM07356. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or NIH. During the course of this work S.F.P. was supported by NIH T32 GM068411 from an Institutional Ruth L. Kirschstein National Research Service Award. This work was also supported by NIH RO1 GM22939. We thank Prof. S.-J. Chen and Dr. S. Cao for help with their pseudoknot parameters.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2619511.
REFERENCES
- Ataide SF, Schmitz N, Shen K, Ke A, Shan SO, Doudna JA, Ban N 2011. The crystal structure of the signal recognition particle in complex with its receptor. Science 331: 881–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, Lipman D 2008. The influenza virus resource at the National Center for Biotechnology Information. J Virol 82: 596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler CF, Reid AH, Dybing JK, Janczewski TA, Fanning TG, Zheng H, Salvatore M, Perdue ML, Swayne DE, García-Sastre A 2001. Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc Natl Acad Sci 98: 2746–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin F, Bach C, Cusack S, Ruigrok RW 1994. Structure of influenza virus RNP. I. Influenza virus nucleoprotein melts secondary structure in panhandle RNA and exposes the bases to the solvent. EMBO J 13: 3158–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhart SH, Hofacker IL, Will S, Gruber AR, Stadler PF 2008. RNAalifold: Improved consensus structure prediction for RNA alignments. BMC Bioinformatics 9: 474–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouloy M, Plotch SJ, Krug RM 1978. Globin mRNAs are primers for the transcription of influenza viral RNA in vitro. Proc Natl Acad Sci 75: 4886–4890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Chen SJ 2006. Predicting RNA pseudoknot folding thermodynamics. Nucleic Acids Res 34: 2634–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Chen SJ 2009. Predicting structures and stabilities for H-type pseudoknots with interhelix loops. RNA 15: 696–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate JH, Gooding AR, Podell ER, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA 1996. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science 273: 1678–1685 [DOI] [PubMed] [Google Scholar]
- Chanturiya AN, Basanez G, Schubert U, Henklein P, Yewdell JW, Zimmerberg J 2004. PB1-F2, an influenza A virus-encoded proapoptotic mitochondrial protein, creates variably sized pores in planar lipid membranes. J Virol 78: 6304–6312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, Basta S, O'Neill R, Schickli J, Palese P 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat Med 7: 1306–1312 [DOI] [PubMed] [Google Scholar]
- Childs JL, Disney MD, Turner DH 2002. Oligonucleotide directed misfolding of RNA inhibits Candida albicans group I intron splicing. Proc Natl Acad Sci 99: 11091–11096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs JL, Poole AW, Turner DH 2003. Inhibition of Escherichia coli RNase P by oligonucleotide directed misfolding of RNA. RNA 9: 1437–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs-Disney JL, Wu M, Pushechnikov A, Aminova O, Disney MD 2007. A small molecule microarray platform to select RNA internal loop–ligand interactions. ACS Chem Biol 2: 745–754 [DOI] [PubMed] [Google Scholar]
- Clever J, Sassetti C, Parslow TG 1995. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J Virol 69: 2101–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compans RW 1972. Structure of the ribonucleoprotein of influenza virus. J Virol 10: 795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dam EBT, Pleij CWA, Bosch L 1990. RNA pseudoknots and translational frameshifting on retroviral, coronaviral and luteoviral RNAs. Virus Genes 4: 121–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirks RM, Pierce NA 2003. A partition function algorithm for nucleic acid secondary structure including pseudoknots. J Comput Chem 24: 1664–1677 [DOI] [PubMed] [Google Scholar]
- Disney MD, Childs JL, Turner DH 2004. New approaches to targeting RNA with oligonucleotides: Inhibition of group I intron self-splicing. Biopolymers 73: 151–161 [DOI] [PubMed] [Google Scholar]
- Disney MD, Labuda LP, Paul DJ, Poplawski SG, Pushechnikov A, Tran T, Velagapudi SP, Wu M, Childs-Disney JL 2008. Two-dimensional combinatorial screening identifies specific aminoglycoside-RNA internal loop partners. J Am Chem Soc 130: 11185–11194 [DOI] [PubMed] [Google Scholar]
- Dreher TW 2009. Role of tRNA-like structures in controlling plant virus replication. Virus Res 139: 217–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dushoff J, Plotkin JB, Viboud C, Earn DJD, Simonsen L 2006. Mortality due to influenza in the United States—an annualized regression approach using multiple-cause mortality data. Am J Epidemiol 163: 181–187 [DOI] [PubMed] [Google Scholar]
- Fodor E, Pritlove DC, Brownlee GG 1994. The influenza virus panhandle is involved in the initiation of transcription. J Virol 68: 4092–4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego J, Varani G 2001. Targeting RNA with small-molecule drugs: Therapeutic promise and chemical challenges. Acc Chem Res 34: 836–843 [DOI] [PubMed] [Google Scholar]
- Gibbs JS, Malide D, Hornung F, Bennink JR, Yewdell JW 2003. The influenza A virus PB1-F2 protein targets the inner mitochondrial membrane via a predicted basic amphipathic helix that disrupts mitochondrial function. J Virol 77: 7214–7224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gog JR, Afonso EDS, Dalton RM, Leclercq I, Tiley L, Elton D, Von Kirchbach JC, Naffakh N, Escriou N, Digard P 2007. Codon conservation in the influenza A virus genome defines RNA packaging signals. Nucleic Acids Res 35: 1897–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AR, Findeiss S, Washietl S, Hofacker IL, Stadler PF 2010. RNAz 2.0: Improved noncoding RNA detection. Pac Symp Biocomput 15: 69–79 [PubMed] [Google Scholar]
- Gultyaev AP, Heus HA, Olsthoorn RCL 2007. An RNA conformational shift in recent H5N1 influenza A viruses. Bioinformatics 23: 272–276 [DOI] [PubMed] [Google Scholar]
- Gultyaev AP, Fouchier RAM, Olsthoorn RCL 2010. Influenza virus RNA structure: Unique and common features. Int Rev Immunol 29: 533–556 [DOI] [PubMed] [Google Scholar]
- Guo F, Gooding AR, Cech TR 2004. Structure of the Tetrahymena ribozyme: Base triple sandwich and metal ion at the active site. Mol Cell 16: 351–362 [DOI] [PubMed] [Google Scholar]
- Hagen M, Chung TD, Butcher JA, Krystal M 1994. Recombinant influenza virus polymerase: Requirement of both 5′ and 3′ viral ends for endonuclease activity. J Virol 68: 1509–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA 2001. BioEdit: A user-friendly biological sequence alignment editor and analysis, version 5.09. Department of Microbiology, North Carolina State University, Raleigh, NC [Google Scholar]
- Harmanci AO, Sharma G, Mathews DH 2007. Efficient pairwise RNA structure prediction using probabilistic alignment constraints in Dynalign. BMC Bioinformatics 8: 130–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu MT, Parvin JD, Gupta S, Krystal M, Palese P 1987. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc Natl Acad Sci 84: 8140–8144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson EC, Curran MD, Read EK, Gog JR, Digard P 2008. Mutational analysis of cis-acting RNA signals in segment 7 of influenza A virus. J Virol 82: 11869–11879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyinskii PO, Schmidt T, Lukashev D, Meriin AB, Thoidis G, Frishman D, Shneider AM 2009. Importance of mRNA secondary structural elements for the expression of influenza virus genes. OMICS 13: 421–430 [DOI] [PubMed] [Google Scholar]
- Jacks T, Madhani HD, Masiarz FR, Varmus HE 1988. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell 55: 447–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang SB, Hung LW, Chi YI, Holbrook EL, Carter RJ, Holbrook SR 1998. Structure of an RNA internal loop consisting of tandem C-A+ base pairs. Biochemistry 37: 11726–11731 [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33: 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka Y, Gorman OT, Ito T, Wells K, Donis RO, Castrucci MR, Donatelli I, Webster RG 1998. Influence of host species on the evolution of the nonstructural (NS) gene of influenza A viruses. Virus Res 55: 143–156 [DOI] [PubMed] [Google Scholar]
- Kieft JS 2008. Viral IRES RNA structures and ribosome interactions. Trends Biochem Sci 33: 274–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo MY, Sharmeen L, Dinter-Gottlieb G, Taylor J 1988. Characterization of self-cleaving RNA sequences on the genome and antigenome of human hepatitis delta virus. J Virol 62: 4439–4444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lee MM, Pushechnikov A, Disney MD 2009. Rational and modular design of potent ligands targeting the RNA that causes myotonic dystrophy 2. ACS Chem Biol 4: 345–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Stombaugh J, Westhof E 2002. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res 30: 3497–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Huang T, Ly H, Parslow TG 2008. Mutational analyses of packaging signals in influenza virus PA, PB1, and PB2 genomic RNA segments. J Virol 82: 229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima S, Hildenbrand J, Korostelev A, Hattman S, Li H 2002. Crystal structure of an RNA helix recognized by a zinc-finger protein: An 18-bp duplex at 1.6 Å resolution. RNA 8: 924–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Shankar N, Turner DH 2009. Fluorescence competition assay measurements of free energy changes for RNA pseudoknots. Biochemistry 49: 623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh GA, Hatami R, Palese P 2007. Specific residues of the influenza A virus hemagglutinin viral RNA are important for efficient packaging into budding virions. J Virol 81: 9727–9736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh GA, Rabadan R, Levine AJ, Palese P 2008. Highly conserved regions of influenza A virus polymerase gene segments are critical for efficient viral RNA packaging. J Virol 82: 2295–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews D 2004. Predicting the secondary structure common to two RNA sequences with Dynalign. Curr Protoc Bioinformatics 12.4.1–12.4.11 [DOI] [PubMed] [Google Scholar]
- Mathews DH, Turner DH 2002. Dynalign: An algorithm for finding the secondary structure common to two RNA sequences. J Mol Biol 317: 191–203 [DOI] [PubMed] [Google Scholar]
- Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH 2004. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci 101: 7287–7292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Moss WN, Turner DH 2010. Folding and finding RNA secondary structure. Cold Spring Harb Perspect Biol doi: 10.1101/cshperspect.a003665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei HY, Cui M, Heldsinger A, Lemrow SM, Loo JA, Sannes-Lowery KA, Sharmeen L, Czarnik AW 1998. Inhibitors of protein–RNA complexation that target the RNA: Specific recognition of human immunodeficiency virus type 1 TAR RNA by small organic molecules. Biochemistry 37: 14204–14212 [DOI] [PubMed] [Google Scholar]
- Muramoto Y, Takada A, Fujii K, Noda T, Iwatsuki-Horimoto K, Watanabe S, Horimoto T, Kida H, Kawaoka Y 2006. Hierarchy among viral RNA (vRNA) segments in their role in vRNA incorporation into influenza A virions. J Virol 80: 2318–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaswamy U, Larios-Sanz M, Hury J, Collins S, Zhang Z, Zhao Q, Fox GE 2002. NCIR: a database of non-canonical interactions in known RNA structures. Nucleic Acids Res 30: 395–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeroff ME, Utans U, Kramer A, Krug RM 1992. Identification of cis-acting intron and exon regions in influenza virus NS1 mRNA that inhibit splicing and cause the formation of aberrantly sedimenting presplicing complexes. Mol Cell Biol 12: 962–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Sagara H, Yen A, Takada A, Kida H, Cheng RH, Kawaoka Y 2006. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature 439: 490–492 [DOI] [PubMed] [Google Scholar]
- Pedersen JS, Meyer IM, Forsberg R, Simmonds P, Hein J 2004. A comparative method for finding and folding RNA secondary structures within protein-coding regions. Nucleic Acids Res 32: 4925–4936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotch SJ, Krug RM 1986. In vitro splicing of influenza viral NS1 mRNA and NS1-beta-globin chimeras: Possible mechanisms for the control of viral mRNA splicing. Proc Natl Acad Sci 83: 5444–5448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotch SJ, Bouloy M, Ulmanen I, Krug RM 1981. A unique cap (m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23: 847–858 [DOI] [PubMed] [Google Scholar]
- Pooggin MM, Ryabova LA, He X, Fütterer J, Hohn T 2006. Mechanism of ribosome shunting in rice tungro bacilliform pararetrovirus. RNA 12: 841–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushechnikov A, Lee MM, Childs-Disney JL, Sobczak K, French JM, Thornton CA, Disney MD 2009. Rational design of ligands targeting triplet repeating transcripts that cause RNA dominant disease: Application to myotonic muscular dystrophy type 1 and spinocerebellar ataxia type 3. J Am Chem Soc 131: 9767–9779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiche K, Stadler PF 2007. RNAstrand: reading direction of structured RNAs in multiple sequence alignments. Algorithms Mol Biol 2: 6–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryabova LA, Hohn T 2000. Ribosome shunting in the cauliflower mosaic virus 35S RNA leader is a special case of reinitiation of translation functioning in plant and animal systems. Genes Dev 14: 817–829 [PMC free article] [PubMed] [Google Scholar]
- Schroeder S 2009. Advances in RNA structure prediction from sequence: New tools for generating hypotheses about viral RNA structure-function relationships. J Virol 83: 6326–6334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro GI, Krug RM 1988. Influenza virus RNA replication in vitro: Synthesis of viral template RNAs and virion RNAs in the absence of an added primer. J Virol 62: 2285–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P, Smith DB 1999. Structural constraints on RNA virus evolution. J Virol 73: 5787–5794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal R, Michener C 1958. A statistical method for evaluating systematic relationships. U Sci Pap Univ Kansas Nat Hist Mus 38: 1409–1438 [Google Scholar]
- Sperschneider J, Datta A 2010. DotKnot: pseudoknot prediction using the probability dot plot under a refined energy model. Nucleic Acids Res 38: e103 doi: 10.1093/nar/gkq021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucheck SJ, Wong CH 2000. RNA as a target for small molecules. Curr Opin Chem Biol 4: 678–686 [DOI] [PubMed] [Google Scholar]
- Theimer CA, Wang Y, Hoffman DW, Krisch HM, Giedroc DP 1998. Non-nearest neighbor effects on the thermodynamics of unfolding of a model mRNA pseudoknot. J Mol Biol 279: 545–564 [DOI] [PubMed] [Google Scholar]
- Tuplin A, Evans DJ, Simmonds P 2004. Detailed mapping of RNA secondary structures in core and NS5B-encoding region sequences of hepatitis C virus by RNase cleavage and novel bioinformatic prediction methods. J Gen Virol 85: 3037–3047 [DOI] [PubMed] [Google Scholar]
- Uzilov AV, Keegan JM, Mathews DH 2006. Detection of non-coding RNAs on the basis of predicted secondary structure formation free energy change. BMC Bioinformatics 7: 173–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S, Hofacker IL, Lukasser M, Hüttenhofer A, Stadler PF 2005a. Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat Biotechnol 23: 1383–1390 [DOI] [PubMed] [Google Scholar]
- Washietl S, Hofacker IL, Stadler PF 2005b. Fast and reliable prediction of noncoding RNAs. Proc Natl Acad Sci 102: 2454–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW Jr, Swanstrom R, Burch CL, Weeks KM 2009. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460: 711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner AM, Maizels N 1987. tRNA-like structures tag the 3′ ends of genomic RNA molecules for replication: Implications for the origin of protein synthesis. Proc Natl Acad Sci 84: 7383–7387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson WD, Li K 2000. Targeting RNA with small molecules. Curr Med Chem 7: 73–98 [DOI] [PubMed] [Google Scholar]
- Xia T, SantaLucia J Jr, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH 1998. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry 37: 14719–14735 [DOI] [PubMed] [Google Scholar]
- Yamano H, Gannon J, Hunt T 1996. The role of proteolysis in cell cycle progression in Schizosaccharomyces pombe. EMBO J 15: 5268–5279 [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Krug RM, Tao YJ 2006. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature 444: 1078–1082 [DOI] [PubMed] [Google Scholar]
- Zamarin D, García-Sastre A, Xiao X, Wang R, Palese P 2005. Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Pathog 1: e4 doi: 10.1371/journal.ppat.0010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamarin D, Ortigoza MB, Palese P 2006. Influenza A virus PB1-F2 protein contributes to viral pathogenesis in mice. J Virol 80: 7976–7983 [DOI] [PMC free article] [PubMed] [Google Scholar]