Summary
Histone deacetylation constitutes an important mechanism for silencing genes. The HDAC-associated mammalian Rpd3S/Sin3S corepressor complex plays key roles in repressing aberrant gene transcription from cryptic transcription initiation sites and mitigating RNA polymerase II progression in intragenic regions of actively transcribed genes. The Sin3 corepressor functions as a molecular adaptor linking HDACs on the one hand, with the chromatin targeting subunits Pf1 and MRG15, on the other. Pf1 also functions as an adaptor by interacting with MRG15 and engaging in multivalent interactions with Sin3 targeting among other domains the two N-terminal PAH domains that serve as sites of interaction with sequence-specific DNA-binding transcription factors. Here, we structurally and functionally evaluate the interaction between the PAH2 domain of mSin3A and the SID1 motif of Pf1 and find the structural aspects to be reminiscent of the interaction between the Mad1/Mxd1 transcription factor and Sin3. Pf1 residues within a highly-conserved sequence motif immediately C-terminal to SID1 appear not to be important for the interaction with Sin3 PAH2. Unexpectedly, the MRG15 subunit competes, rather than collaborate, with Sin3 for the Pf1 segment encompassing the two conserved motifs, implying competition between two subunits for another subunit of the same chromatin-modifying complex.
Keywords: Rpd3S complex, transcription regulation, transcription repression, chromatin-modifying complex assembly, PAH domain, MRG domain, protein-protein interaction, Sin3 corepressor, NMR
Introduction
Post-translational modification of histones is a facile yet powerful mechanism for epigenetic programming.1,2 The reversible nature of these modifications affords reprogramming of any locus with concomitant alterations in gene expression levels. Histone acetylation and deacetylation constitute particularly well-characterized examples and are generally correlated with enhanced or diminished transcript levels, respectively. The enzymes that bring about these epigenetic changes are typically members of multi-protein complexes whose targeting specificity often resides within the non-enzymatic subunits. Comparatively little is known about the precise molecular functions of many of the subunits of these complexes and even less is known about how the subunits assemble to yield a functional complex.
The yeast histone deacetylase Rpd3 and its mammalian homologues HDAC1, HDAC2, and HDAC3 are nuclear proteins found in only a handful of complexes that have distinct functional properties and subunit compositions.3,4 Two related, yet functionally distinct, forms of the Rpd3 complex in yeast and the orthologous Sin3 corepressor complex in mammals play fundamental roles in cellular physiology. The Rpd3L/Sin3L complex is recruited to the promoter regions and represses a broad range of genes involved in cell cycle regulation, differentiation, DNA replication and repair, apoptosis, and mitochondrial metabolism whereas the Rpd3S/Sin3S complex is targeted to the intragenic regions of actively transcribed genes to suppress aberrant transcription initiation from cryptic sites and to mitigate RNA polymerase II progression.4–13 The Rpd3/Sin3 complexes also play key roles in heterochromatin formation at centromeres, telomeres, and ribosomal DNA loci and in DNA replication timing and double-strand break repair.5 Consistent with their fundamental roles, the majority of the subunits of the Rpd3/Sin3 complexes are evolutionarily conserved from yeast to human. The larger, 1.2–2 megadalton Rpd3L/Sin3L complex comprises at least eight subunits whereas the smaller, 0.6 megadalton Rpd3S/Sin3S complex comprises at least five subunits, three of which, including the Sin3, Rpd3/HDAC1/HDAC2, and RbAp46/RbAp48 polypeptides, are shared by both complexes.8,11,14–19
The Pf1 and MRG15 subunits of the mammalian Rpd3S/Sin3S complex as well as their orthologs in budding yeast including Rco1p and Eaf3p, respectively, play critical roles in targeting the complex to the transcribed regions through direct interactions with nucleosomes.9,11,20,21 Pf1 also functions as a molecular adaptor interacting with the MRG domain of MRG15 and linking it with the rest of the complex by engaging multiple domains of Sin3 in direct interactions including the three N-terminal PAH domains and the HID domain.11,18,19 The Sin3 – polypeptide after which the mammalian complex is named – is thought to function as an organizing center or scaffold for the assembly of both Rpd3L/Sin3L and Rpd3S/Sin3S complexes. Sin3 is recruited by a variety of sequence-specific DNA-binding factors through direct protein-protein interactions involving its two N-terminal PAH domains.22–34 The Sin3 PAH3 and HID domains are targeted by various subunits unique to the Rpd3L/Sin3L complex.15,17,35 Thus the multivalent nature of the interactions between Pf1 and Sin3 implicates Pf1 in yet another role – that of limiting access to transcription factors as well as to Rpd3L/Sin3L subunits with disparate functions – thereby preserving the unique specificity of the smaller complex.
The interactions involving the Sin3 PAH1 and PAH2 domains have been structurally and functionally characterized.24,36–40 These studies have shown that PAH domains bind to diverse targets, but do so with a high-degree of specificity. The PAH1 and PAH2 domains recognize distinct sequence motifs (referred to as Sin3 interaction domains or SIDs), yet the directionality of the motifs appears to be unimportant, as motifs running both N-to-C and C-to-N are equally capable of interacting.24,36 This was explained by the SIDs adopting helical conformations in the respective complexes with the precise helical orientation determined by the chain direction of the underlying motif. Sequences analysis of Pf1 orthologs revealed a conserved motif that extended beyond the SID that was shown to be crucial for stable (and PAH2-dependent) association with Sin3. Here, we structurally and functionally characterize the interaction between Pf1 and mammalian Sin3A PAH2 and show that only residues within the SID are important for stable association with Sin3. We also show that rather unexpectedly, MRG15 competes with Sin3 PAH2 for Pf1 SID1, disrupting a presumed important point of contact between Pf1 and Sin3.
Results
Low Resolution Structural Analysis of Pf1 SID1
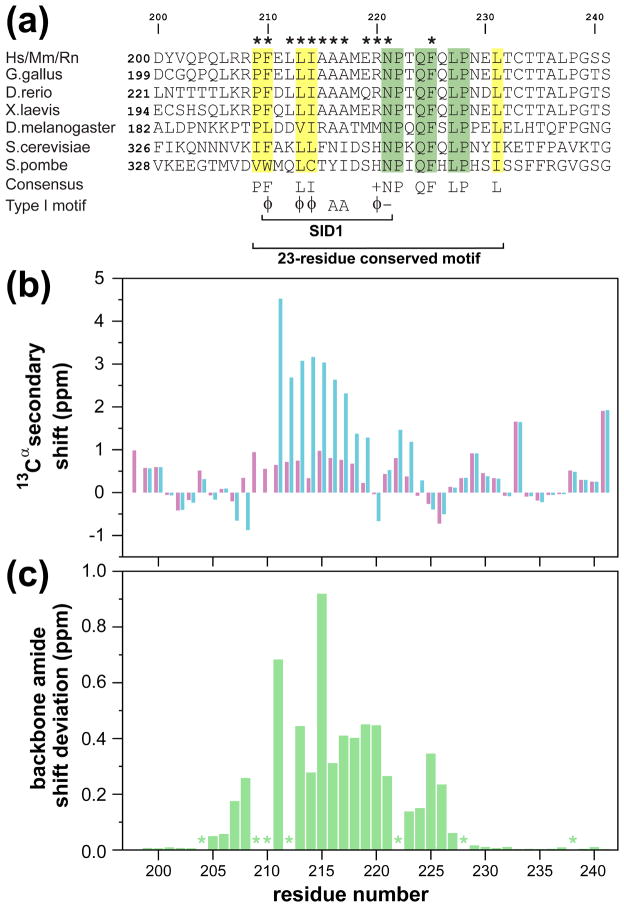
Previous biochemical studies have identified at least three regions within Pf1 as being important for the association with Sin3 including the segments corresponding to SID1 and SID2 in the N- and C-terminal halves of the protein as well as the PHD2 domain.11,18 Comparative analysis of the linker segment connecting the PHD1 and PHD2 domains of various Pf1 orthologs revealed poor conservation of both length and sequence. MEME-based analysis of potential motifs in this region, on the other hand, indicated a conserved segment N-terminal to the PHD2 domain (Figure 1(a)). The 23-residue motif spanning residues 209 to 231 overlaps with the 12-residue SID1 spanning residues 210 to 221, which was classified as a Type I PAH2-interactor (i.e. Mad1/Mxd1-like) based on sequence analysis.24 The high-degree of sequence conservation immediately C-terminal to the SID1 prompted us to evaluate whether these residues might play a role in binding to Sin3 PAH2.
Figure 1.
Conserved sequence motifs and backbone polypeptide conformation in the Pf1 linker region separating the two PHD domains. (a) A MEME-based multiple sequence alignment of Pf1 orthologs (species abbreviations: Hs: Homo sapiens; Mm: Mus musculus; Rn: Rattus norvegicus; G. gallus: Gallus gallus; X. laevis: Xenopus laevis; D. rerio: Danio rerio; D. melanogaster: Drosophila melanogaster; S. cerevisiae: Saccharomyces cerevisiae; S. pombe: Schizosaccharomyces pombe). Invariant and conserved residues are highlighted in green and yellow, respectively. Asterisks identify residues involved in intermolecular contacts in the mSin3A PAH2-Pf1 complex as determined using the program MONSTER.66 Residue abbreviations: φ: Phe, Ile, Leu, Met, or Val; +: His, Lys, or Arg; −: Glu or Asp. (b) 13Cα secondary chemical shifts for the Pf1 polypeptide in the apo (magenta) and mSin3A PAH2-bound (blue) states. (c) Chemical shift deviations for backbone amides between the apo- and mSin3A PAH2-bound states. Asterisks denote prolines or missing data for residues affected by severe resonance broadening.
To this end, we expressed a 42-residue construct of Pf1 spanning residues 200 to 241 (henceforth designated Pf1200–241). The 1H-15N correlated spectrum of Pf1200–241 was characterized by poor amide proton chemical shift dispersion, narrow resonance linewidths, and uniformly intense correlations, characteristic of an unstructured protein (Supplementary Figure S1 (a)). This was confirmed by the absolute magnitude of the 13Cα secondary chemical shifts that were typically <1 ppm (Figure 1(b)). The amide proton chemical shift dispersion of Pf1200–241 increased and correlations belonging to residues in the segment Arg207 to Gln226 were less intense compared to those of residues outside of this region when one equivalent of mSin3A PAH2 was added (Supplementary Figure S1 (b)). The resonances belonging to some of the residues in this segment were considerably broader while those of Phe210 and Leu212 were broadened beyond detection, characteristic of a complex with dissociation kinetics in the slow to intermediate range on the NMR timescale. As expected, the largest changes in backbone amide chemical shifts were observed for residues in the SID1 region. However, residues C-terminal to this motif were also strongly perturbed (Figure 1(c)), suggesting the involvement of this segment in the interaction.
Pf1 SID1 Interacts with mSin3A PAH2 in a Mad1/Mxd1-like Manner
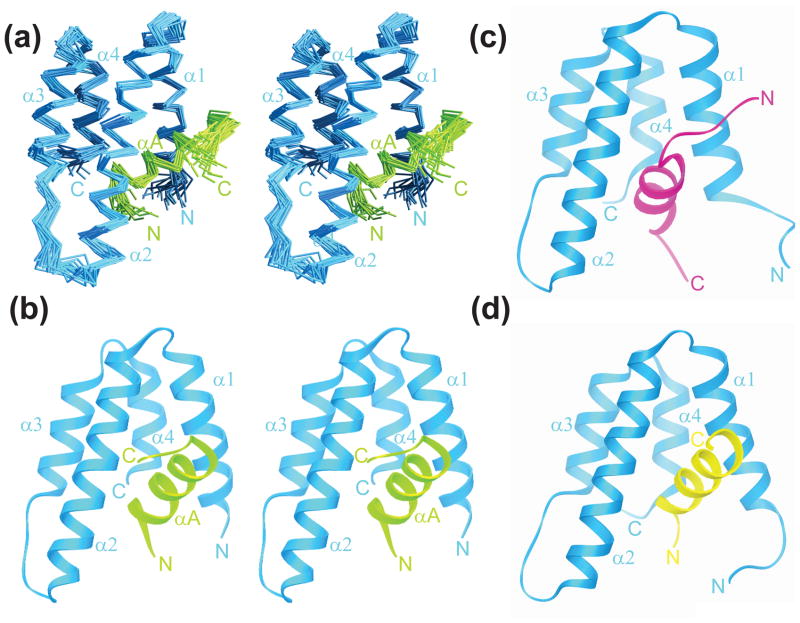
To test the prediction of a Mad1/Mxd1-like interaction for Pf1 with Sin3 PAH2 and to clarify the role of the highly-conserved segment C-terminal to the SID1 motif, we determined the solution structure of the mSin3A PAH2-Pf1200–241 complex using NMR methods. The structure was determined using distance and dihedral angle restraints derived from 1H-1H NOEs and chemical shift data, respectively (see Methods). Conformers of reasonable precision and geometry and in good agreement with experimental data were obtained (Figure 2(a); Table 1). As was observed in the case of the other structurally-characterized mSin3A PAH2 complexes, the PAH2 segment spanning Glu300 to Pro381 is better defined than the regions N- or C-terminal to it. Also consistent with the findings from low-resolution studies of Pf1200–241 (Figure 1(b)), the segment starting with Arg207 through Phe225 comprising SID1 and a portion of the adjacent highly-conserved motif are determined with greater precision than the residues outside of this region.
Figure 2.
Solution structure of the mSin3A PAH2-Pf1 complex and comparison with the Mad1/Mxd1 and HBP1 complexes. Stereoviews of (a) the ensemble of 20 structures following a best-fit superposition of the backbone atoms in the helical regions and (b) a representative structure. Although structures were calculated for residues 295 to 385 and 200 to 241 of mSin3A and Pf1, respectively, for clarity only those residues (300 to 381 and 207 to 225) with average RMSDs <4 Å relative to the average structure are shown. The mSin3A and Pf1 polypeptide segments are shown in blue and green, respectively. Representative structures of the (c) mSin3A PAH2-HBP1 SID (PDB code: 1S5R) and (d) mSin3A PAH2-Mad1/Mxd1 SID (PDB code: 1S5Q) complexes are shown for comparison. The HBP1 and Mad1/Mxd1 peptides are colored in magenta and yellow, respectively. Note the contrasting orientations of the SID helices in these complexes.
Table 1.
NMR Structure Determination Statistics for mSin3A PAH2-Pf1200–241 Complex
| Restraint Statistics | |
| NOE-based distance restraints | 2409 |
| Unambiguous NOE-based restraints | 1890 |
| Intraresidue | 996 |
| Sequential (|i − j| = 1) | 361 |
| Medium-range (1 < |i − j| ≤ 4) | 235 |
| Intramolecular long-range (|i − j| > 4) | 298 |
| Intermolecular | 154 |
| Ambiguous NOE-based restraints | 519 |
| Hydrogen bonding distance restraints | 92 |
| Dihedral angle restraints | 134 (67 φ, 67 ψ) |
| Structure Quality of NMR Ensemble | |
| Restraint satisfaction | |
| Root-mean-square differences for distance restraints (Å) | 0.013 ± 0.002 |
| Root-mean-square differences for torsion angle restraints (°) | 0.187 ± 0.065 |
| Deviations from ideal covalent geometry | |
| Bond lengths (Å) | 0.003 ± 0.000 |
| Bond angles (°) | 0.470 ± 0.015 |
| Impropers (°) | 1.197 ± 0.058 |
| Ramachandran plot statistics (%) | |
| Residues in most favored regions | 79.0 (90.2)a,b |
| Residues in allowed regions | 19.8 (9.3)a,b |
| Residues in disallowed regions | 1.2 (0.5)a,b |
| Average Atomic Root-Mean-Square Deviations from the Average Structure (Å) | |
| All atoms | 5.98 |
| All atoms except disordered regionsb | 1.67 |
| All atoms in helices | 1.32 |
| Backbone atoms (N, Cα, C′) | |
| All residues | 6.14 |
| All residues excluding disordered regionsb | 0.89 |
| All residues in helices | 0.58 |
statistics for ordered regions; deemed to be those residues with backbone RMS deviations less than 4 Å following a best-fit backbone superposition of helical regions
disordered regions include residues 200 to 206 and 226 to 241 of Pf1 SID1 and 295 to 299 and 382 to 385 of mSin3A PAH2
The mSin3A PAH2 domain in the Pf1 complex adopts the well-characterized left-handed, four-helix bundle architecture of PAH domains with helices α2, α3, and α4 packing parallel to each other while helix α1 is splayed open exposing a hydrophobic cleft (Figure 2(b)). The Pf1 SID1 adopts a helical conformation in the complex engaging the cleft through a non-polar surface. Like all previously characterized PAH-interactors including Mad1/Mxd1 and HBP1 (Figure 2), the Pf1 αA helix is the only secondary structural element making contacts with the PAH domain.24 Pf1200–241 and mSin3A PAH2 each bury an average of ~840 Å2 surface area which is comparable to that buried in the HBP1 complex (~800 Å2) but is more than that buried in the Mad1/Mxd1 complex (~700 Å2).
All four PAH2 helices in the Pf1 complex are located at approximately the same locations as in the Mad1/Mxd1 and HBP1 complexes.24 The Pf1 αA helix encompassing residues Pro209 through Glu219 is about the same length as the Mad1/Mxd1 helix (11 versus 13 residues; Figure 2(b) and 2 (d)). The Pf1 helix also adopts the same orientation as the Mad1/Mxd1 helix in complete contrast to the HBP1 helix which adopts an opposite orientation (Figure 2(b), 2 (c) and 2 (d)). A best-fit superposition of the Pf1 and Mad1/Mxd1 complexes yields an RMSD of 1.38 Å with almost all the residues used in this fitting procedure drawn from all five helices. The Pf1 and Mad1/Mxd1 helices superimpose rather well, although a consistent shift in the direction of the N-terminus by ~1.4 Å is detected for the Pf1 helix relative to that of Mad1. The four PAH2 helices in the Pf1 and Mad1/Mxd1 complexes superimpose with an RMSD of 1.24 Å but this drops considerably to 0.46 Å when only the α2 and α3 helices are superimposed; parallel trends are noticed when comparing the Pf1 and HBP1 complexes, implying that the α1 and α4 helices exhibit greater packing diversity and malleability in order to optimize interactions with their targets. In support of this notion, residues that comprise the α1 and α4 helices in the SID-bound states, unlike their counterparts in the α2 and α3 helices, have been found to exhibit substantial conformational diversity in the apo-state by sampling predominantly non-helical conformations.41
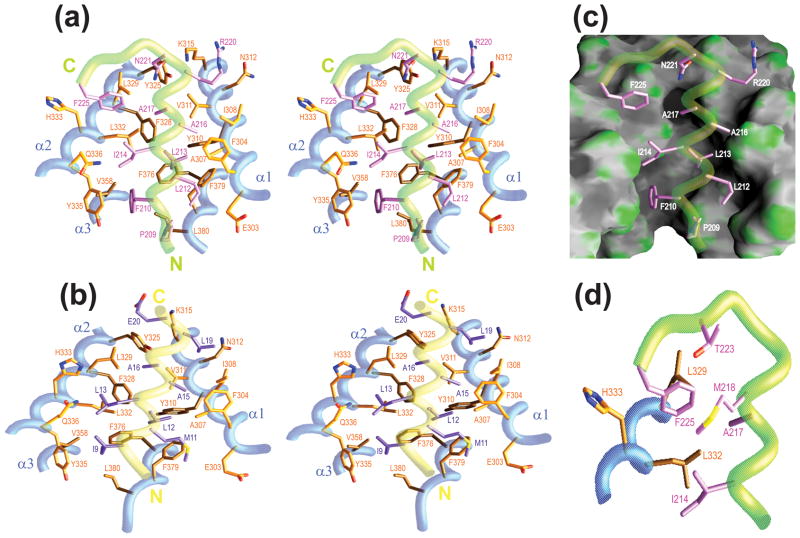
Pf1 SID1 was classified as a Type I (Mad1/Mxd1-like) interaction motif (Figure 1(a)), and like Mad1/Mxd1, the hydrophobic residues in the motif including Phe210, Leu213, Ile214, Ala216 and Ala217 all make contacts with PAH2 residues that comprise the hydrophobic cleft (Figure 3(a)). A key difference between the Pf1 and Mad1/Mxd1 sequences is the presence of a substantially bulkier residue (i.e. Phe210 instead of Ile9) at the first position of the motif (Figure 3(a) and 3 (b)). Molecular modeling suggests that a simple replacement of the isoleucine side chain in the Mad1/Mxd1 structure with phenylalanine would lead to clashes with residues on one side of the cleft including Phe376 and Leu380. We surmise that the origin of the differences in the precise positioning of the Pf1 and Mad1/Mxd1 helices relative to the PAH2 domain as well as the packing diversity noted for residues in the α4 helix and the segment immediately C-terminal to it is due to the rigid and bulky nature of the Phe210 side chain. Leu213, Ile214, Ala216 and Ala217 of Pf1 like Leu12, Leu13, Ala15 and Ala16 of Mad1/Mxd1 target comparable ‘pockets’ on the surface of the PAH2 domain (Figure 3(a) and 3 (b)). By virtue of their short side chains, the alanines in both complexes play a critical role in avoiding steric clashes with the section of the hydrophobic cleft that narrows in their vicinity (Figure 3(c)). The remaining two positions in the Type I motif comprising a hydrophobic and a negatively charged residue in Pf1 are occupied by Arg220 and Asn221, respectively. Since the interactions involving these side chains occur on a more exposed surface, the substantial aliphatic component of the arginine side chain satisfies the former requirement while Asn221 engages in hydrogen bonding interactions with the Lys315 and Tyr325 side chains of PAH2 mimicking to some extent the electrostatic interactions made by Glu21 of Mad1/Mxd1 with Lys315.24,37,42
Figure 3.
Non-covalent interactions in the mSin3A PAH2-Pf1 complex and comparison with the mSin3A PAH2-Mad1/Mxd1 complex. Residues engaging in intermolecular interactions in the (a) Pf1 and (b) Mad1/Mxd1 complexes are shown. The interface in each case is dominated by hydrophobic interactions except for hydrogen bonding interactions involving the side chains of Asn221 of Pf1 with Lys315 and Tyr325 of mSin3A and electrostatic interactions between the Glu20 side chain of Mad1/Mxd1 and the Lys315 side chain of mSin3A. (c) The intermolecular interface in the mSin3A PAH2-Pf1 complex with the PAH2 domain rendered as a molecular surface and the interacting side chains of Pf1 rendered as sticks. (d) A close-up of the medium- and long-range intra- and inter-molecular non-covalent interactions involving Phe225.
Functional Analysis of the mSin3A PAH2-Pf1 SID1 Interaction
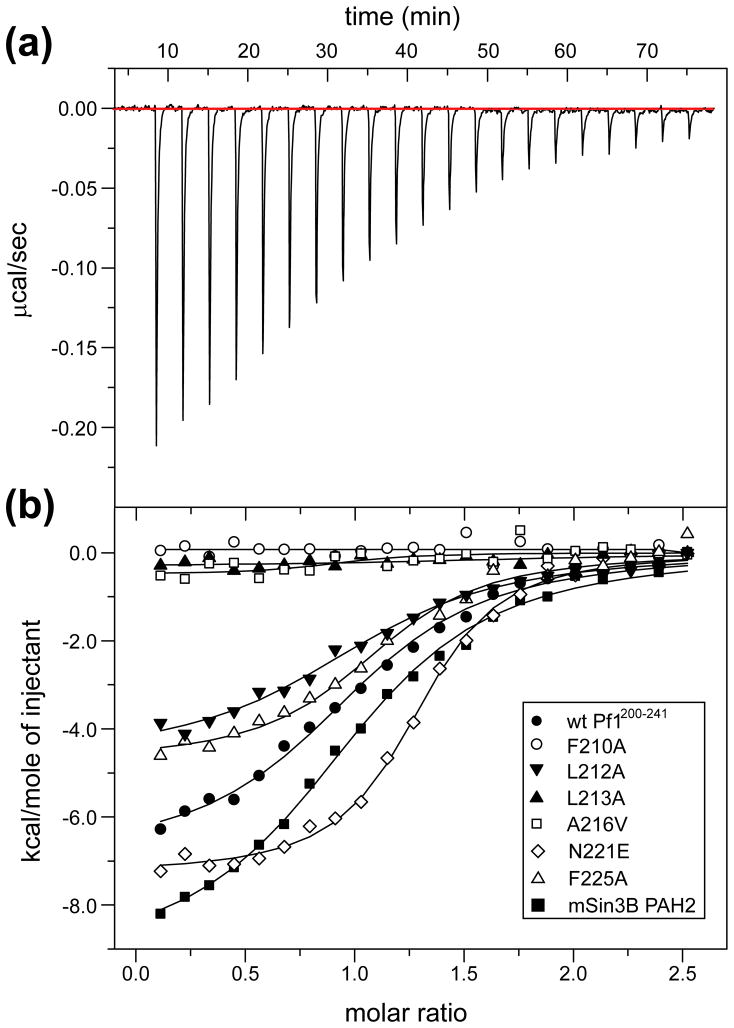
To clarify the roles of individual Pf1 residues at the protein-protein interface, we measured the affinity of a variety of single-site mutants and compared them to the wild-type protein using isothermal titration calorimetry (ITC; Figure 4(a) and 4 (b)). The 2.2 μM equilibrium dissociation constant (KD) measured for the interaction between wild-type Pf1200–241 with mSin3A PAH2 indicates a moderate affinity interaction consistent with the complex being in slow to intermediate exchange on the NMR timescale (cf. above; Supplementary Figure 1(b)). As expected, single-site mutations of Phe210 and Leu213 – both of which interact extensively with the cleft and are shielded from solvent – to alanine adversely affected the interaction. Mutation of Ala216 to the bulkier valine residue also had a similar effect on the interaction likely due to steric clashes with the narrow cleft. Mutation of Leu212, which is not part of the core motif but is partially occluded from solvent and engages in interactions with the peripheral regions of the cleft, to an alanine had a minor effect on binding (Table 2). Interestingly, the binding enthalpy is diminished compared to wild-type but is largely compensated by a corresponding increase in the binding entropy.
Figure 4.
Isothermal titration calorimetric analysis of mSin3A PAH2-Pf1 interactions. (a) A representative titration involving the wild-type Pf1200–241 polypeptide (in the cell) and mSin3A PAH2 (in the syringe) is shown. (b) Overlays of the titration data for wild-type Pf1200–241 (filled circles), Phe210Ala (open circles), Leu212Ala (filled inverted triangles), Leu213Ala (filled triangles), Ala216Val (open squares), Asn221Glu (open diamonds), and Phe225Ala (open triangles) mutants of Pf1200–241 with mSin3A PAH2 are shown. Data for wild-type Pf1200–241 titration with mSin3B PAH2 (filled squares) are shown for comparison. The fitted curves are shown as continuous lines.
Table 2.
Thermodynamic Parameters for Binding Sin3 PAH2 or MRG15 MRG by Wild-type or Mutant Pf1200–241
| Reactants | KD (μM) | N | ΔH (kcal mol−1) | ΔS (cal mol−1 K−1) |
|---|---|---|---|---|
| wild-type Pf1 + mSin3A PAH2 | 2.15 ± 0.45 | 0.99 ± 0.05 | −6.58 ± 0.48 | 3.9 ± 2.0 |
| Pf1 F210A + mSin3A PAH2 | -a | -a | -a | -a |
| Pf1 L212A + mSin3A PAH2 | 2.66 ± 0.46 | 1.00 ± 0.11 | −4.31 ± 0.49 | 11.0 ± 2.0 |
| Pf1 L213A + mSin3A PAH2 | -a | -a | -a | -a |
| Pf1 A216V + mSin3A PAH2 | -a | -a | -a | -a |
| Pf1 N221E + mSin3A PAH2 | 0.63 ± 0.05 | 1.16 ± 0.08 | −7.67 ± 0.43 | 2.6 ± 1.6 |
| Pf1 F225A + mSin3A PAH2 | 1.65 ± 0.43b | 1.12 ± 0.05b | −4.97 ± 0.34b | 9.8 ± 1.7b |
| wild-type Pf1 + mSin3B PAH2 | 2.93 ± 0.41b | 1.02 ± 0.01b | −9.64 ± 0.50b | −6.9 ± 1.8b |
| wild-type Pf1 + MRG15 MRG | 0.015 ± 0.003 | 0.82 ± 0.08 | −13.10 ± 0.40 | −8.0 ± 1.5 |
no detectable binding, hence could not be quantified
average values from two independent measurements
We next asked whether the introduction of a negative charge at the last position of the motif would have an effect on enhancing the affinity of the interaction. Mutation of Asn221 to glutamate indeed enhanced the affinity but only by approximately 3-fold (Table 2). The more than one-order of magnitude difference in affinities between Mad1/Mxd1 (KD ~50 nM)36,37,42 and Pf1 (KD ~2 μM) interactions with mSin3A PAH2 suggests that a phenylalanine residue constitutes a less than an optimal choice at the first position of the Type I interaction motif. Indeed, the resonances of Phe210 and those of residues immediately in its vicinity are most profoundly affected by line broadening in the complex, to a substantially greater extent than those of residues further along the same helix, implying additional internal dynamics at this location. We also asked whether Phe225 – one of the invariant residues C-terminal to SID1 that interacted with both PAH2 and with other residues in Pf1 – was an important affinity determinant for the interaction. Curiously, the Phe225Ala mutant bound with marginally higher affinity than the wild-type protein but, as was noted for the Leu212Ala mutant, with a diminished binding enthalpy compensated by enhanced binding entropy (Figure 4(b); Table 2). This implies that a key distinctive feature of the mSin3A PAH2-Pf1 complex compared with the corresponding mSin3A PAH2-Mad1/Mxd1 complex is dispensable for complex stability.
A recent report showed that the Rpd3S/Sin3S complex was predominantly populated by the mSin3B paralog over mSin3A.11 ITC experiments were conducted to test whether the mSin3B PAH2 bound to Pf1200–241 with higher affinity than mSin3A PAH2. The mSin3B PAH2 bound Pf1200–241 with comparable affinity as mSin3A PAH2 (~3 μM versus ~2 μM) although the underlying enthalpic and entropic contributions were significantly different (Figure 4(b); Table 2). The basis for enthalpy-entropy compensation is unknown considering the close relationship between the proteins at the sequence level, particularly those residues that form the hydrophobic cleft that are virtually identical and hence both PAH2 domains would be expected to interact with Pf1200–241 in a similar fashion. However, we note that the PAH2 domains of mSin3A and mSin3B exhibit very different conformational and oligomerization properties in the apo-state with the former sampling both folded and partially unfolded conformations and existing in monomer-dimer equilibrium whereas the latter is monomeric and predominantly folded.38,41,43 These differences could potentially contribute to the differences in ΔH and ΔS for the two complexes.
Competition between MRG15 and mSin3A PAH2 for Pf1 SID1
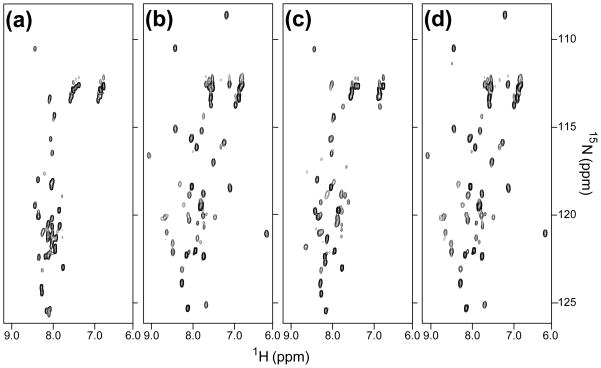
The MRG domain of the MRG15 subunit was previously implicated in interactions with the Pf1 linker segment connecting the two PHD domains.19 Given the poor length and sequence conservation of the linker region among Pf1 orthologs, except for the segment encompassing SID1 and the region immediately C-terminal to it (Figure 1(a)), we asked whether this conserved region might also be targeted by the MRG15 MRG domain. To test this possibility, 15N-labeled Pf1200–241 was titrated with unlabeled MRG15 MRG. The Pf1200–241 spectrum in the presence of an equivalent amount of MRG15 MRG showed a significant enhancement in chemical shift dispersion indicative of a specific interaction (Figure 5(a) and 5 (b)). In comparison, the increase in chemical shift dispersion upon titration of Pf1200–241 with an equivalent amount of mSin3A PAH2 was relatively modest (Figure 5(c)). We then asked whether both mSin3A PAH2 and MRG15 MRG domain could simultaneously interact with Pf1200–241. Surprisingly, the addition of equivalent amounts of both mSin3A PAH2 and MRG15 MRG to Pf1200–241 produced a Pf1 spectrum that was almost identical with the one recorded in the presence of MRG15 MRG alone. Identical results were obtained when MRG15 MRG was added to a mixture of Pf1200–241 and mSin3A PAH2 (data not shown). Consistent with these observations, ITC analysis of the MRG15 MRG-Pf1200–241 interaction revealed a >140-fold increase in affinity for this complex over the corresponding mSin3A PAH2- Pf1200–241 complex (Table 2). Collectively, these results imply that the MRG15 subunit competes, rather than collaborate, with mSin3A for this segment of Pf1.
Figure 5.
NMR titrations of Pf1200–241 with mSin3A PAH2 and MRG15 MRG polypeptides. 1H-15N correlated spectra of (a) apo-Pf1 and of Pf1 recorded in the presence of equimolar amounts of (b) MRG15 MRG, (c) mSin3A PAH2, or (d) both mSin3A PAH2 and MRG15 MRG are shown. All the spectra were recorded under identical solution conditions at pH 7 and 25°C. The NMR data were acquired, processed and displayed with identical parameters.
Discussion
Multi-protein coactivator and corepressor complexes have emerged as important players in eukaryotic transcription control, yet very little is known about how these complexes are assembled and organized. The evolutionarily-conserved Rpd3/Sin3 complexes are excellent model systems for addressing these types of questions although the much smaller size of and fewer subunits in the Rpd3S/Sin3S complex (five subunits but with only two unique subunits) makes it especially attractive for such investigations. In this study, the interactions involving three subunits of this complex were investigated.
Perhaps the most surprising finding to emerge from the present studies is the competition between the MRG15 and mSin3A subunits for a particularly well-conserved segment of Pf1 previously thought to harbor the binding site for the Sin3 PAH2 domain. The loss of contact between Sin3 PAH2 and Pf1 in the presence of MRG15 is unlikely to disrupt the Rpd3S/Sin3S complex, as Pf1-Sin3 interactions are multivalent involving multiple, distinct segments in the two proteins, in contrast to the monovalent interaction between Pf1 and MRG15.11,18 It is conceivable that the conserved segment of Pf1 has evolved to bind both Sin3 and MRG15 but in a mutually exclusive manner, perhaps at different stages of assembly. Given its scaffolding role, Pf1 likely binds to Sin3 in the early stages of Rpd3S/Sin3S complex assembly directing its proper assembly. During this stage, it may be important to occlude Sin3 PAH2 from aberrant recruitment by sequence-specific DNA-binding transcription factors. MRG15 may join the other proteins in the complex at the final step of assembly and at that point, protection of PAH2 from aberrant recruitment may be less important, as the holo-complex might have acquired its innate targeting specificity. We note that MRG15 is also found in multiple, unrelated multi-protein complexes involved in transcription regulation,44,45 recombination repair,46,47 and splicing,48 consistent with its non-scaffolding role.
But why use the same polypeptide sequence for two distinct functions, considering Pf1 is over 1000 residues in length? The answer is unknown but perhaps it might be the Pf1 segment that needs protection from competitors, as it contains a ubiquitous φ-x-x-φ-φ amphipathic helical motif found in transcription factor activation and repression domains.49 Interestingly, the two invariant alanine residues within the Type I PAH2-interaction motif are replaced by bulkier residues in yeast (Figure 1(a)), although the motif itself is preserved from fly to human (we note that the sequence requirements for PAH2-binding are unlikely to be different from yeast to human given the extraordinary level of sequence conservation of Sin3 PAH2 domains particularly those residues that comprise the hydrophobic cleft).37 This implies that the Sin3 PAH2-Pf1 SID1 interaction is most likely absent in yeast. Perhaps, unlike in yeast, the much larger size of the metazoan proteome warrants protection of segments/domains from competitors. Accretion of new functions in orthologous proteins during the course of molecular evolution is not uncommon. For example, the SAP30 subunit of the Rpd3L/Sin3L complex harbors a zinc finger motif that is found in species ranging from fly to human but is notably absent in yeast.50
Two previous studies came to somewhat different conclusions about the importance of the Sin3 PAH2-Pf1 SID1 interaction.11,18 A possible explanation for the seeming disparity between the early and the more recent studies is that the former evaluated the importance of the residues in the Pf1 SID1 region and deemed them to be crucial for Sin3 recruitment whereas the latter tested the requirement of an intact PAH2 domain for complex assembly and found it to be non-essential for this process. Both results are consistent with the results described herein. Our studies point to a role for Pf1 SID1 in not only binding to Sin3 PAH2, possibly in the early steps of complex assembly, but also in MRG15 binding; PAH2 thus may be non-essential for the maturation of the complex.
Our studies also indicate that binding to the MRG15 MRG domain has overlapping sequence and structural requirements as the mSin3A PAH2 domain. However, given the lack of structural similarity between the MRG and PAH2 domains,51,52 the high helical content of these domains notwithstanding, the modes of Pf1 binding are likely to be different. The extraordinary sequence conservation immediately C-terminal to the SID1 motif, suggests that this region might especially play a critical role in MRG binding. An extended protein-protein interface could explain the higher affinity of the MRG15 MRG-Pf1 interaction compared to the mSin3A PAH2-Pf1 interaction. Additional studies are needed to clarify the molecular basis of this interaction.
Intrinsically unstructured/disordered regions are found in a broad range of eukaryotic proteins and are especially common in proteins that regulate basic cellular processes such as transcription.53–55 Pf1 SID1 represents a classic example of a largely unstructured/disordered segment that undergoes folding (into a helix in this case) upon binding to its target. This has also been witnessed for other PAH-binders but what is especially interesting about Pf1 SID1 is that it can bind to two essentially unrelated targets. Indeed, this is viewed as a key advantage enjoyed by polypeptide segments that intrinsically lacked well-defined three-dimensional shapes, as they could instead adopt diverse shapes adapting themselves to their target.
The structure of the mSin3A PAH2-Pf1 complex adds important knowledge regarding how PAH domains, the PAH2 domain in particular, interact with their targets. As expected, Pf1 SID1 adopts a helical conformation and binds to the hydrophobic cleft via residues that comprise the Type I PAH2-interaction motif. However, in spite of the larger solvent-accessible surface buried by the two proteins in this complex compared to the mSin3A PAH2-Mad1/Mxd1 complex, the interaction is of more than 10-fold lower affinity, likely due to a (sub-optimal) phenylalanine residue at the first position of the motif. The conservation of a sub-optimal residue at this position might be to allow the MRG15 MRG domain to compete efficiently with mSin3A PAH2 for Pf1. Finally, Pf1 SID1 is not the only interactor that exhibits conformational plasticity; a subset of the mSin3A PAH2 residues at the interface and beyond, particularly in the α1 and α4 helices, also exhibit side chain packing diversity in the various complexes.24 These are the same regions that also show conformational diversity in apo-mSin3A PAH2.41
Materials and Methods
Production of mSin3A PAH2, mSin3B PAH2 and Pf1200–241 Polypeptides
The coding sequences of mammalian Sin3A PAH2 (residues 295–385) and human Pf1200–241 (residues 200–241) were amplified by PCR and inserted into the pMCSG21 and pMCSG7 expression vectors, respectively. The sub-cloned gene segments were confirmed by DNA sequencing. E. coli BL21(DE3) cells (Novagen, WI) co-transformed with equimolar amounts of the plasmids were grown at 37°C in Terrific Broth (Invitrogen, CA) for improved protein yields. Protein expression was induced with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at 20°C when the OD600 nm reached ~1.2; cells were harvested 12 h thereafter. Cell pellets were resuspended in 20 mM Tris buffer (pH 8) containing 0.2 M NaCl, 5 mM Tris (2-carboxy-ethyl) phosphine hydrochloride (TCEP), 8 M urea, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 μM leupeptin, 1 mM pepstatin, and 0.1% Triton X-100. The cells were lysed using a sonicator and centrifuged and the supernatant was incubated with His-Select Ni2+-resin (Sigma-Aldrich, MO) for 30 min. The resin was washed extensively with 20 mM Tris buffer (pH 8) containing 0.2 M NaCl and 5 mM TCEP followed by and incubated with tobacco etch virus (TEV) protease at 22°C for 4 h and subsequently at 4°C overnight. The cleaved protein was collected and purified to homogeneity via reversed-phase HPLC using a C18 or C8 column (Grace Vydac, CA) and a linear gradient of 0.1% trifluoroacetic acid (TFA) and 0.1% TFA in 80% acetonitrile and the protein-containing fractions were frozen and lyophilized. Uniformly 15N- and/or 13C-labeled proteins were produced using the same procedure except that the cells were grown in M9 minimal medium containing 15N-ammonium sulfate and/or 13C-D-glucose (Cambridge Isotopes, MA), respectively. The identities of the proteins and the extent of isotope enrichment were established by electrospray ionization mass spectrometry (ESI-MS).
Pf1200–241 mutants Phe210Ala, Leu212Ala, Leu213Ala, Ala216Val, Asn221Glu and Phe225Ala, for in vitro binding assays were generated using the QuikChange site-directed mutagenesis protocol (Agilent, CA). All mutations were confirmed by DNA sequencing. Mutant proteins were co-expressed and co-purified with mSin3A PAH2 in an analogous manner to the wild-type Pf1200–241 protein. Mammalian Sin3B PAH2 was expressed and purified as described previously.
Production of the MRG15 MRG Polypeptide
The gene sequence encoding the human MRG15 MRG domain (residues 155–323) was amplified by PCR and inserted into the pMCSG7 expression vector. The sub-cloned gene segment was confirmed by DNA sequencing. The plasmid was transformed in E. coli BL21(DE3) cells and grown at 37°C in Luria-Bertani medium. Protein expression was induced at 20°C by adding 1 mM of IPTG when the OD600 nm reached ~0.8; cells were harvested ~12 hours thereafter. Cell pellets were suspended in 50 mM Tris buffer (pH 8) containing 0.5 M NaCl, 5 mM TCEP, 1 mM PMSF, 1 μM leupeptin, 1 mM pepstatin and 0.1% Triton X-100. The cells were lysed using a sonicator and centrifuged. The supernatant was loaded onto a His-Select Ni2+-resin and incubated at 4°C for 40 min. The resin was washed with 50 mM Tris buffer (pH 8) containing 0.5 M NaCl and 5 mM TCEP followed by incubation with TEV protease at 22°C for 4 h and at 4°C overnight. The cleaved protein was collected, concentrated and further purified by size-exclusion chromatography using a Superdex 75 column (GE Healthcare, NJ) pre-equilibrated with 50 mM Tris buffer (pH 8) containing 5 mM TCEP and 0.2 M NaCl. Fractions containing the purified protein were pooled, concentrated and stored at 4°C for NMR studies. Protein identity and integrity was evaluated by ESI-MS and SDS-PAGE gel electrophoresis.
Isothermal Titration Calorimetry
Isothermal titration calorimetry experiments were performed on a Microcal iTC200 calorimeter (GE Healthcare, NJ). Titrations for each protein pair were performed in triplicate (unless noted otherwise) at 25°C in 20 mM sodium phosphate buffer (pH 7.5). Proteins were dialyzed overnight against the buffer used for the titrations. Protein concentrations were determined spectrophotometrically.56 Wild-type and mutant Pf1200–241 proteins were kept in the cell while the Sin3 PAH2 polypeptides were kept in the syringe at initial concentrations of 20 μM and 0.25 mM, respectively. Titrations between MRG15 MRG and Pf1200–241 were conducted with the former polypeptide in the cell at an initial concentration of 15 μM and the latter polypeptide in the syringe at 0.20 mM. Binding isotherms were analyzed assuming a single-site binding model using the Origin 7.0 software provided by the manufacturer.
mSin3A PAH2-Pf1200–241 Complex Generation and NMR Sample Preparation
NMR samples of mSin3A PAH2 and Pf1200–241 were prepared by dissolving the respective dry, lyophilized protein powders in 20 mM sodium phosphate buffer (pH 6.0) containing 5 mM dithiothreitol-d10, 10% D2O and 0.2% NaN3. The mSin3A PAH2-Pf1200–241 complex was generated by titrating 15N- and/or 13C-labeled mSin3A PAH2 or Pf1200–241 with unlabeled Pf1200–241 or mSin3A PAH2, respectively, until an equimolar ratio was attained. The progress of each titration was monitored by recording 2D 1H-15N HSQC spectra and noting the complete disappearance of correlations arising from the free labeled protein. Protein concentrations were determined spectrophotometrically.56 Samples of the protein-protein complex were lyophilized and re-dissolved in 99.996% D2O for experiments conducted in D2O (Sigma-Aldrich, MO).
NMR Titrations with MRG15 MRG Polypeptide
15N-labeled Pf1200–241 was mixed with equimolar amounts of MRG15 MRG at a concentration of 20 μM at pH 8.0. The sample of the resulting complex was concentrated and exchanged into NMR buffer (20 mM sodium phosphate buffer (pH 7.0) containing 5 mM dithiothreitol-d10, 10% D2O and 0.2% NaN3). Complex formation was confirmed by 2D 1H-15N HSQC spectra. mSin3A PAH2 in dry, lyophilized powder form was dissolved in NMR buffer at equimolar ratio with the MRG15 MRG-Pf1200–241 complex. Sample concentrations used for recording NMR spectra were in the 0.25 mM range. Control spectra for apo-Pf1200–241 and for the mSin3A PAH2-Pf1200–241 complex were recorded under the same solution conditions as for the samples of the MRG15 MRG complexes.
NMR Spectroscopy and Structure Determination
NMR data were acquired on a Varian Inova 600 MHz spectrometer equipped with a pulsed-field-gradient triple-resonance cold probe at 25°C. The concentrations used for the structure determination were in the range of 0.75 to 1.5 mM. NMR data processing and analysis were performed using NMRPipe57 or Felix 98.0 (Accelrys) and Sparky,58 respectively. Backbone 1H, 15N and 13C resonance assignments for apo-Pf1200–241 were obtained by analyzing 3D HNCA, HN(CO)CA, CBCA(CO)NH, HNCACB and HNCO spectra.59,60 Backbone and side chain 1H, 15N and 13C resonance assignments for the mSin3A PAH2-Pf1200–241 complex were obtained by analyzing 3D CBCA(CO)NH, HNCACB, C(CO)NH-TOCSY, HNCO, HCCH-COSY and HCCH-TOCSY spectra.59,60 Aromatic resonances were assigned based on a careful analysis of 2D 15N,13C-double half-filtered NOESY and 1H-13C HSQC spectra.61
Backbone φ and ψ dihedral angle restraints for structure calculations were derived from a combined analysis of the Hα, Cα, Cβ, C′ and backbone 15N chemical shifts using TALOS+.62 Only residues with TALOS+ reliability scores of 10 in helical segments were restrained. NOE-based distance restraints were derived from two sets of four spectra recorded for each protein in the complex including 3D 15N-edited NOESY (τm = 75 ms) recorded in H2O, 3D 13C-filtered, 13C-edited NOESY (τm = 140 ms), 3D 13C-edited aliphatic NOESY (τm = 60 ms) and 2D 15N,13C-double half-filtered NOESY (τm = 60 ms) recorded in D2O.
Structures were determined using ARIA 1.263 in conjunction with CNS 1.1.64 Candidate conformers were calculated via torsion angle dynamics and simulated annealing starting from an initial structure with extended backbone conformations. All NOEs were calibrated automatically and assigned iteratively by ARIA; the assignments were checked manually for errors after every run. Eighty conformers were calculated out of which the 40 conformers with the lowest restraint energies were refined in a shell of water and the 20 conformers with the lowest restraint energies, restraint violations, and RMS deviations from the ideal covalent geometry were selected for further analysis. The final conformers were analyzed using CNS,64 PROCHECK,65 MONSTER,66 DeepView67 and awk scripts written in-house. Molecular images were generated using RIBBONS68 and GRASP.69
Supplementary Material
Acknowledgments
We thank Chetan Velagapudi for contributions towards the development of this work and the members of the Radhakrishnan lab for useful discussions. This work was supported by a grant from the NIH to I.R. (R01 GM064715). Access to instrumentation in the Keck Biophysics Facility and the WCAS Biological NMR Center as well as support for structural biology research from the Robert H. Lurie Comprehensive Cancer Center at Northwestern are gratefully acknowledged.
Abbreviations
- SID
Sin3 interaction domain
- PAH
paired amphipathic helix
- HDAC
histone deacetylase
- MRG
mortality factor on chromosome 4 related gene
Footnotes
Accession numbers. The RCSB PDB accession code for atomic coordinates of the mSin3A PAH2-Pf1200–241 complex is 2L9S and the BMRB code for chemical shifts is 17485.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gardner KE, Allis CD, Strahl BD. OPERating ON Chromatin, a Colorful Language where Context Matters. Journal of Molecular Biology. 2011 doi: 10.1016/j.jmb.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Hayakawa T, Nakayama J. Physiological roles of class I HDAC complex and histone demethylase. J Biomed Biotechnol. 2011;2011:129383. doi: 10.1155/2011/129383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grzenda A, Lomberk G, Zhang JS, Urrutia R. Sin3: master scaffold and transcriptional corepressor. Biochimica et Biophysica Acta. 2009;1789:443–50. doi: 10.1016/j.bbagrm.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silverstein RA, Ekwall K. Sin3: a flexible regulator of global gene expression and genome stability. Curr Genet. 2005;47:1–17. doi: 10.1007/s00294-004-0541-5. [DOI] [PubMed] [Google Scholar]
- 6.McDonel P, Costello I, Hendrich B. Keeping things quiet: roles of NuRD and Sin3 co-repressor complexes during mammalian development. International Journal of Biochemistry and Cell Biology. 2009;41:108–16. doi: 10.1016/j.biocel.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunliffe VT. Eloquent silence: developmental functions of Class I histone deacetylases. Current Opinion in Genetics and Development. 2008;18:404–10. doi: 10.1016/j.gde.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–92. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 9.Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–4. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- 10.Li B, Jackson J, Simon MD, Fleharty B, Gogol M, Seidel C, Workman JL, Shilatifard A. Histone H3 lysine 36 dimethylation (H3K36me2) is sufficient to recruit the Rpd3s histone deacetylase complex and to repress spurious transcription. Journal of Biological Chemistry. 2009;284:7970–6. doi: 10.1074/jbc.M808220200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jelinic P, Pellegrino J, David G. A novel mammalian complex containing Sin3B mitigates histone acetylation and RNA polymerase II progression within transcribed loci. Molecular and Cellular Biology. 2011;31:54–62. doi: 10.1128/MCB.00840-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicolas E, Yamada T, Cam HP, Fitzgerald PC, Kobayashi R, Grewal SI. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat Struct Mol Biol. 2007;14:372–80. doi: 10.1038/nsmb1239. [DOI] [PubMed] [Google Scholar]
- 13.Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Molecular Cell. 2005;20:971–8. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 14.Carrozza MJ, Florens L, Swanson SK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Stable incorporation of sequence specific repressors Ash1 and Ume6 into the Rpd3L complex. Biochim Biophys Acta. 2005;1731:77–87. doi: 10.1016/j.bbaexp.2005.09.005. discussion 75–6. [DOI] [PubMed] [Google Scholar]
- 15.Fleischer TC, Yun UJ, Ayer DE. Identification and characterization of three new components of the mSin3A corepressor complex. Mol Cell Biol. 2003;23:3456–67. doi: 10.1128/MCB.23.10.3456-3467.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Sun ZW, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M, Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol Cell. 1998;1:1021–31. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]
- 17.Laherty CD, Yang WM, Sun JM, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell. 1997;89:349–56. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 18.Yochum GS, Ayer DE. Pf1, a novel PHD zinc finger protein that links the TLE corepressor to the mSin3A-histone deacetylase complex. Mol Cell Biol. 2001;21:4110–8. doi: 10.1128/MCB.21.13.4110-4118.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yochum GS, Ayer DE. Role for the mortality factors MORF4, MRGX, and MRG15 in transcriptional repression via associations with Pf1, mSin3A, and Transducin-Like Enhancer of Split. Molecular and Cellular Biology. 2002;22:7868–76. doi: 10.1128/MCB.22.22.7868-7876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun B, Hong J, Zhang P, Dong X, Shen X, Lin D, Ding J. Molecular basis of the interaction of Saccharomyces cerevisiae Eaf3 chromo domain with methylated H3K36. Journal of Biological Chemistry. 2008;283:36504–12. doi: 10.1074/jbc.M806564200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu C, Cui G, Botuyan MV, Mer G. Structural basis for the recognition of methylated histone H3K36 by the Eaf3 subunit of histone deacetylase complex Rpd3S. Structure. 2008;16:1740–50. doi: 10.1016/j.str.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McArthur GA, Laherty CD, Queva C, Hurlin PJ, Loo L, James L, Grandori C, Gallant P, Shiio Y, Hokanson WC, Bush AC, Cheng PF, Lawrence QA, Pulverer B, Koskinen PJ, Foley KP, Ayer DE, Eisenman RN. The Mad protein family links transcriptional repression to cell differentiation. Cold Spring Harb Symp Quant Biol. 1998;63:423–33. doi: 10.1101/sqb.1998.63.423. [DOI] [PubMed] [Google Scholar]
- 23.Schreiber-Agus N, DePinho RA. Repression by the Mad(Mxi1)-Sin3 complex. Bioessays. 1998;20:808–18. doi: 10.1002/(SICI)1521-1878(199810)20:10<808::AID-BIES6>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 24.Swanson KA, Knoepfler PS, Huang K, Kang RS, Cowley SM, Laherty CD, Eisenman RN, Radhakrishnan I. HBP1 and Mad1 repressors bind the Sin3 corepressor PAH2 domain with opposite helical orientations. Nat Struct Mol Biol. 2004;11:738–46. doi: 10.1038/nsmb798. [DOI] [PubMed] [Google Scholar]
- 25.Zhang JS, Moncrieffe MC, Kaczynski J, Ellenrieder V, Prendergast FG, Urrutia R. A conserved alpha-helical motif mediates the interaction of Sp1-like transcriptional repressors with the corepressor mSin3A. Mol Cell Biol. 2001;21:5041–9. doi: 10.1128/MCB.21.15.5041-5049.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wotton D, Knoepfler PS, Laherty CD, Eisenman RN, Massague J. The Smad transcriptional corepressor TGIF recruits mSin3. Cell Growth Diff. 2001;12:457–63. [PubMed] [Google Scholar]
- 27.Naruse Y, Aoki T, Kojima T, Mori N. Neural restrictive silencer factor recruits mSin3 and histone deacetylase complex to repress neuron-specific target genes. Proc Natl Acad Sci U S A. 1999;96:13691–6. doi: 10.1073/pnas.96.24.13691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Y, Myers SJ, Dingledine R. Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat Neurosci. 1999;2:867–72. doi: 10.1038/13165. [DOI] [PubMed] [Google Scholar]
- 29.Hurlin PJ, Queva C, Eisenman RN. Mnt: a novel Max-interacting protein and Myc antagonist. Curr Top Microbiol Immunol. 1997;224:115–21. doi: 10.1007/978-3-642-60801-8_11. [DOI] [PubMed] [Google Scholar]
- 30.Romm E, Nielsen JA, Kim JG, Hudson LD. Myt1 family recruits histone deacetylase to regulate neural transcription. J Neurochem. 2005;93:1444–53. doi: 10.1111/j.1471-4159.2005.03131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Q, Kong Y, Rothermel B, Garry DJ, Bassel-Duby R, Williams RS. The winged-helix/forkhead protein myocyte nuclear factor beta (MNF-β) forms a co-repressor complex with mammalian sin3B. Biochem J. 2000;345(Pt 2):335–43. [PMC free article] [PubMed] [Google Scholar]
- 32.Melnick AM, Westendorf JJ, Polinger A, Carlile GW, Arai S, Ball HJ, Lutterbach B, Hiebert SW, Licht JD. The ETO protein disrupted in t(8;21)-associated acute myeloid leukemia is a corepressor for the promyelocytic leukemia zinc finger protein. Mol Cell Biol. 2000;20:2075–86. doi: 10.1128/mcb.20.6.2075-2086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim H, Lee JE, Cho EJ, Liu JO, Youn HD. Menin, a tumor suppressor, represses JunD-mediated transcriptional activity by association with an mSin3A-histone deacetylase complex. Cancer Res. 2003;63:6135–9. [PubMed] [Google Scholar]
- 34.Chen X, Bieker JJ. Stage-specific repression by the EKLF transcriptional activator. Mol Cell Biol. 2004;24:10416–24. doi: 10.1128/MCB.24.23.10416-10424.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laherty CD, Billin AN, Lavinsky RM, Yochum GS, Bush AC, Sun JM, Mullen TM, Davie JR, Rose DW, Glass CK, Rosenfeld MG, Ayer DE, Eisenman RN. SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol Cell. 1998;2:33–42. doi: 10.1016/s1097-2765(00)80111-2. [DOI] [PubMed] [Google Scholar]
- 36.Sahu SC, Swanson KA, Kang RS, Huang K, Brubaker K, Ratcliff K, Radhakrishnan I. Conserved themes in target recognition by the PAH1 and PAH2 domains of the Sin3 transcriptional corepressor. Journal of Molecular Biology. 2008;375:1444–56. doi: 10.1016/j.jmb.2007.11.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brubaker K, Cowley SM, Huang K, Loo L, Yochum GS, Ayer DE, Eisenman RN, Radhakrishnan I. Solution structure of the interacting domains of the Mad-Sin3 complex: implications for recruitment of a chromatin-modifying complex. Cell. 2000;103:655–65. doi: 10.1016/s0092-8674(00)00168-9. [DOI] [PubMed] [Google Scholar]
- 38.van Ingen H, Lasonder E, Jansen JF, Kaan AM, Spronk CA, Stunnenberg HG, Vuister GW. Extension of the binding motif of the Sin3 interacting domain of the Mad family proteins. Biochemistry. 2004;43:46–54. doi: 10.1021/bi0355645. [DOI] [PubMed] [Google Scholar]
- 39.Spronk CA, Tessari M, Kaan AM, Jansen JF, Vermeulen M, Stunnenberg HG, Vuister GW. The Mad1-Sin3B interaction involves a novel helical fold. Nature Structural Biology. 2000;7:1100–4. doi: 10.1038/81944. [DOI] [PubMed] [Google Scholar]
- 40.Nomura M, Uda-Tochio H, Murai K, Mori N, Nishimura Y. The neural repressor NRSF/REST binds the PAH1 domain of the Sin3 corepressor by using its distinct short hydrophobic helix. J Mol Biol. 2005;354:903–15. doi: 10.1016/j.jmb.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Zhang Z, Demeler B, Radhakrishnan I. Coupled unfolding and dimerization by the PAH2 domain of the mammalian Sin3A corepressor. J Mol Biol. 2006;360:7–14. doi: 10.1016/j.jmb.2006.04.069. [DOI] [PubMed] [Google Scholar]
- 42.Cowley SM, Kang RS, Frangioni JV, Yada JJ, DeGrand AM, Radhakrishnan I, Eisenman RN. Functional analysis of the Mad1-mSin3A repressor-corepressor interaction reveals determinants of specificity, affinity, and transcriptional response. Mol Cell Biol. 2004;24:2698–709. doi: 10.1128/MCB.24.7.2698-2709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He Y, Radhakrishnan I. Solution NMR studies of apo-mSin3A and mSin3B reveal that the PAH1 and PAH2 domains are structurally independent. Protein Sci. 2008;17:171–5. doi: 10.1110/ps.073097308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doyon Y, Selleck W, Lane WS, Tan S, Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Molecular and Cellular Biology. 2004;24:1884–96. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pardo PS, Leung JK, Lucchesi JC, Pereira-Smith OM. MRG15, a novel chromodomain protein, is present in two distinct multiprotein complexes involved in transcriptional activation. Journal of Biological Chemistry. 2002;277:50860–6. doi: 10.1074/jbc.M203839200. [DOI] [PubMed] [Google Scholar]
- 46.Hayakawa T, Zhang F, Hayakawa N, Ohtani Y, Shinmyozu K, Nakayama J, Andreassen PR. MRG15 binds directly to PALB2 and stimulates homology-directed repair of chromosomal breaks. Journal of Cell Science. 2010;123:1124–30. doi: 10.1242/jcs.060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sy SM, Huen MS, Chen J. MRG15 is a novel PALB2-interacting factor involved in homologous recombination. Journal of Biological Chemistry. 2009;284:21127–31. doi: 10.1074/jbc.C109.023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Plevin MJ, Mills MM, Ikura M. The LxxLL motif: a multifunctional binding sequence in transcriptional regulation. Trends Biochem Sci. 2005;30:66–9. doi: 10.1016/j.tibs.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 50.He Y, Imhoff R, Sahu A, Radhakrishnan I. Solution structure of a novel zinc finger motif in the SAP30 polypeptide of the Sin3 corepressor complex and its potential role in nucleic acid recognition. Nucleic Acids Res. 2009;37:2142–52. doi: 10.1093/nar/gkp051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang P, Zhao J, Wang B, Du J, Lu Y, Chen J, Ding J. The MRG domain of human MRG15 uses a shallow hydrophobic pocket to interact with the N-terminal region of PAM14. Protein Science. 2006;15:2423–34. doi: 10.1110/ps.062397806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bowman BR, Moure CM, Kirtane BM, Welschhans RL, Tominaga K, Pereira-Smith OM, Quiocho FA. Multipurpose MRG domain involved in cell senescence and proliferation exhibits structural homology to a DNA-interacting domain. Structure. 2006;14:151–8. doi: 10.1016/j.str.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 53.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. Journal of Molecular Biology. 1999;293:321–31. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 54.Wright PE, Dyson HJ. Linking folding and binding. Current Opinion in Structural Biology. 2009;19:31–8. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nature Reviews Molecular Cell Biology. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 56.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–26. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 57.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 58.Goddard TD, Kneller DG. Sparky. 2004;3 http://www.cgl.ucsf.edu/home/sparky/
- 59.Ferentz AE, Wagner G. NMR spectroscopy: a multifaceted approach to macromolecular structure. Q Rev Biophys. 2000;33:29–65. doi: 10.1017/s0033583500003589. [DOI] [PubMed] [Google Scholar]
- 60.Bax A, Grzesiek S. Methodological advances in protein NMR. Accounts Chem Res. 1993;26:131–138. [Google Scholar]
- 61.Otting G, Wüthrich K. Heteronuclear filters in two-dimensional [1H,1H]-NMR spectroscopy: combined use with isotope labelling for studies of macromolecular conformation and intermolecular interactions. Q Rev Biophys. 1990;23:39–96. doi: 10.1017/s0033583500005412. [DOI] [PubMed] [Google Scholar]
- 62.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. Journal of Biomolecular NMR. 2009;44:213–23. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Linge JP, Habeck M, Rieping W, Nilges M. ARIA: automated NOE assignment and NMR structure calculation. Bioinformatics. 2003;19:315–6. doi: 10.1093/bioinformatics/19.2.315. [DOI] [PubMed] [Google Scholar]
- 64.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 65.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–86. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 66.Salerno WJ, Seaver SM, Armstrong BR, Radhakrishnan I. MONSTER: inferring non-covalent interactions in macromolecular structures from atomic coordinate data. Nucleic Acids Res. 2004;32:W566–8. doi: 10.1093/nar/gkh434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–23. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 68.Carson M. Macromolecular Crystallography. Pt B. Vol. 277. Academic Press Inc; San Diego: 1997. Ribbons; pp. 493–505. [Google Scholar]
- 69.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–96. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.