Abstract
Aim of the study.
Ethnobotanically-driven drug-discovery programs include data related to many aspects of the preparation of botanical medicines, from initial plant collection to chemical extraction and fractionation. The Traditional Medicine-Collection Tracking System (TM-CTS) was created to organize and store data of this type for an international collaborative project involving the systematic evaluation of commonly used Traditional Chinese Medicinal plants.
Materials and Methods.
The system was developed using domain-driven design techniques, and is implemented using Java, Hibernate, PostgreSQL, Business Intelligence and Reporting Tools (BIRT), and Apache Tomcat.
Results.
The TM-CTS relational database schema contains over 70 data types, comprising over 500 data fields. The system incorporates a number of unique features that are useful in the context of ethnobotanical projects such as support for information about botanical collection, method of processing, quality tests for plants with existing pharmacopoeia standards, chemical extraction and fractionation, and historical uses of the plants. The database also accommodates data provided in multiple languages and integration with a database system built to support high throughput screening based drug discovery efforts. It is accessed via a web-based application that provides extensive, multi-format reporting capabilities.
Conclusions.
This new database system was designed to support a project evaluating the bioactivity of Chinese medicinal plants. The software used to create the database is open source, freely available, and could potentially be applied to other ethnobotanically-driven natural product collection and drug-discovery programs.
Keywords: Database, Traditional Chinese Medicine, High Throughput Screening, Ethnobotany, Drug Discovery
1. Introduction
Recent survey results have demonstrated that 83 million adults in the United States use some form of complementary and alternative medicine, spending a total of $14.8 billion dollars per year on nonmineral, nonvitamin natural products, the majority of which consist of herbal medicines (Nahin et al., 2009). In light of these trends, an international collaborative project was initiated to create a screening library of more than 200 species of plants and fungi commonly used in Traditional Chinese Medicine (Eisenberg et al., 2011). The primary goal of the project was to evaluate this group of medicinal plants and fungi for their pharmacological properties and potential as novel therapeutics using high throughput screening techniques (“The Academic Pursuit of Screening,” 2007). The Traditional Medicine Collection Tracking System (TM-CTS) was created to support some of the data requirements of this project. The main objectives of the TM-CTS were to establish an information structure to support the inventory and tracking needs of the project, to ensure that the work was recorded in such a way that plants, plant extracts, and fractions could be prepared as reproducibly as possible should recollection be required in the future, and to share the data among an international group of collaborators. This paper briefly describes the rationale, content, methods, and implementation of the TM-CTS.
Botanical medicines, like the natural populations of plants they come from, can be variable in many aspects. Consequently, drug-discovery programs based on traditional botanical medicines should incorporate data from all aspects of medicinal plant preparation with the dual goals of ensuring transparency and reproducibility to the maximum extent feasible. Considerable chemical variation exists in natural populations of plants and fungi (e.g., Binns et al., 2002) and that variation can have pharmacological consequences (e.g., Toselli et al., 2009). Additional variation in ethnobotanical medicines and plant-based products may be introduced as a result of differences in methods of cultivation, harvesting, and processing (e.g., Jeffery et al., 2003). Also, the production of ethnobotanical medicines may be guided by existing standards, such as those in published pharmacopoeias (e.g., Chinese Pharmacopoeia Commission, 2005) and can have additional accessory information, such as a detailed history of use in multiple languages (e.g., Traditional Chinese Medicine, Ayurvedic medicine). As a result, drug-discovery programs based on traditional botanical knowledge have particular data requirements in addition to those shared with projects involving plant collection or biodiversity-based natural product screening.
During the initial development phase of the TM-CTS, several existing database solutions were evaluated for their suitability. These included databases that support natural products research (e.g., NAPIS, White Point Systems), biological collections (e.g., Specify Software Project), and information about ethnobotanical uses (e.g., Ji et al., 2005). However, results of an early assessment indicated that some key-feature requirements would be best addressed by the creation of a custom database for the project. In particular, the requirement for detailed records of the processing of specimens, tracking of shipments, as well as incorporation of a number of different types of quality assessment and authentication analyses led to the decision to develop a custom solution. In addition, the project required Chinese textual data and English translations to be stored together. Finally, because the project was done in collaboration with investigators using high throughput screening technology, there was a need to federate the TM-CTS database with an existing database used for project-related screening activities and experimental result data. The Screensaver database system used to support the high throughput screening platform is described in Tolopko et al. (2010).
2. Database Content
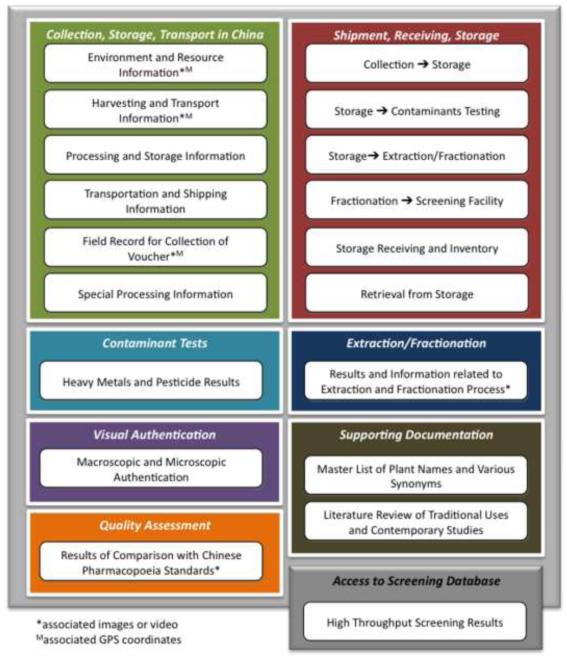
The TM-CTS was created to support information requirements related to project activities from plant cultivation and collection through to fractionation of plant extracts. The Chinese medicinal plant project ultimately included more than 550 plant samples representing 202 species collected from locations throughout China. In most cases up to three samples per species were collected for the project to account for variation in plant quality. Through the course of the project, bulk samples of the medicinal material as well as voucher specimens (Hildreth et al., 2007) were collected in areas traditionally known for the production of the medicinal material. Bulk medicinal materials were processed according to traditional methods, and tested for quality according to standards specified by the 2005 edition of the Pharmacopoeia of the People’s Republic of China (Chinese Pharmacopoeia Commission, 2005). Plants were authenticated according to microscopic and macroscopic visual characteristics (e.g. Zhao et al., 2007), and in some cases by DNA barcoding (CBOL Plant Working Group, 2009). These multiple authentication steps (visual, chemical, microscopic) were done to ensure both taxonomic identity and quality. The authentication status (pass/fail) and details of the results were recorded, and only plants that passed these authentication steps were sent for further testing. Plants were then shipped to the United States where they were placed in storage and tested for the presence of heavy metal and pesticide contaminants. Following the tests for contaminants, appropriate plant samples were selected for extraction and fractionation. All fractions were then aliquoted into 96- and 384-well microplate format for high throughput screening. In addition to these data, literature reviews were also created during the course of the project to provide detailed background information on the traditional uses of each plant. All text-based data were needed in both English and Chinese to support use by research collaborators in the United States and China. The content of the TM-CTS is graphically summarized in Fig. 1. A more detailed summary of the various aspects of the project is provided in Eisenberg et al. (2011).
Fig. 1.
Summary of data content included in the Traditional Medicine Collection Tracking System (TM-CTS). TM-CTS supports data related to all aspects of an ethnobotanically-driven drug discovery program from plant cultivation and collection through to fractionation of plant extracts. The database currently includes data from plant collections, authentication and quality tests, shipment, extraction, and fractionation. The database is designed to work in conjunction with a screening database (Screensaver; Tolopko et al., 2010) that stores information related to high throughput screens of the plant fractions.
3. Software Design and Implementation
3.1 Domain Model
The TM-CTS was created using domain-driven design techniques (Evans, 2003), producing a software-based domain model that reflects the real-world entities and activities that comprise the research project. This domain model defines the relationships, constraints, and behaviors of these entities and processes, and is used to generate the underlying relational database schema that is used to store the system’s data.
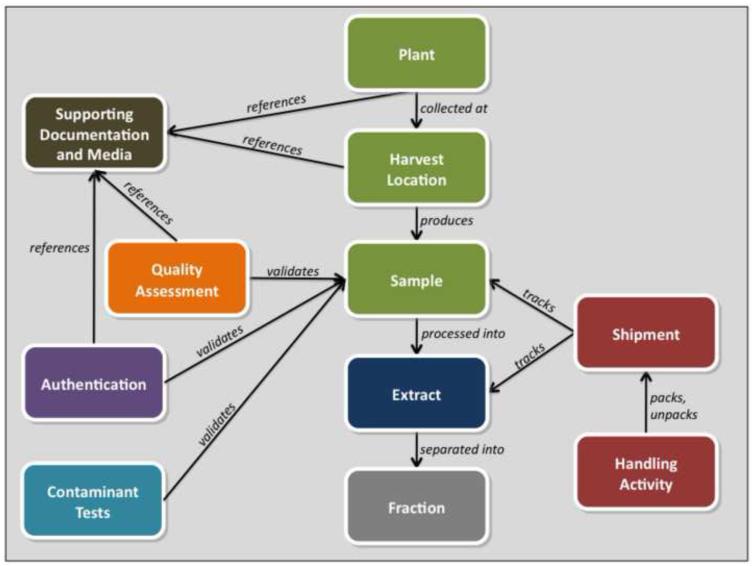
A simplified domain model diagram shows the high-level domain concepts that form the basis of the TM-CTS design (Fig. 2). A Plant serves as the primary entity in the model and identifies a particular species that is to be collected, tracked, processed, and screened. A plant is collected from a Harvest Location, which records the precise geographic location and environmental conditions of the harvesting activity. A harvested plant is divided into Samples that can then be independently shipped, tracked, and inventoried by the system. Quality Assessment, Authentication, and Contaminant Tests are associated with samples. Samples are processed into Extracts that are separated into Fractions.
Fig. 2.
Simplified object model for the TM-CTS indicating the high-level domain concepts used in database design and their relationships with one another. For example, the Plant entity is collected at a Harvest Location and produces a Sample that can be extracted and fractionated. Supporting Documentation and Media can be referenced by a number of different domain entities, including the Plant, Harvest Location, Quality Assessment, and Visual Authentication. Further explanation of these relationships is provided in the text. The color scheme in this model correlates to the colors used in Fig. 1.
These fractions are aliquoted into libraries for use in high throughput screening based drug-discovery efforts. The Shipment and Handling Activity entities record detailed tracking and storage information for the samples and the extracts. Supporting Documentation and Media, in the form of document, image, or video files that can be uploaded into TM-CTS, may be referenced by plant, harvest locations, quality assessment and authentication records. Over 500 data fields detailing plant specimen acquisition, verification, tracking and fractionation are maintained across the entire domain model. Many data fields are stored in duplicate in order to record the original Chinese textual data and their associated English translations.
3.2 Technologies
TM-CTS uses free, open source software technologies that were chosen because of their flexibility and extensibility. TM-CTS is implemented as a Java-based web application that can be deployed to an Apache Tomcat (http://tomcat.apache.org/) servlet container. Data are stored in a PostgreSQL (8.3) database (http://www.postgresql.org/) and can therefore be accessed from any external software system that can achieve database connectivity. The Hibernate framework (http://www.hibernate.org/) is used for the system’s object-relational mapping layer and provides persistence operations for the Java-implemented domain model. The Eclipse Business Intelligence and Reporting Tools (BIRT) package (http://www.eclipse.org/birt/phoenix/) is used to create user-customizable reports of the data stored in the database.
3.3 Data Import
Data are recorded in spreadsheets that are batch-imported into the system. The spreadsheet data are parsed and verified against the requirements of the domain model. Admissible data are stored in a relational database. If errors are identified in the spreadsheet, an output spreadsheet is generated that references the specific line of each error in the input data. The user may then review and correct each error and resubmit the input data. Supporting documentation and media files are imported into the system using spreadsheets to reference the location of the files to be uploaded.
3.4 Generating Reports from the Database
To access data in the TM-CTS, a large number of reports drawing from all elements of the database can be generated by users via the TM-CTS Report Navigator. Two types of reports are available: commonly requested fixed-format reports, and user-configurable reports. The “Single Species Overview” report (Supplementary Material S1) is an example of the fixed-format report. It displays all information relevant to a single plant species in the collection. A user-configurable report is set up using a parameter selection form (Supplementary Material S2). Reports may contain hyperlinks to other reports, allowing users access to the underlying details of data items shown in a report. Reports are also capable of displaying multimedia data such as images and videos (Supplementary Material S3) and can be linked to maps provided by geographic information systems (GIS) via the web (Supplementary Material S4). A user may export a report to a variety of different file formats, including PDF, CSV, and Excel formats. User access to reports is controlled by database administrators, and different levels of access can be specified for different users. More detail on the administration and creation of reports in the TM-CTS is provided in Supplementary Material S5.
Currently one of the most common use-scenarios for the database is to search information from plant fractions that have been identified as “positives” in high throughput screening campaigns. Using fraction information from the high throughput screening database (Tolopko et al., 2010), a user can search for information through a user-configurable report in the TM-CTS. The user can then access any available information about the plant or fraction, such as various names of the plant (e.g., scientific, pharmaceutical, Chinese), details about the fractionation process, traditional uses of the plant, as well as any collection-related details of interest.
4. Concluding Remarks
A number of the features supported by the TM-CTS are generally applicable to ethnobotanical projects. For example, all ethnobotanical projects typically involve the general activities of plant collection, quality assessment, tests for contaminants, information about traditional uses, and chemical extraction (Malone, 1983). Although the data for the project described in this paper (Eisenberg et al., 2011) is not in the public domain, the software described herein is open source (GNU GPL2 license) and freely available (http://tm-cts.sourceforge.net). Therefore, other ethnobotanical projects with similar informatics requirements may be able to adapt the TM-CTS to their particular needs. In this way, it is hoped that the TM-CTS and its underlying domain model will help in general efforts to evaluate and standardize traditional medicines derived from plants and fungi (Berman and Straus, 2004).
Supplementary Material
Acknowledgements
This work was supported primarily by a grant from the National Institutes of Health (U19 CA128534) and, in part, by the Osher Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Appendix A. Supplementary data
Supplementary data associated with this article can be found in the online version.
Current Address for ESJH: Harvard University Herbaria, 22 Divinity Avenue, Cambridge, MA 02138 USA.
References
- Berman JD, Straus SE. Implementing a research agenda for Complementary and Alternative Medicine. Annual Review of Medicine. 2004;55:239–254. doi: 10.1146/annurev.med.55.091902.103657. [DOI] [PubMed] [Google Scholar]
- Binns SE, Arnason JT, Baum BR. Phytochemical variation within populations of Echinacea angustifolia (Asteraceae) Biochemical Systematics and Ecology. 2002;30:837–854. [Google Scholar]
- CBOL Plant Working Group A DNA barcode for land plants. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinese Pharmacopoeia Commission . Pharmacopoeia of the Peoples Republic of China. 2005 ed. Vol. 1. People’s Medical Publishing House; Beijing: 2005. [Google Scholar]
- Eisenberg DM, Harris ESJ, Littlefield BA, Cao SG, Craycroft JA, Scholten R, Fu YL, Wang WQ, Liu Y, Zhao ZZ, Kaptchuk T, Shamu CE, Clardy J. Developing a library of authenticated Traditional Chinese Medicinal (TCM) plants for systematic biological evaluation – Rationale, methods and preliminary results from a Sino-American collaboration. Fitoterapia. 82:17–33. doi: 10.1016/j.fitote.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E. Domain-driven design: Tackling complexity in the heart of software. Addison-Wesley Longman Publishing; Boston: 2003. [Google Scholar]
- Hildreth J, Hrabeta-Robinson E, Applequist W, Betz J, Miller J. Standard operating procedure for the collection and preparation of voucher plant specimens for use in the nutraceutical industry. Analytical and Bioanalytical Chemistry. 2007;389:13–17. doi: 10.1007/s00216-007-1405-x. [DOI] [PubMed] [Google Scholar]
- Jeffery EH, Brown AF, Kurilich AC, Keck AS, Matusheski N, Klein BP, Juvik JA. Variation in content of bioactive components in broccoli. Journal of Food Composition and Analysis. 2003;16:323–330. [Google Scholar]
- Ji ZL, Zhou H, Wang JF, Han LY, Zheng CJ, Chen YZ. Traditional Chinese medicine information database. Journal of Ethnopharmacology. 2006;103:501. doi: 10.1016/j.jep.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Malone M. The pharmacological evaluation of natural products - general and specific approaches to screening ethnopharmaceuticals. Journal of Ethnopharmacology. 1983;8:127–147. doi: 10.1016/0378-8741(83)90050-8. [DOI] [PubMed] [Google Scholar]
- Nahin RL, Barnes PM, Stussman BJ, Bloom B. Costs of complementary and alternative medicine (CAM) and frequency of visits to CAM practitioners: United States, 2007. National Health Statistics Reports. 2009;18:1–14. [PubMed] [Google Scholar]
- “The Academic Pursuit of Screening,” Editorial. Nature Chemical Biology. 2007;3:433. doi: 10.1038/nchembio0807-433. [DOI] [PubMed] [Google Scholar]
- Tolopko AN, Sullivan JP, Erickson SD, Wrobel D, Chiang SL, Rudnicki K, Rudnicki S, Nale J, Selfors LM, Greenhouse D, Muhlich JL, Shamu CE. Screensaver: an open source lab information management system (LIMS) for high throughput screening facilities. BMC Bioinformatics. 2010;11:260. doi: 10.1186/1471-2105-11-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toselli F, Matthias A, Gillam EMJ. Echinacea metabolism and drug interactions: The case for standardization of a complementary medicine. Life Sciences. 2009;85:97–106. doi: 10.1016/j.lfs.2009.04.023. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Xiao P, Xiao Y, Yuen JPS. Quality assurance of Chinese Herbal Medicines (CHMs) Journal of Food and Drug Analysis. 2007;15:337–346. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.