Abstract
We have identified a novel tRNA methyltransferase in Saccharomyces cerevisiae that we designate Trm9. This enzyme, the product of theYML014w gene, catalyzes the esterification of modified uridine nucleotides, resulting in the formation of 5-methylcarbonylmethyluridine in tRNAArg3 and 5-methylcarbonylmethyl-2-thiouridine in tRNAGlu. In intact yeast cells, disruption of the TRM9 gene results in the complete loss of these modified wobble bases and increased sensitivity at 37°C to paromomycin, a translational inhibitor. These results suggest a role for this potentially reversible methyl esterification reaction when cells are under stress.
In the last few years, it has become increasingly evident that S-adenosylmethionine (AdoMet)-dependent methyltransferases play a wide range of roles in cell physiology. Unlike other covalent modifications such as phosphorylation that occur mainly on proteins, methylation has been observed in DNA, RNA, proteins, lipids, polysaccharides, and small molecules (15, 35, 36, 38). As a result of such diversity, identifying the methyltransferases and their substrates has been challenging. Although more than 25 methylated nucleotides have been documented for eukaryotic tRNAs alone, only a few of the corresponding enzymes and their genes have been identified (16, 47).
In 1999, using conserved AdoMet binding motifs, we identified 26 putative methyltransferases (designated F1 to F26) encoded by the genome of the yeast Saccharomyces cerevisiae (40). Biochemically, we have been able to show that two of these species are in fact methyltransferases; F3 (YDR465c) catalyzes the transfer of a methyl group to the delta-nitrogen atom of arginine residues in a novel protein posttranslational reaction (40, 58), while F9 (YER175c) catalyzes the methyl esterification of the small molecule trans-aconitate (9). Recently, F1 (YDL201w) was found to be responsible for formation of 7-methylguanosine at position 46 of tRNA (3).
One major approach in identifying novel enzymes has been the comparison of the methylation spectra of strains lacking the putative methyltransferase and those of their isogenic wild-type parents by separation techniques such as sodium dodecyl sulfate (SDS) gel electrophoresis (26, 59). In the yeast S. cerevisiae, one can study biological methylation in vivo by incubating cells with S-adenosyl-l-[methyl-3H]methionine ([3H]AdoMet), the major biological methyl donor (26). As a result, all methyl-accepting species such as RNAs, proteins, and small molecules can become radiolabeled and the fate of the methylated species can be followed biochemically. One can then look for differences in the methylation spectra between a mutant strain and its parent.
In all domains of life, tRNAs are highly modified posttranscriptionally (46, 47). Methylation reactions account for the majority of these modifications. In the yeast S. cerevisiae, eight tRNA methyltransferases have been identified so far. The TRM1 gene product forms N2,N2-dimethylguanosine at position 26 (14). TRM2 encodes a protein that forms 5-methyluridine at position 54 (41). TRM3 encodes a protein that catalyzes methylation on the 2′-O-ribose moiety of guanosine 18 (11). TRM4 encodes a methyltransferase that forms 5-methylcytosine at positions 34, 40, 48, and 49 (37). The TRM5 gene product forms 1-methylguanosine (m1G37) at position 37 and 1-methylinosine (m1I) and participates in Y-base (yW) formation (8). The GCD14 gene product is part of a complex that forms 1-methyladenosine (4). Trm7 was identified as a methyltransferase that forms 2′-O-methylribose in the anticodon loop (43). Recently, Trm8, working in a complex, was found to be required for formation of 7-methylguanosine at position 46 of tRNA (3).
In this paper, we present evidence for the identification of the F6 (YML014w) candidate methyltransferase as the enzyme responsible for the esterifications of the modified 5-methylcarbonylmethyluridine (mcm5U) and 5-methylcarbonylmethyl-2-thiouridine (mcm5s2U) wobble bases in tRNA. We show that deletion of the F6 gene can lead to hypo-methyl esterification of tRNA at these bases in vivo. In addition, we show that the F6 gene product can methyl esterify tRNA in vitro, and we have now designated the F6 gene TRM9. Finally, we show that trm9 deletion mutants are hypersensitive to the translational inhibitor paromomycin at elevated temperatures, suggesting the importance of the methyl-esterified bases during heat shock.
MATERIALS AND METHODS
S. cerevisiae strains.
Strain BY4742 (MATα hisΔ1 leu2Δ0 lys2Δ0 ura3Δ0) and the F6 (YML014w) kanamycin insert deletion strain in the BY4742 background were obtained from ResGen/Invitrogen (Carlsbad, Calif.). In this work, we designated the F6 deletion strain HKY101 (BY4742 trm9::Kanr). The CEN.PK2-1C strain (MATα ura3 his3 leu2 trp1) was a gift from Cathy Clarke, University of California—Los Angeles. Strain HKY102 (CEN.PK2-1c trm9::TRP1) was made using a PCR-based methodology with forward primer 5′-ATGGAGATAAACCAAGCGGCTGAAAAAGAACAGGAGTATGCCGCGGTGGCCGCTCTA-3′, reverse primer 5′-TCATCTCTTCTGGGCCACCACCCACCAATTGTCGCGGCATCGATAAGCTTGATATCGA-3′, and template plasmid pBluescript KS(+)-TRP1 (6). To generate the expression vector pRS316-TRM9, PCR methodology was used to amplify a 1,292-bp genomic fragment that contains the entire coding sequence of TRM9 and 452 bp upstream of the TRM9 gene. The forward primer (5′-GCCGGATCCGGGACTTTGTTGTTGATAGAGTCCGG-3′) contains BamHI site-flanking sequences and the reverse primer (5′-TAAGTCGACTCATCTCTTCTGGGCCACCACCAC-3′) contains SalI site-flanking sequences. The HKY111 strain, in which the wild-type TRM9 gene is replaced in the yeast genome by a fusion of TRM9 with an N-terminal hemagglutinin (HA) epitope-encoding segment under the control of the GAL1 promoter, was prepared as described previously (26). Briefly, specific primers included a forward primer containing nucleotides −90 to −51 of the promoter region of YML014w incorporated into the 5′ end of the F1 sequence and a reverse primer containing nucleotides +50 to +11 of YML014w incorporated into the 5′ end of the R1 sequence (26).
In vivo labeling and preparation of cell extracts.
Strains of S. cerevisiae were grown to early log phase (optical density at 600 nm [OD600] of between 0.6 and 0.8) in 50 ml of medium (1% yeast extract [Difco, Detroit, Mich.], 2% peptone [Difco], and 2% of either d-glucose for yeast extract-peptone-dextrose [YPD] medium or d-galactose [Sigma Chemical Co., St Louis, Mo.] for yeast extract-peptone-glucose [YPG] medium).An aliquot of 5 OD600 units of cells was collected by centrifugation at 110 × g for 5 min at 4°C, and the cells were washed three times with 10 ml of YPD or YPG medium. The cell pellet was resuspended in 820 μl of YPD or YPG medium and 180 μl of [3H]AdoMet (80 Ci/mmol in hydrochloric acid-ethanol diluted 9:1 [pH 2.0 to 2.5; Amersham Biosciences, Piscataway, N.J.]) to give a final [3H]AdoMet concentration of 2 μM. Cells were incubated in a gyratory shaker at 225 rpm for 30 min at 30°C, pelleted as described above, and washed twice with 1 ml of water.
The cell pellet was then resuspended in 50 μl of lysis buffer (1% SDS [wt/vol] and 0.67 mM phenylmethylsulfonyl fluoride). Glass beads (0.2 g and 0.5 mm in diameter; Biospec Products, Inc., Bartlesville, Okla.) were added to the cell suspension, and the mixture was vortexed for 1 min, followed by incubation on ice for another 1 min. The vortexing step was repeated seven times. The extract was collected into a new tube, and another 50 μl of lysis buffer was added to the beads and vortexed for 30 s to wash the remaining protein from the beads. This washed extract was then pooled with the original extract.
SDS gel electrophoresis and analysis of 3H-methylated species.
Approximately 50 μl of the extract was mixed with an equal volume of concentrated gel electrophoresis sample buffer (3.5% [vol/vol] β-mercaptoethanol, 6% [wt/vol] SDS, 0.18 M Tris-HCl [pH. 6.8], 10% glycerol, 0.005% [wt/vol] bromophenol blue), incubated at 100°C for 5 min, and loaded onto a 1.5-mm-thick slab gel containing a stacking gel and a 10.5-cm-long resolving gel. The resolving gel was made from 10% (wt/vol) acrylamide and 0.34% (wt/vol) N,N-methylenebisacrylamide. Molecular mass standards (20-μl aliquots of Bio-Rad low-molecular-weight standard, no. 161-0304, each at 2 mg/ml) were mixed with 20 μl of sample buffer; 10 μl was loaded into wells. Electrophoresis was performed at 20 mA until the dye front ran off the end of the resolving gel (32). Gels were stained for 15 min in 0.1% Coomassie brilliant blue in 50% (vol/vol) methanol and 10% (vol/vol) acetic acid in water and then destained overnight at room temperature in 5% (vol/vol) methanol and 10% (vol/vol) acetic acid in water. The gels were vacuum-dried at 65°C onto Whatman 3MM chromatography paper.
Dried gels were cut into 3-mm-thick slices that were 8 mm wide. To analyze gel slices for 3H radioactivity in methyl ester linkages, 150 μl of 1.5 M Na2CO3 (pH 12) was added to each dried gel slice in a 1.5-ml polypropylene microcentrifuge tube. The tubes were gently placed into 20-ml scintillation vials containing 5 ml of Safety Solve counting fluid (Research Products International, Mt. Prospect, Ill.) so that no mixing occurred, and the vials were tightly capped. Vials were incubated at 37°C for 24 h to allow [3H]methanol derived from base hydrolysis of methyl esters to diffuse in the vapor phase from the microcentrifuge tubes to the scintillation fluid. Scintillations in vialswere counted using a Beckman LS6500 scintillation counter. To determine the total level of 3H radioactivity present in each gel slice, 1 ml of 30% hydrogen peroxide was added gently to each microcentrifuge tube containing the gel slice after the scintillations in the vials were counted to measure [3H]methanol levels. Vials were capped loosely and incubated at 37°C for an additional 24 h to allow for the digestion of the gel slice. After the vials were shaken to mix completely the contents of the microcentrifuge tube with the scintillation fluid, scintillations in the vials were recounted.
tRNA extraction and digestion.
Yeast cells grown in 50 or 500 ml of YPD to log phase (OD600 of 0.5 to 1.0) at 30°C were harvested by centrifugation for 5 min at 1,300 × g at 4°C. The cells were resuspended in 300 μl of 0.9% NaCl per 10 OD units of cells, and 2 volumes of phenol were then added to the suspension. The mixture was rotated gently at room temperature for 30 min. Chloroform (0.1 volume) was then added to the mixture, and the samples were incubated for an additional 15 min at room temperature. The samples were spun down at 10,600 × g for 20 min. The aqueous phase was collected and mixed with 2.5 volumes of ethanol and 0.1 volume of 20% potassium acetate to precipitate RNA. tRNA was purified with the 2 M LiCl extraction method as described previously (5). The tRNA pellet was washed twice with 80% ethanol and finally dissolved in water or 2.4 M tetraethylammonium chloride. tRNA was digested as previously described (18, 44). Briefly, 50 μl (100 to 200 μg of RNA as determined by A260) of tRNA was heat denatured at 90°C for 2 min and 5 μl of 10 mM zinc sulfate and 10 μl (200 U per ml) of nuclease P1 (Boehringer-Mannheim, Mannheim, Germany) were added. The mixture was incubated at 37°C for 16 h. Ten microliters of 0.5 M Tris buffer (pH 8.0) and 10 μl (100 U per ml) of alkaline phosphatase (bacterial type III; Sigma) were added. The mixture was incubated at 37°C for 2 h.
Purification of specific tRNAs.
To isolate specific tRNA species, the total tRNA extract described above was hybridized to matrix-bound oligonucleotides. Synthetic nucleotides containing biotin at the 5′ end were designed to specifically hybridize to S. cerevisiae tRNAArg3 or tRNAGlu. Biotinylated primers were synthesized that were specific for tRNAArg3 and tRNAGlu and contained 35 nucleotides complementary to the 5′ ends up to position 34, at which the mcm5U or mcm5s2U nucleotides are present. The probe (0.5 nmol) was bound to Dynabeads M-280 with streptavidin covalently attached to the surface, according to the recommendation of the manufacturer (Dynal A.S., Olso, Norway). Briefly, the beads were washed three times in the high salt buffer (5 mM Tris-HCl, 0.5 mM EDTA, 1 M NaCl). The beads were resuspended in 250 μl of the high salt buffer and 10 μl (1 nmol) of biotinylated synthetic probes. The mixture was incubated at room temperature for 15 min. Beads were washed three times with the high salt buffer. The hybridization method used here is described elsewhere (49). Briefly, tRNA (50 to 200 μg), dissolved in 2.4 M tetraethylammonium chloride, was denatured at 60°C for 3 min and then mixed with the streptavidin-bound oligonucleotide beads at 15°C for 30 min. The beads were washed three times with 2.4 M tetraethylammonium chloride, and then bound tRNA was eluted after heating at 60°C for 3 min in 100 μl of 2.4 M tetraethylammonium chloride. tRNA was concentrated and desalted by centrifugation by using Centricon 10 columns (Amicon, Beverly, Mass.).
High-pressure liquid chromatography (HPLC).
In a modification of the method of Pomerantz and McCloskey (44), digested tRNA was injected onto a C18 reverse-phase column (Supelcosil; 5-μm-diameter beads, 4.6-mm inside diameter, and 250-mm length) equilibrated in solvent A (5 mM ammonium acetate, pH 6.0) for 15 min and eluted at 1 ml/min at 25°C with increasing solvent B (40% acetonitrile in 60% water) concentrations. Amounts of time (in minutes) and percentages of A and B were as follows: 0, 100, and 0; 3, 100, and 0; 5.8, 98, and 2; 7.2, 97, and 3; 10, 95, and 5; 25, 75, and 25; 30, 50, and 50; 34, 25, and 75; 37, 25, and 75; 43, 0, and 100; and 48, 0, and 100, respectively. The nucleotides were detected using a UV detector (Waters Lambda-Max model 489) set at 254 nm. The fractions were collected at either at 1- or 0.5-min intervals. The collected fractions were used directly for the base-labile assay (described below) or dried overnight under a vacuum for mass spectrometry.
HPLC fractions were analyzed for [3H]methyl esters by mixing 100 to 150 μl with NaOH to give a 1 M solution in a 200-μl final volume to form [3H]methanol. The hydrolysate was spotted onto a 1.5- by 8-cm piece of Whatman 3MM paper and suspended in the neck of a 20-ml scintillation vial above 5 ml of Safety Solve scintillation fluid. After 2 h at room temperature, the paper was removed and the scintillations in the vial were counted to measure the level of [3H]methanol that had transferred to the fluid in the vapor phase.
Mass spectrometry.
A Perkin-Elmer Sciex API III triple quadrupole mass spectrometer was used as previously described (27). HPLC fractions containing nucleosides were analyzed by direct injection (15 μl) into a mixture of water-acetonitrile-formic acid (50/50/0.1, vol/vol/vol). Normal spectra were obtained by scanning from an m/z of 200 to an m/z of 600 (0.3-Da step size, 30-msec dwell time, 6.0 s/scan, and orifice voltage of 60). Ion series were transformed using version 3.3 of MacSpec software. For tandem mass spectrometry (MS/MS) analysis, positive ion spectra of Q1 preselected parent ions were generated by collisionally induced dissociation (10% nitrogen in argon) with a collision gas thickness instrument setting (CGT) of 100 and an R0-R2 offset of 20 V via scanning of Q3 from an m/z of 50 to an m/z of 400 (step size, 0.3 Da; dwell time, 30 ms; scan time, 5.02 s; orifice voltage, 60).
Preparation of saponified tRNA.
Yeast tRNA was saponified as described previously (29). Briefly, 1 ml of 0.5% of a commercial preparation of yeast tRNA (Boehringer-Mannheim) was mixed with 125 μl of 1 M NaOH and the mixture was incubated at room temperature for 10 min. The solution was neutralized with 125 μl of 1 M acetic acid. This preparation was used directly in the in vitro reactions.
In vitro methylation of immunoprecipitated HA-tagged protein.
Genomically HA-tagged TRM9 gene product was immunoprecipitated from extracts of HKY111 cells (100 OD600 units) as described previously (26). Briefly, the protein-bound beads were resuspended in 150 μl of buffer containing 50 mM Tris-HCl (pH 7.5) and 150 mM NaCl. An in vitro reaction mixture containing 5 μl of [3H]AdoMet (final concentration of 1.4 μM in hydrochloric acid-ethanol diluted 9:1 [pH 2.0 to 2.5]), 40 μl of beads, and various amounts of commercially purchased baker's yeast tRNA (Boehringer-Mannheim) was incubated at 37°C for 30 min. The reaction was stopped using 2× gel electrophoresis sample buffer, and the [3H]methyl ester counts were measured as described previously (26).
Preparation of a GST-Trm9 fusion protein.
Plasmid pAN105 was made by Agnieszka Niewmierzycka in our laboratory to express a glutathione-S-transferase (GST)-Trm9 fusion protein (39). Briefly, DNA including the open reading frame of YML014w (TRM9) was amplified by PCR from yeast genomic DNA by using primers containing a BamHI site in the 5′ end (CTAGGATCCAACATGGAGATAAACC) and an EcoRI site in the 3′ end (CTAGAATTCACCTTCATCTCTTCTG). The amplified products and the plasmid pGEX-2T (Amersham Biosciences) were then each digested with EcoRI and BamHI. The digested insert was ligated into the digested vector in frame with the GST coding region to encode a GST-Trm9 fusion protein. Escherichia coli DH5α cells were transformed with the ligation reaction, and the correct clone was selected by restriction analysis. The GST fusion protein was expressed by IPTG (isopropyl-β-d-thiogalactopyranoside; 1 mM final concentration) induction of a 2-liter culture of cells in Luria-Bertani broth containing 100 μg of ampicillin/ml at an OD600 of 0.4. After 3 h, the cells were spun down and washed twice, and the pellet was resuspended in 25 ml of a mixture of 140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4 (pH 7.3) containing a dissolved protease cocktail tablet (Roche, Inc.). The resuspended cells were sonicated for 5 min, and the mixture was spun down by centrifugation at 18,600 × g for 15 min at 4°C. The supernatant was transferred to 0.5 ml of a 50% glutathione-Sepharose 4B slurry (Amersham Biosciences) and shaken gently at 4°C for 1 h. Beads were spun down and washed three times with the lysis buffer. The fusion protein was eluted using 1 ml of 10 mM reduced glutathione-50 mM Tris-HCl (pH 8.0).
RESULTS
In vivo identification of a novel RNA methyltransferase.
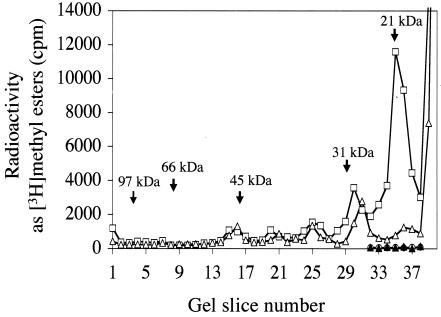
We labeled methylated cellular components of strains of the yeast S. cerevisiae by incubating intact cells with the biological methyl donor [3H]AdoMet. We then fractionated extracts by SDS gel electrophoresis, measuring [3H]methyl-esterified species with an assay in which gel slices are treated with base to release [3H]methanol, which is then detected in a vapor phase assay for volatile radioactivity (26, 59). In Fig. 1, we summarize a comparison of [3H]methyl-esterified species of the candidate methyltransferase F6 (YML014w) from a wild-type parent and a knockout strain. A significant reduction of methylation was observed in the 21-kDa region of the mutant compared to that in its isogenic parent, suggesting that the substrate of the F6 methyltransferase eluted at this position. It has been shown previously that the majority of methyl-esterified species in the 21-kDa region actually are modified RNA species rather than proteins (23).
FIG. 1.
[3H]methyl ester spectra of yeast extracts separated by SDS gel electrophoresis. Yeast cells were labeled in vivo with [3H]AdoMet as explained in Materials and Methods. Subsequently, the cellular extract was fractionated by SDS-10% polyacrylamide gel electrophoresis and the [3H]methanol resulting from hydrolysis of methyl esters was measured as described previously (26). Polypeptide size standards are as follows: phosphorylase b, 97 kDa; bovine serum albumin, 66 kDa; ovalbumin, 45 kDa; carbonic anhydrase, 31 kDa; and soybean trypsin inhibitor, 21 kDa. For RNase treatment, extracts containing approximately 100 μg of protein were incubated with 40 μg of bovine pancreatic RNase A (preincubated by heating at 100°C for 15 min). Symbols: □, wild-type strain BY4742; ▵, F6 (YML014w) deletion mutant HKY101; ○, RNase-treated wild-type strain BY4742; ▴, RNase-treated F6 (YML014w) deletion mutant HKY101.
In order to confirm that the F6-dependent methylation in the 21-kDa region is that of an RNA species, we treated cellular extracts of in vivo-radiolabeled yeast cells with either proteinase K or RNase A. Proteinase K treatment eliminated almost all of the methyl ester radioactivity in the gel except in the 21-kDa region, where much of the radioactivity was preserved (data not shown). On the other hand, when extracts were treated with RNase A, almost all the methyl ester radioactivity at the 21-kDa position disappeared (Fig. 1). We conclude that the methyl esterification reduced in the F6 deletion mutant is present in RNA.
F6 is a tRNA methyltransferase (Trm9) responsible for the methylation of mcm5U and mcm5s2U nucleosides.
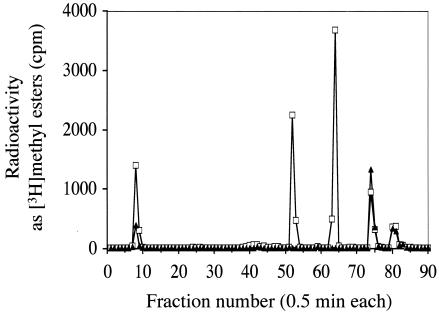
We focused on tRNA methylation because tRNAs would be expected to migrate in the 21-kDa region of SDS gels. tRNA was extracted from in vivo [3H]AdoMet-radiolabeled cells and was subsequently hydrolyzed to nucleosides. The resulting nucleosides were separated by HPLC, and the fractions were analyzed either by direct counting for total radioactivity or by the vapor phase assay for [3H]methyl esters. No large differences were observed in the patterns of total radioactivity in F6 deletion strains, suggesting that the F6 gene product is not one of the major tRNA methylated species (data not shown). However, when the methyl ester radioactivity levels were compared, it was found that two of the major species were completely missing in the F6 deletion strain (Fig. 2). One of these missing species eluted at 26 min (fraction 52) and the other at 32 min (fraction 64). Two candidate modified nucleosides that would be expected to elute at these positions are mcm5U and mcm5s2U (18, 44). When the elution profiles of authentic mcm5U and mcm5s2U (both generous gifts of Darrell R. Davis, University of Utah) were examined under our HPLC conditions, mcm5U eluted exactly with the 26-min peak missing in the F6 deletion mutant and mcm5s2U eluted exactly with the 32-min peak also absent in the F6 deletion mutant (data not shown). We thus designate the F6 (YML014w) gene TRM9 (for tRNA methyltransferase).
FIG. 2.
HPLC fractionation of methyl-esterified tRNA nucleosides. Total tRNA was extracted from in vivo-radiolabeled cells (10 OD units) as described in Materials and Methods. Subsequently, tRNA was digested with P1 nuclease and alkaline phosphatase to generate free nucleosides and then fractionated by HPLC as described in Materials and Methods. Methyl ester radioactivity was measured by adding 100 μl of 2 N NaOH to 100 μl of each fraction and measuring vapor phase [3H]methanol as described in Materials and Methods. Symbols: □, wild-type strain BY4742; ▴, F6 deletion mutant HKY101.
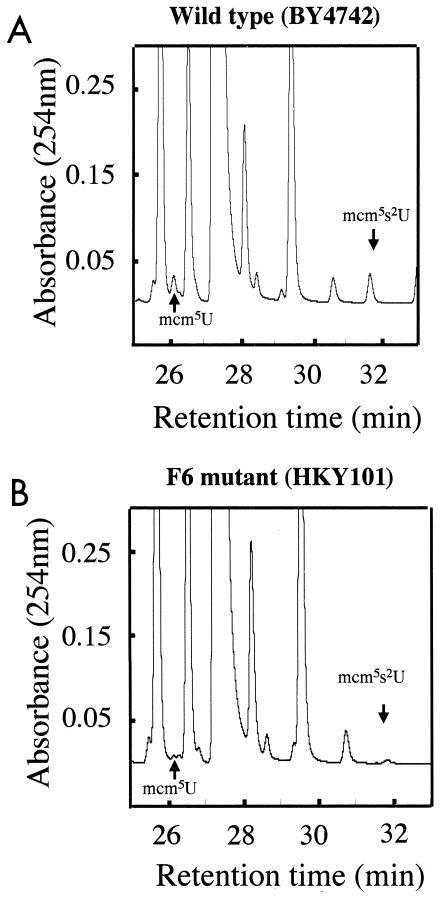
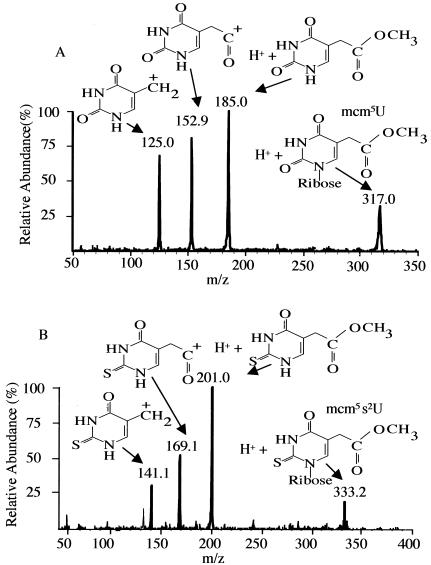
Mass spectrometry was used to directly determine the structure of the modified nucleosides that were not present in the trm9 deletion strain. tRNA was purified in large quantities from both the wild-type and the mutant strains, digested into nucleosides, and fractionated by HPLC (Fig. 3). The digested tRNA samples were mixed with tracer amounts of radiolabeled modified nucleosides (prepared as indicated in Fig. 2) to identify the nonradiolabeled materials in the bulk tRNA preparations. Two small peaks, present in the wild-type preparation at 26 and 32 min (Fig. 3), corresponded with radioactivity; both the A254 peaks were missing in the corresponding fractions from the trm9 deletion strain (Fig. 3). Fractions containing the methyl ester and fractions that eluted immediately before or after were analyzed in an electrospray quadrupole mass spectrometer. In the analysis of the 26-min fraction, the only marked difference found between samples from the wild-type and mutant strains was an ion of m/z of 316.9 corresponding to that expected for the M + H+ species of mcm5U (317.0). This ion was present only in the 26-min fraction eluting with the tracer radioactivity; it was not observed in adjacent fractions (data not shown). To confirm that this species is mcm5U, the fractions were subjected to MS/MS to examine the pattern of ions after fragmentation. The pattern of ions observed was exactly consistent with the mcm5U structure (Fig. 4A). The same type of analysis was done for the 32-min fraction. In this case, TRM9-dependent peaks of m/z 332.9 and 355.1 were found corresponding to the M + H+ and M + Na+ forms of mcm5s2U (data not shown). The product ions of the 333.0-m/z precursor ion were consistent with the mcm5s2U chemical structure (Fig. 4B). We conclude that the deletion strain lacking the TRM9 gene does not have the two highly modified nucleosides mcm5U and mcm5s2U.
FIG. 3.
Large-scale HPLC fractionation of tRNA-derived nucleosides. Total tRNA from yeast cells (500 OD units) was extracted and treated to generate nucleosides as described in the legend to Fig. 2, and chromatography was performed as described in Materials and Methods. (A) Wild-type parent BY4742. (B) F6 deletion mutant HKY101.
FIG. 4.
Mass spectral identification of modified nucleosides missing in the trm9 deletion mutant. HPLC fractions corresponding to the mcm5U and mcm5s2U peaks shown in Fig. 3 were analyzed as described in Materials and Methods. The MS/MS spectrum of the ion of m/z 317.0 from the 26-min peak is shown in the upper panel (A); the MS/MS spectrum of the ion of m/z 333.0 from the 32-min peak is shown in the lower panel (B).
The Trm9 methyltransferase modifies yeast tRNAArg3 and tRNAGlu.
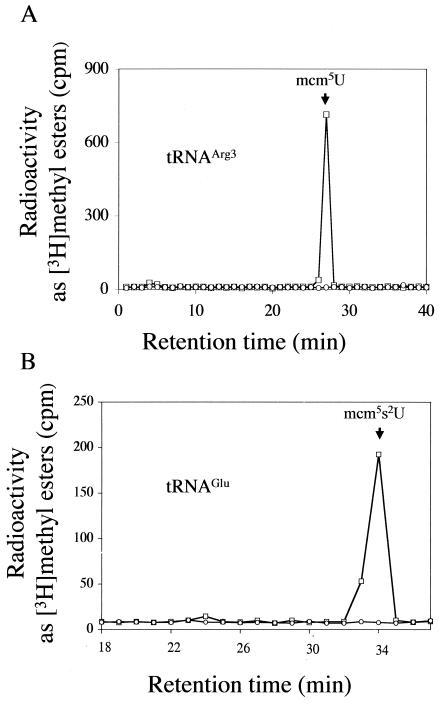
We asked which yeast tRNAs contain methyl-esterified nucleotides and whether these would be absent in the trm9 deletion strain. It has previously been reported that tRNAArg3 and tRNAGlu of S. cerevisiae contain mcm5U and mcm5s2U modifications, respectively, at the wobble position 34 (28, 30, 31). Therefore, each of these specific tRNAs was purified from in vivo [3H]AdoMet-labeled wild-type and mutant strains by using complementary oligonucleotides coupled to magnetic beads. After the purified tRNAs were digested into nucleosides, the mixture was separated by HPLC and fractions were analyzed for [3H]methyl esters (Fig. 5). For digested tRNAArg3, a peak of [3H]methyl esters was seen only at 26.5 min and was absent in the trm9 mutant (Fig. 5A). When purified tRNAGlu from wild-type cell digests was examined, only one peak of methyl ester radioactivity was found in the position corresponding to mcm5s2U at 33 min in digests of wild-type cells and no radioactive peak was found in digests of the trm9 deletion strain (Fig. 5B). Thus, we conclude that the tRNAArg3 and tRNAGlu species from the trm9 deletion mutant lack the methyl-esterified nucleosides at the wobble position 34.
FIG. 5.
Isolation and characterization of tRNAArg3 and tRNAGlu from in vivo-radiolabeled yeast cells. Total tRNA was isolated from 15 OD600 units of radiolabeled cells as described in Materials and Methods. Biotinylated primers specific for either tRNAArg3 or tRNAGlu were used to purify the tRNAs from either the wild-type parent BY4742 or the HKY101 trm9 deletion strain. Isolated tRNA was digested with P1 nuclease and alkaline phosphatase to generate free nucleosides that were fractionated by HPLC as described in the legend to Fig. 2. One hundred microliters of each fraction was used for measuring the methyl ester radioactivity as described in the legend to Fig. 2. (A) Methyl ester radioactivity from digested tRNAArg3 from the parent (□) and the trm9 deletion (○) strains. (B) Methyl ester radioactivity from digested tRNAGlu from the parent (□) and the trm9 deletion (○) strains.
In vitro analysis shows that Trm9 catalyzes the methyl esterification reaction.
We then questioned whether the methyl esterification reactions that are dependent upon the TRM9 gene in vivo could be catalyzed by the TRM9 gene product in vitro. With the use of a saponified preparation of tRNA enriched with carboxylic acid-methyl-accepting substrates, it was found that both wild-type and Trm9-overexpressing extracts of yeast catalyzed significant incorporation of methyl esters into tRNA whereas trm9 mutant extracts demonstrated no significant incorporation compared to background controls in which extract or tRNA was absent (Table 1). Importantly, the level of Trm9-dependent methyl esterification observed with nonsaponified tRNA was much lower. This indicates that a free carboxylate group generated by saponification is the substrate for the Trm9-catalyzed methylation reaction (Table 1).
TABLE 1.
In vitro methyl esterification catalyzed by the Trm9 methyltransferasea
| Reaction component(s) | Radioactivity as 3H ester (cpm ± SD) |
|---|---|
| Expt 1 | |
| trm9 mutant extract and saponified tRNA | 270 ± 71 |
| Wild-type extract and saponified tRNA | 3,546 ± 530 |
| Trm9-overexpressing extract and saponified tRNA | 5,138 ± 1,100 |
| trm9 mutant extract and tRNA | 428 ± 187 |
| Wild-type extract and tRNA | 668 ± 261 |
| Trm9-overexpressing extract and tRNA | 846 ± 257 |
| Saponified tRNA alone | 340 ± 141 |
| tRNA alone | 236 ± 13 |
| trm9 mutant extract alone | 314 ± 159 |
| Wild-type extract alone | 340 ± 173 |
| Trm9-overexpressing extract alone | 334 ± 122 |
| Expt 2 | |
| Trm9 immunoprecipitate and tRNA | 623 |
| Trm9 immunoprecipitate alone | 11 |
| tRNA alone | 80 |
| Expt 3 | |
| Control immunoprecipitate (YPD) and tRNA | 51 |
| Control immunoprecipitate (YPD) alone | 39 |
In experiment 1, reaction mixtures containing 3 μl of [3H]AdoMet (final concentration, 0.8 μM), 50 μg of the designated protein extract, and 50 μg of either a commercial preparation of tRNA from baker's yeast or its saponified derivative (prepared as described in Materials and Methods) were incubated at 30°C for 30 min in a final volume of 50 μl containing 100 mM Tris-HCl, pH 8.0. Reactions were stopped by adding 1 volume of phenol and 1 volume of chloroform. tRNA was precipitated from the aqueous phase by adding 2 volumes of ethanol, and its [3H]methyl ester content was assayed as described in Materials and Methods. In experiment 2, in vitro reaction mixtures were prepared as described in Materials and Methods. The enzyme source was the monoclonal anti-HA immunoprecipitated protein (26) from strain HKY111 cells grown in YPG medium to express the HA-Trm9 fusion protein. In experiment 3, in vitro reaction mixtures were prepared as in experiment 2 except that the HKY111 cells were grown in YPD medium so that the HA-Trm9 fusion protein was not expressed.
To confirm that the TRM9 gene product is the enzyme responsible for the methyl esterification, this gene product was purified using monoclonal antibodies to immunoprecipitate Trm9-HA-tagged protein from extracts of cells grown in galactose in which the protein was expressed. We subsequently asked whether this preparation could methylate the commercial mixture of tRNA species as described above. When volatile [3H]methyl esters were measured, significant radioactivity was found only when the enzyme and tRNA substrates were present (Table 1, experiment 2). When cells were grown in glucose in which the Trm9-HA protein would not be expressed, no significant radioactivity was found (Table 1, experiment 3).
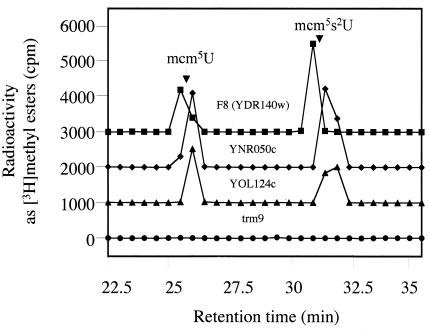
Genome-wide analysis of yeast protein complexes has suggested that the Trm9 protein is present in a complex with another putative methyltransferase, F8 (YDR140w), as well as three other gene products (YNR050c, YOL124c, and YNR046w) (17). The deletion mutants lacking these interacting proteins were examined, with the exception of the YNR046w deletion mutant, since deletion of YNRO46w is lethal. All deletion mutants were found to contain the mcm5U and mcm5s2U modified nucleosides (Fig. 6).
FIG. 6.
HPLC fractionation of methyl-esterified tRNA nucleosides from mutant yeast cells deficient in products potentially found in a complex with Trm9 (17). Total tRNA was extracted from in vivo-radiolabeled cells with mutations in the F8 (YDR140w), YNR050c, and YOL124c genes as well as in the TRM9 gene as described in the legend to Fig. 2. Subsequently, tRNA was digested with P1 nuclease and subjected to alkaline phosphatase to generate free nucleosides, which were fractionated by HPLC. Methyl ester radioactivity was measured by adding 100 μl of 2 N NaOH to 100 μl of each fraction and measuring vapor phase [3H]methanol as described in Materials and Methods. The elution positions of mcm5U and mcm5s2U methyl esters are shown with arrows. For clarity, the baseline has been adjusted; there is no significant radioactivity seen except in the region of mcm5U and mcm5s2U.
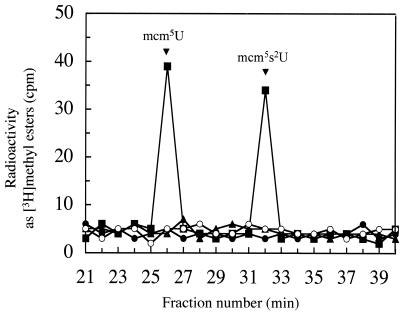
To directly show that the Trm9 protein can catalyze methyl ester formation in the absence of other yeast proteins, we generated a GST-Trm9 fusion protein in E. coli cells that have no homolog of the TRM9 gene and that lack mcm5U and mcm5s2U modified nucleosides. In Fig. 7, we show that the purified fusion protein catalyzes the formation of methyl esters in both mcm5U and mcm5s2U. No methyl ester formation is seen when the GST-Trm9 fusion protein is incubated in the absence of tRNA or when an unrelated GST fusion protein is used (Fig. 7). At this point, it is unclear whether potential protein partners may enhance the catalytic activity. However, it is clear that the Trm9 polypeptide can itself catalyze the methyl esterification reactions leading to the two modified nucleosides.
FIG. 7.
In vitro formation of mcm5U and mcm5s2U catalyzed by GST-Trm9. Saponified yeast tRNA (50 μg) prepared as described in Materials and Methods was incubated with [3H]AdoMet (final concentration of 0.8 μM) and the purified GST-Trm9 fusion protein (5 μg of protein, prepared as described in Materials and Methods) in a final volume of 50 μl of 100 mM Tris-HCl, pH 8.0. Samples were incubated for 60 min at 30°C. Reactions were stopped by adding 1 volume of phenol and 1 volume of chloroform. tRNA was precipitated from the aqueous phase by adding 2 volumes of ethanol. Samples were enzymatically digested and fractionated by HPLC as described in Materials and Methods. Methyl ester radioactivity in fractions was measured by adding 50 μl of 4 N NaOH to 150 μl of each fraction and measuring vapor phase [3H]methanol as described in Materials and Methods (▪). The elution positions of mcm5U and mcm5s2U are indicated. Control experiments were performed in which no GST fusion protein was added (•), in which GST-Trm9 was incubated without tRNA (○), or in which an unrelated GST fusion protein, GST-Rmt1, was used (▴).
Sequence analysis of Trm9.
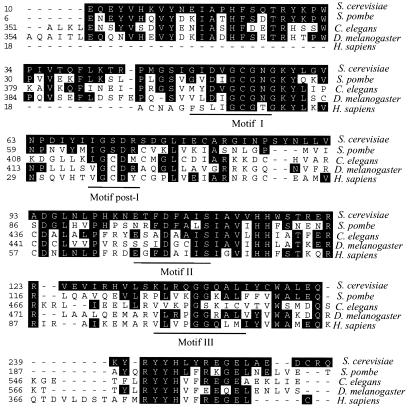
A BLAST search of the amino acid sequence encoded by the TRM9 gene of S. cerevisiae revealed that homologs are present in all eukaryotes for which the complete genomes have been sequenced, including fungi, worms, flies, plants, and mammals (Fig. 8). A multialignment of sequences from the species S. cerevisiae, Schizosaccharomyces pombe, Caenorhabditis elegans, Drosophilia melanogaster, and Homo sapiens demonstrates strong sequence similarity in the region containing the conserved AdoMet binding motifs (40) and in a C-terminal region including the S. cerevisiae sequence RYYHLYRELEL at residues 244 to 251. The sequence identities in the region of the yeast protein at residues 244 to 251 are especially interesting in that they may define residues involved in specific interactions with the substrates (tRNA).
FIG. 8.
Protein sequence identity of the Trm9 methyltransferase from S. cerevisiae with potential orthologs from four other eukaryotes. Deduced sequences were aligned using the Clustal method in the MegAlign program. Amino acid residues identical to those in the S. cerevisiae sequence are shaded. GenBank accession numbers are as follows: S. cerevisiae, S55105; Schizosaccharomyces pombe, spac13d.o3; Caenorhabditis elegans, CAB63431; Drosophila melanogaster, CG17807; and Homo sapiens, KIAA1456.
Sensitivity of a trm9 deletion strain to an inhibitor of protein translation at elevated temperatures.
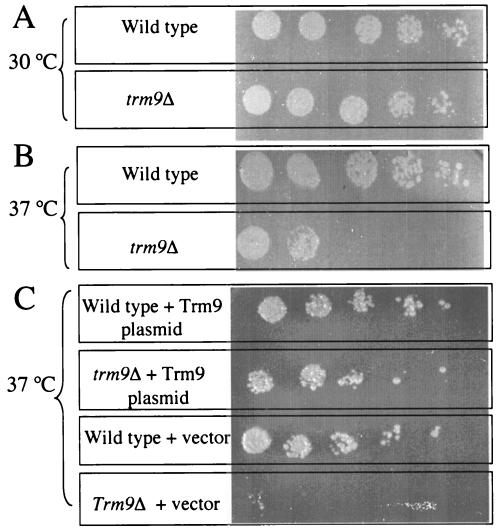
To understand the physiological roles of Trm9, the deletion mutant strain HKY101 was subjected to adverse conditions such as elevated temperatures and UV exposure, and no major difference was observed between the mutant strain and its isogenic parent BY4742 (data not shown). The growth of the trm9 deletion strain was also examined in various media containing nonfermentable sugars such as ethanol and glycerol as the sole carbon source, in media lacking the amino acid arginine, in media containing the arginine analog l-canavanine, and in media containing the protein translational inhibitor paromomycin, but again no significant phenotypic difference from the parent strain was observed (data not shown). However, the presence of the kanamycin resistance cassette in the HKY101 trm9 mutant would complicate the analysis for related protein synthesis inhibitors such as paromomycin. We therefore created a new trm9 knockout strain (designated HKY102) in which the TRM9 gene is disrupted by TRP1 rather than the kanamycin cassette. Upon examination of the new deletion strain for phenotypic differences, the HKY102 strain mutant appears to be much more sensitive than its isogenic parent to paromomycin at 37°C but not at 30°C (Fig. 9). To determine whether the sensitivity phenotype is based solely on the lack of the TRM9 gene product, a rescue experiment was done in which the TRM9 gene, under its native promoter, was expressed in the HKY102 strain via a yeast centromer plasmid. The lack of sensitivity to paromomycin in this strain shows that the effect is due to the TRM9 gene product (Fig. 9).
FIG. 9.
Sensitivity of trm9 deletion strains to paromomycin. (A) Yeast strains (wild-type parent CEN.PK2-1CHKY102 and the trm9Δ HKY102 strain) were grown to log phase at 30°C in the presence of 300 μg of paromomycin/ml. Cells (0.5 OD600 unit) were collected and resuspended in 200 μl of sterilized water. Ten microliters of the suspension was diluted in fivefold serial dilutions and plated onto YPD plates, and colonies were allowed to grow for 3 days. (B) Experimental conditions were the same as those described for panel A except that strains were grown at 37°C. (C) Yeast strains containing either the vector pRS316 or the TRM9 expression vector pRS316-TRM9 were grown at 37°C and plated as described above but in a synthetic dextrose medium lacking uracil.
DISCUSSION
In this work, we demonstrate that the Trm9 methyltransferase is responsible for the methylation of wobble bases at position 34. It is the first described enzyme that catalyzes the potentially reversible methyl esterification of tRNA species.
Uridine residues at the wobble position 34 are generally modified. In E. coli the modification is either of the xo5U (5-hydroxyuridine derivative) type or the xm5(s2) U(m) (5-methyluridine, 5-methyl-2-thiouridine, or 5-2′-O-methyluridine) type. The xo5U modification is found in tRNAs in which the wobble base can recognize all four nucleotides in the codon (53). Modified nucleosides of the xm5(s2) U(m) type include 5-methylaminomethyl-2-thiouridine (mnm5s2U) and 5-carboxymethylaminomethyl-2′-O-methyluridine (cmnm5Um) in E. coli. They are found in tRNA families that can read A or G in the third position of the codon (12, 50). In eukaryotic tRNA, methyl-esterified derivatives of the xm5(s2) U(m) family (mcm5U and mcm5s2U) have been found in the wobble positions of tRNAArg3 and tRNAGlu of yeast (28, 30, 31) and mcm5s2U has been reported in human tRNALys3 as well as in tRNAs of other mammalian species (42, 45). These methyl-esterified modifications have been found only in eukaryotes to date. Their apparent absence in eubacteria and archaebacteria suggests additional functions for these modifications as organisms became more complex.
Crick's original wobble hypothesis proposed that U in position 34 of the anticodon recognizes both A and G in the third position of the codon (13). With knowledge of the modifications of uridine residues in the wobble position, this hypothesis has now been revised to propose that the unmodified U at position 34 can recognize all four bases while the modified uridine residues are more restrictive and limit the recognition to only A and G, or to only one of these residues, at this position (33, 56). As a result, disruption of the modification of this uridine residue may lead to misreading in the third position of the codon and ultimately the incorporation of the wrong amino acid into the protein (57).
In vitro studies have shed some light onto the possible functions of the methyl esterification reaction in relation to these modified nucleotides. It has been shown that the mcm5U modification restricts the pairing of the anticodon UCU so that it recognizes the AGA codon only; yeast tRNAArg3 lacking this modification can recognize both AGA and AGG codons (52). Results of other in vitro binding and translation studies support the hypothesis that the 2-thiouridine (s2U34) modification and the 5-methylaminoethyl (mnm5U34) modification at position 34 restrict the reading of noncognate or near-cognate codons ending in U and C that specify different amino acids but allow recognition of their cognate codons ending in A or G (2, 20, 34, 54). With nuclear magnetic resonance, it has been shown that an mnm5s2U34-hypermodifiedtRNALys stabilizes a U turn structure whereas the s2U34 modification alone cannot bring about the same structural change (48). It is interesting, however, that in vivo studies using E. coli mutants lacking mnm5s2U34 or s2U34 actually demonstrate reduced rather than increased misreading of asparagine codons (22). It is clear that further work is needed to fully understand the role of these modifications.
What is the consequence of the loss of the Trm9 methyltransferase identified in yeast cells? Mutant cells are clearly viable under a number of conditions tested. However, we have been able to demonstrate sensitivity of trm9 mutants to paromomycin at elevated temperatures (Fig. 9). Paromomycin is an aminoglycoside antibiotic that has been found to interact with the ribosomal A site (10, 51). It is possible that the trm9 mutant lacking the methyl ester at the wobble position incorporates incorrect amino acids at a significant rate and that this leads to its sensitivity to paromomycin at the higher temperatures. Strains with TRM9 mutations were also found in a genome-wide screen of viable diploid deletion mutants of S. cerevisiae for gamma-ray sensitivity (7). These mutants also showed sensitivity to bleomycin (a DNA-damaging agent) and resistance to camptothecin (a topoisomerase I inhibitor) and hydroxyurea (an inhibitor of DNA replication). Although we have not confirmed these phenotypes in our haploid deletion mutants, pleiotropic effects may be expected from the disruption of the function of two (or more) tRNA species. In humans, we note that a mutation in a tRNALys species results in a defect in the modification of its wobble 2-thiouridine residue that disturbs the codon-anticodon interaction and is associated with an epilepsy linked to abnormal “ragged-red” muscle fibers (55). Additionally, in at least one host-pathogen interaction, the modified nucleoside mcm5s2U found in human tRNALys3 plays a key role in controlling the primer-template interaction in the reverse transcription initiation complex of human immunodeficiency virus type 1 (24, 25).
Among the eight tRNA methyltransferases in yeast described previously, only the Gcd10/Gcd14 methyltransferase has been shown to be essential (4). However, a deletion strain lacking TRM5, which is responsible for modification of m1G37, m1I, and yW, was found to grow very poorly (8). In addition, a deletion strain lacking TRM7, responsible for 2′-O-methylribose in the anticodon loop, showed slow growth and sensitivity to paromomycin (43). Although the tRNA methyltransferases are evolutionarily conserved among many organisms, which points to essential roles, the generally nonlethal mutant phenotypes suggest subtle roles in fine-tuning of tRNA function under a variety of physiological conditions.
We examined the subcellular localization of Trm9 in yeast cells by using a green fluorescent protein fusion construct and found Trm9 to be present in both nuclei and cytoplasm (data not shown). For those tRNA methyltransferases of which the subcellular localization patterns have been determined, the enzymes are found in either the nucleus or the cytoplasm. Trm1, Trm4, and Gcd10/Gcd14 are found in the nucleus (2, 12, 32), whereas Trm7 is found to be cytoplasmic (43).
In two yeast tRNA methyltransferases (Gcd10/Gcd14 and Trm8/Trm82), the cellular function requires more than one polypeptide chain in a complex (3, 4), and it has been suggested that this situation may be more general (1). It was of interest to us that at least two steps are required to form mcm5U and mcm5s2U from the precursor U. These reactions include the addition of the sulfur atom, the formation of the carboxyl-methyl group on carbon-5, and the methyl esterification step performed by Trm9. It is not clear whether Trm9 can also perform some or all of the other steps mentioned above. The TRM9 gene product has been reported to be associated with four other gene products in a high-throughput pull-down screen (17). However, we show here that deletion mutants lacking one of three of these genes all show the same modification of mcm5U and mcm5s2U as the wild-type parent strain. It is possible that the fourth potentially interacting product, that of YNR046w, of which deletion proves lethal, is a subunit required for one or more of these steps. Nevertheless, YNR046w has no methyltransferase signature motifs and has not been associated with Trm9 in a two-hybrid screen (data not shown).
Our study has described the first enzyme that can catalyze the methyl esterification of a nucleobase, in this case mcm5U and mcm5s2U in yeast tRNA. In gram-negative bacteria, a structurally related methyl-esterified nucleobase, uridine 5-oxyacetic acid methyl ester (mcmo5U), containing an ether moiety is found in several tRNA species (21). A pathway for the biosynthesis of mcmo5U has been suggested in which hydroxylation of C-5 is followed by a methylation step and one or more reactions resulting in the addition of a carboxyl group (21). It is unclear whether the biosynthetic pathways of mcm5U and mcm5s2U may also involve a similar methylated precursor, in this case 5-methyluridine. Determination of the biosynthetic pathways may be complicated by specific structural requirements of the tRNA substrate as well (19). In any case, neither the gene nor the protein for the methyl esterification reaction has been identified in this bacterial system and we observe no prokaryotic homologs of the Trm9 protein.
Acknowledgments
This work was supported by grants GM26020 and AG18000 from the National Institutes of Health.
We thank Darrell R. Davis (Department of Medicinal Chemistry, University of Utah) for his generous gifts of the modified nucleosides. We also thank Kym Faull and Joseph Loo for their insightful suggestions on mass spectrometry experiments.
REFERENCES
- 1.Agris, P. F., D. Setzer, and C. W. Gehrke. 1977. Characterization of a unique enzyme complex composed of S-adenosyl-l-methionine-tRNA-methyltransferase and aminoacyl-tRNA synthetase activities. Nucleic Acids Res. 4:38-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agris, P. F., D. Soll, and T. Seno. 1973. Biological function of 2-thiouridine in Escherichia coli glutamic acid transfer ribonucleic acid. Biochemistry 12:4331-4337. [DOI] [PubMed] [Google Scholar]
- 3.Alexandrov, A., M. R. Martzen, and E. M. Phizicky. 2002. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA 8:1253-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, J., L. Phan, and A. G. Hinnebusch. 2000. The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97:5173-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avital, S., and D. Elson. 1969. A convenient procedure for preparing transfer ribonucleic acid from Escherichia coli. Biochim. Biophys. Acta. 179:297-307. [DOI] [PubMed] [Google Scholar]
- 6.Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett, C. B., L. K. Lewis, G. Karthikeyan, K. S. Lobachev, Y. H. Jin, J. F. Sterling, J. R. Snipe, and M. A. Resnick. 2001. Genes required for ionizing radiation resistance in yeast. Nat. Genet. 29:426-434. [DOI] [PubMed] [Google Scholar]
- 8.Björk, G. R., K. Jacobsson, K. Nilsson, M. J. Johansson, A. S. Bystrom, and O. P. Persson. 2001. A primordial tRNA modification required for the evolution of life? EMBO J. 20:231-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai, H., D. Dumlao, J. E. Katz, and S. Clarke. 2001. Identification of the gene and characterization of the activity of the trans-aconitate methyltransferase from Saccharomyces cerevisiae. Biochemistry 40:13699-13709. [DOI] [PubMed] [Google Scholar]
- 10.Carter, A. P., W. M. Clemons, D. E. Brodersen, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407:340-348. [DOI] [PubMed] [Google Scholar]
- 11.Cavaille, J., F. Chetouani, and J. P. Bachellerie. 1999. The yeast Saccharomyces cerevisiae YDL112w ORF encodes the putative 2′-O-ribose methyltransferase catalyzing the formation of Gm18 in tRNAs. RNA 5:66-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakraburtty, K. 1975. Primary structure of tRNA Arg II of E. coli. Nucleic Acids Res. 2:1787-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crick, F. H. 1966. Codon-anticodon pairing: the wobble hypothesis. J. Mol. Biol. 19:548-555. [DOI] [PubMed] [Google Scholar]
- 14.Ellis, S. R., M. J. Morales, J. M. Li, A. K. Hopper, and N. C. Martin. 1986. Isolation and characterization of the TRM1 locus, a gene essential for the N2,N2-dimethylguanosine modification of both mitochondrial and cytoplasmic tRNA in Saccharomyces cerevisiae. J. Biol. Chem. 261:9703-9709. [PubMed] [Google Scholar]
- 15.Fujioka, M. 1992. Mammalian small molecule methyltransferases: their structural and functional features. Int. J. Biochem. 24:1917-1924. [DOI] [PubMed] [Google Scholar]
- 16.Garcia, G. A., and D. M. Goodenough-Lashua. 1998. Appendix III: general properties of RNA-modifying and -editing enzymes, p. 555-565. In H. Grosjean and R. Benne (ed.), Modification and editing of RNA. ASM Press, Washington, D.C.
- 17.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 18.Gehrke, C. W., and K. C. Kuo. 1989. Ribonucleoside analysis by reversed-phase high-performance liquid chromatography. J. Chromatogr. 471:3-36. [DOI] [PubMed] [Google Scholar]
- 19.Grosjean, H., J. Edqvist, K. B. Straby, and R. Giege. 1996. Enzymatic formation of modified nucleosides in tRNA: dependence on tRNA architecture. J. Mol. Biol. 255:67-85. [DOI] [PubMed] [Google Scholar]
- 20.Grosjean, H. J., S. de Henau, and D. M. Crothers. 1978. On the physical basis for ambiguity in genetic coding interactions. Proc. Natl. Acad. Sci. USA 75:610-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagervall, T. G., Y. H. Jönsson, C. G. Edmonds, J. M. McCloskey, and G. R. Björk. 1990. Chorismic acid, a key metabolite in modification of tRNA. J. Bacteriol. 172:252-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagervall, T. G., S. C. Pomerantz, and J. A. McCloskey. 1998. Reduced misreading of asparagine codons by Escherichia coli tRNALys with hypomodified derivatives of 5-methylaminomethyl-2-thiouridine in the wobble position. J. Mol. Biol. 284:33-42. [DOI] [PubMed] [Google Scholar]
- 23.Hrycyna, C. A., M. C. Yang, and S. Clarke. 1994. Protein carboxyl methylation in Saccharomyces cerevisiae: evidence for STE14-dependent and STE14-independent pathways. Biochemistry 33:9806-9812. [DOI] [PubMed] [Google Scholar]
- 24.Isel, C., J. M. Lanchy, S. F. Le Grice, C. Ehresmann, B. Ehresmann, and R. Marquet. 1996. Specific initiation and switch to elongation of human immunodeficiency virus type 1 reverse transcription require the post-transcriptional modifications of primer tRNA3Lys. EMBO J. 15:917-924. [PMC free article] [PubMed] [Google Scholar]
- 25.Isel, C., R. Marquet, G. Keith, C. Ehresmann, and B. Ehresmann. 1993. Modified nucleotides of tRNA(3Lys) modulate primer/template loop-loop interaction in the initiation complex of HIV-1 reverse transcription. J. Biol. Chem. 268:25269-25272. [PubMed] [Google Scholar]
- 26.Kalhor, H. R., K. Luk, A. Ramos, P. Zobel-Thropp, and S. Clarke. 2001. Protein phosphatase methyltransferase 1 (Ppm1p) is the sole activity responsible for modification of the major forms of protein phosphatase 2A in yeast. Arch. Biochem. Biophys. 395:239-245. [DOI] [PubMed] [Google Scholar]
- 27.Kalhor, H. R., A. Niewmierzycka, K. F. Faull, X. Yao, S. Grade, S. Clarke, and P. A. Rubenstein. 1999. A highly conserved 3-methylhistidine modification is absent in yeast actin. Arch. Biochem. Biophys. 370:105-111. [DOI] [PubMed] [Google Scholar]
- 28.Keith, G., and G. Dirheimer. 1980. Reinvestigation of the primary structure of brewer's yeast tRNA 3 Arg. Biochem. Biophys. Res. Commun. 92:116-119. [DOI] [PubMed] [Google Scholar]
- 29.Kennedy, T. D., and B. G. Lane. 1975. Selective labelling of the methyl carboxylate substituents found in the anticodon sequences of some species of yeast transfer RNA. Can. J. Biochem. 53:1-10. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi, T., T. Irie, M. Yoshida, K. Takeishi, and T. Ukita. 1974. The primary structure of yeast glutamic acid tRNA specific to the GAA codon. Biochim. Biophys. Acta. 366:168-181. [DOI] [PubMed] [Google Scholar]
- 31.Kuntzel, B., J. Weissenbach, R. E. Wolff, T. D. Tumaitis-Kennedy, B. G. Lane, and G. Dirheimer. 1975. Presence of the methylester of 5-carboxymethyl uridine in the wobble position of the anticodon of tRNAIII Arg from brewer's yeast. Biochimie 57:61-70. [DOI] [PubMed] [Google Scholar]
- 32.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 33.Lim, V. I., and J. F. Curran. 2001. Analysis of codon:anticodon interactions within the ribosome provides new insights into codon reading and the genetic code structure. RNA 7:942-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lustig, F., P. Elias, T. Axberg, T. Samuelsson, I. Tittawella, and U. Lagerkvist. 1981. Codon reading and translational error. Reading of the glutamine and lysine codons during protein synthesis in vitro. J. Biol. Chem. 256:2635-2643. [PubMed] [Google Scholar]
- 35.McBride, A. E., and P. A. Silver. 2001. State of the arg: protein methylation at arginine comes of age. Cell 106:5-8. [DOI] [PubMed] [Google Scholar]
- 36.Meehan, R. R., and I. Stancheva. 2001. DNA methylation and control of gene expression in vertebrate development. Essays Biochem. 37:59-70. [DOI] [PubMed] [Google Scholar]
- 37.Motorin, Y., and H. Grosjean. 1999. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA 5:1105-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nau, F. 1976. The methylation of tRNA. Biochimie 58:629-645. [DOI] [PubMed] [Google Scholar]
- 39.Niewmierzycka, A. 1998. Ph.D. thesis. University of California-Los Angeles, Los Angeles.
- 40.Niewmierzycka, A., and S. Clarke. 1999. S-Adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. Identification of a novel protein arginine methyltransferase. J. Biol. Chem. 274:814-824. [DOI] [PubMed] [Google Scholar]
- 41.Nordlund, M. E., J. O. Johansson, U. von Pawel-Rammingen, and A. S. Bystrom. 2000. Identification of the TRM2 gene encoding the tRNA(m5U54)methyltransferase of Saccharomyces cerevisiae. RNA 6:844-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ortwerth, B. J., and J. V. Carlson. 1977. Lysine transfer RNA from liver: a sulfur-containing species that codes for AAG. Arch. Biochem. Biophys. 178:278-284. [DOI] [PubMed] [Google Scholar]
- 43.Pintard, L., F. Lecointe, J. M. Bujnicki, C. Bonnerot, H. Grosjean, and B. Lapeyre. 2002. Trm7p catalyses the formation of two 2′-O-methylriboses in yeast tRNA anticodon loop. EMBO J. 21:1811-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pomerantz, S. C., and J. A. McCloskey. 1990. Analysis of RNA hydrolyzates by liquid chromatography-mass spectrometry. Methods Enzymol. 193:796-824. [DOI] [PubMed] [Google Scholar]
- 45.Raba, M., K. Limburg, M. Burghagen, J. R. Katze, M. Simsek, J. E. Heckman, U. L. Rajbhandary, and H. J. Gross. 1979. Nucleotide sequence of three isoaccepting lysine tRNAs from rabbit liver and SV40-transformed mouse fibroblasts. Eur. J. Biochem. 97:305-318. [DOI] [PubMed] [Google Scholar]
- 46.Rozenski, J., P. F. Crain, and J. A. McCloskey. 1999. The RNA modification database: 1999 update. Nucleic Acids Res. 27:196-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sprinzl, M., C. Horn, M. Brown, A. Ioudovitch, and S. Steinberg. 1998. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26:148-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sundaram, M., P. C. Durant, and D. R. Davis. 2000. Hypermodified nucleosides in the anticodon of tRNALys stabilize a canonical U-turn structure. Biochemistry 39:12575-12584. [DOI] [PubMed] [Google Scholar]
- 49.Tsurui, H., Y. Kumazawa, R. Sanokawa, Y. Watanabe, T. Kuroda, A. Wada, K. Watanabe, and T. Shirai. 1994. Batchwise purification of specific tRNAs by a solid-phase DNA probe. Anal. Biochem. 221:166-172. [DOI] [PubMed] [Google Scholar]
- 50.Uziel, M., and A. J. Weinberger. 1975. Sequence of E. coli tRNA-Glu1 by automated sequential degradation. Nucleic Acids Res. 2:469-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vicens, Q., and E. Westhof. 2001. Crystal structure of paromomycin docked into the eubacterial ribosomal decoding A site. Structure (Cambridge) 9:647-658. [DOI] [PubMed] [Google Scholar]
- 52.Weissenbach, J., and G. Dirheimer. 1978. Pairing properties of the methylester of 5-carboxymethyl uridine in the wobble position of yeast tRNA3Arg. Biochim. Biophys. Acta. 518:530-534. [DOI] [PubMed] [Google Scholar]
- 53.Williams, R. J., W. Nagel, B. Roe, and B. Dudock. 1974. Primary structure of E. coli alanine transfer RNA: relation to the yeast phenylalanyl tRNA synthetase recognition site. Biochem. Biophys. Res. Commun. 60:1215-1221. [DOI] [PubMed] [Google Scholar]
- 54.Yarian, C., H. Townsend, W. Czestkowski, E. Sochacka, A. J. Malkiewicz, R. Guenther, A. Miskiewicz, and P. F. Agris. 2002. Accurate translation of the genetic code depends on tRNA modified nucleosides. J. Biol. Chem. 277:16391-16395. [DOI] [PubMed] [Google Scholar]
- 55.Yasukawa, T., T. Suzuki, N. Ishii, S. Ohta, and K. Watanabe. 2001. Wobble modification defect in tRNA disturbs codon-anticodon interaction in a mitochondrial disease. EMBO J. 20:4794-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yokoyama, S., T. Watanabe, K. Murao, H. Ishikura, Z. Yamaizumi, S. Nishimura, and T. Miyazawa. 1985. Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. Proc. Natl. Acad. Sci. USA 82:4905-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yokoyama, S., and S. Nishimura. 1995. Modified nucleosides and codon recognition, p. 207-223. In D. Söll and U. L. RajBhandary (ed.), tRNA: structure, biosynthesis, and function. ASM Press, Washington, D.C.
- 58.Zobel-Thropp, P., J. D. Gary, and S. Clarke. 1998. Delta-N-methylarginine is a novel posttranslational modification of arginine residues in yeast proteins. J. Biol. Chem. 273:29283-29286. [DOI] [PubMed] [Google Scholar]
- 59.Zobel-Thropp, P., M. C. Yang, L. Machado, and S. Clarke. 2000. A novel post-translational modification of yeast elongation factor 1A. Methylesterification at the C terminus. J. Biol. Chem. 275:37150-37158. [DOI] [PubMed] [Google Scholar]