Abstract
Background
The emergence of artesunate-mefloquine (AS+MQ)–resistant Plasmodium falciparum in the Thailand-Cambodia region is a major concern for malaria control. Studies indicate that copy number increase and key alleles in the pfmdr1 gene are associated with AS+MQ resistance. In the present study, we investigated evidence for a selective sweep around pfmdr1 because of the spread of adaptive mutation and/or multiple copies of this gene in the P. falciparum population in Cambodia.
Methods
We characterized 13 microsatellite loci flanking (± 99 kb) pfmdr1 in 93 single-clone P. falciparum infections, of which 31 had multiple copies and 62 had a single copy of the pfmdr1 gene.
Results
Genetic analysis revealed no difference in the mean (± standard deviation) expected heterozygosity (He) at loci around single (0.75 ± 0.03) and multiple (0.76 ± 0.04) copies of pfmdr1. Evidence of genetic hitchhiking with the selective sweep of certain haplotypes was seen around mutant (184F) pfmdr1 allele, irrespective of the copy number. There was an overall reduction of 28% in mean He (± SD) around mutant allele (0.56 ± 0.05), compared with wild-type allele (0.84 ± 0.02). Significant linkage disequilibrium was also observed between the loci flanking mutant pfmdr1 allele.
Conclusion
The 184F mutant allele is under selection, whereas amplification of pfmdr1 gene in this population occurs on multiple genetic backgrounds.
The Thailand-Cambodia border has been an epicenter for drug-resistant malaria and is where resistance to chloroquine (CQ) emerged in the early 1960s [1]. Sulfadoxine-pyrimethamine (SP) officially became the first-line treatment for falciparum malaria in 1989, followed by mefloquine (MQ) in 1994, although both drugs were likely available in the private sector much earlier. Resistance to MQ exceeded 25% (RI–RIII) in western Cambodia in the mid-1990s, and artemisinin-based combination therapy (ACT) consisting of artesunate (AS) and MQ, was adopted as the first-line treatment against Plasmodium falciparum countrywide in 2000 [2]. There is growing evidence that efficacy of AS+MQ is decreasing along the Thailand-Cambodia border [3–7], as well as in southwestern Cambodia [8].
One of the most widely used molecular markers for predicting MQ or AS+MQ early treatment failure and/or recrudescence is an increase in copy number in the pfmdr1 gene [3, 8–10]. The pfmdr1 gene amplification has been found to be associated with resistance to MQ, AS, quinine (QN), and halofantrine (HAL) in in vitro–selected parasite lines [11–13] and genetically modified parasites [14], as well as in clinical isolates [9, 15–18]. In addition to gene amplification, pfmdr1 alleles 86N, 1034S, 1042N, and 1246D have also been associated with in vitro resistance to MQ and AS in genetically modified parasite lines [19, 20] and clinical isolates [17, 21]. Field studies have also shown selection of the 86N allele in recurrent infections after treatment with AS+MQ or artemether plus lumefantrine (AL) [9, 22–24], which suggests that 86N could be a potential marker of MQ and lumefantrine resistance in vivo. However, no clear association between pfmdr1 184F mutation and MQ failure has been established, although this allele is widespread in Cambodia [25]. Western Cambodia has a higher prevalence of MQ resistance than does eastern Cambodia, and this is consistent with an increased frequency of multiple copies of pfmdr1 and the 184F mutation in the western part of the country, compared with the eastern part of the country [25, 26]. Although AS+MQ treatment policy for uncomplicated falciparum malaria applies for the whole of Cambodia, the difference in drug pressure between western and eastern Cambodia is well known. The available data indicate high efficacy (~100%) of AS+MQ in eastern Cambodia, compared with Pailin in western Cambodia, where a reduced efficacy (79%) of AS+MQ has been reported [6, 25].
Microsatellites are important markers to identify regions in the genome that have been under selection. During the process of selection, a beneficial mutation spreads through the population that results in a reduction of heterozygosity at both the selected locus and neutral flanking microsatellite loci (genetic hitchhiking) [27]. Under continuous selection, the mutation eventually gets fixed in the population, and sequence diversity is reduced around the selected locus (selective sweep). Evidence of selective sweeps around drug-resistant pfcrt and dhfr, dhps alleles due to CQ and SP drug pressure, respectively, have been described in several P. falciparum populations [28–32]. Microsatellite analysis around these alleles also revealed that CQ and SP resistance originated independently at only a few places and later spread to other parts of the world, which suggests the role of gene flow in the evolution of drug resistance [29, 30, 32–34]. Hence, it is important to investigate whether evolving MQ resistance has a similar genetic basis.
Previous studies have addressed the genetic basis of pfmdr1 gene amplification and found that the amplification involves a wide chromosomal region (up to 100 kb) and contains multiple copies of the pfmdr1 gene [11, 15, 35]. Recently, these findings were further confirmed in P. falciparum isolates from the Thailand-Myanmar border, which showed that a 15–49 kb region of the chromosome 5 is amplified and contains 2–4 copies of pfmdr1 [36]. It is not known whether the amplification of the pfmdr1 gene in Cambodian parasites is similar.
Here, we have attempted to understand the nature of selective sweeps occurring around pfmdr1 caused by the spread of adaptive mutation and/or multiple copies of this gene in the P. falciparum population in Cambodia. This study also addresses geographical differences in the pfmdr1 haplotype pattern between eastern and western Cambodia.
MATERIALS AND METHODS
Sample collection, DNA isolation, pfmdr1 genotyping, and copy number estimation
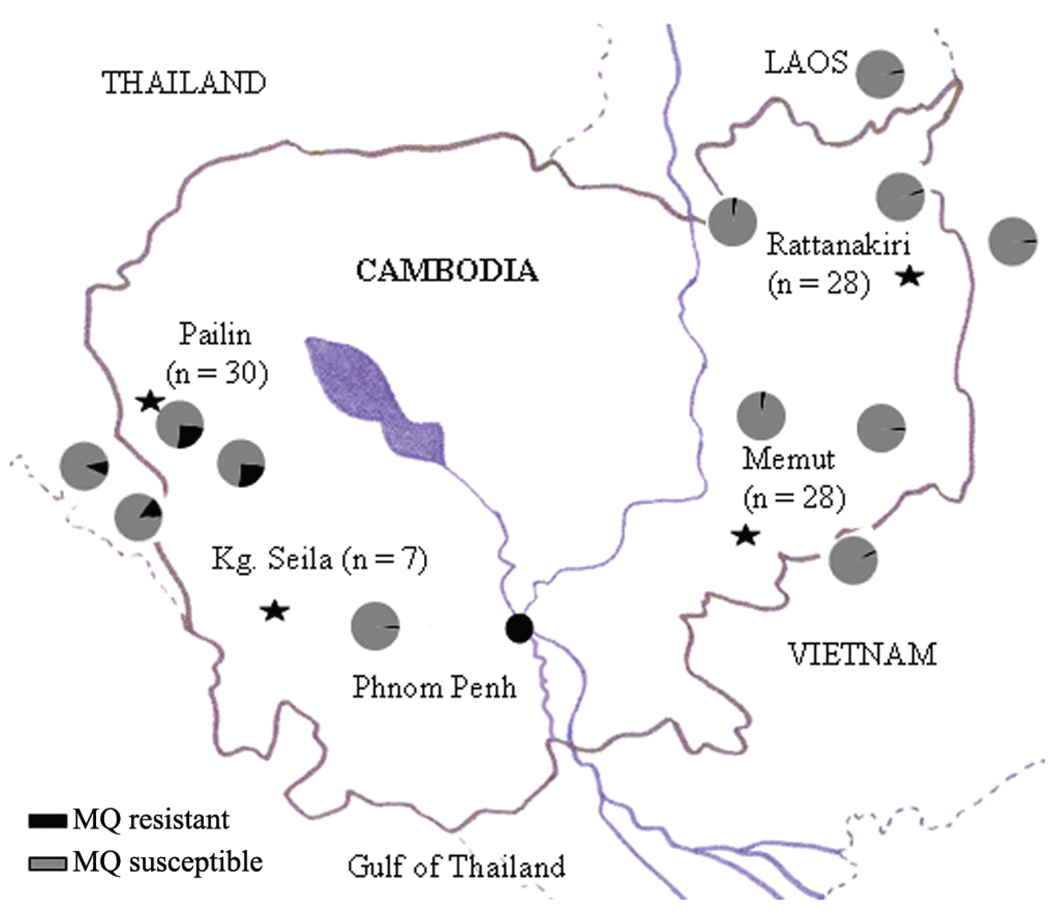
Clinical isolates of P. falciparum were collected from patients with uncomplicated falciparum malaria from 4 sites across Cambodia: Pailin and Kampong Seila in western Cambodia and Memut and Rattanakiri in eastern Cambodia, as described earlier [25]. The location of the study sites and distribution of MQ resistance is shown in Figure 1. Epidemiologically, malaria incidence is much lower in western Cambodia than it is in eastern Cambodia. Eastern Cambodia is less developed, has a greater population of ethnic minorities, and has a very poor public health system. In eastern Cambodia, local transmission within villages is more common, and all age groups are affected. In western Cambodia, malaria is more predominant among adults, who are usually occupationally exposed in the jungles (ie, outside of their villages), than it is among other age groups.
Figure 1.
A map of Cambodia showing the location of the 4 sites (Pailin, Kampong Seila, Memut, and Rattanakiri) from which the isolates used in this study were obtained. The proportion of mefloquine (MQ) resistance in Cambodia and bordering areas (Thailand to the west, Vietnam to the east, and Laos to the north) is indicated by pie charts.
This study was approved by the Institutional Review Boards of the Cambodia National Ethics Committee for Health Research, the US Naval Medical Research Unit No.2 (Jakarta, Indonesia), and the University of North Carolina at Chapel Hill (Chapel Hill, NC).Written informed consent was obtained from each participant before blood samples were collected. The patients were treated with AS+MQ in accordance with current national antimalarial drug policy.
DNA was extracted from filter paper blood spots using QIAamp Mini kit (Qiagen). The pfmdr1 copy number estimation and genotyping of pfmdr1 codons 86, 184, 1034, 1042, and 1246 was done previously using real-time polymerase chain reaction (PCR) [25]. These coded DNA samples were sent to the Centers for Disease Control and Prevention laboratory at Atlanta, Georgia, for further analysis. Of 158 DNA samples, 62 had multiple copies (≥2) of pfmdr1, whereas the remaining 96 samples had single copies of pfmdr1 [25]. We also performed direct sequencing of partial fragments of the pfmdr1 (for codons 86–184 and 1034–1246) on these samples to analyze any additional mutations. The pfmdr1 fragments were amplified in 2 separate PCR reactions. The first fragment (~799 base pairs [bp]) covered codons 86–184 was amplified using AL6875F (5′-CCGTTTAAATGTTTACCTGCAC-3′) and AL6876R (5′-TGGGGTATTGATTCGTTGCAC-3′) primers with the following PCR cycling parameters: initial denaturation at 94°C for 10 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 57°C for 1 min, extension at 72°C for 1 min, and a final extension at 72°C for 10 min. The second fragment (~909 bp) that covered codons 1034–1246 was amplified using AL6794F (5′-TATGCATACTGTTATTAATTATGG-3′) and AL6795R (5′-TTCGATAAATTCATCTATAGCAG-3′) primers. The PCR cycling parameters for these primers were also the same as for the first fragment, except that an annealing temperature of 56°C was used. The PCR product was sequenced on an ABI 3130xl Genetic Analyzer using BigDye Terminator, version 3.1 (Applied Biosystems). The internal primers AL6877F (5′-GTATGTGCTGTATTATCAGGAG-3′) and AL6878R (5′-AGCCTCTTCTATAATGGACATG-3′) were used to sequence the first fragment, whereas the second fragment was sequenced using AL6794F and AL6795R primers.
Microsatellite allele scoring and statistical analysis
All 158 isolates were first assayed for 8 neutral microsatellite loci on chromosomes 2 and 3, unlinked to any gene under selection [37]. Of 158 samples, 65 had multiple alleles at ≥1 neutral loci and thus were excluded from the study. The remaining 93 isolates were further analyzed for 13 microsatellite loci (−99, −54, −29.5, −9.3, −4.2, and −3.3 kb upstream and 0, 0.16, 0.45, 3.6, 9.1, 23.3, 89 kb downstream) flanking the pfmdr1 gene on chromosome 5. Primer sequences and PCR cycling parameters for all 8 neutral loci and 13 pfmdr1 loci have been described elsewhere [31, 36, 37]. PCR products were separated on ABI 3130xl Genetic Analyzer and analyzed using Gene-Mapper software, version 3.7 (Applied Biosystems).
The genetic variation at each locus was measured as expected heterozygosity (He) and number of alleles per locus (A). He was calculated using the formula , where n is the number of samples genotyped for that locus and pi is the frequency of the ith allele. The Microsatellite ToolKit was used to compute He, pi as well as A [38]. The sampling variance for He was calculated as . The closest 8 loci (± 9 kb) around pfmdr1 (−9.3, −4.2, −3.3, 0 [within gene], 0.16, 0.45, 3.6 and 9.1 kb) were used for grouping isolates or parasites into haplotypes. The haplotypes were considered to be unique if they differed at even 1 of the 8 loci analyzed.
Significant associations between pairs of loci were determined on the basis of an exact test of linkage disequilibrium performed in Arlequin, version 3.01 [39]. To correct for multiple testing, Bonferroni correction was used. The genetic differentiations between groups (single copy vs multiple copies and wild-type [184Y] vs mutant [184F] alleles) were determined through Wright’s fixation index (FST) using Arlequin, version 3.01 [39].
We used the same 8 loci (± 9 kb) to make a median-joining haplotypes network (Network, version 4.5.1.0.) to predict the genetic relationship among the haplotypes. Median joining networks are used for reconstructing the phylogeny of regions with reticulate evolution [40].
RESULTS
pfmdr1 Mutant alleles and copy number in Cambodia
We sequenced the pfmdr1 region covering codons 86–184 and 1034–1246 in Cambodian P. falciparum isolates. We grouped the isolates from all 4 sites on the basis of whether they harbored a single copy or multiple (≥2) copies of pfmdr1. Overall, the pfmdr1 sequencing results for 93 isolates (from patients with single-clone infections) revealed that only a minority (2.15%) contained mutations at codon 86 (86Y), whereas ~54% of the isolates harbored a mutation at codon 184 (184F). None of the isolates contained mutation at codon 1246. Mutations at codons 1034 and 1042 were only observed in conjunction with mutation at codon 184 (Table 1). Two major differences in the distribution of pfmdr1 alleles and copy number were observed between isolates from western and eastern Cambodia. First, we observed an unbiased distribution of 184F mutation in isolates from western Cambodia (Pailin and Kampong Seila), where both isolates with a single copy and isolates with multiple copies of pfmdr1 carried the 184F mutation. This was in striking contrast to the eastern Cambodian (Memut and Rattanakiri) population, where the 184F mutation was completely absent in isolates with multiple copies of pfmdr1. Second, the 86Y mutation and the recently described mutations 130K and 1109I [41] were observed exclusively in the eastern population, albeit at low frequencies (3.5%, 3.5%, and 7.1%, respectively). However, both populations contained the double mutant 184F plus 1042D, although this was found exclusively in isolates that contained a single copy of pfmdr1.
Table 1.
Distribution of Mutations at Codons 86, 130, 184, 1034, 1042, and 1109 in pfmdr1 Gene in Cambodian Plasmodium falciparum Isolates
| Study site, copy number | No. of isolates (n = 93) |
Wild- type pfmdr1 |
Single mutation | Double mutation | |||||
|---|---|---|---|---|---|---|---|---|---|
| 86Y | 130K | 184F | 1109I | 184F plus 1034C |
184F plus 1042D |
130K plus 1109I |
|||
| Western Cambodia | |||||||||
| Pailin | |||||||||
| SC | 16 | 1 | … | … | 12 | … | 1 | 2 | … |
| MC | 14 | 4 | … | … | 10 | … | … | … | … |
| Kampong Seila | |||||||||
| SC | 4 | … | … | … | 4 | … | … | … | … |
| MC | 3 | … | … | … | 3 | … | … | … | … |
| Eastern Cambodia | |||||||||
| Rattanakiri | |||||||||
| SC | 16 | 9 | … | … | 7 | … | … | … | … |
| MC | 12 | 8 | 1 | … | … | 3 | … | … | … |
| Memut | |||||||||
| SC | 26 | 12 | 1 | 1 | 8 | … | … | 3 | 1 |
| MC | 2 | 2 | … | … | … | … | … | … | … |
| Overall | |||||||||
| SC | 62 | 22 | 1 | 1 | 31 | … | 1 | 5 | 1 |
| MC | 31 | 14 | 1 | … | 13 | 3 | … | … | … |
NOTE. No singleton mutations were found at pfmdr1 codons 1034 and 1042. MC, isolates bearing multiple (≥2) copies of pfmdr1; SC, isolates bearing a single copy of pfmdr1.
Genetic variation at microsatellite loci flanking pfmdr1 and selective sweep of 184F allele
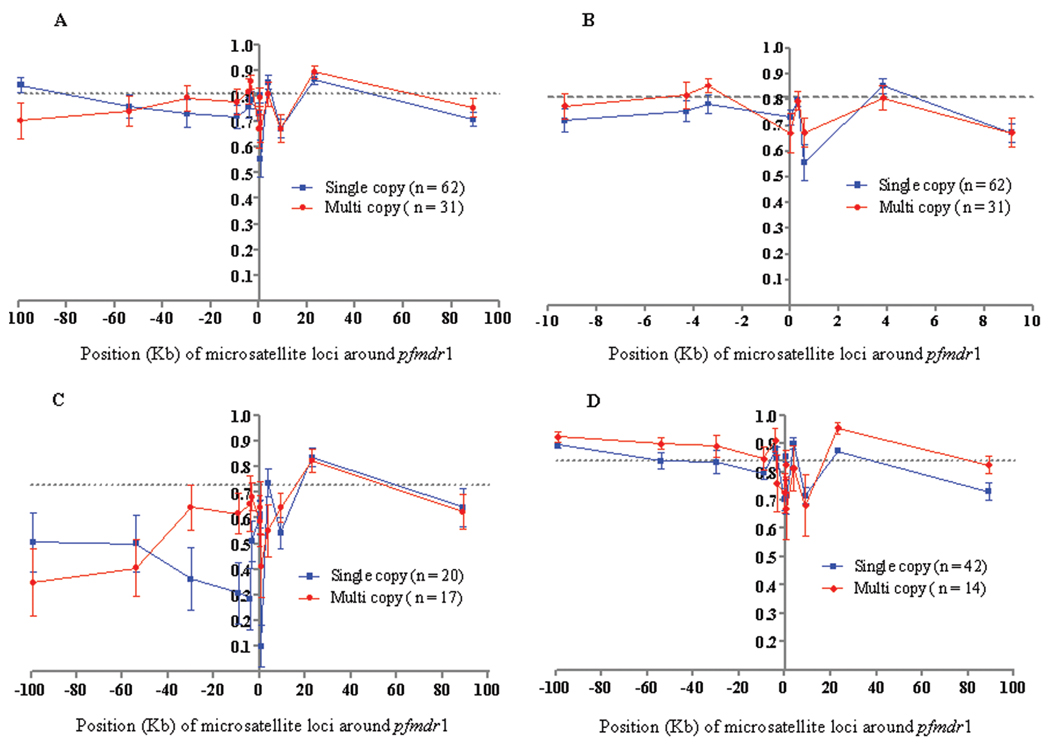
We attempted to study the genetic hitchhiking of flanking pfmdr1 loci resulting from the spread of multiple copies of pfmdr1 and/or mutant 184F allele among these isolates. In this regard, we compared the level of heterozygosity (He) and number of alleles per locus (A) on the basis of single versus multiple copies of pfmdr1 and wild-type (184Y) versus mutant (184F) groups (Figures 2 and 3 and Table 2). Overall, fewer alleles and less genetic diversity were found in the loci surrounding pfmdr1 than were found in the neutral loci on chromosomes 2 and 3 (Table 2). However, there were no obvious differences in the heterozygosities for any of the 13 loci between samples with single copies and those with multiple copies of pfmdr1. The single-copy and multiple-copy groups had almost identical mean He values (± standard deviation [SD]) of 0.75 ± 0.03 and 0.76 ± 0.04, respectively (Figure 2A and 2B). However, interpopulation analysis revealed markedly reduced He at almost all of the loci in the western Cambodian population, compared with the eastern Cambodian population (Figure 2C and 2D).
Figure 2.
The expected heterozygosity (He) at 13 microsatellite loci around pfmdr1. Comparison of He between groups of isolates with a single copy or multiple copies of pfmdr1. The mean He at neutral loci on chromosomes 2 and 3 is shown by a dotted line. The error bars indicate ± 1 standard deviation. A, isolates from both western and eastern Cambodia; B, detail showing the closest 8 microsatellite loci flanking (± 9 kb) pfmdr1 for isolates from western and eastern Cambodia; C, isolates from western Cambodia; D, isolates from eastern Cambodia.
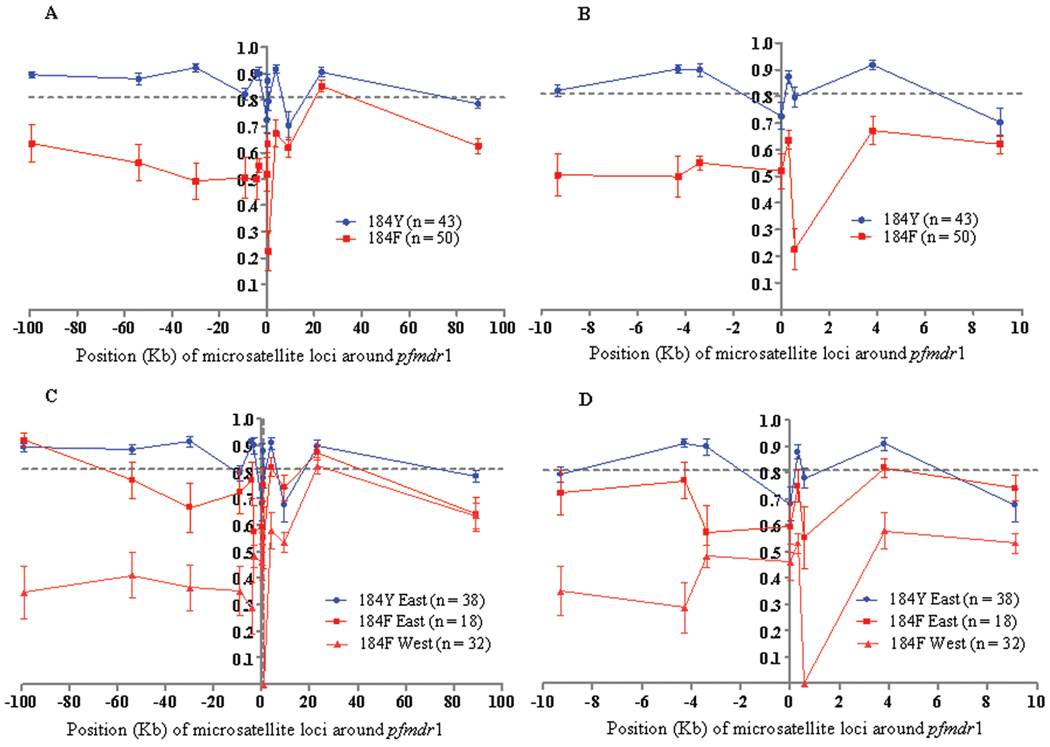
Figure 3.
The expected heterozygosity (He) at 13 loci around wild-type (184Y) and mutant (184F) pfmdr1. Comparison of He between 184Y and 184F pfmdr1 groups of isolates from both western and eastern Cambodia (A) and the closest 8 microsatellite loci flanking (± 9 kb) pfmdr1 (B), and comparison of 184F group of isolates from western Cambodia with 184F and 184Y groups of isolates from eastern Cambodia (C) and the closest 8 microsatellite loci flanking (± 9 kb) pfmdr1 (D). The mean He at neutral loci on chromosomes 2 and 3 is shown by a dotted line. The error bars indicate ± 1 standard deviation.
Table 2.
Genetic Characteristics of the 13 Microsatellite Loci around pfmdr1 on Chromosome 5 and 8 Neutral Microsatellite Loci (4 Each on Chromosomes 2 and 3) Based on pfmdr1 Copy Number and 184F Mutation
| Loci | Based on copy number | Based on Y184F | ||||||
|---|---|---|---|---|---|---|---|---|
| Single copy (n = 62) |
Multiple copies (n = 31) |
184Y (n = 43) |
184F (n = 50) |
|||||
| A | He ± SD | A | He ± SD | A | He ± SD | A | He ± SD | |
| pfmdr1 | ||||||||
| −99 kb | 13 | 0.84 ± 0.03 | 8 | 0.70 ± 0.07 | 11 | 0.89 ± 0.01 | 10 | 0.63 ± 0.07 |
| −54 kb | 12 | 0.75 ± 0.04 | 7 | 0.74 ± 0.06 | 12 | 0.88 ± 0.02 | 7 | 0.56 ± 0.07 |
| −29.5 kb | 14 | 0.73 ± 0.05 | 9 | 0.79 ± 0.05 | 14 | 0.92 ± 0.01 | 7 | 0.49 ± 0.07 |
| −9.3 kb | 7 | 0.72 ± 0.04 | 7 | 0.77 ± 0.05 | 8 | 0.82 ± 0.02 | 6 | 0.50 ± 0.07 |
| −4.2 kb | 11 | 0.76 ± 0.04 | 11 | 0.81 ± 0.05 | 13 | 0.90 ± 0.01 | 7 | 0.49 ± 0.07 |
| −3.3 kb | 12 | 0.78 ± 0.03 | 10 | 0.85 ± 0.02 | 14 | 0.90 ± 0.02 | 4 | 0.55 ± 0.02 |
| 0 kb | 8 | 0.73 ± 0.02 | 8 | 0.67 ± 0.07 | 10 | 0.72 ± 0.05 | 5 | 0.51 ± 0.06 |
| 0.16 kb | 10 | 0.81 ± 0.02 | 7 | 0.79 ± 0.03 | 12 | 0.87 ± 0.02 | 5 | 0.63 ± 0.03 |
| 0.45 kb | 10 | 0.56 ± 0.07 | 6 | 0.67 ± 0.05 | 9 | 0.79 ± 0.03 | 5 | 0.22 ± 0.07 |
| 3.6 kb | 14 | 0.85 ± 0.02 | 9 | 0.80 ± 0.04 | 13 | 0.91 ± 0.01 | 7 | 0.67 ± 0.05 |
| 9.1 kb | 5 | 0.67 ± 0.03 | 6 | 0.67 ± 0.05 | 7 | 0.70 ± 0.05 | 5 | 0.61 ± 0.03 |
| 23.3 kb | 11 | 0.86 ± 0.02 | 11 | 0.89 ± 0.02 | 12 | 0.90 ± 0.01 | 10 | 0.85 ± 0.02 |
| 89 kb | 5 | 0.71 ± 0.02 | 5 | 0.75 ± 0.03 | 6 | 0.78 ± 0.01 | 4 | 0.62 ± 0.03 |
| Mean value | 10.15 | 0.75 ± 0.03 | 8 | 0.76 ± 0.04 | 10.84 | 0.84 ± 0.02 | 6.30 | 0.56 ± 0.05 |
| Chr 2a | ||||||||
| C2M27 | 13 | 0.91 ± 0.02 | 12 | 0.91 ± 0.02 | 14 | 0.92 ± 0.01 | 12 | 0.85 ± 0.03 |
| C2M29 | 10 | 0.68 ± 0.06 | 9 | 0.77 ± 0.07 | 10 | 0.74 ± 0.06 | 11 | 0.71 ± 0.06 |
| C2M34 | 17 | 0.91 ± 0.02 | 17 | 0.95 ± 0.01 | 15 | 0.92 ± 0.01 | 17 | 0.91 ± 0.02 |
| C2M33 | 15 | 0.89 ± 0.02 | 11 | 0.89 ± 0.02 | 14 | 0.92 ± 0.01 | 14 | 0.88 ± 0.02 |
| Mean value | 13.75 | 0.85 ± 0.05 | 12.25 | 0.88 ± 0.03 | 13.25 | 0.87 ± 0.04 | 13.50 | 0.84 ± 0.04 |
| Chr 3a | ||||||||
| C3M40 | 16 | 0.89 ± 0.02 | 14 | 0.93 ± 0.02 | 15 | 0.91 ± 0.02 | 14 | 0.87 ± 0.02 |
| C3M88 | 16 | 0.91 ± 0.01 | 11 | 0.92 ± 0.01 | 12 | 0.91 ± 0.01 | 13 | 0.91 ± 0.01 |
| C3M69 | 11 | 0.85 ± 0.02 | 8 | 0.82 ± 0.03 | 11 | 0.88 ± 0.02 | 10 | 0.82 ± 0.03 |
| C3M39 | 6 | 0.44 ± 0.07 | 3 | 0.31 ± 0.11 | 6 | 0.59 ± 0.07 | 3 | 0.19 ± 0.07 |
| Mean value | 12.25 | 0.77 ± 0.11 | 12.25 | 0.75 ± 0.14 | 11 | 0.82 ± 0.07 | 10 | 0.71 ± 0.16 |
NOTE. The isolates were considered as multiple copy isolates if they contained ≥2 copies of pfmdr1 gene as determined by real-time quantitative polymerase chain reaction. A, number of alleles per locus; He, expected heterozygosity; SD, standard deviation.
Interestingly, evidence of genetic hitchhiking was seen that was due to selective sweep of 184F pfmdr1 allele in the population. There was an overall reduction of 28% in mean He (± SD) observed for the isolates carrying the 184F allele (0.56 ± 0.05), compared with those harboring the wild-type 184Y allele (0.84 ± 0.02) (Figure 3A and 3B and Table 2). The difference in He between wild-type (0.86) and mutant (0.53) alleles was greater (33% reduction) in the upstream region of pfmdr1. On the other hand, the difference in He between wild-type (0.82) and mutant (0.60) alleles was relatively less (22% reduction) in the downstream region (Figure 3A). To investigate the role of 184F-driven selective sweep, we compared the heterozygosities around the 184Y and 184F alleles in isolates from eastern Cambodia. The results revealed a reduction in heterozygosity around 184F, compared with the wild-type allele, in the eastern Cambodian population (Figure 3C and 3D). The reduction around the 184F allele was greater in the isolates from western Cambodia (Figure 3C and 3D). We could not compare heterozygosities around the 184Y allele in western Cambodia, because there were only 4 isolates that harbored this allele.
Linkage disequilibrium between loci flanking pfmdr1
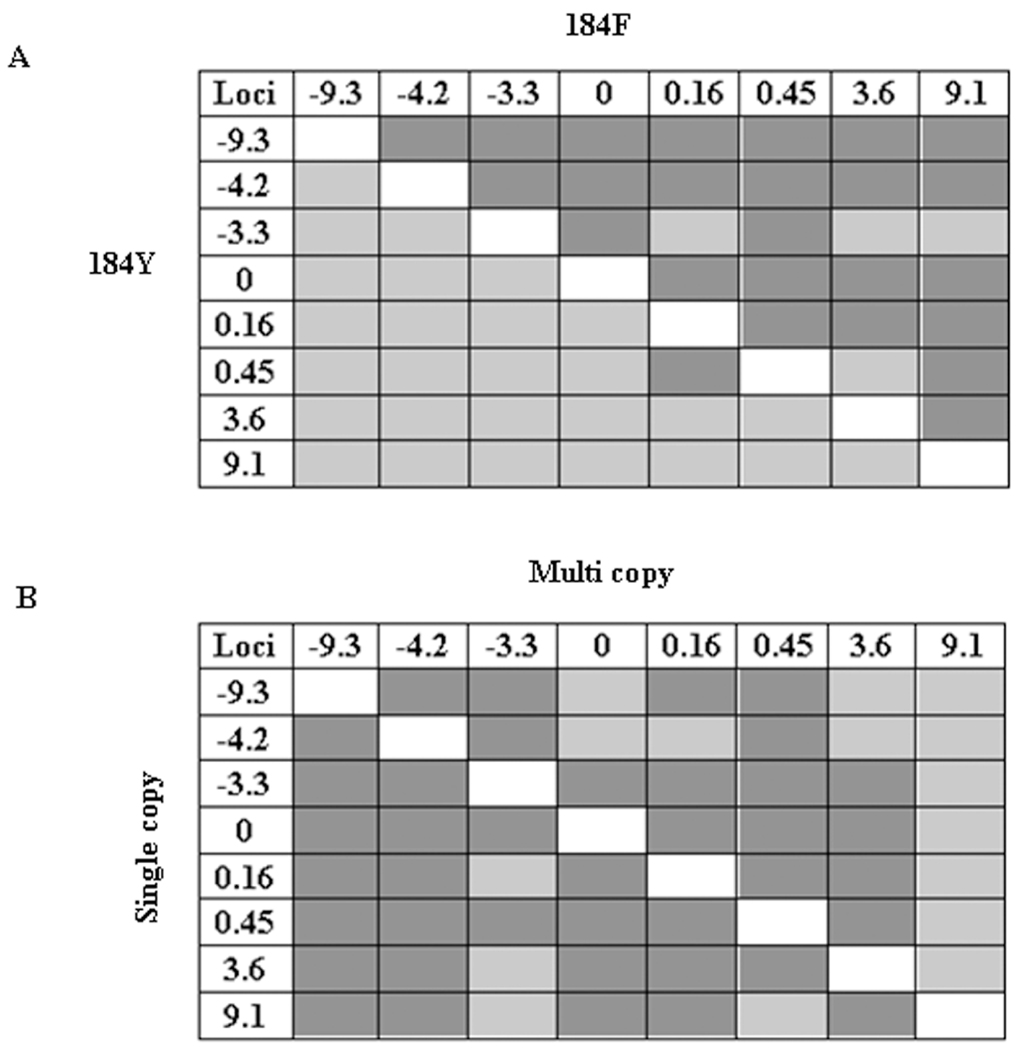
The pattern of linkage disequilibrium was also deduced to explore the extent of 184F-induced selective sweeps operating in the population. The pair-wise linkage disequilibrium distribution between the closest (± 9 kb) pfmdr1 microsatellite loci is shown in Figure 4. Significant linkage disequilibrium was observed between loci close to pfmdr1 in the group with the mutant 184F allele, but no association was observed between loci in the group with the 184Y allele (Figure 4A). On the other hand, we found significant linkage disequilibrium between these loci in both groups with single or multiple copies of the pfmdr1 gene (Figure 4B).
Figure 4.
Pair-wise linkage disequilibrium around pfmdr1 (± 9 kb). A, linkage disequilibrium between loci within the wild-type group (lower half of the diagonal) and mutant group (upper half of the diagonal). B, linkage disequilibrium between loci within the group of isolates with a single copy of pfmdr1 (lower half of the diagonal) and those with multiple copies of pfmdr1 (upper half of the diagonal). Each cell shows a comparison between polymorphic pairs of loci. P < .006 was considered to be significant after Bonferroni correction. Light gray cells represent P values that are not significant (>.006), and dark gray cells indicate statistically significant P values (<.006).
Genetic differentiation and relationships among pfmdr1 microsatellite haplotypes
The genetic differentiation between the 184Y (wild-type) and 184F (mutant) groups using the closest 8 microsatellite loci (± 9 kb) around the pfmdr1 gene was measured employing Wright’s fixation index, FST. Significant FST (FST, 0.03; P < .001) was observed between the wild-type (184Y) and mutant (184F) groups, as would be expected because of the decrease in genetic diversity within this subpopulation. Unsurprisingly, a low but insignificant level of genetic differentiation was observed between subpopulations with a single copy and those with multiple copies of pfmdr1 (FST, 0.003; P > .05).
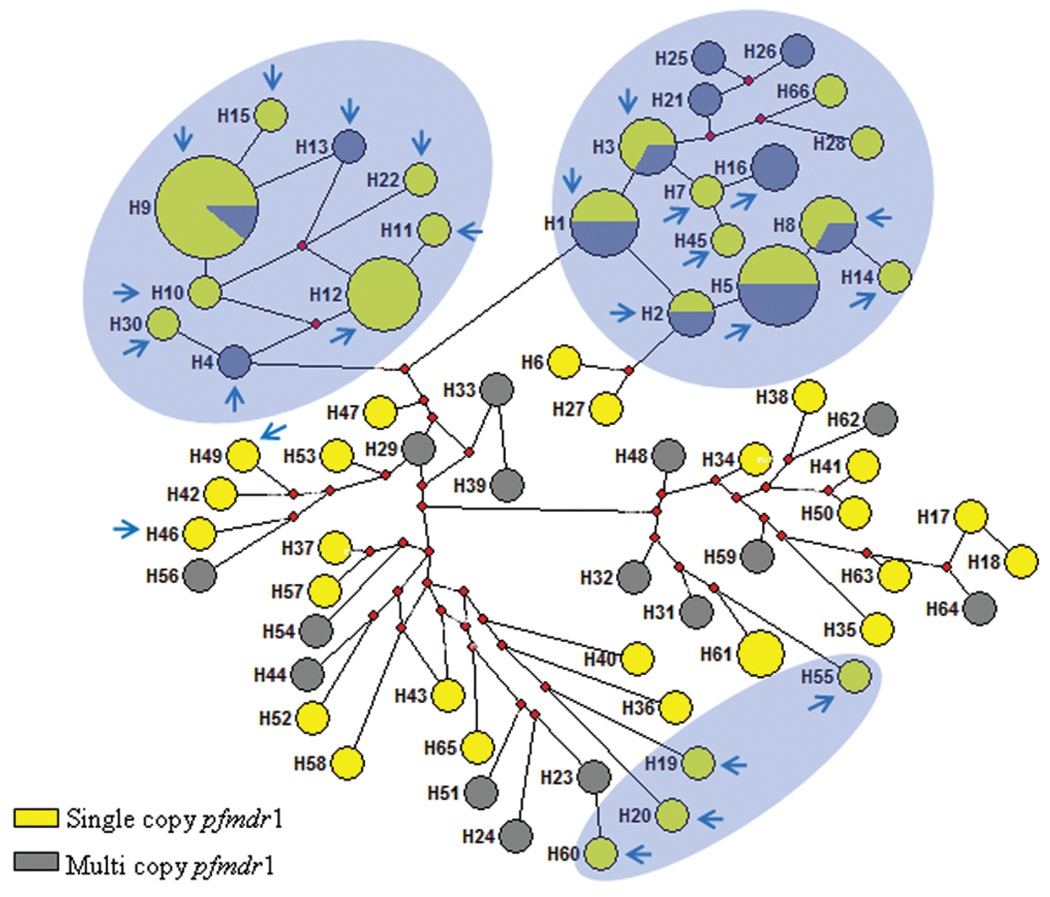
The same 8 loci (± 9 kb) were also used for classifying the 93 isolates into 66 haplotypes (H1–H66) (Figure 5). When we looked at the distribution of haplotypes irrespective of pfmdr1 copy number by geography, we found fewer haplotypes in western Cambodia (19 haplotypes among 37 isolates) than in eastern Cambodia (51 haplotypes among 56 isolates), which suggests less genetic diversity in western Cambodia. Only 4 haplotypes (H1, H8, H9, and H12) were shared between eastern and western isolates, including isolates that bore a single copy and those that bore multiple copies of pfmdr1 (Figure 5). Overall, the subpopulation with multiple copies of pfmdr1 displayed a relatively smaller number of haplotypes (27) than did the subpopulation with single copies (45) (Figure 5). Only 6 haplotypes (H1, H2, H3, H5, H8, and H9) were shared by subpopulations with both single and multiple pfmdr1 copies, and all of them had the pfmdr1 184F mutation (Figure 5). We also found that wild-type and mutant pfmdr1 alleles did not share any microsatellite haplotypes. The haplotypes with the mutant 184F allele (on genetic backgrounds of both single and multiple pfmdr1 copies) were found to form 3 main independent clusters (Figure 5).
Figure 5.
Median-joining network showing genetic relationships among pfmdr1 haplotypes in Cambodia. The closest 8 loci (± 9 kb) were used for constructing the network. Each circle in this network represents a unique haplotype, with the size of the circle being proportional to the number of isolates harboring that haplotype. The circles shown in yellow and gray represent isolates carrying single and multiple copies of pfmdr1, respectively. The haplotype shared by both isolates with single and isolates with multiple pfmdr1 copies (proportion indicated in pie charts) are shown as a circle with both yellow and gray shading. The red dots are the median vectors that imply software-generated “hypothetical haplotypes,” joining 2 haplotypes in the diagram from which single and/or multiple pfmdr1 copy–containing haplotypes may have originated. The isolates with 184F are marked by blue arrows enclosed in the shaded clusters.
DISCUSSION
Resistance to AS+MQ has developed on the Thailand–Cambodia border. This study provides comprehensive data on the distribution of pfmdr1 alleles and the pattern of selective sweeps in 4 sites in Cambodia with different levels of transmission and different drug resistance profiles. As has been shown elsewhere [25], our results from pfmdr1 sequencing also confirmed the high prevalence (~86%) of the 184F allele in western Cambodia (Table 1), where a significant level of MQ resistance has been reported. On the other hand, the prevalence of 184F allele in eastern Cambodia was low (~32%), which also correlates with the reduced level of MQ resistance in this region (Table 1). In western Cambodia, the pfmdr1 amplification was found to occur on both 184Y and 184F backgrounds, in contrast with eastern Cambodia, where all of the amplification was found on the 184Y background (Table 1). A possible explanation for this is that the pfmdr1 amplification is independent of the acquisition of the 184F mutation by the parasites. Although an increased copy number in the pfmdr1 gene has been associated with MQ resistance, the significance of the 184F mutation remains less well understood [9, 42]. A study on the Southeast Asian P. falciparum population showed that isolates that fall under category I (all wild-type pfmdr1 codons: 86N, 184Y, 1034S, and 1042N) and category III (mutation at only 184 codon: 86N, 184F, 1034S, and 1042N) exhibited increased resistance to MQ and AS [17]. Also, the isolates with amplified pfmdr1 fell into category I or III and were significantly more resistant to these drugs. A recent study from Cambodia demonstrated that isolates with single 184F mutation had a significantly elevated 50% inhibitory concentration for MQ [41]. Similar results were obtained in studies from Tanzania and Uganda, where treatment with another widely used ACT (AL) selected for reinfecting parasites with 86N, 184F, and 1246D pfmdr1 alleles [24, 43, 44]. The wild-type 86N allele has been found to be associated with increased 50% inhibitory concentrations to MQ and AS [9, 18, 42, 45].
We characterized microsatellites flanking (± 99 kb) the pfmdr1 gene to estimate the strength of genetic hitchhiking resulting from the spread of the 184F mutant allele and/or amplification events in pfmdr1. As evident from Figure 3, 184F mutant allele appears to be under strong selection with reduced variation spanning from 99 kb upstream to 88 kb downstream of the gene. Generally, downstream loci exhibited a lower reduction in He, except at the 0.45-kb locus, where we found a sharp reduction of 57%, compared with wild-type pfmdr1 (Table 2 and Figure 3). We could not find in the PlasmoDB any other potential gene under selection around this locus that could be responsible for such a sharp reduction in He. Present data seems to suggest that the 184F allele has undergone a selective sweep, but it is not clear whether MQ or other drug pressure is the cause for this selection. Although a previous study by Nair et al [36] reported that a significant proportion (30%; n = 326) of the Thailand isolates harbor the 184F allele, the authors did not mention any genetic hitchhiking resulting from the spread of this allele.
Amplification of the pfmdr1 gene was found to occur on multiple haplotype backgrounds based on the typing of 13 microsatellite loci (Figure 5). Our observation indicating multiple origins for the development of strains with multiple copies of pfmdr1 in Cambodia is in agreement with previous observations in Thailand and other studies [35, 36]. Nair et al [36] have described a “soft selective sweep” in the isolates with multiple copies of pfmdr1 from the border of Thailand and Myanmar. They have shown a “limited” reduction (~42%; range, 21%–63%) in heterozygosities in the 16 loci surrounding (± 18 kb) pfmdr1 in isolates bearing multiple copies, compared with those carrying a single copy. Our results are also consistent with the soft selective sweep model, because it is evident that putatively advantageous alleles with multiple copies of pfmdr1 have entered the population recurrently (Figure 5). Thus, we may not have detected a decrease in heterozygosity simply because the strength of that pattern is affected by the number and neutral divergence among adaptive equivalent alleles [46]. The amplification of pfmdr1 has been suggested to be a frequent event, and an increase from a single-copy state to having 2 copies of pfmdr1 has been found to occur in every 108 parasites [47]. The rate of occurrence of gene duplication is much higher than that of mutation within codons, which occurs at a much lower frequency (1:1014) [48]. In the presence of MQ drug pressure, an increase in pfmdr1 copy number has been observed, whereas deamplification occurs under CQ pressure in laboratory isolates [12–14, 49].
We attempted to study the genetic relationships among haplotypes using Network to visualize whether the multiple pfmdr1 copy–bearing haplotype emerged from the single-copy haplotype (Figure 5). Indeed, the multiple pfmdr1 copy group was found to share some haplotypes with the single-copy group. Possible explanations for not finding the progenitor haplotype from which single or multiple pfmdr1 copies may have emerged in our sample set could be attributable to a recent selective sweep (ie, not enough time has passed to detect selective changes) or attributable to recurring pfmdr1 copy number changes that promote duplication events on multiple genetic backgrounds. The negligible FST value and similarity between the amount of linkage disequilibrium between single and multiple pfmdr1 copy sub-populations suggests that the frequency of duplication events is independent of the genetic background. Other studies similarly suggest multiple and independent origins of pfmdr1 gene amplification [15, 35, 36].
In conclusion, this study provides evidence that rapid changes in pfmdr1 copy number are evolving on multiple genetic backgrounds. It remains to be seen whether the amplified pfmdr1 gene will spread in the population as rapidly as CQ-and SP-resistant genotypes. The 184F allele appears to be under selective pressure, but it remains to be established to what extent this mutation influences resistance to MQ. Moreover, it will be important to monitor whether the dissemination of the 184F could also assist in spreading strains with multiple copies of the pfmdr1 gene in the population. Therefore, continuing molecular surveillance using genetic markers may help track the spread of MQ resistant parasites and allow better containment of MQ resistance.
Acknowledgments
We thank Dr Duong Socheat, for his support, and Dr Laurence Slutsker, for his critical review of the manuscript and support for the project.
Financial support: The Atlanta Research and Education Foundation, Atlanta Veterans Affairs Medical Center, American Society for Microbiology and Coordinating Center for Infectious Diseases (postdoctoral fellowship to M.T.A.), Centers for Disease Control and Prevention Antimicrobial Resistance Working Group, US Navy Medical In-House Laboratory Independent Research Program at Naval Health Research Center/Naval Medical Research Center, Department of Defense Global Emerging Infections Systems, the International Atomic Energy Agency (CRP E1.50.19), and the National Institutes of Health (grant R01GM084320 to A.A.E.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention or the US Department of the Navy or the US Department of Defense.
Footnotes
Potential conflicts of interest: none reported.
References
- 1.Eyles DE, Hoo CC, Warren M, Sandosham AA. Plasmodium falciparum resistant to chloroquine in Cambodia. Am J Trop Med Hyg. 1963;12:840–843. doi: 10.4269/ajtmh.1963.12.840. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. Geneva, Switzerland: World Health Organization; 2002. Development of South-Asia surveillance network for malaria drug resistance. Report on informal consultative meeting. [Google Scholar]
- 3.Lim P, Alker AP, Khim N, et al. Pfmdr1 copy number and arteminisin derivatives combination therapy failure in falciparum malaria in Cambodia. Malar J. 2009;8:11. doi: 10.1186/1475-2875-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 5.Wongsrichanalai C, Meshnick SR. Declining artesunate-mefloquine efficacy against falciparum malaria on the Cambodia-Thailand border. Emerg Infect Dis. 2008;14:716–719. doi: 10.3201/eid1405.071601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denis MB, Tsuyuoka R, Poravuth Y, et al. Surveillance of the efficacy of artesunate and mefloquine combination for the treatment of uncomplicated falciparum malaria in Cambodia. Trop Med Int Health. 2006;11:1360–1136. doi: 10.1111/j.1365-3156.2006.01690.x. [DOI] [PubMed] [Google Scholar]
- 7.Vijaykadga S, Rojanawatsirivej C, Cholpol S, Phoungmanee D, Nakavej A, Wongsrichanalai C. In vivo sensitivity monitoring of mefloquine monotherapy and artesunate-mefloquine combinations for the treatment of uncomplicated falciparum malaria in Thailand in 2003. Trop Med Int Health. 2006;11:211–219. doi: 10.1111/j.1365-3156.2005.01557.x. [DOI] [PubMed] [Google Scholar]
- 8.Rogers WO, Sem R, Tero T, et al. Failure of artesunate-mefloquine combination therapy for uncomplicated Plasmodium falciparum malaria in southern Cambodia. Malar J. 2009;8:10. doi: 10.1186/1475-2875-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Price RN, Uhlemann AC, Brockman A, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alker AP, Lim P, Sem R, et al. Pfmdr1 and in vivo resistance to artesunate-mefloquine in falciparum malaria on the Cambodian-Thai border. Am J Trop Med Hyg. 2007;76:641–647. [PubMed] [Google Scholar]
- 11.Cowman AF, Galatis D, Thompson JK. Selection for mefloquine resistance in Plasmodium falciparum is linked to amplification of the pfmdr1 gene and cross-resistance to halofantrine and quinine. Proc Natl Acad Sci U S A. 1994;91:1143–1147. doi: 10.1073/pnas.91.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peel SA, Bright P, Yount B, Handy J, Baric RS. A strong association between mefloquine and halofantrine resistance and amplification, overexpression, and mutation in the P-glycoprotein gene homolog (pfmdr) of Plasmodium falciparum in vitro. Am J Trop Med Hyg. 1994;51:648–658. doi: 10.4269/ajtmh.1994.51.648. [DOI] [PubMed] [Google Scholar]
- 13.Wilson CM, Serrano AE, Wasley A, et al. Amplification of a gene related to mammalian mdr genes in drug-resistant Plasmodium falciparum. Science. 1989;244:1184–1186. doi: 10.1126/science.2658061. [DOI] [PubMed] [Google Scholar]
- 14.Sidhu AB, Uhlemann AC, Valderramos SG, Valderramos JC, Krishna S, Fidock DA. Decreasing pfmdr1 copy number in Plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J Infect Dis. 2006;194:528–535. doi: 10.1086/507115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaiyaroj SC, Buranakiti A, Angkasekwinai P, Looressuwan S, Cowman AF. Analysis of mefloquine resistance and amplification of pfmdr1 in multidrug-resistant Plasmodium falciparum isolates from Thailand. Am J Trop Med Hyg. 1999;61:780–783. doi: 10.4269/ajtmh.1999.61.780. [DOI] [PubMed] [Google Scholar]
- 16.Wilson CM, Volkman SK, Thaithong S, et al. Amplification of pfmdr1 associated with mefloquine and halofantrine resistance in Plasmodium falciparum from Thailand. Mol Biochem Parasitol. 1993;57:151–160. doi: 10.1016/0166-6851(93)90252-s. [DOI] [PubMed] [Google Scholar]
- 17.Pickard AL, Wongsrichanalai C, Purfield A, et al. Resistance to anti-malarials in Southeast Asia and genetic polymorphisms in pfmdr1. Antimicrob Agents Chemother. 2003;47:2418–2423. doi: 10.1128/AAC.47.8.2418-2423.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price RN, Cassar C, Brockman A, et al. The pfmdr1 gene is associated with a multidrug-resistant phenotype in Plasmodium falciparum from the western border of Thailand. Antimicrob Agents Chemother. 1999;43:2943–2949. doi: 10.1128/aac.43.12.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. Pgh1 modulates sensitivity and resistance to multiple anti-malarials in Plasmodium falciparum. Nature. 2000;403:906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- 20.Sidhu AB, Valderramos SG, Fidock DA. pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol Microbiol. 2005;57:913–926. doi: 10.1111/j.1365-2958.2005.04729.x. [DOI] [PubMed] [Google Scholar]
- 21.Ngo T, Duraisingh M, Reed M, Hipgrave D, Biggs B, Cowman AF. Analysis of pfcrt, pfmdr1, dhfr, and dhps mutations and drug sensitivities in Plasmodium falciparum isolates from patients in Vietnam before and after treatment with artemisinin. Am J Trop Med Hyg. 2003;68:350–356. [PubMed] [Google Scholar]
- 22.Sisowath C, Stromberg J, Martensson A, et al. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem) J Infect Dis. 2005;191:1014–1017. doi: 10.1086/427997. [DOI] [PubMed] [Google Scholar]
- 23.Martensson A, Stromberg J, Sisowath C, et al. Efficacy of artesunate plus amodiaquine versus that of artemether-lumefantrine for the treatment of uncomplicated childhood Plasmodium falciparum malaria in Zanzibar, Tanzania. Clin Infect Dis. 2005;41:1079–1086. doi: 10.1086/444460. [DOI] [PubMed] [Google Scholar]
- 24.Sisowath C, Ferreira PE, Bustamante LY, et al. The role of pfmdr1 in Plasmodium falciparum tolerance to artemether-lumefantrine in Africa. Trop Med Int Health. 2007;12:736–742. doi: 10.1111/j.1365-3156.2007.01843.x. [DOI] [PubMed] [Google Scholar]
- 25.Shah NK, Alker AP, Sem R, et al. Molecular surveillance for multidrug-resistant Plasmodium falciparum, Cambodia. Emerg Infect Dis. 2008;14:1637–1640. doi: 10.3201/eid1410.080080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Socheat D, Denis MB, Fandeur T, et al. Mekong malaria. II. Update of malaria, multi-drug resistance and economic development in the Mekong region of Southeast Asia. Southeast Asian J Trop Med Public Health. 2003;34 Suppl 4:1–102. [PubMed] [Google Scholar]
- 27.Smith JM, Haigh J. The hitch-hiking effect of a favourable gene. Genet Res. 1974;23:23–35. [PubMed] [Google Scholar]
- 28.Maiga O, Djimde AA, Hubert V, et al. A shared Asian origin of the triple-mutant dhfr allele in Plasmodium falciparum from sites across Africa. J Infect Dis. 2007;196:165–172. doi: 10.1086/518512. [DOI] [PubMed] [Google Scholar]
- 29.McCollum AM, Poe AC, Hamel M, et al. Antifolate resistance in Plasmodium falciparum: multiple origins and identification of novel dhfr alleles. J Infect Dis. 2006;194:189–197. doi: 10.1086/504687. [DOI] [PubMed] [Google Scholar]
- 30.McCollum AM, Mueller K, Villegas L, Udhayakumar V, Escalante AA. Common origin and fixation of Plasmodium falciparum dhfr and dhps mutations associated with sulfadoxine-pyrimethamine resistance in a low-transmission area in South America. Antimicrob Agents Chemother. 2007;51:2085–2091. doi: 10.1128/AAC.01228-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nair S, Williams JT, Brockman A, et al. A selective sweep driven by pyrimethamine treatment in southeast Asian malaria parasites. Mol Biol Evol. 2003;20:1526–1536. doi: 10.1093/molbev/msg162. [DOI] [PubMed] [Google Scholar]
- 32.Wootton JC, Feng X, Ferdig MT, et al. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 2002;418:320–323. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]
- 33.Anderson TJ, Roper C. The origins and spread of antimalarial drug resistance: lessons for policy makers. Acta Trop. 2005;94:269–280. doi: 10.1016/j.actatropica.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 34.Ariey F, Fandeur T, Durand R, et al. Invasion of Africa by a single pfcrt allele of South East Asian type. Malar J. 2006;5:34. doi: 10.1186/1475-2875-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Triglia T, Foote SJ, Kemp DJ, Cowman AF. Amplification of the multidrug resistance gene pfmdr1 in Plasmodium falciparum has arisen as multiple independent events. Mol Cell Biol. 1991;11:5244–5250. doi: 10.1128/mcb.11.10.5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nair S, Nash D, Sudimack D, et al. Recurrent gene amplification and soft selective sweeps during evolution of multidrug resistance inmalaria parasites. Mol Biol Evol. 2007;24:562–573. doi: 10.1093/molbev/msl185. [DOI] [PubMed] [Google Scholar]
- 37.Su X, Ferdig MT, Huang Y, et al. A genetic map and recombination parameters of the human malaria parasite Plasmodium falciparum. Science. 1999;286:1351–1353. doi: 10.1126/science.286.5443.1351. [DOI] [PubMed] [Google Scholar]
- 38.Park SDE. PhD thesis. Dublin, Ireland: University of Dublin; 2001. Trypanotolerance in West African cattle and the population genetic effects of selection. [Google Scholar]
- 39.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 40.Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 41.Khim N, Bouchier C, Ekala MT, et al. Countrywide survey shows very high prevalence of Plasmodium falciparum multilocus resistance genotypes in Cambodia. Antimicrob Agents Chemother. 2005;49:3147–3152. doi: 10.1128/AAC.49.8.3147-3152.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duraisingh MT, Jones P, Sambou I, von Seidlein L, Pinder M, Warhurst DC. The tyrosine-86 allele of the pfmdr1 gene of Plasmodium falciparum is associated with increased sensitivity to the anti-malarials mefloquine and artemisinin. Mol Biochem Parasitol. 2000;108:13–23. doi: 10.1016/s0166-6851(00)00201-2. [DOI] [PubMed] [Google Scholar]
- 43.Dokomajilar C, Nsobya SL, Greenhouse B, Rosenthal PJ, Dorsey G. Selection of Plasmodium falciparum pfmdr1 alleles following therapy with artemether-lumefantrine in an area of Uganda where malaria is highly endemic. Antimicrob Agents Chemother. 2006;50:1893–1895. doi: 10.1128/AAC.50.5.1893-1895.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Humphreys GS, Merinopoulos I, Ahmed J, et al. Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob Agents Chemother. 2007;51:991–997. doi: 10.1128/AAC.00875-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duraisingh MT, Roper C, Walliker D, Warhurst DC. Increased sensitivity to the antimalarials mefloquine and artemisinin is conferred by mutations in the pfmdr1 gene of Plasmodium falciparum. Mol Microbiol. 2000;36:955–961. doi: 10.1046/j.1365-2958.2000.01914.x. [DOI] [PubMed] [Google Scholar]
- 46.Pennings PS, Hermisson J. Soft sweeps II—molecular population genetics of adaptation from recurrent mutation or migration. Mol Biol Evol. 2006;23:1076–1084. doi: 10.1093/molbev/msj117. [DOI] [PubMed] [Google Scholar]
- 47.Preechapornkul P, Imwong M, Chotivanich K, et al. Plasmodium falciparum pfmdr1 amplification, mefloquine resistance, and parasite fitness. Antimicrob Agents Chemother. 2009;53:1509–1515. doi: 10.1128/AAC.00241-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White NJ, Pongtavornpinyo W. The de novo selection of drug-resistant malaria parasites. Proc Biol Sci. 2003;270:545–554. doi: 10.1098/rspb.2002.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barnes DA, Foote SJ, Galatis D, Kemp DJ, Cowman AF. Selection for high-level chloroquine resistance results in deamplification of the pfmdr1 gene and increased sensitivity to mefloquine in Plasmodium falciparum. EMBO J. 1992;11:3067–3075. doi: 10.1002/j.1460-2075.1992.tb05378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]