Abstract
A wide range of mammalian signaling and stress pathways are mediated by nitric oxide (NO), which is synthesized in vivo by the nitric oxide synthase (NOS) family of enzymes. Experimental manipulations of NO are frequently achieved by either inhibition or activation of endogenous NOS or via providing exogenous NO sources. On the contrary, many microbes consume NO via flavohemoglobin (FlavoHb), a highly efficient NO-dioxygenase that protects from nitrosative stress. Here we report a novel resource for studying NO in mammalian cells by heterologously expressing E. coli FlavoHb within a lentiviral delivery system. This technique boosts endogenous cellular consumption of NO, thus providing a simple and efficacious approach to studying mammalian NO-biology that can be employed as both a primary experimental and confirmatory tool.
INTRODUCTION
The diatomic gas nitric oxide (NO) plays a multitude of roles in mammalian, plant and microbial biology (1–4). In models of infection, mammalian-derived NO facilitates microbial killing (5,6), indicating that NO is a central element of innate immunity. In return, bacteria and fungi possess two major systems to metabolize NO: the flavorubredoxin NO-reductase (7) and flavohemoglobin (FlavoHb) NO-dioxygenase (8–10) enzymes, which operate under strictly anaerobic and aerobic/microaerophilic conditions, respectively. The importance of NO-metabolism in the prokaryotic life cycle is evidenced by the evolution of the FlavoHb family nearly 2 billion years ago (11), suggesting that microbes have been coping with nitrosative stress long before mammals existed.
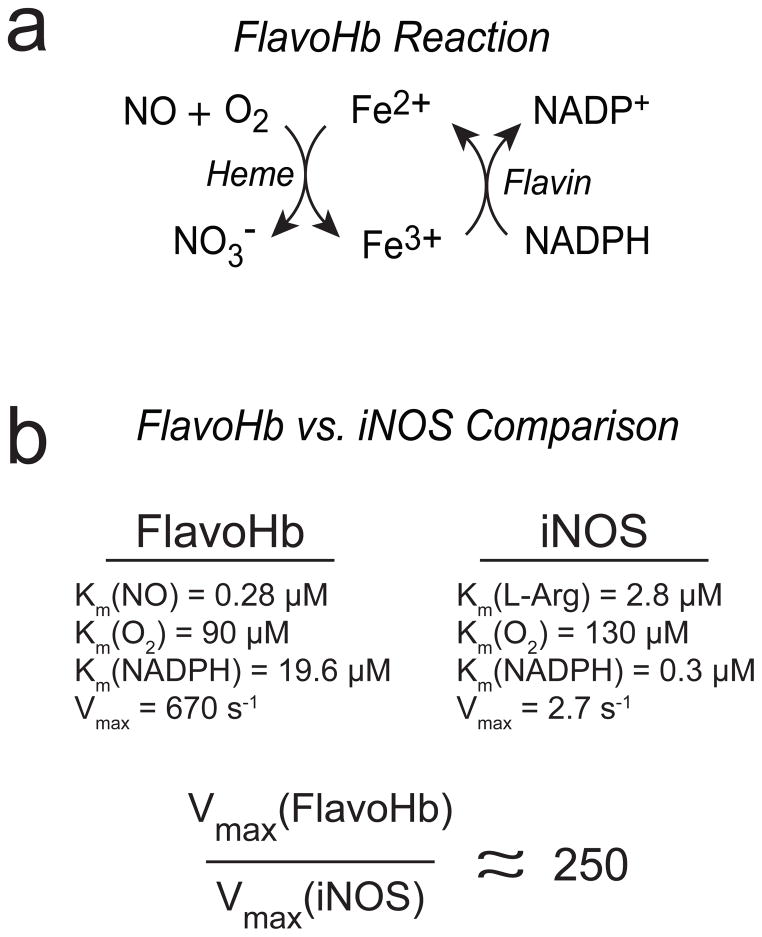
A wealth of studies over the past decade has elucidated the mechanism and function of FlavoHb in bacteria and fungi. Under aerobic/microaerophilic conditions, FlavoHb converts NO and O2 into nontoxic nitrate (NO3−) with concomitant oxidation of ferrous (Fe2+) to ferric (Fe3+) heme within FlavoHb (9). The active site flavin adenine dinucleotide (FAD) supports one-electron reduction back to the ferrous state, driven by electrons from NADH or NADPH (Figure 1A). The enzyme is not known to react with substrates other than NO. Further, FlavoHb is transcriptionally induced by NO in many bacterial and fungal pathogens, and has been shown to play a central role in protection from NO in multiple pathogenic microbes (5,10,12–19).
Figure 1.
Schematic of the FlavoHb-catalyzed reaction and a kinetic comparison to human iNOS. (A) The heme domain converts NO and O2 to nitrate (NO3−), with concomitant oxidation of the ferrous to ferric states. The flavin-containing reductase domain uses electrons from NADPH to reduce the ferric back to ferrous heme, thus allowing enzymatic turnover. (B) A kinetic comparison of FlavoHb and iNOS (iNOS was chosen for comparison because it is the most active NOS isoform (i.e., highest Vmax), and therefore the most conservative comparison to FlavoHb).
In contrast to microbes, the FlavoHb gene is not present in metazoans or mammals. On the contrary, mammals synthesize NO via three conserved nitric oxide synthase (NOS) isoforms: (“inducible”) iNOS/NOS2, (“endothelial”) eNOS/NOS3 and (“neuronal”) nNOS/NOS1, each of which play distinct biological roles (20). Though the overall importance of NO is widely appreciated, techniques to determine the cellular roles of NO have relied predominantly on manipulating NOS expression or activity, most frequently via arginine-based NOS inhibitors. While this approach is undoubtedly powerful, there are several drawbacks to NOS inhibitors: 1) they rarely exhibit strong isoform selectivity, with the exception of some iNOS inhibitors such as 1400W (21) and BYK191023 (22), 2) they are typically arginine analogues, and several studies have suggested they may perturb arginine uptake or metabolism (23–25) and 3) NOS-independent sources of NO are unaffected. On the contrary, NOS overexpression or administration of NO-donor compounds may result in supraphysiologic levels of NO. Reliance on such methods might therefore lead to aberrant cellular effects, thus confounding the interpretation of NO’s roles in mammalian cells. A technique to selectively deplete NO – independently of NO source – would therefore be of significant utility in studies of NO biology.
Given the remarkable specificity and catalytic efficiency of bacterial NO-consuming enzymes, we hypothesized that heterologous expression of such an enzyme in mammalian cells might be a useful tool to interrogate the biological role(s) of NO. While the flavorubredoxin is a potent NO reductase, it requires exquisitely anaerobic conditions. On the contrary, FlavoHb operates under a wide range of O2 concentrations, suggesting it might also operate in mammalian cells. Further, flavorubredoxin consists of two separate polypeptides (the NO reductase and flavorubredoxin reductase), whereas FlavoHb is a 44 kD protein encoded by a single gene, thus simplifying the strategy of heterologous expression. As compared in Figure 1B, the kinetic parameters of E. coli FlavoHb (26) are far superior to even the most robust mammalian NOS isoform, iNOS (27). We therefore chose to focus our efforts on FlavoHb.
MATERIALS AND METHODS
Materials
All materials were from Sigma unless otherwise indicated. LPS was from E. coli strain 026:B6. Protease inhibitor (PI) cocktail was from Roche. “NONOate” NO donors and 1400W were from Cayman Chemical and freshly prepared in 10 mM NaOH. Sources of antibodies were: α-Txnip/Vdup1 mouse mAb (MBL International, #K0205-3), α-GAPDH mouse mAb (Millipore, #MAB374), α-COX2 rabbit pAb (Cell Signaling, #4842), α-VASP rabbit pAb (Cell Signaling, #3112), α-phospho-VASP rabbit pAb (Cell Signaling, #3111), α-iNOS mouse mAb (BD Biosciences, #610328), α-Flag M2 mouse mAb for immunoblotting (Sigma, #F1804), α-Flag rabbit pAb for immunofluorescence (Cell Signaling, #2368), α-catalase mouse mAb (Sigma, #C0979), α-PCNA mouse mAb (Cell Signaling, #2586).
Cell culture
All cell lines were obtained from ATCC. The RAW264.7 and HEK293 cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS), 100 U/ml penicillin and 0.1 mg/ml streptomycin. For transient transfection experiments, HEK293 cells were transfected in 6 well dishes with 5 μg total DNA and 20 μl Superfect (Qiagen, 1:4 DNA:Superfect ratio).
Cloning and DNA manipulation
All PCRs were performed with Advantage DNA polymerase (Clontech). The E. coli gene for FlavoHb (hmpA) was amplified from genomic DNA (Strain BW25113) via PCR with the following primers: 5′-AATAGAATTCACCATGGACTACAAAGACGATGACGACAAGCTTGACGCTCAAACCA TCGCTAC-3′ and 5′-AAATGGATCCTTACAGCACCTTATGCGG-3′. The 1.2 kb PCR product was digested with EcoRI and BamHI, and ligated into the lentiviral expression plasmid pCDH-CMV-MCS-EF1-copGFP (System Biosciences) digested at the same sites. All products were verified by DNA sequencing (Duke DNA Sequencing Facility). Human iNOS was cloned into pCDH-CMV-MCS-EF1-copGFP at the XbaI and NotI sites using the following PCR primers: 5′-CTTATATCTAGAACCATGGCCTGTCCTTGG-3′ and 5′-ATTATAGCGGCCGCTTAGAGCGCTGACA-3’.
Lentiviral production
Lentiviral particles were generated by co-transfecting subconfluent (60–70%) 293FT cells with empty pCDH vector or pCDH-CMV-FlavoHb-EF1-copGFP plasmid DNA and the packaging and enveloping constructs, pCI-VSVG and ps-PAX2, respectively (Addgene). For a 10 cm dish, 6 μg plasmid DNA was co-transfected with 2 μg pCI-VSVG and 3 μg ps-PAX2 using 33 μL Lipofectamine 2000. Media were changed 8–12 hours after transfection to fresh DMEM with 10% FBS, and the virus-laden supernatant was collected at 24 and 48 hours. The supernatant was filtered and employed for experiments or frozen at −80 °C in aliquots.
Generation and isolation of cells stably infected with empty vs. FlavoHb lentivirus
Stable FlavoHb-expressing or vector control cells were generated by treating HEK293 or RAW264.7 macrophages twice with virus prepared as described above. Twenty-four hours after the second infection, HEK293 or RAW264.7 cells were dissociated using trypsin or cell scraping, respectively, and viable GFP-positive cells were single-cell sorted into 96-well cell culture plates containing DMEM using a FACS AriaII Cell Sorter (BD Biosciences). Single cell sorting was verified 24 hours after sorting and media changed on amplifying clones every 3 days until cell growth warranted splitting cells to a larger culture vessel. Amplified stable GFP-expressing cultures were evaluated for FlavoHb-expression by anti-Flagimmunoblotting.
Measurement of FlavoHb activity in cellular extracts via NO-dependent NADPH consumption
A 15 cm dish of HEK293 cells stably infected with either control lentivirus or FlavoHb was lysed via repeated passage through a 28 g needle in 50 mM PO4, 0.5 mM EDTA, 2 μM FAD, pH 7.4 containing 1 mM DTT and protease inhibitor cocktail (Roche). A cytosolic-enriched sample was obtained via clarification at 20,000 g × 30 min. Protein was measured via BCA assay. For measurement of NADPH consumption, 94 μl of lysate was mixed with either 2 μl of buffer or 10 mM NADPH, and absorbance at 340 nm was measured on a Beckman DU650 UV-Vis Spectrophotometer at 37 °C. Spermine-NONOate (final 4 mM, stock solution in 10 mM NaOH) was added to initiate the reaction, and absorbance at 340 nm was followed over time.
Subcellular fractionation for determination of FlavoHb localization in cells
Basic fractionation was achieved via differential centrifugation as briefly described. HEK293 cells were lysed via repeated passage through a 28 g needle in 25 mM Tris, 25 mM NaCl, 0.1 mM EDTA, pH 7.4 containing PI cocktail. Lysis efficiency was verified > 99% by trypan blue staining. The lysates were centrifuged at 500 g × 10 min to obtain a nuclei-enriched pellet. The supernatant was removed and centrifuged at 20,000 g × 30 min to obtain a membrane/organelle-enriched pellet and cytosol-enriched supernatant. To facilitate solubilization, lysis buffer was added to each fraction along with 0.2% Triton X-100 (v/v). Protein was measured by BCA assay, and equal amounts of material (40 μg each) were employed for immunoblotting analysis.
Measurement of NO2− and NO3− in culture media
Following the indicated iNOS transfection(s) or LPS-stimulations, culture media was collected and clarified by centrifugation at 5,000 g × 5 min. The supernatant was saved, and NO2− was measured by the standard Griess reaction. In brief, 40 μl of media or NO2− standard in culture media was transferred to a 96 well dish and 100 μl of 1% sulfanilamide (w/v) in 1 M HCl was added. The mixture was incubated at room temperature for 5 min, and 50 μl 0.1% N-1-napthylethylenediamine dihydrochloride (NEDD) in 0.5 M HCl was added to each well. Absorbance at 540 nm was measured via spectrophotometry and all samples were normalized to standard NO2− solutions. For measurement of NO3−, 40 μl of media or NO3− standard in culture media was transferred to two wells of a 96 well dish (labeled “A” and “B”). To all wells was added 40 μl of PBS containing 4 μM FAD, then 10 μl of 4 mM NADPH. To “A” and “B” wells was added 10 μl PBS or NO3− reductase (2 mU/μl in PBS), respectively. Plates were incubated at 37 °C for 1 h, and 100 μl of 1% sulfanilamide (w/v) in 1 M HCl was added. The mixture was incubated at room temperature for 5 min, and 50 μl 0.1% N-1-napthylethylenediamine dihydrochloride (NEDD) in 0.5 M HCl was added to each well. Absorbance at 540 nm was measured via spectrophotometry. The value of “B” minus “A” (i.e., NO3−-derived signal) was calculated for all samples/standards, and normalized accordingly.
Immunofluorescence of FlavoHb in mammalian cells
HEK293 cells were plated onto poly-D-lysine-coated coverslips in 6 well dishes. Upon reaching 50–60% confluence, cells were transfected with 2 μg pCDH-Flag-FlavoHb for 18 h, washed with PBS × 2, and fixed in 4% paraformaldehyde in PBS for 30 min at room temperature. Cells were again washed with PBS and permeabilized with 0.1% Triton X-100 in PBS for 15 min on ice. Cells were blocked for 1 hour in PBS containing 5% goat serum, and stained overnight at 4 °C with PBS containing 5% goat serum and anti-flag rabbit pAb (1:50). Similar results were obtained with anti-Flag M5 mAb (data not shown) at 1:400 dilution. Cells were washed again with PBS and secondary anti-rabbit antibody with an Alexafluor-568 conjugate (Invitrogen) was added at 1:1000 dilution for 40 min at room temperature. Cells were washed repeatedly with PBS, mounted for imaging using mounting media and coverslips affixed to glass slides with nail polish adhesive. Cells were imaged on a Zeiss Axio Imager widefield fluorescence microscope (Duke Microscopy Core Facility) at 63x magnification.
For confocal imaging, HEK293 cells were plated onto tissue-culture treated chamber slides (Lab-Tek). Twenty-four hours after plating, cells were washed with PBS × 2 and fixed in paraformaldehyde (4% in PBS) for 15 minutes at room temperature. Cells were washed three times with PBS, and were permeabilized with 0.1% Triton X-100 in PBS and blocked with normal goat serum (10% in PBS) for 1 hour at room temperature. Slides were stained overnight at 4 °C with anti-Flag rabbit pAb (1:100), washed three times with PBS and incubated with Alexafluor-568 conjugate (Invitrogen) at 1:250 dilution for 20 min at room temperature. Cells were washed three times with PBS, were stained with Hoechst 33258 at 5 μg/mL for 10 min at room temperature before washing three times with PBS. Cells were coverslipped using FluorSave mounting media (Calbiochem) and imaged on a Leica SP5 confocal microscope, using sequential scanning technique with a 63x oil immersion objective.
Assessment of nitrosative stress by 3H-thymidine uptake, cell counting and colony formation assays
For evaluation of 3H-thymidine uptake, HEK293 cells were stably infected with either empty lentivirus or FlavoHb and viable cells were plated in equal numbers in 12-well plates (30,000 cells per well). Cells were treated with: 1) 0, 200, 500 or 1000 μM DETA-NO for 24 hours, or 2) were transfected with pCDH-CMV-iNOS-EF1-copGFP or vector control (pCDH-CMV-MCS-EF1-copGFP) for 12 hours before the media was changed to fresh DMEM + 10% FBS for 48 hours. At this point, cells were incubated for 5 h with 4 μCi of 3H-thymidine (Perkin Elmer), washed once with PBS, fixed for 1 hour in 10% trichloroacetic acid, and incubated overnight in 0.01 M NaOH before assessing radioactivity incorporation of the lysed cellular contents by scintillation counting (Beckman 156000).
For cell counting, HEK293 cells were stably infected with either empty lentivirus or FlavoHb and viable cells were plated in equal numbers in 6-well plates (100,000 cells per well). Cells were treated with 0, 200, 500 or 1000 μM DETA-NO for 48 hours, at which point cells were dissociated by trypsin, resuspended in a defined volume, and were mixed 1:1 with trypan blue for two minutes before viable cells were counted on a hemacytometer.
For colony formation assays, cells stably expressing either vector control or Flag-FlavoHb were plated at a density of 500 cells/well of a 6 well plate. DETA-NONOate was added at either 0, 0.1 or 0.5 mM final concentration and colonies were grown for 7 days before visualization by staining with 1% methylene blue in methanol (wt/vol) for 2 minutes.
RESULTS AND DISCUSSION
The E. coli FlavoHb gene (hmpA) was amplified by PCR from genomic DNA with a forward 5’-primer encoding an N-terminal Flag epitope. This PCR product was subcloned into a pCDH plasmid (Systems Biosciences), which employs both a lentiviral vector system and EGFP driven by a distinct promoter. Therefore, this system allows for enrichment or identification of positive cells (i.e., transfected, infected) by EGFP-based selection. It is worth noting that FlavoHb and EGFP are expressed as separate polypeptides, thus EGFP is simply an indicator of FlavoHb presence, not localization.
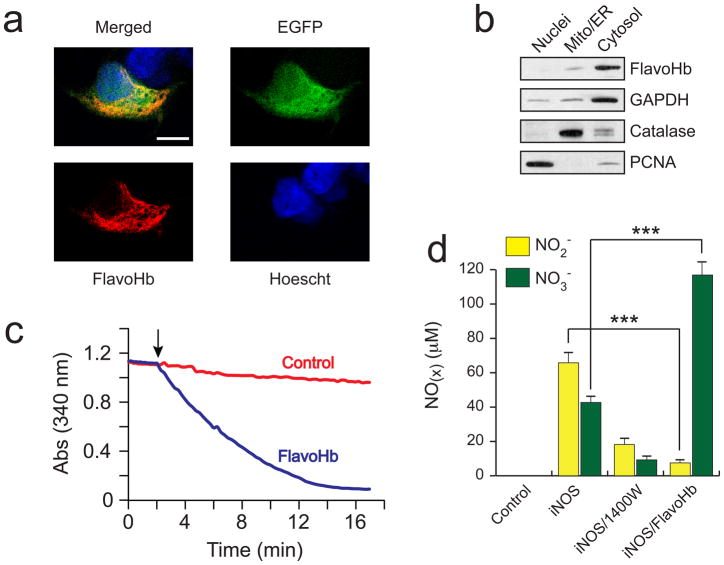
The cellular localization of FlavoHb was determined in HEK293 cells via confocal immunofluorescence (Figure 2A, Supplementary Figure 1), as well as subcellular fractionation (Figure 2B). Flavohemoglobin displayed diffuse subcellular localization with a cytosolic predominance, as expected since FlavoHb lacks any known human localization signal. To determine whether E. coli FlavoHb is enzymatically active in human cells, cytosolic extracts from transfected HEK293 cells were assayed by measuring NO-dependent NADPH consumption (Figure 2C). Expression of FlavoHb resulted in markedly increased NADPH consumption, which was dependent on the addition of exogenous NO. These features are characteristic of FlavoHb activity (8–10). Further, expression of FlavoHb did not exhibit any noticeable toxicity or growth suppression in several tested mammalian cell types (data not shown).
Figure 2.
E. coli FlavoHb is a functional NO-consuming enzyme when expressed in mammalian cells. (A) Confocal immunofluorescence reveals that Flag-FlavoHb exhibits diffuse cytosolic localization in transfected HEK293 cells. The white bar indicates 10 μm. (B) Subcellular fractionation demonstrates that Flag-FlavoHb partitions with cytosolic markers (e.g. GAPDH), and is therefore localized predominantly to the cytosol. (C) HEK293 cells were transfected for 24 h with either empty pCDH or pCDH-Flag-FlavoHb, and cytosolic extracts were analyzed for NADPH consumption via spectrophotometry at 340 nm. Reactions were conducted in a phosphate buffer at 37 °C, pH 7.4, and initiated by adding 5 mM spermine-NONOate (indicated by arrow). (D) The presence of FlavoHb shifts the balance of NO oxidation from uncatalyzed auto-oxidation (NO2-) to FlavoHb-catalyzed NO dioxygenation (NO3−), as measured by the Griess reaction in the media of HEK293 cells transfected −/+ iNOS and FlavoHb (***, p < 0.001).
Endogenously synthesized NO is normally oxidized to NO2− (nitrite) and NO3− (nitrate) in an approximately 60:40 ratio via auto-oxidation (28,29). Since FlavoHb converts NO to NO3− (not NO2−), the efficacy of FlavoHb to metabolize endogenous NO was examined via measuring NO2− and NO3− in the media of FlavoHb-transfected HEK293 cells. As shown in Figure 2D, the iNOS inhibitor 1400W led to decreased NO2− and NO3− due to inhibition of NO synthesis, whereas FlavoHb heavily shifted the ratio towards NO3−. Similar results were obtained in lipopolysaccharide (LPS)-stimulated RAW264.7 macrophages stably expressing FlavoHb (Supplementary Figure 2). These data demonstrate that FlavoHb indeed metabolizes endogenously synthesized NO to NO3−in mammalian cells without affecting NO synthesis itself.
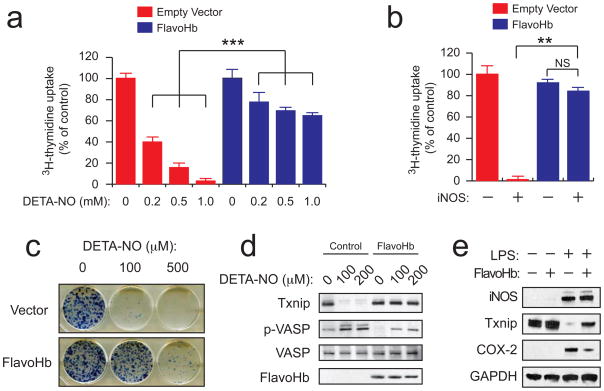
FlavoHb is known to play a major role in protection from nitrosative stress in bacteria and fungi. As high levels of NO drive nitrosative stress and toxicity in mammalian cells, we evaluated the ability of FlavoHb to protect mammalian cells from nitrosative stress. In HEK293 cells, FlavoHb blocked the growth suppressive effects of both exogenous NO (Figure 3A) and endogenous iNOS-derived NO (Figure 3B), as assayed by 3H-thymidine uptake. Diethylenetriamine NONOate (DETA-NO) was chosen as the exogenous NO source, as its long half life (t1/2 ~ 20 h) provides a sustained level of NO over the course of the experiment.
Figure 3.
FlavoHb can be used to study NO-dependent stress and signaling in mammalian cells. (A) HEK293 cells expressing either empty vector or FlavoHb were treated with DETA-NO for 18 h, then subjected to the 3H-thymidine uptake assay to assess cell proliferation. (B) As in (A), except cells were co-transfected with either empty vector or iNOS for 24 h, then subjected to the 3H-thymidine uptake assay (**, p < 0.01; ***, p < 0.001 by ANOVA; NS, not significant). (C) Colony formation assay with HEK293 cells expressing either empty vector or FlavoHb. Cells were plated into 6 well dishes (500 cells per well) and incubated with DETA-NO at the indicated concentrations for one week. Colonies were then fixed and visualized by methylene blue staining. (D) HEK293 cells expressing either empty vector or FlavoHb were treated with DETA-NO for 18 h, then subjected to immunoblotting as shown. Txnip suppression and VASP phosphorylation are two established NO-dependent signaling pathways, and are blocked by FlavoHb. (E) RAW264.7 macrophages expressing either empty vector or FlavoHb were stimulated with 500 ng/ml LPS for 18 h, and analyzed for Txnip suppression and COX-2 induction by immunoblotting.
Confirmatory data were obtained via cell counting (Supplementary Figure 3) and colony formation assays (Figure 3C and Supplementary Figure 4). Collectively, these experiments demonstrate that FlavoHb – the predominant NO-protective system in microbial systems – exhibits a similar ability to protect mammalian cells from both exogenous and endogenous nitrosative stresses. Further, they emphasize that FlavoHb is enzymatically active in human cells, which is remarkable given the disparities between these two cellular environments.
As NO is frequently studied in the context of cell signaling, the utility of FlavoHb for interrogating NO-dependent signaling pathways was examined. In HEK293 cells, FlavoHb blunted both NO-dependent Txnip suppression (30) and VASP phosphorylation (31) (Figure 3D), two established downstream mediators of NO in epithelial cells. FlavoHb also inhibited COX-2 induction (32) and Txnip suppression (30) in LPS-stimulated RAW264.7 macrophages (two established NO-dependent events) (Figure 3E). These findings suggest that FlavoHb expression within mammalian cells can be used to interrogate the role of NO in various physiological or pathophysiological processes.
Here we provide evidence that E. coli FlavoHb is enzymatically active in mammalian cells, thereby protecting them from nitrosative stress and inhibiting NO-signaling pathways. Importantly, the FlavoHb protein does not exhibit any apparent toxicities in the tested mammalian cell types (HEK293 epithelial cells, RAW264.7 macrophages), and there exists no evidence why it should in other mammalian cells (with the exception of cells that exhibit NO-dependent growth or survival). Heterologous expression of FlavoHb is therefore a novel and useful strategy to probe the effects of NO in mammalian cells independently of NO synthesis. This tool will likely find application in a range of NO-related studies and may provide a novel route to therapeutic depletions of NO in vivo.
Supplementary Material
Acknowledgments
This work was supported by grants F30NS063496 from the National Institute of Neurological Disorders and Stroke (C.E.E.), and NIH grants NS054276, CA116659, and CA129958 (J.N.R.). Additional support was provided by the Goldhirsh Foundation, Damon Runyon Cancer Research Foundation, and the Brain Tumor Society of the James S. McDonnell Foundation. We thank Patrick J. Casey (Duke University) for providing laboratory support and Rainbo C. Hultman for help with immunofluorecence experiments.
Footnotes
AUTHOR CONTRIBUTIONS
M.T.F. designed and performed experiments, analyzed data and wrote the manuscript. C.E.E. designed and performed experiments, and analyzed data. J.N.R. analyzed data and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Stamler JS, Singel DJ, Loscalzo J. Biochemistry of nitric oxide and its redox-activated forms. Science. 1992;258:1898–1902. doi: 10.1126/science.1281928. [DOI] [PubMed] [Google Scholar]
- 2.Jaffrey SR, Snyder SH. Nitric oxide: a neural messenger. Annu Rev Cell Dev Biol. 1995;11:417–440. doi: 10.1146/annurev.cb.11.110195.002221. [DOI] [PubMed] [Google Scholar]
- 3.Spiro S. Regulators of bacterial responses to nitric oxide. FEMS Microbiol Rev. 2007;31:193–211. doi: 10.1111/j.1574-6976.2006.00061.x. [DOI] [PubMed] [Google Scholar]
- 4.Moreau M, Lindermayr C, Durner J, Klessig DF. NO synthesis and signaling in plants--where do we stand? Physiol Plant. 2010;138:372–383. doi: 10.1111/j.1399-3054.2009.01308.x. [DOI] [PubMed] [Google Scholar]
- 5.Fang FC. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat Rev Microbiol. 2004;2:820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 6.Nathan C, Shiloh MU. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc Natl Acad Sci U S A. 2000;97:8841–8848. doi: 10.1073/pnas.97.16.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardner AM, Helmick RA, Gardner PR. Flavorubredoxin, an inducible catalyst for nitric oxide reduction and detoxification in Escherichia coli. J Biol Chem. 2002;277:8172–8177. doi: 10.1074/jbc.M110471200. [DOI] [PubMed] [Google Scholar]
- 8.Gardner PR, Gardner AM, Martin LA, Salzman AL. Nitric oxide dioxygenase: an enzymic function for flavohemoglobin. Proc Natl Acad Sci U S A. 1998;95:10378–10383. doi: 10.1073/pnas.95.18.10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hausladen A, Gow AJ, Stamler JS. Nitrosative stress: metabolic pathway involving the flavohemoglobin. Proc Natl Acad Sci U S A. 1998;95:14100–14105. doi: 10.1073/pnas.95.24.14100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crawford MJ, Goldberg DE. Role for the Salmonella flavohemoglobin in protection from nitric oxide. J Biol Chem. 1998;273:12543–12547. doi: 10.1074/jbc.273.20.12543. [DOI] [PubMed] [Google Scholar]
- 11.Vinogradov SN, Hoogewijs D, Bailly X, Mizuguchi K, Dewilde S, Moens L, Vanfleteren JR. A model of globin evolution. Gene. 2007;398:132–142. doi: 10.1016/j.gene.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 12.Richardson AR, Dunman PM, Fang FC. The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol Microbiol. 2006;61:927–939. doi: 10.1111/j.1365-2958.2006.05290.x. [DOI] [PubMed] [Google Scholar]
- 13.Sebbane F, Lemaitre N, Sturdevant DE, Rebeil R, Virtaneva K, Porcella SF, Hinnebusch BJ. Adaptive response of Yersinia pestis to extracellular effectors of innate immunity during bubonic plague. Proc Natl Acad Sci U S A. 2006;103:11766–11771. doi: 10.1073/pnas.0601182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Svensson L, Poljakovic M, Save S, Gilberthorpe N, Schon T, Strid S, Corker H, Poole RK, Persson K. Role of flavohemoglobin in combating nitrosative stress in uropathogenic Escherichia coli--implications for urinary tract infection. Microb Pathog. 2010;49:59–66. doi: 10.1016/j.micpath.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Turrion-Gomez JL, Eslava AP, Benito EP. The flavohemoglobin BCFHG1 is the main NO detoxification system and confers protection against nitrosative conditions but is not a virulence factor in the fungal necrotroph Botrytis cinerea. Fungal Genet Biol. 2010;47:484–496. doi: 10.1016/j.fgb.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 16.de Jesus-Berrios M, Liu L, Nussbaum JC, Cox GM, Stamler JS, Heitman J. Enzymes that counteract nitrosative stress promote fungal virulence. Curr Biol. 2003;13:1963–1968. doi: 10.1016/j.cub.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 17.Goncalves VL, Nobre LS, Vicente JB, Teixeira M, Saraiva LM. Flavohemoglobin requires microaerophilic conditions for nitrosative protection of Staphylococcus aureus. FEBS Lett. 2006;580:1817–1821. doi: 10.1016/j.febslet.2006.02.039. [DOI] [PubMed] [Google Scholar]
- 18.Ullmann BD, Myers H, Chiranand W, Lazzell AL, Zhao Q, Vega LA, Lopez-Ribot JL, Gardner PR, Gustin MC. Inducible defense mechanism against nitric oxide in Candida albicans. Eukaryot Cell. 2004;3:715–723. doi: 10.1128/EC.3.3.715-723.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hromatka BS, Noble SM, Johnson AD. Transcriptional response of Candida albicans to nitric oxide and the role of the YHB1 gene in nitrosative stress and virulence. Mol Biol Cell. 2005;16:4814–4826. doi: 10.1091/mbc.E05-05-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stuehr DJ, Santolini J, Wang ZQ, Wei CC, Adak S. Update on mechanism and catalytic regulation in the NO synthases. J Biol Chem. 2004;279:36167–36170. doi: 10.1074/jbc.R400017200. [DOI] [PubMed] [Google Scholar]
- 21.Garvey EP, Oplinger JA, Furfine ES, Kiff RJ, Laszlo F, Whittle BJ, Knowles RG. 1400W is a slow, tight binding, and highly selective inhibitor of inducible nitric-oxide synthase in vitro and in vivo. J Biol Chem. 1997;272:4959–4963. doi: 10.1074/jbc.272.8.4959. [DOI] [PubMed] [Google Scholar]
- 22.Strub A, Ulrich WR, Hesslinger C, Eltze M, Fuchss T, Strassner J, Strand S, Lehner MD, Boer R. The novel imidazopyridine 2-[2-(4-methoxy-pyridin-2-yl)-ethyl]-3H-imidazo[4,5-b]pyridine (BYK191023) is a highly selective inhibitor of the inducible nitric-oxide synthase. Mol Pharmacol. 2006;69:328–337. doi: 10.1124/mol.105.017087. [DOI] [PubMed] [Google Scholar]
- 23.Bogle RG, MacAllister RJ, Whitley GS, Vallance P. Induction of NG-monomethyl-L-arginine uptake: a mechanism for differential inhibition of NO synthases? Am J Physiol. 1995;269:C750–756. doi: 10.1152/ajpcell.1995.269.3.C750. [DOI] [PubMed] [Google Scholar]
- 24.DeGeorge GL, Heck DE, Laskin JD. Arginine metabolism in keratinocytes and macrophages during nitric oxide biosynthesis: multiple modes of action of nitric oxide synthase inhibitors. Biochem Pharmacol. 1997;54:103–112. doi: 10.1016/s0006-2952(97)00144-5. [DOI] [PubMed] [Google Scholar]
- 25.MacAllister RJ, Fickling SA, Whitley GS, Vallance P. Metabolism of methylarginines by human vasculature; implications for the regulation of nitric oxide synthesis. Br J Pharmacol. 1994;112:43–48. doi: 10.1111/j.1476-5381.1994.tb13026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gardner AM, Martin LA, Gardner PR, Dou Y, Olson JS. Steady-state and transient kinetics of Escherichia coli nitric-oxide dioxygenase (flavohemoglobin). The B10 tyrosine hydroxyl is essential for dioxygen binding and catalysis. J Biol Chem. 2000;275:12581–12589. doi: 10.1074/jbc.275.17.12581. [DOI] [PubMed] [Google Scholar]
- 27.Stuehr DJ, Cho HJ, Kwon NS, Weise MF, Nathan CF. Purification and characterization of the cytokine-induced macrophage nitric oxide synthase: an FAD- and FMN-containing flavoprotein. Proc Natl Acad Sci U S A. 1991;88:7773–7777. doi: 10.1073/pnas.88.17.7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stuehr DJ, Marletta MA. Mammalian nitrate biosynthesis: mouse macrophages produce nitrite and nitrate in response to Escherichia coli lipopolysaccharide. Proc Natl Acad Sci U S A. 1985;82:7738–7742. doi: 10.1073/pnas.82.22.7738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stuehr DJ, Marletta MA. Synthesis of nitrite and nitrate in murine macrophage cell lines. Cancer Res. 1987;47:5590–5594. [PubMed] [Google Scholar]
- 30.Forrester MT, Seth D, Hausladen A, Eyler CE, Foster MW, Matsumoto A, Benhar M, Marshall HE, Stamler JS. Thioredoxin-interacting protein (Txnip) is a feedback regulator of S-nitrosylation. J Biol Chem. 2009;284:36160–36166. doi: 10.1074/jbc.M109.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oelze M, Mollnau H, Hoffmann N, Warnholtz A, Bodenschatz M, Smolenski A, Walter U, Skatchkov M, et al. Vasodilator-stimulated phosphoprotein serine 239 phosphorylation as a sensitive monitor of defective nitric oxide/cGMP signaling and endothelial dysfunction. Circ Res. 2000;87:999–1005. doi: 10.1161/01.res.87.11.999. [DOI] [PubMed] [Google Scholar]
- 32.Habib A, Bernard C, Lebret M, Creminon C, Esposito B, Tedgui A, Maclouf J. Regulation of the expression of cyclooxygenase-2 by nitric oxide in rat peritoneal macrophages. J Immunol. 1997;158:3845–3851. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.