Abstract
Nkx2.5 (also known as Csx) is an evolutionarily conserved cardiac transcription factor of the homeobox gene family. Nkx2.5 is required for early heart development, since Nkx2.5-null mice die before completion of cardiac looping. To identify genes regulated by Nkx2.5 in the developing heart, we performed subtractive hybridization by using RNA isolated from wild-type and Nkx2.5-null hearts at embryonic day 8.5. We isolated a mouse cDNA encoding myocardin A, which is an alternative spliced isoform of myocardin and the most abundant isoform in the heart from embryo to adult. The expression of myocardin A and myocardin was markedly downregulated in Nkx2.5-null mouse hearts. Transient-cotransfection analysis showed that Nkx2.5 transactivates the myocardin promoter. Inhibition of myocardin function in the teratocarcinoma cell line P19CL6 prevented differentiation into cardiac myocytes after dimethyl sulfoxide treatment. Myocardin A transactivated the promoter of the atrial natriuretic factor gene through the serum response element, which was augmented by bone morphogenetic protein 2 and transforming growth factor β-activated kinase 1. These results suggest that myocardin expression is regulated by Nkx2.5 and that its function is required for cardiomyogenesis.
Nkx2.5 (also referred to as Csx) is a member of the NK2 class of homeoproteins (20, 22). NK2 class homeoproteins are expressed in a tissue-specific manner, suggesting a role in tissue specification, differentiation, and patterning (13). Murine Nkx2.5 was identified as a homologue of the Drosophila tinman gene (5), which is required for subdivision of the dorsal mesoderm into the cardiac and visceral mesoderm (20, 22). In mice, expression of Nkx2.5 begins as early as embryonic day (ED) 7.5 and continues through adulthood (17, 20, 22). Nkx2.5-null mice die before ED 11 with an arrest of cardiac development (23, 36).
Myocardin is expressed in cardiac myocytes and smooth muscle cells, and its expression begins in the cardiac crescent at ED 7.75 (42). Myocardin is a transcription cofactor and transactivates the serum response element (SRE) (also referred to as CArG box) dependent promoters by forming a ternary complex with serum response factor (SRF). Expression of a dominant-negative mutant of myocardin in Xenopus embryos prevents heart formation (42). However, regulation of myocardin gene expression is unknown.
Bone morphogenetic proteins (BMPs) are members of the transforming growth factor β (TGF-β) superfamily. TGF-β-activated kinase 1 (TAK1) is a member of the mitogen-activated protein kinase kinase kinase family and a mediator of TGF-β and BMP signal transduction (43). By using the teratocarcinoma P19 cells and their clonal derivative P19CL6 cells in an in vitro system, BMPs have been shown to be essential for cardiac myocyte differentiation (15, 28). BMP signaling regulates Nkx2.5 activity during cardiomyogenesis in P19 cells (15). In P19CL6 cells, TAK1, Nkx2.5, and GATA-4 play a pivotal role in the cardiogenic BMP signaling pathway (28). In addition, overexpression of a dominant-negative mutant of Nkx2.5 inhibited cardiomyogenesis in both P19 cells and P19CL6 cells (16, 38).
We and others have previously reported that several genes, including atrial natriuretic factor (ANF), B-type natriuretic peptide, CARP, eHAND, MEF2C, myosin light chain 2v (MLC2v), SM22, N-myc, Msx2, Irx4, and Chisel are downregulated in the Nkx2.5-null heart (3, 4, 6, 9, 23, 29, 36, 47). To identify other genes regulated by Nkx2.5 in the developing heart, we performed suppression subtractive hybridization with RNAs isolated from the hearts of wild-type and Nkx2.5-null mice at ED 8.5. We describe here that myocardin and myocardin A are downregulated in Nkx2.5-null hearts. Nkx2.5 transactivates the myocardin promoter. A dominant-negative mutant of myocardin inhibited cardiomyogenesis in P19CL6 cells. BMP2 and TAK1 augmented myocardin A transcriptional activation at the SRE.
MATERIALS AND METHODS
RNA preparation, cDNA synthesis, and subtractive hybridization.
Total RNA (150 ng) from the hearts of wild-type and Nkx2.5-null embryos at ED 8.5 was treated with RNase-free DNase I and used to generate cDNA by using the SMART cDNA synthesis kit (Clontech). Subtractive hybridization was performed by using the PCR-Select cDNA subtraction kit (Clontech). Briefly, two pools of RsaI-digested cDNA from wild-type hearts were used as tester and ligated to unique adapters. RsaI-digested cDNA from Nkx2.5-null hearts was used as the driver without adapters. Hybridization of the tester population with excess driver and amplification of subtracted species with the two unique adapters in the tester cDNA produced a pool of PCR-amplified fragments theoretically present in wild-type hearts but not present in Nkx2.5-null hearts.
Cloning of myocardin A cDNA and the promoter region.
cDNA clones representing myocardin A were isolated from ED 10 mouse heart cDNA libraries (Stratagene) by using standard methodology. 5′ RNA ligase-mediated RACE (rapid amplification of cDNA ends) was performed according to the manufacturer's protocol (Ambion, Inc.). A 2-kb fragment of the myocardin promoter (positions −1961 to +39 in the myocardin gene) was isolated from a bacterial artificial chromosome clone (RP23-298L23; Research Genetics).
In situ hybridization.
In situ hybridization was performed with paraformaldehyde-fixed, paraffin-embedded mouse embryos at ED 9.5. A common cDNA fragment for myocardin A and myocardin was made by PCR with mouse heart cDNA as a template with the following primers: a forward primer (5′-CAACTGTCACCTTTCCTGTCACG-3′) and a reverse primer (5′-CCACCAGCATCTTGTCCTTCTC-3′). The fragment was cloned into pCRII (Invitrogen) and used to generate a 35S-labeled probe by in vitro transcription. Hybridization, washing, and probe detection were performed as previously described (36).
Northern blot analysis.
Total RNA was isolated by using Trizol reagent (Invitrogen) or an RNeasy Mini kit (Qiagen). mRNA was isolated from total RNA by using NucleoTrap mRNA kit (Clontech). Total RNA or mRNA was size fractionated on 1.3% agarose gel containing 2.2 M formaldehyde and transferred to nylon membranes. The membranes were then hybridized with a probe radiolabeled with [32P]dCTP by random priming. Blots were hybridized at 42°C for 16 h in the presence of 50% formamide. After hybridization, the membranes were washed at 65°C in the presence of 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% sodium dodecyl sulfate. Visualization was achieved by exposure to Kodak Biomax MS film (Eastman Kodak Co.). A specific probe for myocardin A was made by PCR with myocardin A cDNA as a template with a forward primer (5′-AACTCAGGGGCACACGAAGG-3′) and a reverse primer (5′-CTTTGGATGAATACTTGGGC-3′). A PstI-EcoRV fragment of N-cadherin was used as a probe as previously described (36). A PflMI-EcoRI fragment of Nkx2.5 was used as a probe. α-Myosin heavy chain (MHC) 3′-untranslated region cDNA was used as a probe for MHC (kindly provided by Margaret Buckingham, Institut Pasteur, Paris, France). MEF2C cDNA was used as a probe.
Reverse transcriptase PCR (RT-PCR).
Total RNA from embryonic (ED 10.5), neonatal, and adult hearts were treated with RNase-free DNase I. First-strand cDNA synthesis was performed with 1 μg of total RNA with Superscript II RT, and random hexamer. PCR amplification was performed with primers that anneal to both myocardin A and myocardin cDNAs and that flank the myocardin A-specific exon (forward primer [5′-TCACTGTGTGGAGTCCTCAGGTC-3′] and reverse primer [5′-TGGCATCGGCTGGCATTTC-3′]). PCR conditions were as follows: 94°C for 2 min and then 30 cycles at 94°C for 15 s, 58°C for 30 s, and 72°C for 1 min. PCR products were electrophoresed on 2% agarose gels and visualized by ethidium bromide staining.
Primer extension analysis.
One microgram of mouse heart mRNA and 105 cpm end-labeled oligonucleotide (antisense for mouse myocardin A cDNA nucleotides 27 to 50,5′-TGAGGGTGGCCGGGAACAGCCGCA-3′) were hybridized in hybridization buffer (150 mM KCl, 10 mM Tris [pH 8.3], 1 mM EDTA) at 65°C for 90 min and cooled slowly to room temperature. The sample was mixed with reaction mix (20 mM Tris [pH 8.3], 5.5 mM dithiothreitol, 10 mM MgCl2, 150 μM deoxynucleoside triphosphate, 150 μg of actinomycin D/ml, 40 U of Superscript II RT [Invitrogen]) and then incubated at 42°C for 60 min. The sample was treated with 14 μg of RNase A/ml at 37°C for 15 min and extracted with phenol-chloroform. The ethanol precipitated sample was resupended with formamide loading buffer. After heating the mixture at 65°C for 5 min, the sample was loaded onto an 8% acrylamide-8 M urea gel. After electrophoresis, the gel was dried and exposed to film.
Plasmid construct.
A cDNA encoding myocardin A with a C-terminal hemagglutinin (HA) epitope was cloned into pcDNA3 (Invitrogen) to generate pcDNA3-myocardin A. A deletion mutant of myocardin A-tagged HA (MC129-509) was made by PCR with pcDNA3-myocardin A as a template with a forward primer (5′-CCCCAAGCTTCCCACCATGGATTCTTCCGTGAAAGAGGCT-3′) and a reverse primer (5′-CCGGAATTCTTAAGCGTAATCTGGAACATCGTATGGGTAGCCCGAGCCCCCATTCAGGCTGCTCA-3′). MC129-509 was ligated into pcDNA3 to construct pcDNA3-MC129-509. pcDNA3-myocardin was made by replacing the myocardin A fragment with a myocardin fragment. The myocardin fragment was obtained by PCR with mouse neonatal heart cDNA as a template with the following primers: a forward primer (5′-TCACTGTGTGGAGTCCTCAGGTC-3′) and a reverse primer (5′-TGGCATCGGCTGGCATTTC-3′). The PCR fragments were then digested with Bsu36I and EcoRV and ligated with Bsu36I-EcoRV-digested pcDNA3-myocardin A to construct pcDNA3-myocardin. All of the PCR fragments were confirmed by DNA sequencing. A 2-kb fragment of the myocardin promoter was ligated with pGL2-Basic vector (Promega) to construct myocardin-Luc. Promoter mutations were constructed by site-directed mutagenesis with the Quickchange kit (Stratagene). The TCAAGTG nucleotide sequence in the NKE was changed to a TCCCTCG sequence. The AACTTGA nucleotide sequence in the NKEr2 was changed to a AGAGGGA sequence. Mutations of the promoter constructs were checked by sequencing.
Transfection and reporter assay.
COS cells and P19CL6 cells were plated at densities of 4.0 × 105 and 1.0 × 105 cells/well, respectively, in six-well plates. The following day, the cells were transfected with 3 μg of myocardin-Luc, ANF luciferase reporter construct (−638 ANF Luc; kindly provided by Kenneth R. Chien, University of California, San Diego), or various SRE mutants in the ANF luciferase reporter construct (kindly provided by Andrew Thorburn, Wake Forest University School of Medicine). The cells were also transfected with 1 μg of expression vectors containing Nkx2.5, Nkx2.5(I183P), myocardin A, myocardin, MC129-509, SRF, TAK1, or TAK1 mutants and 1 μg of pTKβ-Gal by the calcium phosphate method. The total plasmid amount was adjusted to 6 μg with an empty vector plasmid. Expression vectors containing TAK1 and TAK1 mutants were kindly provided by Hiroshi Shibuya (Tokyo Medical and Dental University, Tokyo, Japan). An SRF expression vector was kindly provided by Ron Prywes (Columbia University). At 6 h after transfection, cells were washed with phosphate-buffered saline (PBS), and the medium was changed. Cells were cultured for another 24 to 48 h, lysed with 200 μl of reporter lysis buffer (Promega), and assayed for luciferase activity (by a Promega assay) and β-galactosidase activity. Luciferase activity was normalized against β-galactosidase activity.
Electrophoretic mobility shift assay.
Nkx2.5 protein was prepared by coupled in vitro transcription/translation of a T7-driven Nkx2.5 plasmid in reticulocyte lysate by using a TNT kit (Promega). Labeled DNA probes were incubated with 1 μl of programmed lysate in 2 μg of bovine serum albumin and 2 μg of poly(dG-dC) in 10 mM HEPES (pH 7.9)-50 mM KCl-1 mM EGTA-10% glycerol-2.5 mM dithiothreitol-7 mM MgCl2 in a 20-μl reaction volume for 30 min at room temperature and then separated in 4% polyacrylamide gel by using a Tris-glycine buffer. All probes were double stranded and were radiolabeled with [γ-32P]ATP. The single-stranded sequences of the probes were as follows (the putative DNA-binding sites are underlined): NKE (GAATTCCAAGATTCAAGTGCATTGGTGCAACC) and NKEr2 (GATGTAATAAGAACTTGACCCACAATTTATG).
Replication-defective recombinant adenovirus and gene transfer.
HA-tagged myocardin A cDNA and LacZ cDNA were used to generate recombinant adenoviruses expressing myocardin A (AdMCA) and LacZ (AdLacZ), respectively, by using an Adeno-X Expression System (Clontech). At 24 h after seeding, cardiac myocytes were infected with AdMCA or AdLacZ diluted in the culture media at the multiplicity of infection of 10 and incubated for 2 h. The viral suspension was removed, and cardiac myocytes were cultured with the serum-depleted culture medium for 48 h.
Cell culture and differentiation.
Rat neonatal cardiac myocyte culture from 1-day-old Wister rats was prepared as previously described with some modifications (32). Mouse neonatal cardiac myocyte culture from 1-day-old C57BL/6 mice was prepared as described above in rat with some modifications. P19CL6 cells were kindly provided by Issei Komuro (Chiba University Graduate school of Medicine). P19CL6 cells were cultured as described previously (12, 27, 28). Briefly, cells were cultured in a-minimal essential medium (Invitrogen), supplemented with 10% fetal bovine serum (Invitrogen), penicillin (100 U/ml), and streptomycin (100 μg/ml). To induce differentiation under adherent conditions, cells were plated at a density of 3.7 × 105 cells in a 60-mm culture dish with 1% dimethyl sulfoxide (DMSO). The medium was changed every 2 days. Days of differentiation were numbered consecutively, with the first day of DMSO treatment as day 0.
Stable transformants.
Stable P19CL6 cell lines expressing cDNAs (P19CL6[pcDNA3] and P19CL6[MC129-509]) were generated by using Lipofectamine 2000 reagent according to the manufacturer's protocol (Invitrogen). Stable transformants were selected with 600 μg of G418 (Invitrogen)/ml, and 12 independent cell lines were cloned.
Immunofluorescence.
After a 10-min fixation in methanol on ice, cells were rehydrated in PBS for 15 min at room temperature. The fixed cells were incubated for 1 h at room temperature in the presence of MF20, a mouse anti-sarcomeric MHC monoclonal antibody. After three washes with PBS containing 0.5% Tween 20, the cells were incubated for 1 h at room temperature in the dark with TO-PRO-3 (Molecular Probes) and anti-mouse immunoglobulin G conjugated with tetramethyl rhodamine isothiocyanate (TRITC) as the secondary antibody (Jackson Immunoresearch Laboratories). Next, the cells were washed three times and mounted in a solution of 90% glycerol, 10% PBS, and 1 mg of p-phenylenediamine/ml onto glass slides.
RESULTS
Myocardin is downregulated in Nkx2.5-null hearts.
To identify potential downstream target genes in the Nkx2.5-dependent pathway in the developing heart, we performed suppression subtraction hybridization by using RNAs isolated from hearts of wild-type and Nkx2.5-null embryos at ED 8.5. One cDNA fragment isolated in this screening appeared to encode the myocardin gene. To examine whether myocardin mRNA is downregulated in Nkx2.5-null hearts, we performed in situ hybridization and Northern blotting. In situ hybridization revealed that myocardin mRNA expression in the heart of Nkx2.5-null embryos at ED 9.5 was downregulated compared to that of wild-type embryos (Fig. 1A). We then performed Northern blotting to compare myocardin mRNA expression quantitatively using total RNA isolated from hearts at ED 9.5 and 10.5. Since GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA was upregulated in Nkx2.5-null hearts, myocardin mRNA expression was normalized to 28S RNA loading. Myocardin mRNA expression in Nkx2.5-null hearts was decreased by 60 to 65% compared to that in wild-type hearts (Fig. 1B).
FIG. 1.
Myocardin mRNA expression in Nkx2.5-null hearts. Myocardin mRNA expression was detected by in situ hybridization (A) and Northern blot analysis (B). (A) 35S-labeled common probe for myocardin A and myocardin was hybridized to a wild-type embryo (+/+) and a Nkx2.5-null embryo (−/−) at ED 9.5. Bar, 100 μm. (B) Blots were made with total RNA (1 μg) isolated from hearts of wild-type embryos (+/+), Nkx2.5-null embryos (−/−), and Nkx2.5 heterozygote embryos (+/−) at ED 9.5 and 10.5. Myocardin expression was downregulated in Nkx2.5-null hearts. 28S RNA was used as a control for assessing RNA loading.
We also examined N-cadherin mRNA expression. N-cadherin is expressed in the notochord, otic and optic vesicles, and heart at ED 9.5 in mice (30). In N-cadherin-null mice, heart looping is arrested at the same stage as in Nkx2.5-null mice. As shown in Fig. 1B, N-cadherin mRNA expression was not affected in Nkx2.5-null hearts as previously reported (36). Together, these results suggest that downregulation of myocardin mRNA expression in Nkx2.5-null hearts is specific.
Cloning of myocardin A.
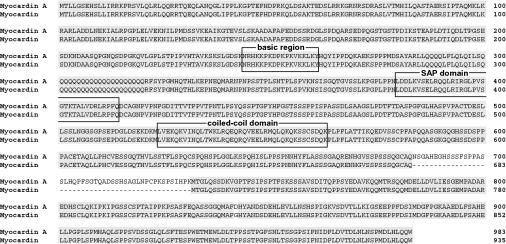
Northern blotting revealed multiple transcripts that hybridized to the myocardin cDNA probe and the major hybridized band was ca. 8 kb (Fig. 1B and 3). However, a previous report showed the major hybridized bands of myocardin to be ca. 3 to 5 kb (42). Therefore, we suspected that the major band that we identified was a distinct myocardin isoform. We cloned this isoform by using cDNA libraries of mouse embryonic hearts at ED 10. The sequence of the cDNA encoded a protein of 983 amino acids that was identical to myocardin except for the presence of a 48-amino-acid insertion (Fig. 2). During the course of the present study, a cDNA that has the same insertion site as in our study but starts from the 129th amino acid, as determined in our study, was independently cloned and named myocardin A (GenBank NM_145136). Therefore, we also refer to our gene as myocardin A. Examination of a database of mouse genomic DNA revealed that myocardin A was located on chromosome 11. Myocardin A consists of 14 exons, and the myocardin A-specific exon is located between exon 10 and 11 of myocardin, indicating that myocardin A is an alternative spliced isoform of myocardin.
FIG. 3.
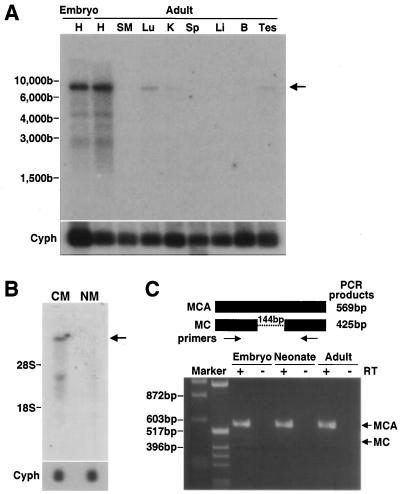
Myocardin A expression in murine tissues. (A) Blots were made with mRNA (1 μg) isolated from hearts of embryos at ED 10.5 and tissues of 8-week-old mice. An 8-kb transcript (arrow) hybridized to the myocardin A specific probe in both embryo and adult heart. Weaker signals were observed with mRNA from lung, kidney, and testis in adult. A cyclophilin (Cyph) probe was used as a control for assessing RNA loading. (B) Blots were made with total RNA (10 μg) isolated from mouse cultured neonatal cardiac myocytes (CM) and nonmyocytes (NM). An 8-kb myocardin A transcript was observed in cardiac myocytes (arrow), but not in nonmyocytes. A cyclophilin (Cyph) probe was used as a control for assessing RNA loading. (C) RT-PCR was performed with cDNAs from embryonic (ED 10.5), neonatal, and adult hearts as templates with common primers for myocardin A and myocardin that sandwich the myocardin A-specific exon. RT was included or omitted from samples as indicated. Expected sizes of PCR products from myocardin A and myocardin are 569 and 425 bp, respectively. The myocardin A transcript but not the myocardin transcript was detected in this experimental condition.
FIG. 2.
Primary sequence of mouse myocardin A. Alignment and amino acid sequence comparison of mouse myocardin A (AY303755) and myocardin (NM_146386). Identical amino acids are shaded in gray, gaps are represented by a “-”. Positions in the amino acid sequence are given by numbers. The sequence of the myocardin A cDNA encoded a protein of 983 amino acids. Myocardin A and myocardin contain a basic region, a SAP domain, and a coiled-coil domain.
Northern blot analysis was performed with a variety of tissues to examine the pattern of myocardin A mRNA expression with the myocardin A-specific exon as a probe. Analysis of adult mouse tissues revealed a band representing an 8-kb mRNA in the heart. Very weak signals were found in the lung, kidney, and testis (Fig. 3A). Other, weaker signals (ca. 4.5 and 3 kb) were observed with RNA from the hearts of embryos and adult mice. When cardiac myocytes were separated from nonmyocytes by using primary culture, myocardin A was found to be expressed in myocytes but not in nonmyocytes (Fig. 3B). We also examined the overall mRNA expression of myocardin and myocardin A by using a common probe for myocardin and myocardin A. The tissue distribution and pattern of hybridized bands was the same as that obtained with the myocardin A-specific probe (data not shown). In addition, RT-PCR with RNA from embryo, neonate, and adult hearts revealed that the myocardin A transcript is much more abundant than the myocardin transcript (Fig. 3C). In fact, the myocardin transcript was not readily visualized by 30 cycles RT-PCR. Therefore, myocardin A is the most abundant isoform in the heart.
Of note, the myocardin A sequence revealed that the probes that we used for in situ hybridization and Northern blotting in Fig. 1 recognize both myocardin and myocardin A transcripts. Therefore, the results of Fig. 1 indicate that overall mRNA expression of myocardin and myocardin A is downregulated in Nkx2.5-null hearts.
Nkx2.5 transactivated the myocardin promoter.
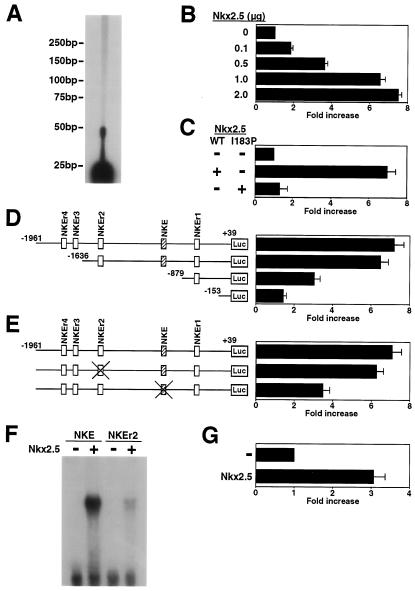
To examine whether myocardin expression is regulated by Nkx2.5, we performed a reporter assay using the myocardin promoter. We determined the 5′ end of myocardin A cDNA by primer extension analysis (Fig. 4A). We then isolated a 2-kb fragment of the myocardin promoter (positions −1961 to +39 in the myocardin gene). Wild-type Nkx2.5 transactivated the myocardin promoter in a dose-dependent manner (Fig. 4B). On the other hand, a point mutant of Nkx2.5(I183P [Ile183Pro]) that failed to bind DNA (18) did not transactivate the myocardin promoter (Fig. 4C).
FIG. 4.
Transactivation of the myocardin promoter by Nkx2.5. (A) Primer extension analysis was performed with mouse heart mRNA and an antisense oligonucleotide for mouse myocardin A cDNA (nucleotides 27 to 50). A band was seen at 50 bp. Positions in the nucleotide are given by numbers from mouse myocardin A (AY303755). (B) COS cells were transiently cotransfected with myocardin-Luc and the indicated amounts of wild-type Nkx2.5 expression vector. Nkx2.5 transactivated the myocardin promoter in a dose-dependent manner. (C) COS cells were transiently cotransfected with myocardin-Luc and with wild-type Nkx2.5 (WT) or Nkx2.5(I183P) expression vectors. Wild-type Nkx2.5 activated myocardin-Luc, but Nkx2.5(I183P) did not. (D) COS cells were transiently cotransfected with either myocardin-Luc or deletion mutant constructs and wild-type Nkx2.5 expression vector. A deletion between −1636 and −879 markedly reduced the promoter activity. Consensus and similar binding motifs for Nkx2.5 are indicated by boxes. (E) COS cells were transiently cotransfected with myocardin-Luc, mutated NKE in myocardin-Luc, or mutated NKEr2 in myocardin-Luc, and wild-type Nkx2.5 expression vector. Mutated NKE in the myocardin promoter markedly reduced responsiveness to Nkx2.5. (F) NKE and NKEr2 were incubated with in vitro-translated Nkx2.5 protein. NKE had a high binding affinity for Nkx2.5 protein, whereas NKEr2 had a lower binding affinity for Nkx2.5 protein. (G) Cardiac myocytes were transiently cotransfected with myocardin-Luc and wild-type Nkx2.5 expression vector. Nkx2.5 transactivated the myocardin promoter in cardiac myocytes.
We then made deletion mutants of myocardin-Luc and examined the role of the Nkx2.5-responsive element in the myocardin promoter. Deletion of the myocardin promoter region from −1961 to −1636 slightly reduced the promoter activation by Nkx2.5. However, deletion between −1636 and −879 markedly reduced the promoter activity (Fig. 4D), indicating that the Nkx2.5-responsive element is located between −1636 and −879 of the myocardin promoter region. The myocardin promoter has one consensus binding motif for Nkx2.5 (TCAAGTG) at −1069. We designated this site NKE (Fig. 4C). Four additional sequences similar to the consensus motif (i.e., NKEr1, -2, -3, and -4) were found within the myocardin promoter from −1961 to −1. We speculated that NKE is important for the transcriptional activation of the myocardin promoter by Nkx2.5. To examine this possibility, the mutated NKE and NKEr2 constructs were cotransfected with the expression vector of Nkx2.5. Mutated NKE in the myocardin promoter markedly reduced responsiveness to Nkx2.5, whereas mutated NKEr2 did not (Fig. 4E).
The result described above suggested that NKE is necessary for the full activation of the myocardin promoter by Nkx2.5. To confirm that Nkx2.5 binds to NKE, an electrophoretic mobility shift assay was performed with in vitro-translated Nkx2.5 protein and oligonucleotide probes corresponding to NKE and NKEr2. As shown in Fig. 4F, Nkx2.5 bound to NKE, whereas it bound to NKEr2 with much lower affinity.
In cardiac myocytes, Nkx2.5 also transactivated the myocardin promoter (Fig. 4G). Together, these results suggest that Nkx2.5 regulates myocardin gene transcription in vitro.
Myocardin A transactivates the ANF promoter through the SRE.
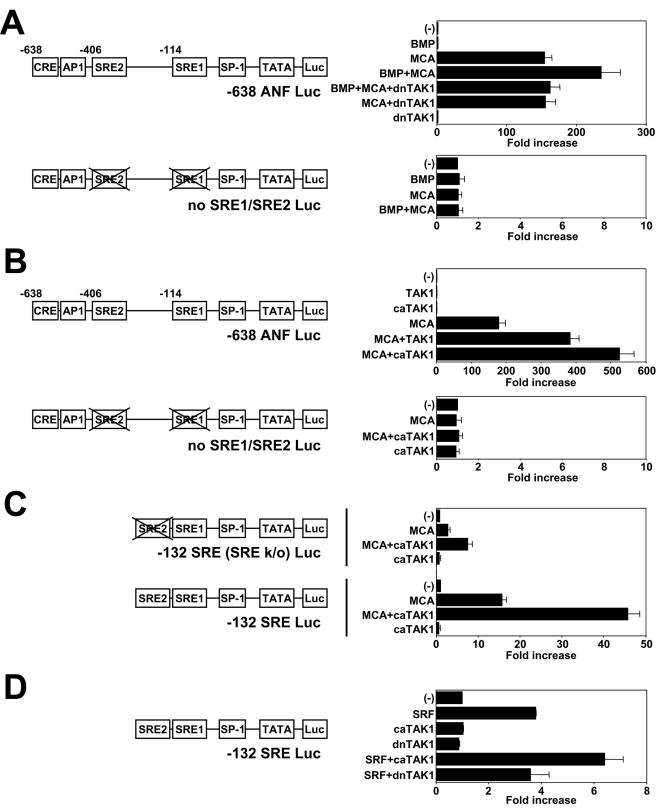
Myocardin activates SRE-dependent promoters such as the ANF promoter and the SM22 promoter (42). We examined the transcriptional activity of myocardin A by using the ANF promoter (−638 ANF Luc), which has two SRF binding sites (14). Myocardin A transactivated −638 ANF Luc by >100-fold, and the extent of activation by myocardin A was the same as that caused by myocardin (Fig. 5). We then examined the requirement of the SRE sites in the ANF promoter for responsiveness to myocardin A. Mutation of the SRE2 (no SRE2 Luc) reduced responsiveness to myocardin A by 75%. In contrast, mutation of the SRE1 (no SRE1 Luc) markedly reduced responsiveness to myocardin A, and mutation of both SREs (no SRE1/SRE2 Luc) completely abolished the activation by myocardin A. The requirement of the SRE sites in the ANF promoter for responsiveness to myocardin A was the same as that of myocardin. These results indicate that although there is a difference in the requirement between the SRE1 and the SRE2, the transcriptional activities of myocardin A and myocardin are mediated though the SRE sites.
FIG. 5.
Transactivation of the ANF promoter by myocardin A and myocardin. (A) COS cells were transiently cotransfected with −638 ANF Luc or −638 ANF Luc with mutations in SRE1 (no SRE1 Luc), SRE2 (no SRE2 Luc), or both SRE1 and SRE2 (no SRE1/SRE2 Luc) and with or without myocardin A or myocardin expression vector. Myocardin A and myocardin activated −638 ANF Luc to the same degree. Mutation of the SRE2 reduced responsiveness to myocardin A. Mutation of the SRE1 almost completely reduced responsiveness to myocardin A, and mutation of both SREs completely abolished activation by myocardin A. (B) Blots were made with total RNA (5 μg) isolated from cardiac myocytes infected with AdLacZ or AdMCA. Overexpression of myocardin A induced endogenous ANF mRNA expression. A GAPDH probe was used as a control for assessing RNA loading.
We then made a recombinant adenovirus expressing myocardin A (AdMCA) and examined whether myocardin A induces endogenous ANF mRNA expression. As shown in Fig. 5B, myocardin A induced ANF mRNA expression in cardiac myocytes.
Myocardin is required for cardiomyogenesis in P19CL6 cells.
We then examined myocardin expression during cardiomyogenesis in P19CL6 cells. In P19CL6 cells, spontaneously beating cells were first observed in small patches on day 10 after treatment with 1% DMSO, and subsequently the majority of cells beat synchronously, a finding consistent with previous reports (12, 27, 28). As shown in Fig. 6A, myocardin mRNA expression was faintly detectable on day 6 and became maximal on day 8 and 10. The expression pattern of MEF2C mRNA was similar to that of myocardin mRNA because MEF2C mRNA was detectable on day 6 and became maximal on day 8. On the other hand, Nkx2.5 mRNA expression was also first detectable on day 6, but its expression became maximal after day 10. MLC2v mRNA expression was detected on day 10 and increased through day 16.
FIG. 6.
Inhibition of cardiomyogenesis by overexpression of a dominant-negative mutant of myocardin. (A) Blots were made with total RNA (10 μg) isolated from P19CL6 cells on the indicated days. Myocardin (arrow) and Nkx2.5 mRNA expressions were first detectable at day 6. 28S RNA was used as a control for assessing RNA loading. (B) Schematic representation of myocardin A, myocardin, and MC129-509. Abbreviations: B, basic region; S, SAP domain; C, coiled-coil domain. P19CL6 cells were transiently cotransfected with the ANF truncated promoter with a functional SRE2 fused to the end (−132 SRE Luc), with or without myocardin A or myocardin expression vector and with or without MC129-509 expression vector. MC129-509 inhibited both actions of myocardin A and myocardin on this promoter. (C) P19CL6[pcDNA3] cells and P19CL6[MC129-509] cells were treated with 1% DMSO. At 16 days after treatment, cells were stained by immunofluorescence with MF20 (upper panels) or TO-PRO-3 (lower panels). About 80% of the P19CL6[pcDNA3] cells were differentiated into cardiac myocytes. In contrast, <10% of P19CL6[MC129-509] cells were differentiated into cardiac myocytes. Bar, 100 μm. (D) P19CL6[pcDNA3] cells and P19CL6[MC129-509] cells were treated with 1% DMSO. At 16 days after treatment, cells were harvested. Blots were made with total RNA (10 μg) isolated from P19CL6 cells on day 16. mRNA expressions of MLC2v, Nkx2.5, MHC, and MEF2C were reduced in three independent P19CL6[MC129-509] cells (lanes 1, 2, and 3). 28S RNA was used as a control for assessing RNA loading.
Since Nkx2.5 activity has been reported to be essential for cardiomyogenesis in P19 cells and P19CL6 cells (16, 38), we examined whether myocardin also plays an important role during cardiomyogenesis with P19CL6 cells. The deletion mutant of myocardin A (MC129-509) per se did not affect the basal transcriptional activity of the truncated promoter containing both SRE1 and SRE2 (−132 SRE Luc). When MC129-509 was cotransfected with myocardin A or myocardin, it inhibited the transcriptional activity of both myocardin A and myocardin on −132 SRE Luc in P19CL6 cells (Fig. 6B), indicating that it works as a dominant negative. We tried to make stable cell lines expressing myocardin A and MC129-509. We obtained cell lines expressing MC129-509 (P19CL6[MC129-509]), but we could not obtain cell lines constitutively expressing wild-type myocardin A, suggesting that overexpression of myocardin A may terminally differentiate cells or reduce their proliferating capacity. To determine the effect of myocardin activity on cardiomyogenesis, we sought to determine whether stable cell lines expressing MC129-509 had reduced expression of sarcomeric MHC, as determined previously by staining with the anti-MHC antibody, MF20 (27, 28, 38). About 80% of the P19CL6[pcDNA3] cells were differentiated into cardiac myocytes in the presence of 1% DMSO at day 16. In contrast, <10% of P19CL6[MC129-509] cells were differentiated into cardiac myocytes at day 16 (Fig. 6C). Furthermore, we performed Northern blotting to compare mRNA expression of cardiac markers in 1% DMSO-treated P19CL6[pcDNA3] and three independently cloned P19CL6[MC129-509] cell lines at day 16. mRNA expressions of MEF2C, Nkx2.5, MHC, and MLC2v were markedly reduced in P19CL6[MC129-509] cell lines at day 16 (Fig. 6D). These results demonstrate that myocardin function is required for differentiation into cardiac myocytes in P19CL6 cells.
Myocardin A action is modulated by BMP/TAK1 signaling at the SRE.
In P19CL6 cells BMPs are required for cardiac myocyte differentiation (28). BMPs act through Smad and TAK1 pathways (40). Smads, TAK1, and their common target ATF2 play pivotal roles in the cardiogenic BMP signaling pathway (27, 28). In cardiac myocytes TGF-β and TAK1 activate transcription through the SRE (24, 39, 44). Therefore, we examined whether BMP/TAK1 signaling modulates myocardin A action on using the ANF promoter in P19CL6 cells. BMP2 alone did not activate the ANF promoter under our experimental conditions, whereas BMP2 augmented myocardin A-induced ANF promoter activation (Fig. 7A). To examine the effect of TAK1 on BMP signaling, we used a dominant-negative mutant of TAK1 (dnTAK1) in which Lys-63 of the ATP-binding site was replaced by tryptophan (43). dnTAK1 inhibited BMP2 action on myocardin A-induced ANF promoter activation. These results indicate that BMP2 augments responsiveness to myocardin A through TAK1 on the ANF promoter. In addition, wild-type TAK1 augmented myocardin A-induced ANF promoter activation, and a constitutively active TAK1 (caTAK1) which lacks the N-terminal 22 amino acids (43) caused more transactivation (Fig. 7B).
FIG. 7.
Augmentation of myocardin A-induced ANF promoter activity by BMP/TAK1 signaling. P19CL6 cells were transiently cotransfected with various luciferase reporter constructs, with or without myocardin A expression vector, or with or without SRF expression vector and with or without various TAK1 expression vectors as indicated. (A) The transfected cells were treated with vehicle or BMP2 (100 ng/ml) for 24 h. BMP2 augmented myocardin A-induced ANF promoter activation, and dnTAK1 inhibited BMP2 action on myocardin A-induced ANF promoter activity. Mutation of both SREs (no SRE1/SRE2 Luc) completely abolished BMP action. (B) Wild-type of TAK1 or caTAK1 augmented myocardin A-induced ANF promoter activation. Mutation in both SRE1 and SRE2 (no SRE1/SRE2 Luc) abolished myocardin A action and augmentation by caTAK1. (C) caTAK1 augmented the myocardin A action on −132 SRE Luc. Responsiveness to myocardin A was reduced on −132 SRE (SREk/o) Luc, but TAK1 still augmented myocardin A action on this promoter. (D) SRF transactivated the truncated promoters containing both SRE1 and SRE2 (−132 SRE Luc), and caTAK1 augmented the SRF action on −132 SRE Luc.
To further analyze the mechanism of BMP/TAK1 signaling to the ANF promoter, we used a mutant promoter in both SREs. As shown in Fig. 7A and B, mutation of both SREs (no SRE1/SRE2 Luc) abolished not only activation by myocardin A but also BMP2 and TAK1 action, indicating that SREs in the ANF promoter are necessary for BMP/TAK1 action on myocardin A. We then examined whether the SRE is sufficient to confer responsiveness to TAK1. As shown in Fig. 7C, myocardin A transactivated the truncated promoters containing SRE1 (−132 SRE [SREk/o] Luc), and both SRE1 and SRE2 (−132 SRE Luc). caTAK1 augmented responsiveness to myocardin A on −132 SRE (SREk/o) Luc and −132 SRE Luc (Fig. 7C).
We then examined whether TAK1 phosphorylates myocardin A. TAK1 did not phosphorylate myocardin A (data not shown). We also examined whether TAK1 augments SRF action at the SRE. As shown in Fig. 7D, caTAK1 augmented responsiveness to SRF on −132 SRE Luc, indicating that TAK1 signaling regulates SRF function at the SRE site. Together, these results indicate that TAK1 action and myocardin A action are integrated at the SRE sites in P19CL6 cells.
DISCUSSION
We have shown here that myocardin A and myocardin are downregulated in Nkx2.5-null hearts. In addition, at least five consensus and similar binding motifs for Nkx2.5 (TYAAGTG and CWTAATTG) (7) were found within the mouse myocardin promoter from −1961 to −1, and Nkx2.5 directly regulates this promoter in vitro, at least in part through the NKE site. These findings suggest that Nkx2.5 directly regulated myocardin expression during embryonic development. On the other hand, the Nkx2.5 promoter has been reported to be activated by myocardin (42). We showed that Nkx2.5 mRNA expression was reduced during differentiation into cardiac myocytes in P19CL6[MC129-509] cells. Therefore, it is likely that Nkx2.5 and myocardin regulate their expression on each other in the embryonic heart.
We showed that mutation of both SREs (no SRE1/SRE2 Luc) completely abolished activation by myocardin A (Fig. 5). These results suggest that both SRE sites on the −638 ANF promoter are necessary for myocardin A-dependent transactivation, a finding consistent with previous results showing that myocardin functions as an SRF cofactor at SRE sites (42). However, other sequences in the −638 ANF promoter are also required for full activation of the −638 ANF promoter by myocardin A, because responsiveness to myocardin A on the truncated ANF promoter containing both SRE1 and SRE2 (−132 SRE Luc) was reduced compared to that of the −638 ANF promoter (Fig. 7C). The region deleted in −132 SRE Luc contains binding sites for other transcription factors, including the ATF/CREB family, Nkx2.5, and GATA-4. Hines et al. proposed a model suggesting that SRF at the SRE1 site of the ANF promoter functions as an “activator bridge” to promote productive interactions between activated transcription factors at the other sites and the basal transcription machinery (14). ATF6 has been shown to interact with SRF and be involved in the activation of transcription by SRF (37, 46). SRF also interacts with Nkx2.5 and GATA-4, and the recruitment of Nkx2.5 and GATA-4 by SRF enhances SRF DNA-binding activity (34). Consistent with these data, our results indicate that full ANF promoter activation requires interactions between myocardin and SRF at SRE sites, and other transcription factors at other sites distal to SRE1.
Overexpression of a dominant-negative mutant of Nkx2.5 inhibited cardiomyogenesis in both P19 cells and P19CL6 cells (16, 38). These findings demonstrate that dominant-negative strategies can bypass potential functional redundancies with other family members and allow for the determination of the role of certain genes. To examine the effect of myocardin function on cardiomyogenesis, we used the dominant-negative strategy using the deletion mutant of myocardin A (MC129-509). We showed here that MC129-509 worked as a dominant-negative on the truncated ANF promoter containing SREs and that inhibition of myocardin function prevents differentiation into cardiac myocytes in P19CL6 cells. Expression of a dominant-negative mutant of myocardin in Xenopus embryos interfered with differentiation of myocardial cells (42). Our findings indicate that myocardin function is required for cardiomyogenesis not only in amphibians but also in cultured mammalian cells. The SRF gene was demonstrated to be necessary for cardiac mesoderm formation (1). Our findings indicate that in the presence of SRF, P19CL6 cells cannot differentiate into cardiac myocytes without myocardin function. On the other hand, we could not obtain a cell line expressing wild-type myocardin A. This suggests that timing and the amount of myocardin expression are important for the maintenance of cell function.
BMPs are required for cardiac myocyte differentiation in P19 cells and P19CL6 cells (15, 28). In Drosophila, dpp, the BMP homologue, is required to maintain tinman expression in the mesoderm (10). In chick and Xenopus, disruption of BMP signaling with noggin or dominant-negative BMP receptors can prevent cardiomyogenesis (21, 33, 35, 41). In mouse, BMP2-deficient embryos die before ED 10.5 with a defect in cardiac development (45). These results suggest that BMP signaling is essential for cardiomyogenesis in a number of species.
BMPs function via binding to two types of serine/threonine kinase receptors, type I and type II receptors (19). Among the type I receptors for BMPs, activin receptor-like kinase 3 (ALK3)/BMP type I-A receptor (BMPR-IA) mutant embryos die by ED 9.5 due to a defect in mesoderm formation (26). Development is also arrested in ALK2 mutant embryos at the early gastrulation stage, and these mutants display disruption of mesoderm formation (11, 25). In addition, BMPR-II-deficient embryos die by ED9.5 and lacked mesoderm (2). These results indicate that BMP receptors are important for cardiac development, including mesoderm formation.
BMPs are expressed in tissues adjacent to the precardiac mesoderm, and treatment of BMPs can induce Nkx2.5, GATA-4, and ventricular MHC expression and differentiation into cardiac myocytes of nonprecardiac mesodermal cells in chick (33). However, BMPs can induce heart formation in limited regions of the nonprecardiac mesoderm. These findings suggest that BMP signaling and additional factor(s) in the specified region cooperatively regulate cardiac specificity.
The SRE site is important for expression of early response genes such as c-fos, and cardiac, skeletal, and smooth muscle specific genes (31). Although SRF is highly expressed in muscle cell lineages during embryogenesis, it is not muscle specific. Myocardin has been reported to be expressed in cardiac myocytes and smooth muscle cells (8, 42). We showed here that myocardin A mRNA is highly expressed in heart. However, it was also weakly detected in lung, kidney, and testis. This raises the possibility that expression of SRF and myocardin alone may not account for cardiac myocyte specificity of its target genes and that additional factor(s) are also required for cardiac specificity. Consistent with this viewpoint, the results of the present study showed that BMP/TAK1 signaling augments myocardin function at the SRE sites. Although we have not examined whether the interaction of myocardin with SRF is enhanced by TAK1 or not, our results at least indicate that myocardin A action and TAK1 action are integrated at the SRE site. This may be one of the mechanisms of cardiac myocyte specificity of the SRE-dependent genes.
Acknowledgments
We thank W. Pu, J. R. McMullen, and E. Höcht for critical reading of the manuscript. We also thank I. Chen for preparing the figures.
This study was supported by Japan Heart Foundation and Bayer Yakuhin Research Grant Abroad, a postdoctoral fellowship from the American Heart Association (AHA; New England affiliate) to T.U., an AHA (Massachusetts Affiliate) Beginning-Grant-in-Aid to H.K., and SCOR in congenital Heart Disease grant from the NIH (P50-HL61036) to S.I.
REFERENCES
- 1.Arsenian, S., B. Weinhold, M. Oelgeschlager, U. Ruther, and A. Nordheim. 1998. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J. 17:6289-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beppu, H., M. Kawabata, T. Hamamoto, A. Chytil, O. Minowa, T. Noda, and K. Miyazono. 2000. BMP type II receptor is required for gastrulation and early development of mouse embryos. Dev. Biol. 221:249-258. [DOI] [PubMed] [Google Scholar]
- 3.Biben, C., and R. P. Harvey. 1997. Homeodomain factor Nkx2-5 controls left/right asymmetric expression of bHLH gene eHand during murine heart development. Genes Dev. 11:1357-1369. [DOI] [PubMed] [Google Scholar]
- 4.Biben, C., S. Palmer, D. A. Elliott, and R. P. Harvey. 1997. Homeobox genes and heart development. Cold Spring Harbor Symp. Quant. Biol. 62:395-403. [PubMed] [Google Scholar]
- 5.Bodmer, R. 1993. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development 118:719-729. [DOI] [PubMed] [Google Scholar]
- 6.Bruneau, B. G., Z. Z. Bao, M. Tanaka, J. J. Schott, S. Izumo, C. L. Cepko, J. G. Seidman, and C. E. Seidman. 2000. Cardiac expression of the ventricle-specific homeobox gene Irx4 is modulated by Nkx2-5 and dHand. Dev. Biol. 217:266-277. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C. Y., and R. J. Schwartz. 1995. Identification of novel DNA binding targets and regulatory domains of a murine tinman homeodomain factor, Nkx-2.5. J. Biol. Chem. 270:15628-15633. [DOI] [PubMed] [Google Scholar]
- 8.Chen, J., C. M. Kitchen, J. W. Streb, and J. M. Miano. 2002. Myocardin: a component of a molecular switch for smooth muscle differentiation. J. Mol. Cell Cardiol. 34:1345-1356. [DOI] [PubMed] [Google Scholar]
- 9.Christoffels, V. M., P. E. Habets, D. Franco, M. Campione, F. de Jong, W. H. Lamers, Z. Z. Bao, S. Palmer, C. Biben, R. P. Harvey, and A. F. Moorman. 2000. Chamber formation and morphogenesis in the developing mammalian heart. Dev. Biol. 223:266-278. [DOI] [PubMed] [Google Scholar]
- 10.Frasch, M. 1995. Induction of visceral and cardiac mesoderm by ectodermal Dpp in the early Drosophila embryo. Nature 374:464-467. [DOI] [PubMed] [Google Scholar]
- 11.Gu, Z., E. M. Reynolds, J. Song, H. Lei, A. Feijen, L. Yu, W. He, D. T. MacLaughlin, J. van den Eijnden-van Raaij, P. K. Donahoe, and E. Li. 1999. The type I serine/threonine kinase receptor ActRIA (ALK2) is required for gastrulation of the mouse embryo. Development 126:2551-2561. [DOI] [PubMed] [Google Scholar]
- 12.Habara-Ohkubo, A. 1996. Differentiation of beating cardiac muscle cells from a derivative of P19 embryonal carcinoma cells. Cell Struct. Funct. 21:101-110. [DOI] [PubMed] [Google Scholar]
- 13.Harvey, R. P. 1996. NK-2 homeobox genes and heart development. Dev. Biol. 178:203-216. [DOI] [PubMed] [Google Scholar]
- 14.Hines, W. A., J. Thorburn, and A. Thorburn. 1999. A low-affinity serum response element allows other transcription factors to activate inducible gene expression in cardiac myocytes. Mol. Cell. Biol. 19:1841-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jamali, M., C. Karamboulas, P. J. Rogerson, and I. S. Skerjanc. 2001. BMP signaling regulates Nkx2-5 activity during cardiomyogenesis. FEBS Lett. 509:126-130. [DOI] [PubMed] [Google Scholar]
- 16.Jamali, M., P. J. Rogerson, S. Wilton, and I. S. Skerjanc. 2001. Nkx2-5 activity is essential for cardiomyogenesis. J. Biol. Chem. 276:42252-42258. [DOI] [PubMed] [Google Scholar]
- 17.Kasahara, H., S. Bartunkova, M. Schinke, M. Tanaka, and S. Izumo. 1998. Cardiac and extracardiac expression of Csx/Nkx2.5 homeodomain protein. Circ Res. 82:936-946. [DOI] [PubMed] [Google Scholar]
- 18.Kasahara, H., A. Usheva, T. Ueyama, H. Aoki, N. Horikoshi, and S. Izumo. 2001. Characterization of homo- and heterodimerization of cardiac Csx/Nkx2.5 homeoprotein. J. Biol. Chem. 276:4570-4580. [DOI] [PubMed] [Google Scholar]
- 19.Kawabata, M., T. Imamura, and K. Miyazono. 1998. Signal transduction by bone morphogenetic proteins. Cytokine Growth Factor Rev. 9:49-61. [DOI] [PubMed] [Google Scholar]
- 20.Komuro, I., and S. Izumo. 1993. Csx: a murine homeobox-containing gene specifically expressed in the developing heart. Proc. Natl. Acad. Sci. USA 90:8145-8149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ladd, A. N., T. A. Yatskievych, and P. B. Antin. 1998. Regulation of avian cardiac myogenesis by activin/TGFβ and bone morphogenetic proteins. Dev. Biol. 204:407-419. [DOI] [PubMed] [Google Scholar]
- 22.Lints, T. J., L. M. Parsons, L. Hartley, I. Lyons, and R. P. Harvey. 1993. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development 119:419-431. (Erratum, 119:969.) [DOI] [PubMed] [Google Scholar]
- 23.Lyons, I., L. M. Parsons, L. Hartley, R. Li, J. E. Andrews, L. Robb, and R. P. Harvey. 1995. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 9:1654-1666. [DOI] [PubMed] [Google Scholar]
- 24.MacLellan, W. R., T. C. Lee, R. J. Schwartz, and M. D. Schneider. 1994. Transforming growth factor-beta response elements of the skeletal alpha-actin gene. Combinatorial action of serum response factor, YY1, and the SV40 enhancer-binding protein, TEF-1. J. Biol. Chem. 269:16754-16760. [PubMed] [Google Scholar]
- 25.Mishina, Y., R. Crombie, A. Bradley, and R. R. Behringer. 1999. Multiple roles for activin-like kinase-2 signaling during mouse embryogenesis. Dev. Biol. 213:314-326. [DOI] [PubMed] [Google Scholar]
- 26.Mishina, Y., A. Suzuki, N. Ueno, and R. R. Behringer. 1995. Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes Dev. 9:3027-3037. [DOI] [PubMed] [Google Scholar]
- 27.Monzen, K., Y. Hiroi, S. Kudoh, H. Akazawa, T. Oka, E. Takimoto, D. Hayashi, T. Hosoda, M. Kawabata, K. Miyazono, S. Ishii, Y. Yazaki, R. Nagai, and I. Komuro. 2001. Smads, TAK1, and their common target ATF-2 play a critical role in cardiomyocyte differentiation. J. Cell Biol. 153:687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monzen, K., I. Shiojima, Y. Hiroi, S. Kudoh, T. Oka, E. Takimoto, D. Hayashi, T. Hosoda, A. Habara-Ohkubo, T. Nakaoka, T. Fujita, Y. Yazaki, and I. Komuro. 1999. Bone morphogenetic proteins induce cardiomyocyte differentiation through the mitogen-activated protein kinase kinase kinase TAK1 and cardiac transcription factors Csx/Nkx-2.5 and GATA-4. Mol. Cell. Biol. 19:7096-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmer, S., N. Groves, A. Schindeler, T. Yeoh, C. Biben, C. C. Wang, D. B. Sparrow, L. Barnett, N. A. Jenkins, N. G. Copeland, F. Koentgen, T. Mohun, and R. P. Harvey. 2001. The small muscle-specific protein Csl modifies cell shape and promotes myocyte fusion in an insulin-like growth factor 1-dependent manner. J. Cell Biol. 153:985-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radice, G. L., H. Rayburn, H. Matsunami, K. A. Knudsen, M. Takeichi, and R. O. Hynes. 1997. Developmental defects in mouse embryos lacking N-cadherin. Dev. Biol. 181:64-78. [DOI] [PubMed] [Google Scholar]
- 31.Reecy, L., N. Belaguli, and R. Schwartz. 1999. Serum response factor-NK homeodomain interaction: role in cardiac development, p. 273-287. In R. Harvey and N. Rosenthal (ed.), Heart development. Academic Press, Inc., New York, N.Y.
- 32.Sadoshima, J., L. Jahn, T. Takahashi, T. J. Kulik, and S. Izumo. 1992. Molecular characterization of the stretch-induced adaptation of cultured cardiac cells: an in vitro model of load-induced cardiac hypertrophy. J. Biol. Chem. 267:10551-10560. [PubMed] [Google Scholar]
- 33.Schultheiss, T. M., J. B. Burch, and A. B. Lassar. 1997. A role for bone morphogenetic proteins in the induction of cardiac myogenesis. Genes Dev. 11:451-462. [DOI] [PubMed] [Google Scholar]
- 34.Sepulveda, J. L., S. Vlahopoulos, D. Iyer, N. Belaguli, and R. J. Schwartz. 2002. Combinatorial expression of GATA4, Nkx2-5, and serum response factor directs early cardiac gene activity. J. Biol. Chem. 277:25775-25782. [DOI] [PubMed] [Google Scholar]
- 35.Shi, Y., S. Katsev, C. Cai, and S. Evans. 2000. BMP signaling is required for heart formation in vertebrates. Dev. Biol. 224:226-237. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka, M., Z. Chen, S. Bartunkova, N. Yamasaki, and S. Izumo. 1999. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development 126:1269-1280. [DOI] [PubMed] [Google Scholar]
- 37.Thuerauf, D. J., N. D. Arnold, D. Zechner, D. S. Hanford, K. M. DeMartin, P. M. McDonough, R. Prywes, and C. C. Glembotski. 1998. p38 Mitogen-activated protein kinase mediates the transcriptional induction of the atrial natriuretic factor gene through a serum response element: a potential role for the transcription factor ATF6. J. Biol. Chem. 273:20636-20643. [DOI] [PubMed] [Google Scholar]
- 38.Toko, H., W. Zhu, E. Takimoto, I. Shiojima, Y. Hiroi, Y. Zou, T. Oka, H. Akazawa, M. Mizukami, M. Sakamoto, F. Terasaki, Y. Kitaura, H. Takano, T. Nagai, R. Nagai, and I. Komuro. 2002. Csx/Nkx2-5 is required for homeostasis and survival of cardiac myocytes in the adult heart. J. Biol. Chem. 277:24735-24743. [DOI] [PubMed] [Google Scholar]
- 39.Ueyama, T., S. Kawashima, T. Sakoda, K. Hirata, Y. Ohashi, W. Yamochi, H. Akita, and M. Yokoyama. 1998. Transforming growth factor-β1 and protein kinase C synergistically activate the c-fos serum response element in myocardial cells. J. Mol. Cell Cardiol. 30:551-562. [DOI] [PubMed] [Google Scholar]
- 40.von Bubnoff, A., and K. W. Cho. 2001. Intracellular BMP signaling regulation in vertebrates: pathway or network? Dev. Biol. 239:1-14. [DOI] [PubMed] [Google Scholar]
- 41.Walters, M. J., G. A. Wayman, and J. L. Christian. 2001. Bone morphogenetic protein function is required for terminal differentiation of the heart but not for early expression of cardiac marker genes. Mech. Dev. 100:263-273. [DOI] [PubMed] [Google Scholar]
- 42.Wang, D., P. S. Chang, Z. Wang, L. Sutherland, J. A. Richardson, E. Small, P. A. Krieg, and E. N. Olson. 2001. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 105:851-862. [DOI] [PubMed] [Google Scholar]
- 43.Yamaguchi, K., K. Shirakabe, H. Shibuya, K. Irie, I. Oishi, N. Ueno, T. Taniguchi, E. Nishida, and K. Matsumoto. 1995. Identification of a member of the MAPKKK family as a potential mediator of TGF-β signal transduction. Science 270:2008-2011. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, D., V. Gaussin, G. E. Taffet, N. S. Belaguli, M. Yamada, R. J. Schwartz, L. H. Michael, P. A. Overbeek, and M. D. Schneider. 2000. TAK1 is activated in the myocardium after pressure overload and is sufficient to provoke heart failure in transgenic mice. Nat. Med. 6:556-563. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, H., and A. Bradley. 1996. Mice deficient for BMP2 are nonviable and have defects in amnion/chorion and cardiac development. Development 122:2977-2986. [DOI] [PubMed] [Google Scholar]
- 46.Zhu, C., F. E. Johansen, and R. Prywes. 1997. Interaction of ATF6 and serum response factor. Mol. Cell. Biol. 17:4957-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zou, Y., S. Evans, J. Chen, H. C. Kuo, R. P. Harvey, and K. R. Chien. 1997. CARP, a cardiac ankyrin repeat protein, is downstream in the Nkx2-5 homeobox gene pathway. Development 124:793-804. [DOI] [PubMed] [Google Scholar]