Abstract
Coat protein complex I (COPI) coated vesicles, one of three major types of vesicular carriers in the cell, mediate the early secretory pathway and retrograde transport from the Golgi to the endoplasmic reticulum. COPI vesicles are generated through activation of the regulatory GTPase Arf1 at the donor membrane and the subsequent recruitment of coatomer, a coat protein complex consisting of seven stably associated components. Coatomer functions in binding and sequestering cargo molecules and assembles into a polymeric protein shell that encompasses the surface of COPI vesicles. Little is known about the structural properties of this heptameric complex. We have isolated native yeast coatomer and examined its structure and subunit organization by single-particle electron microscopy (EM). Our analyses provide the first three-dimensional picture of the complete coatomer, and reveal substantial conformational flexibility likely to be critical for its scaffolding function.
Transport vesicles shuttle proteins and lipids between different subcellular compartments. These cargo carriers are generated at the donor compartment by a highly regulated, multi-step process that involves several essential factors including coat proteins. Coat proteins are a set of cytoplasmic proteins or protein complexes that are recruited to the membrane of the donor compartment by a regulatory GTPase of the Arf family upon its activation by GDP-to-GTP exchange1. Coat proteins play critical roles in sorting and concentrating cargo molecules and assemble into a polymeric “coat” structure that deforms the donor membrane, a process that ultimately leads to vesicle budding2. It has been suggested that coat proteins, which cover the surface of transport vesicles, might also contribute to vesicle targeting specificity by serving as a molecular identification code for recognition by the vesicle tethering machinery located at the acceptor membrane3.
Clathrin, coat protein complex I (COPI), and coat protein complex II (COPII) are the three major types of coats found on transport vesicles2,4. Clathrin-coated vesicles mediate endocytic uptake and post-endoplasmic reticulum (ER) sorting at the Golgi, while COPI-coated vesicles participate in both intra-Golgi trafficking and retrograde transport from the Golgi to the ER. COPII-coated vesicles are responsible for exporting cargo from the ER to the Golgi. These three coat proteins are divergent in composition but are highly analogous in overall design. In particular, they all consist of an “adaptor” module that interacts directly with the cargo and the donor membrane, and a “cage” module that mediates intersubunit interaction and formation of a polymeric coat structure2,4.
For clathrin and COPII, the adaptor and cage modules exist as separate protein complexes, which integrate into a two-layer coat structure during biogenesis of the transport vesicle2. The molecular architecture and organization of the clathrin and COPII coats have been extensively studied. In particular, pseudo-atomic models of the clathrin and COPII coats have been generated based on cryo-EM reconstructions of in vitro assembled coats and crystal structures of individual components or subcomplexes5,6,7. These structures revealed how clathrin and COPII coat proteins, specifically their cage modules, use extended α solenoid and β propeller domains to mediate intersubunit interactions.
By contrast, the adaptor and cage modules of the COPI coat form a single complex known as coatomer. Coatomer consists of seven components (α, β, β’, γ, δ, ε, ζ), and is recruited en bloc to the donor membrane by activated Arf18,9. Biochemical analyses suggested that the α, β’, and ε components of coatomer form the cage module or the B subcomplex, while β’, γ, δ, and ζ, four components that share limited sequence similarities with subunits of the canonical clathrin adaptors AP-1 and AP-2, constitute the corresponding adaptor module or the F subcomplex10. Although high-resolution structures of two small subunits (ε and ζ), and fragments of three larger subunits (α, β’, γ) have recently been determined11,12,13,14, crystallographic analysis of the complete heptameric coatomer has not yet been successful, and little is known about the overall structure of coatomer and its subunit organization. Here, we report the structural characterization of native yeast coatomer by single-particle electron microscopy (EM). Our analyses provide insights into the overall architecture and subunit organization of the full coatomer complex, and reveal substantial conformational flexibility likely to be critical for its scaffolding function.
Purification of native coatomer from yeast
To obtain sufficient quantities of coatomer for structural studies, we first developed a reproducible purification method for this complex. The fact that coatomer contains seven subunits, four of which are larger than 90 kDa, has precluded the use of recombinant expression system for this purpose. The traditional method of isolating native coatomer involves repeated ion exchange chromatography and high salt elution steps15,16, and these more stringent procedures could compromise the structural integrity of coatomer. Tandem affinity purification (TAP) is a proven method for isolating native complexes from yeast extracts17. The TAP tag consists of a calmodulin-binding peptide separated from a protein A tag by a TEV protease cleavage site17. The two-step TAP procedure, involving IgG sepharose and calmodulin sepharose, is simple and uses mild buffers and elution conditions. To evaluate the feasibility of using the TAP method to isolate coatomer, we carried out conventional two-step TAP purification from the extracts of six different yeast strains. Each of these strains encodes a TAP tag fused to the C terminus of the essential α, β, β’, γ, δ, subunits or the non-essential ε subunit. The addition of the tag does not appear to interfere with the function of coatomer, as all six strains exhibited wild type growth rates in YPD medium. We were able to recover a 7-component complex from the six independent purifications (Fig. 1a), although the subunit stoichiometry varied to some degree. Mass spectrometry confirmed that the bands on the SDS-PAGE gels corresponded to the seven coatomer subunits (Table S1).
Figure 1.
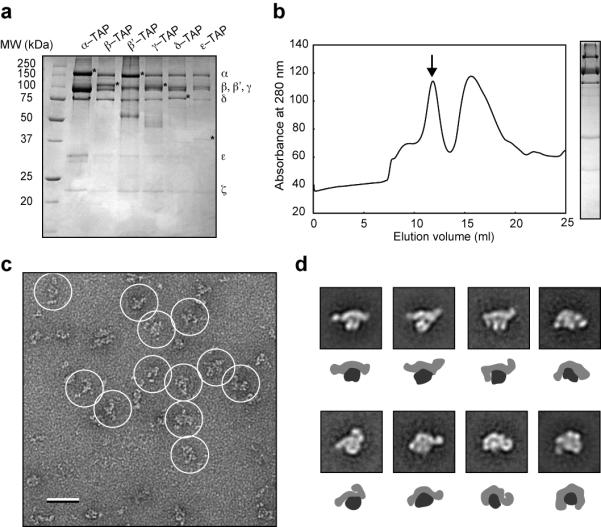
Purification and negative-stain EM analysis of yeast coatomer. (a) Coomassie-stained 12.5% SDS-PAGE gel of coatomer purified by the conventional two-step TAP procedure17. The TAP-tagged strains are shown on the top, with the identities of the individual subunits denoted on the right. Asterisks in each lane indicate the TAP-tagged subunit, which contains only the calmodulin-binding peptide (CBP) part of the tag as the protein A module is proteolytically removed during TAP purification. (b) Gel filtration chromatography. The arrow indicates the coatomer peak, with the corresponding silver-stained gel shown on the right. The second peak corresponds to contaminants and dissociated or degraded complexes. (c) A raw image of negatively stained coatomer with individual particles circled. Scale bar represents 50 nm. (d) Eight representative class averages of cryo-negatively stained coatomer with schematic representations underneath each panel showing the two major domains of the coatomer (dark gray for globular domain, light gray for extended domain). Side length of each panel is 38 nm.
We next tested whether we could modify the standard TAP procedure to improve the purity of the isolated coatomer. In particular, we substituted the second affinity purification step (calmodulin-sepharose) with gel filtration chromatography, which is more effective than calmodulin-sepharose in separating and removing aggregated protein, subcomplexes, and the TEV protease used to elute the complexes from the IgG-sepharose resin. We carried out the two-step IgG-sepharose-gel filtration procedure for all six TAP-tagged strains and observed one major peak in the gel filtration profile of all samples (α-TAP is shown in Fig. 1b). SDS-PAGE analysis confirmed that the peak fractions contained all seven coatomer subunits and showed that the new procedure resulted in improved purity of the complex (α-TAP is shown in Fig. 1b). Coatomer eluted from the gel filtration column at a volume similar to that of the mass standard thyroglobulin (669 kDa), indicating that the purified complex (calculated mass of 570 kDa) is likely a “monomer”, with a large hydrodynamic radius (Fig. 1b). We also found that addition of glycerol and the detergent NP40 was necessary to prevent protein aggregation. Furthermore, we noticed that the detergent CHAPS, although successfully used in the purification of the Nup8418 and TRAPPII19 complexes, compromised the structural integrity of coatomer.
Negative stain 2D analysis and 3D reconstruction of yeast coatomer
With the ability to isolate relatively pure yeast coatomer, we next characterized its structure by single-particle EM methods. We first used uranyl formate to prepare negatively stained specimens from the peak fraction of the gel filtration column (α-TAP), and then examined these specimens on a CM10 electron microscope operated at 100 kV. The raw EM images showed mono-dispersed particles with different overall shapes (Fig. 1c). To better resolve the structural details of coatomer, we collected more images on imaging plates at a nominal magnification of 67,000x and a defocus value of −1.5 μm, using a Tecnai T12 electron microscope operated at 120 kV. We selected 12,515 particles from the digitized images using BOXER, the display program associated with the EMAN software package20, and classified these particle images into 100 classes using the SPIDER software package21. From the averaged images of these classes, we deduced that coatomer consists of two major domains: a more globular domain connected to the mid section of a more extended domain (Fig. S1a). The observation of these two domains is consistent with the notion that coatomer consists of two subcomplexes, which represent the cage and the adaptor modules, respectively10. These two domains appear to adopt multiple conformations, resulting in striking differences in the overall morphology seen in the calculated class averages (Fig. S1a).
The substantial structural variability of the coatomer population revealed by the 2D averages (Fig. S1a) ruled out cryo-EM analysis of the coatomer structure, as it is currently virtually impossible to use this technique for very heterogeneous specimens. Negative staining provides not only higher image contrast but also often induces the molecules to adsorb in one or a few preferred orientations to the carbon support film, making it possible to classify the imaged particles into structurally homogeneous groups according to their conformation. However, negative staining also introduces preparation artifacts, most notably flattening of the particles upon sample dehydration. Cryo-negative staining has been shown to minimize preparation artifacts, and when combined with the random conical tilt approach, provides a powerful method to determine accurate 3D reconstructions de novo22. We therefore prepared cryo-negatively stained specimens by embedding purified coatomer (α-TAP) in a layer of uranyl formate sandwiched between two carbon films on Quantifoil grids. The specimens were frozen in liquid nitrogen and maintained at liquid nitrogen temperature during data collection. We collected pairs of images (50° tilted/untilted) from these specimens at a magnification of 50,000x and a defocus value of −2 μm, using an Oxford CT-3500 cryo-transfer holder and a Tecnai F20 electron microscope operated at 200 kV (Fig. S1b). We selected 20,561 pairs of particles from 56 corresponding pairs of images using WEB, the display program associated with SPIDER21, and classified the particles from the images of the untilted specimen into 100 classes using SPIDER. The resulting class averages showed a lack of stain accumulation around the particles and clear structural features, indicating that the purified complexes were well preserved during specimen preparation and imaging (Fig. 1d, Fig. S1c). The majority of the 100 class averages resemble those seen in the class averages obtained with the negatively stained specimen (Fig. S1b & S1c). Using the corresponding particles from the images of the tilted specimen, we calculated independent 3D reconstructions using the random conical tilt approach23, a procedure implemented in SPIDER21. We only selected classes with at least 200 particles to ensure that the 3D reconstructions we calculated would represent a significant percentage of the coatomer population. We also omitted classes that showed diffuse densities in their averaged images, as these indicate poor alignment and/or substantial remaining heterogeneity in the class.
We determined 3D reconstructions of 20 classes. The density maps show a similar two-domain architecture but display considerable differences in the structural features and orientation of the two domains (Fig. S1d, Fig. S2a-e). In particular, the extended domain, as shown by the 2D class averages (Fig. 1d) and substantiated by the 3D reconstructions, is highly flexible and leads to an overall shape of coatomer that ranges from a “V” to a “horseshoe” (Fig. 1d). The terminal regions of the extended domain project to different directions, resulting in even more structural diversity. The inherent flexibility of the extended domain may be important for coatomer to make the necessary adjustments needed to form coats on vesicles of varying sizes. This structural property may also allow coatomer to overcome the geometrical constraints imposed by having the cage and the adaptor modules in a single entity. We also found that the size of the globular domain varies between different reconstructions when we displayed the maps at the contour level consistent with the calculated mass of the coatomer. We attribute this problem to the more flexible parts being averaged out in the reconstruction process. Inspection of each density map at different contour levels showed that the 3D reconstruction from class 36 was least affected by this issue (Fig. 2a), indicating that it was the class with the least heterogeneity. We therefore chose this particular reconstruction as the reference structure for our interpretation and further discussion. Based on its strong resemblance to other class averages, this density map also seems to capture most of the key structural elements of coatomer. With an estimated resolution of 36 Å according to the Fourier shell correlation = 0.5 criterion (Fig. S1d), the class 36 reconstruction has an overall T shape with two well-defined domains (Fig. 2a). The extended domain is triangular and slightly curved, and spans a distance of approximately 250 Å, with an ellipsoidal density at one end (Fig. 2a). In projection, the extended domain resembles the Sec13-Sec31 complex, the cage module of the COPII coat24. The globular domain, with dimension of about 100 × 110 × 60 Å, connects to the extended domain at an angle, and contains a central groove reminiscent of that in AP1 and AP2, two clathrin adaptor complexes that are believed to share structural homology with the adaptor module of the coatomer25,26.
Figure 2.
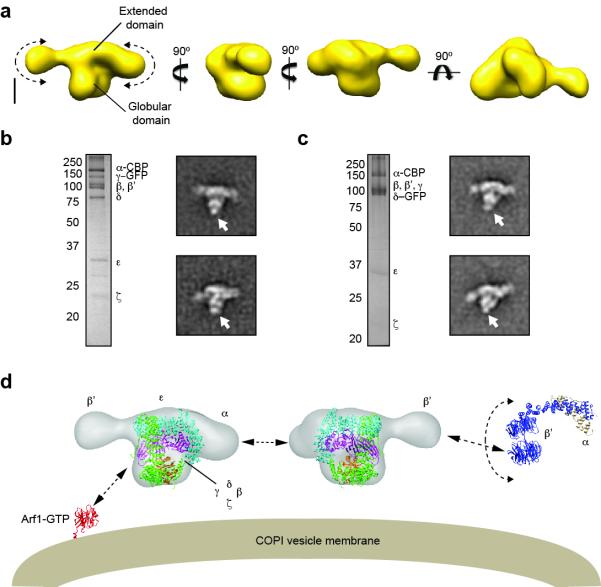
3D reconstruction and subunit organization of yeast coatomer. (a) Different views of the 3D reconstruction of coatomer corresponding to the class average shown in the top left panel of Figure 1a. The arrows indicate the flexibility of the terminal parts of the extended domain. Scale bar represents 5 nm. (b) A silver-stained gel of purified coatomer with GFP fused to the γ subunit (γ–GFP) (left) with two representative averages (right). The arrow indicates the GFP density. Side length of each panel is 43 nm. (c) A silver stained gel of purified coatomer with GFP fused to the δ subunit (δ–GFP) (left) with two representative averages (right). The arrow indicates the GFP density. Side length of each panel is 43 nm. (d) Placement of the crystal structure of the clathrin AP-2 core complex (PDB code: 2JKR)25 into the coatomer EM density map. The labels indicate the proposed locations of the seven subunits. A putative model for coatomer recruitment and intersubunit interaction is shown. Upon its activation by GTP exchange, Arf1 (PDB code: 2KSQ)30 recruits coatomer by binding to its β and γ subunits. Coatomer units interact with each other through the terminal regions of the B subcomplex, which contains the β propeller domains of the α and β’ subunits. In particular, the tandem β propeller domains of the β’ subunits from neighboring complexes interact with each other to form the vertex element of the COPI coat (PDB code of core B subcomplex: 3MKQ)12.
Subunit organization of the coatomer
To validate our assignment of the two modules, we carried out EM-based green fluorescent protein (GFP) localization studies to investigate the subunit organization of coatomer. In this approach, we take advantage of the globular fold instead of the fluorescence property of GFP, which is sufficiently large to be observed by EM27. We generated three yeast strains, in which GFP was fused to the C terminus of components of the B subcomplex, α, β’, and ε, and two additional strains, in which GFP was fused to the C terminus of components of the F subcomplex, γ and δ. We purified the GFP-tagged complexes from these strains based on TAP-tagged α or, in the case of α-GFP, TAP-tagged δ. Although the overall yields varied across the different strains, all five GFP-tagged complexes could be isolated at a purity comparable to that of the non-GFP-tagged complex (Fig. 2b, c, Fig. S2f-h). Furthermore, the GFP tag did not seem to interfere with complex assembly, as all five complexes eluted at volumes from the gel filtration column as the one without GFP (data not shown). We prepared negatively stained specimens from the purified GFP-labeled complexes, and collected images using a Tecnai T12 electron microscope. For each of the five samples, we selected at least 3,000 particles from the digitized images, which we classified into 200 classes using SPIDER. For γ-GFP and δ-GFP, we could unambiguously identify an additional density in the class averages positioned directly underneath the globular domain (Fig. 2b, c). By contrast, we could not detect any additional density in the class averages obtained for α-GFP, β’-GFP, or ε-GFP (Fig. S2f-h). A possible explanation is that the GFP tags are positioned within the core density and are thus obscured. Alternatively, the positions of the GFPs could be too variable, causing them to be averaged out. Nevertheless, the GFP labeling data provide evidence supporting our interpretation that the globular domain represents the F subcomplex or adaptor module, and implies that the extended domain corresponds to the B subcomplex or cage module.
Using the position of the central groove as a guide, we were able to fit the crystal structure of the clathrin AP-2 core into the globular domain of the class 36 density map. According to this docking, coatomer is positioned with its globular domain facing the donor membrane (Fig. 2d), which would place the adaptor module subunits β and γ in close proximity to Arf1. In agreement with this proposal, previous studies have shown that Arf1, upon activation by GTP exchange, localizes to the donor membrane and recruits coatomer by binding to these two proteins28,29 (Fig. 2d). We were, however, unable to fit the crystal structure of the core cage module (β’ in complex with a fragment of α) into the class 36 density map12. One possible explanation is that this particular 3D map captured a conformation of coatomer different from the one found in the crystal structure, but the uncertainty concerning the appropriate threshold for contouring prevented us from using our other density maps for docking experiments. Our 3D reconstruction also does not have sufficient resolution and structural detail to allow unambiguous docking of the crystal and NMR structures of the ε and ζ components into the density map. We did notice, however, that the N-terminal β propeller domains of the β’ subunit, which engage in a 3-way junction hypothesized to mediate inter-subunit interactions at the vertex of the COPI cage12, are flexible and could potentially represent the ellipsoid subdomain in our EM map. Future experiments will focus on developing methods to reconstitute COPI coats in vitro to not only validate this proposal, but also to further delineate the molecular details of the interaction between different subunits within coatomer and between different coatomer complexes within an assembled coat.
Supplementary Material
Acknowledgements
We thank Zongli Li for assistance with microscopy and image processing. C.K.Y acknowledges fellowships from the Jane Coffin Childs Memorial Fund and the Canadian Institutes for Health Research. T.W. is an investigator in the Howard Hughes Medical Institute. The molecular electron microscopy facility at Harvard Medical School was established with a generous donation from the Giovanni Armenise Harvard Center for Structural Biology and is maintained with funds from NIH grant PO1 GM62580 (to Stephen C. Harrison).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kahn RA, Volpicelli-Daley L, Bowzard B, Shrivastava-Ranjan P, Li Y, Zhou C, Cunningham L. Arf family GTPases: roles in membrane traffic and microtubule dynamics. Biochem Soc Trans. 2005;33:1269–72. doi: 10.1042/BST0331269. [DOI] [PubMed] [Google Scholar]
- 2.McMahon HT, Mills IG. COP and clathrin-coated vesicle budding: different pathways, common approaches. Curr Opin Cell Biol. 2004;16:379–91. doi: 10.1016/j.ceb.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 3.Cai H, Yu S, Menon S, Cai Y, Lazarova D, Fu C, Reinisch K, Hay JC, Ferro-Novick S. TRAPPI tethers COPII vesicles by binding the coat subunit Sec23. Nature. 2007;445:941–4. doi: 10.1038/nature05527. [DOI] [PubMed] [Google Scholar]
- 4.Harrison SC, Kirchhausen T. Structural biology: Conservation in vesicle coats. Nature. 2010;466:1048–9. doi: 10.1038/4661048a. [DOI] [PubMed] [Google Scholar]
- 5.Fotin A, Cheng Y, Sliz P, Grigorieff N, Harrison SC, Kirchhausen T, Walz T. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 2004;432:573–9. doi: 10.1038/nature03079. [DOI] [PubMed] [Google Scholar]
- 6.Stagg SM, Gurkan C, Fowler DM, LaPointe P, Foss TR, Potter CS, Carragher B, Balch WE. Structure of the Sec13/31 COPII coat cage. Nature. 2006;439:234–8. doi: 10.1038/nature04339. [DOI] [PubMed] [Google Scholar]
- 7.Stagg SM, LaPointe P, Razvi A, Gurkan C, Potter CS, Carragher B, Balch WE. Structural basis for cargo regulation of COPII coat assembly. Cell. 2008;134:474–84. doi: 10.1016/j.cell.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beck R, Rawet M, Wieland FT, Cassel D. The COPI system: molecular mechanisms and function. FEBS Lett. 2009;583:2701–9. doi: 10.1016/j.febslet.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 9.Hsu VW, Lee SY, Yang JS. The evolving understanding of COPI vesicle formation. Nat Rev Mol Cell Biol. 2009;10:360–4. doi: 10.1038/nrm2663. [DOI] [PubMed] [Google Scholar]
- 10.Lowe M, Kreis TE. In vitro assembly and disassembly of coatomer. J Biol Chem. 1995;270:31364–71. doi: 10.1074/jbc.270.52.31364. [DOI] [PubMed] [Google Scholar]
- 11.Hsia KC, Hoelz A. Crystal structure of alpha-COP in complex with epsilon-COP provides insight into the architecture of the COPI vesicular coat. Proc Natl Acad Sci U S A. 2010;107:11271–6. doi: 10.1073/pnas.1006297107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee C, Goldberg J. Structure of coatomer cage proteins and the relationship among COPI, COPII, and clathrin vesicle coats. Cell. 2010;142:123–32. doi: 10.1016/j.cell.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu W, Lin J, Jin C, Xia B. Solution structure of human zeta-COP: direct evidences for structural similarity between COP I and clathrin-adaptor coats. J Mol Biol. 2009;386:903–12. doi: 10.1016/j.jmb.2008.12.083. [DOI] [PubMed] [Google Scholar]
- 14.Hoffman GR, Rahl PB, Collins RN, Cerione RA. Conserved structural motifs in intracellular trafficking pathways: structure of the gammaCOP appendage domain. Mol Cell. 2003;12:615–25. doi: 10.1016/j.molcel.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Hosobuchi M, Kreis T, Schekman R. SEC21 is a gene required for ER to Golgi protein transport that encodes a subunit of a yeast coatomer. Nature. 1992;360:603–5. doi: 10.1038/360603a0. [DOI] [PubMed] [Google Scholar]
- 16.Waters MG, Serafini T, Rothman JE. ‘Coatomer’: a cytosolic protein complex containing subunits of non-clathrin-coated Golgi transport vesicles. Nature. 1991;349:248–51. doi: 10.1038/349248a0. [DOI] [PubMed] [Google Scholar]
- 17.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–2. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 18.Kampmann M, Blobel G. Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex. Nat Struct Mol Biol. 2009;16:782–8. doi: 10.1038/nsmb.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yip CK, Berscheminski J, Walz T. Molecular architecture of the TRAPPII complex and implications for vesicle tethering. Nat Struct Mol Biol. 2010;17:1298–304. doi: 10.1038/nsmb.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 21.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–9. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 22.Cheng Y, Wolf E, Larvie M, Zak O, Aisen P, Grigorieff N, Harrison SC, Walz T. Single particle reconstructions of the transferrin-transferrin receptor complex obtained with different specimen preparation techniques. J Mol Biol. 2006;355:1048–65. doi: 10.1016/j.jmb.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 23.Radermacher M. Three-dimensional reconstruction of single particles from random and nonrandom tilt series. J Electron Microsc Tech. 1988;9:359–94. doi: 10.1002/jemt.1060090405. [DOI] [PubMed] [Google Scholar]
- 24.Fath S, Mancias JD, Bi X, Goldberg J. Structure and organization of coat proteins in the COPII cage. Cell. 2007;129:1325–36. doi: 10.1016/j.cell.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 25.Collins BM, McCoy AJ, Kent HM, Evans PR, Owen DJ. Molecular architecture and functional model of the endocytic AP2 complex. Cell. 2002;109:523–35. doi: 10.1016/s0092-8674(02)00735-3. [DOI] [PubMed] [Google Scholar]
- 26.Heldwein EE, Macia E, Wang J, Yin HL, Kirchhausen T, Harrison SC. Crystal structure of the clathrin adaptor protein 1 core. Proc Natl Acad Sci U S A. 2004;101:14108–13. doi: 10.1073/pnas.0406102101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lees JA, Yip CK, Walz T, Hughson FM. Molecular organization of the COG vesicle tethering complex. Nat Struct Mol Biol. 2010;17:1292–7. doi: 10.1038/nsmb.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao L, Helms JB, Brunner J, Wieland FT. GTP-dependent binding of ADP-ribosylation factor to coatomer in close proximity to the binding site for dilysine retrieval motifs and p23. J Biol Chem. 1999;274:14198–203. doi: 10.1074/jbc.274.20.14198. [DOI] [PubMed] [Google Scholar]
- 29.Zhao L, Helms JB, Brugger B, Harter C, Martoglio B, Graf R, Brunner J, Wieland FT. Direct and GTP-dependent interaction of ADP ribosylation factor 1 with coatomer subunit beta. Proc Natl Acad Sci U S A. 1997;94:4418–23. doi: 10.1073/pnas.94.9.4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Kahn RA, Prestegard JH. Dynamic structure of membrane-anchored Arf*GTP. Nat Struct Mol Biol. 2010;17:876–81. doi: 10.1038/nsmb.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.