Abstract
Melanoma is a highly aggressive and deadly skin cancer. Early intervention correlates with nearly 100% patient survival, but greater than 80% mortality is associated with advanced disease. Currently, few treatment options are available for patients with metastatic melanoma, and the global incidence of melanoma is increasing faster than that of other cancers. Therefore, it is vitally important to uncover and use genetic and epigenetic regulatory mechanisms at work during the development and progression of melanoma for better prevention, diagnosis, and clinical management. MicroRNA (miRNA) is a set of small, single-stranded, noncoding RNAs that target the 3′-untranslated region of an estimated 30% of all human genes to inhibit their expression. Our understanding of miRNA-mediated regulation of cancers has grown immensely over the past decade. Here we review currently available data on melanoma-associated miRNAs, highlighting those deregulated miRNAs targeting important genes and signaling pathways involved in the progression of melanocytes to primary and metastatic melanoma. Understanding the important roles of miRNAs in melanoma progression and metastasis development will contribute to the development of miRNA-targeted therapy in the future.
Keywords: Epigenetics, melanoma, metastasis, microRNA
INTRODUCTION
Melanoma is currently the sixth most common cancer in white men and women in the United States. According to the American Cancer Society, 68,720 new cases of melanoma and 8,650 deaths were predicted for 2009.1 Although not the most common of skin cancers, comprising only 5% of all skin cancers, it is by far the deadliest, responsible for 75% of skin cancer-related deaths. Incidence rates for white men and women in the United States are currently at 1 in 45 and 1 in 58, respectively,1 with the incidence rate of melanoma increasing faster than any other cancer in the world. Those at high risk for developing melanoma may have any combination of fair skin and eye color, familial melanoma, presence of common or dysplastic/atypical nevi (moles), high density of freckles, intermittent sun exposure with burns, and/or other current or previous skin cancer lesions. Primary cutaneous melanoma diagnosed early in its course has an excellent outcome with early surgical intervention, but regional and distant metastatic disease has a much more dismal prognosis. Current 5-year survival rates for all races equal 91% for all disease sites (significantly higher than 1975-1977 at 82%), 99% for local disease, 65% for regional disease (including regional lymph node involvement), and only 16% for those with distant metastasis (median survival is only 7.5 months). Unfortunately, for those patients who will develop locally advanced and metastatic melanoma, current treatment options are limited. Metastatic melanoma is highly resistant to therapy; thus, the overall poor mortality rates for patients with regional and distant disease emphasize the need for better treatment modalities and recognition of early neoplasia.
Melanocytes and Melanoma
Cutaneous melanoma arises from epidermal melanocytes of neural-crest developmental origin. Neural-crest cells, induced at the time of gastrulation, are highly invasive and undergo an epithelial-to-mesenchymal cell-type transition (EMT), as they must migrate through embryonic tissues during early development to form cartilage, bone, connective tissue, neural and glial cells, sympathoadrenal cells, and smooth muscle and pigment-producing melanocytes, among other cell types.2 Some of these multipotent cells differentiate to melanocytes and/or the basal layer of the epidermis, as well as hair follicles, meninges, the eye, and cochlea.2 They produce melanin pigment in response to ultraviolet (UV) radiation in melanosome organelles that may either remain in the cell or, more often, be deposited into neighboring keratinocytes via extending dendritic processes for protecting the skin from UV radiation and reactive oxygen species. Importantly, metastatic melanoma cells have been shown to overexpress proteins associated with neural-crest cell invasion and differentiation relative to their nonmetastatic counterparts as well as to downregulate proteins characteristic of normal melanocytes, leading to the idea of tumor development and metastasis being a cell “lineage reversion” to a more neural-crest cell phenotype.3,4
The conversion of normal melanocytes to melanoma cells has thereby been attributed to a number of factors. Notably, UV exposure has been associated with the development of skin cancer, including melanoma.5 Indeed, focused efforts on decreasing UV exposure appear to result in decreased melanoma incidence, and high sun protection factor (SPF) sunscreens afford a protective advantage against UV radiation.6,7 Direct DNA damage by UV radiation can deregulate normal DNA repair mechanisms and result in unimpeded proliferation.
Melanocyte integrity is also highly influenced by its microenvironment. In the epidermis, melanocytes exist at roughly 1/5 the number of basal-level keratinocytes present and 1/36 of total keratinocytes. Paracrine hormone communication, direct cell-to-cell adhesion communication, and autocrine regulation serve to direct melanocyte homeostatic mechanisms, including differentiation, quiescence, and proliferative and apoptotic responses.8 Deregulation of these processes can lead to the formation of benign and dysplastic nevi and melanoma.9 For example, by way of the transmembrane protein, Notch, melanocyte precursors (melanoblasts) maintain their nondifferentiated status, and Notch1 has been described as overexpressed in melanoma relative to melanocytes and benign nevi.10-12 In a recent study, transfection of an active truncated Notch1 transgene construct into a normal melanocyte resulted in induction of tumorigenic character, including anchorage-independent growth, increased proliferation in limited media, and increased migration and invasive capacity. The authors thus described Notch itself as the first melanoma-transforming oncogene.12
Understanding the gene expression character of early melanocytic transformation, such as Notch signaling, and further metastatic processes and their regulatory mechanisms is vital for the future of targeted melanoma therapy. Interest in epigenetics, defined as all heritable changes in gene expression not involving DNA coding, has grown significantly over the past decade because of its recognized influence on normal cellular processes and disease states; in particular, cancer. Epigenetic modes of gene expression regulation include DNA methylation (promoter methylation inhibits transcription factor binding), histone modifications (influences histone binding affinity for DNA), and RNA-associated silencing, including microRNA (miRNA).13,14 Of these epigenetic mechanisms, messenger RNA (mRNA) silencing by miRNA may be the most complex and promising field for future cancer research and therapy.
MicroRNA
MiRNAs are a set of small [∼22 nucleotides (nt)], single-stranded, non−protein-coding RNA molecules, which can recognize and bind 3′-untranslated regions (UTRs) of mRNA, blocking translation of the gene or inducing cleavage of the mRNA.15,16 Currently, more than 700 miRNAs have been discovered in humans, ∼800 are predicted as remaining, and more than 4,000 in all eukaryotic species have been examined.17
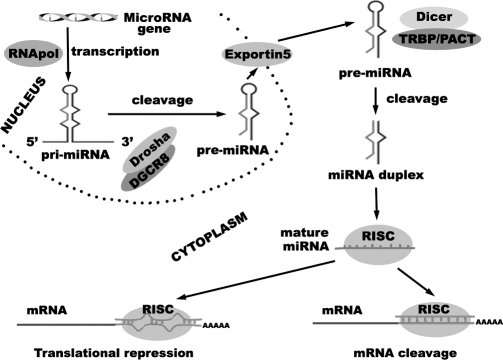
A primary miRNA is the result of transcription as either an independent transcript or as coexpressed from an intron of another gene by RNA polymerase II and may include multiple regions that will become mature miRNAs (multiple miRNAs that are transcribed together are termed clusters, ie, miR-17-92 cluster). The primary miRNA that was coexpressed undergoes further splicing before it and the individual transcript primary miRNA are bound in the nucleus by the microprocessor complex, which consists of the RNase III-type endonuclease Drosha and its cofactor Pasha (DGCR8). This complex then crops the primary miRNA into a hairpin loop, cleaving off 3′ and 5′ regions of excess mRNA, to give a precursor miRNA of ∼70 nt in length. Precursor miRNA is then actively transported to the cytoplasm by exportin-5, where it is bound by the RNAse III-type endonuclease Dicer, which removes the loop and results in a mature miRNA duplex of complementary sequences. One strand is bound by the RNA-induced silencing complex (RISC), which guides mature miRNA to target mRNA for subsequent silencing. The remaining strand is usually degraded but may be bound by the RISC and target its own mRNAs, denoted with an asterisk (eg, miR-373 and miR-373*).18,19 MiRNA synthesis and function are summarized in Figure 1.
Figure 1.
MicroRNA synthesis and function.
MiRNAs mediate gene expression at the posttranscriptional and translational levels in both plants and animals.20,21 Because of their critical function in gene regulation and expression, it is important to understand their roles and significance in tumor cell development, differentiation, proliferation, and apoptosis.22-25 Interestingly, a single miRNA potentially binds hundreds of its cognate mRNA 3′-UTR sequences. It is thus predicted that miRNAs may regulate the expression of more than 30% of all mammalian genes, acting as tumor suppressors or oncogenes, depending on which they dominantly bind and repress.16
The first indication that miRNAs might function as tumor suppressor genes was derived from a study by Calin et al,26 who found that miR-15a and miR-16-1 were commonly deleted in more than 65% of patients with B-cell chronic lymphocytic leukemia. This study proved that miR-15a and miR-16-1 negatively regulated Bcl2, which is an antiapoptotic protein that is often overexpressed in a variety of tumor histologies.26 Recent reports have also demonstrated that more than 50% of miRNA genes are located in cancer-associated genomic regions or within fragile sites.27 This suggests that miRNAs might play a more important role in the pathogenesis of human cancer than was previously realized.
MICRORNA IN MELANOMA
Overview
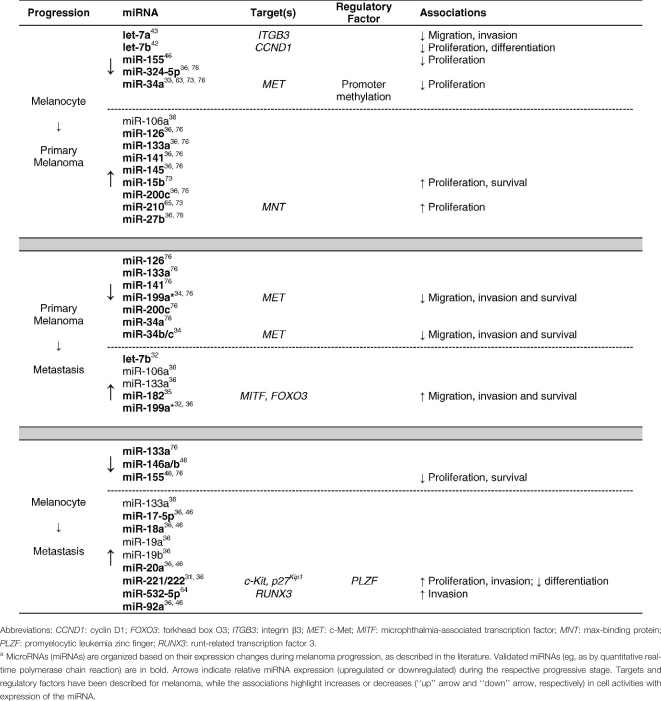
Compared with studies in other solid tumors, relatively few studies have focused on the role of miRNAs in the pathogenesis and development of melanoma. Zhang et al28 published one of the first studies on miRNA in melanoma, documenting that 86% of primary melanoma cell lines had DNA copy number alterations in genomic loci containing miRNA genes. They also identified 243 miRNA genes (83 with gains vs 160 with losses) that were unique to melanoma. Using highly sensitive miRNA quantitative real-time polymerase chain reaction (qRT-PCR) techniques, Gaur et al29 identified a uniquely clustered gene group that was able to distinguish melanoma from all other tumor histologies. Moreover, miR-137 was identified as a trigger for micropthalmia-associated transcription factor (MITF), a master regulator of melanocyte development and survival of significant functional importance in melanoma.30 MiR-221 and miR-222 (miR-221/222) possess an oncogenic role in melanoma and other cancers when their expression is deregulated.31 Worley et al32 identified several miRNA profiles associated with a “good” prognostic signature in the progression of uveal melanoma, with the overexpression of miR-199a and let-7b found primarily in metastatic lesions. It should be noted that uveal and cutaneous melanomas do have important clinical and molecular differences; for instance, uveal melanoma favors hematogenous spread rather than lymphatic, has a high propensity for metastasis to the liver and lungs, and demonstrates only rare instances of BRAF and CDKN2A mutation compared to cutaneous melanoma. MiR-34a appears to act as a tumor suppressor in uveal melanoma cell proliferation and migration through the downregulation of c-Met.33 Furthermore, it is likely that miR-34b, miR-34c, and miR-199a* have the capacity to impair c-Met−mediated invasive growth.34 MiR-182 ectopic expression stimulated migration of melanoma cells in vitro and increased their metastatic potential in vivo, whereas miR-182 downregulation impeded invasion and triggered cellular apoptosis.35 Mueller et al36 recently identified a large cohort of miRNAs associated with the malignant transformation and progression of melanoma using in vitro models. Taken together, these results suggest that miRNAs play an important role in the biology and progression of melanoma. Clearly, additional studies are required to validate a distinct profile of miRNA markers with the capacity to predict the potential for developing metastatic melanoma. The Table lists notable miRNAs involved in the progression from melanocyte to metastatic melanoma, their validated or proposed targets, regulatory factors, and effects of miRNA upregulation or downregulation during the respective stages of progression.
Table.
Representative miRNAs involved in the progression of melanoma
MITF and Associated miRNAs in Melanoma
MITF is a recognized oncogene in melanoma that regulates cell proliferation and apoptosis and is overexpressed in 10% to 20% of human melanomas.37 It is a member of the Myc supergene family of basic helix-loop-leucine-zipper transcription factors, necessary for functional melanocyte formation.38 Because of the critical role of MITF in melanoma progression, several recent studies have explored the impact of miRNAs on melanoma through MITF-mediated pathways.
MicroRNAs Targeting MITF. MicroRNA.org, an online database for miRNA target prediction, provides more than 300 miRNA candidates that putatively target MITF, although only a few of them have been validated thus far. MiR-137 is located at the chromosomal region 1p22, which is known to harbor an allele for melanoma susceptibility. Bioinformatic and in vitro analyses verified miR-137 targeting of MITF in melanoma cells.30 Segura et al35 described miR-182 as another negative regulator of MITF expression. MiR-182 is located in 7q31-34, a chromosomal region frequently altered in melanoma. MiR-182 was demonstrated to increase survival and invasive potential of melanoma cells by repressing MITF and FOXO3, a forkhead family transcription factor. Importantly, 7q31-34 also harbors c-Met (encoding hepatocyte growth factor receptor with tyrosine-kinase activity) and BRAF (member of the raf/mil family of serine/threonine protein kinases), two important regulators in the MAPK/ERK signaling pathway.39 They found that miR-182 was overexpressed not only in human melanoma cell lines but also in tissue specimens. These results were inversely correlated with MITF and FOXO3 expression in predicting melanoma progression and development. Moreover, ectopic expression of miR-182 in melanoma cells stimulated anchorage-independent growth and invasion, using an in vitro extracellular matrix assay, and promoted melanoma lung metastasis in a mouse model, whereas miR-182 downregulation impeded invasion and triggered apoptosis of melanoma cells.
MiRNAs Regulated by MITF at the Transcriptional Level. Ozsolak et al40 identified a number of miRNAs to be regulated by MITF in melanoma cells using nucleosome mapping and linker sequence analyses. These miRNAs include some members of the let-7 family (let-7a-1, -7d, -7f-1, and -7i), miR-221/222, miR-17-92 cluster, miR-106-363 cluster, miR-29, miR-146a, miR-148b, and miR-125b.40
The let-7 family is highly conserved across species in sequence and function, the first miRNAs validated to be involved in tumorigenesis.41 Schultz et al42 revealed 5 members of the let-7 family (let-7a, -7b, -7d, -7e, and -7g) to be significantly downregulated in primary melanoma compared to benign nevi, suggesting the let-7 family might act as tumor suppressors in melanoma. The ectopic expression of let-7b diminished the anchorage-independent growth ability of melanoma cells and inhibited cell-cycle progression. Overexpression of let-7b eventually repressed cyclins (D1, D3, and A) and cyclin-dependent kinase 4 (CDK4), all of which have been described as playing a role in melanoma development.
Another study found that let-7a was lost in melanoma when comparing primary melanocytes and malignant melanoma cell lines, with sequence analysis suggesting that it interacts with the 3′-UTR of integrin β3 mRNA.43 Integrin β3 is highly related to melanoma progression and leads to enhanced migratory and invasive potential of melanoma cells.8 Transfection of melanoma cells with let-7a precursor miRNA molecules resulted in the downregulation of integrin β3 mRNA and protein expression, which suggested that the loss of let-7a expression might be an essential regulatory mechanism leading to increased integrin β3 expression in melanoma cells.43 Müller and Bosserhoff43 also proved that the overexpression of let-7a in melanoma cells reduced their invasive potential by approximately 75%, while transfection with antisense oligonucleotides that directly bind and inhibit the actions of miRNAs resulted in the induction of integrin β3 expression and induced migration of anti-let-7a−transfected melanocytes. These findings revealed let-7a to be an important regulator of integrin β3, the loss of let-7a thus being involved in the development and progression of malignant melanoma.
The miR-17-92 cluster locates to chromosome 13 and contains 6 members (miR-17, -18a, -19a, -20a, -19b-1, and -92a-1), while another miRNA cluster, miR-106-363, which shares many similarities with the miR-17-92 cluster, locates to the X chromosome and also consists of 6 members (miR-106a, -18b, -20b, -19b-2, -92a-2, and -363). Both miRNA clusters are described as oncogenic and found highly expressed in a variety of cancers.44,45 Mueller et al36 compared the profiles of normal human melanocytes and well-characterized melanoma cell lines derived from primary tumors and melanoma metastases. They showed that all members of the miR-17-92 cluster were upregulated in primary tumor cell lines compared to normal melanocytes. The expression of the miR-17-92 cluster was even higher in metastatic cell lines, with an approximately twofold upregulation compared to primary melanoma cell lines. The expression of the miR-106-363 cluster was similar to the expression of the miR-17-92 cluster in melanocytes and melanoma cells. The researchers detected a strong upregulation of miR-106a expression in primary tumor cells and a further increase in expression level in metastatic melanoma cells.36 In addition to finding miR-17-5p, miR-18a, miR-20a, and miR-92a overexpressed and miR-146a, miR-146b, and miR-155 downregulated in the majority of melanoma cell lines with respect to melanocytes, Levati et al46 found that ectopic expression of miR-155 in melanoma cells inhibits their proliferation. These results imply involvement of the miR-17-92 cluster in melanoma progression.
Both miR-221 and miR-222 are regulated by MITF at the transcriptional level.31 These 2 miRNAs are clustered on the X chromosome, transcribed as a common precursor, and overexpressed in a variety of cancers with the function of repressing the c-Kit receptor. In normal melanocytes, stem cell factor-dependent, c-Kit−mediated signaling supports proliferation, migration, and differentiation of cells47; however, constitutive activation of the c-Kit receptor tyrosine kinase alone is insufficient to induce tumorigenic transformation of melanocytes in vitro or in vivo.48 Cutaneous melanomas are often characterized with a loss of c-Kit expression.49 Inhibition of the c-Kit receptor tyrosine kinase in c-Kit−positive melanoma cells showed increased apoptosis and G1-phase cell-cycle arrest,49 while the re-expression of c-Kit in c-Kit−negative melanoma cells restored c-Kit−mediated apoptosis and resulted in a loss of tumorigenic potential.50 In accordance with these observations, Felicetti et al31 found that upregulation of miR-221/222 repressed the expression of the c-Kit receptor and p27Kip1 (cyclin-dependent kinase inhibitor 1B) tumor suppressor during melanoma progression from a weakly invasive primary tumor to a more invasive phenotype. Overexpression of miR-221/222 in melanoma cells led to increased proliferation and invasion in vitro and accelerated tumor growth in a mouse melanoma model. Conversely, treatment with anti-miRNAs against both miRNAs resulted in a reduced rate of proliferation and ability to migrate in melanoma cells with a high level of miR-221/222. The researchers also found that the elevated expression of miR-221/222 in melanoma cells was caused by the loss of a transcription factor, promyelocytic leukemia zinc finger (PLZF). PLZF binds to the miR-221/222 promoter and inhibits their transcription in normal melanocytes. Igoucheva and Alexeev51 recently described a similar result. They confirmed that c-Kit was downregulated by miR-221/222 and revealed that c-Kit regulation was mainly based on miRNA-dependent posttranscriptional mechanisms instead of an AP-2-dependent transcriptional mechanism.
Phosphatase and tensin homolog (PTEN) is an important tumor suppressor that is either mutated or deregulated in a variety of cancers, including melanoma.52-54 It inhibits the phosphorylation/activation of Akt3 (p-Akt), a member of the serine/threonine kinase family, and the phosphoinositide 3-kinase/protein kinase B (PI3K/AKT) pathway, known to promote proliferation, survival, invasion, and angiogenesis in melanoma. Akt3 becomes more active (p-Akt) toward advanced, metastatic melanoma with a concomitant loss of PTEN expression. No miRNAs are currently described as targeting PTEN in melanoma, although recent reports highlighted miR-141 in nasopharyngeal carcinoma and miR-221/222 in aggressive non-small cell lung cancer and hepatocarcinoma as oncogenic miRNAs (oncomirs) capable of directly targeting and inhibiting the expression of the tumor suppressor PTEN.55,56
MiR-221/222 are increased from primary to metastatic melanoma samples (Table), where PTEN mutation is relatively rare (5%-20%).31,52 These miRNAs act in melanoma to increase proliferation, invasion, migration, and anchorage-independent growth and to reduce differentiation and melanogenesis by downregulating the c-Kit receptor and p27Kip1. Felicetti et al31 described miR-221/222 overexpression in melanoma cells as being the result of a decrease in the PLZF transcription factor, a direct inhibitory factor of miR-221/222. Cyclin-dependent kinase 2 (CDK2) has been reported to phosphorylate PLZF, triggering its ubiquitination and subsequent degradation.57 Furthermore, p27Kip1, which is induced to coexpress with PTEN, is important for the efficient induction of G1 cell-cycle arrest by PTEN and is necessary for PTEN-induced downregulation of CDK2.58,59 Therefore, PTEN itself may be an important regulator of miR-221/222 in melanoma as its coexpression with p27Kip1 in turn inhibits CDK2-mediated phosphorylation of PLZF, thereby maintaining PLZF levels to bind miR-221/222 promoters, preventing their transcription. Additionally, PTEN is important for Ha-ras–mediated astrocyte elevated gene-1 (AEG-1) promoter activation.60 AEG-1 directly binds PLZF, preventing it from binding its target promoters,61 including those of miR-221/222. As a result, there exists a putative positive feedback loop for miR-221/222 expression, promoting melanoma progression through the joint inhibition of PTEN and p27Kip1 and blocking PTEN/AEG-1/PLZF and/or p27Kip1/CDK2/PLZF-mediated repression of miR-221/222 (Figure 2).
Figure 2.
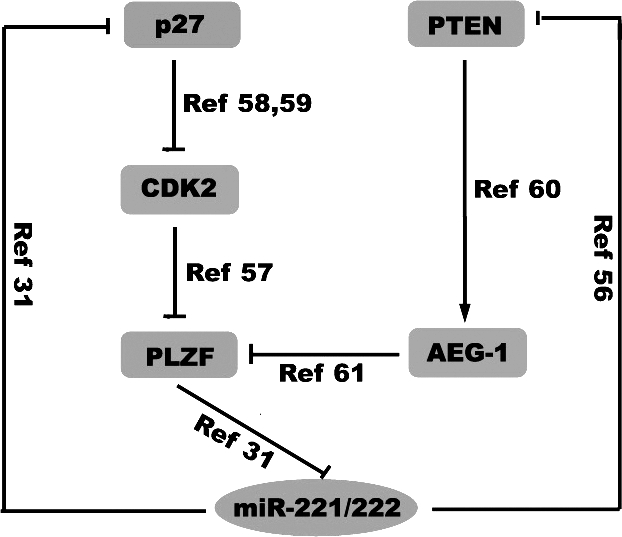
The regulatory network of miR-221/222.
MiR-34 Family and Melanoma
Recently, miR-34 was identified as a target and a potential key effector of the tumor suppressor gene product p53. Ectopic expression of miR-34a induced a G1 cell-cycle arrest, senescence, and apoptosis, suggesting that miR-34 may act as a tumor suppressor.62 The altered expression of miR-34 was also found during melanoma progression.33,34,63 Lodygin et al63 reported that miR-34a expression is silenced in several types of cancer because of aberrant CpG methylation of its promoter. Indeed, 43.2% of melanoma cell lines and 62.5% of primary melanoma samples displayed CpG methylation of the miR-34a promoter and loss of miR-34a expression, whereas the 2 samples of normal melanocytes included in the study showed little to no promoter methylation. Migliore et al34 identified 3 miRNAs–miR-34b, miR-34c, and miR-199a*–in melanoma cells that negatively regulate the expression of MET, an oncogene that encodes the tyrosine kinase receptor for hepatocyte growth factor. MET is frequently overexpressed in many human tumors and promotes the “invasive growth” that results from stimulation of cell motility and protection from apoptosis. Exogenous expression of these miRNAs in primary melanoma cells led to a decreased MET protein expression and resulted in the impairment of MET-mediated motility in these cells.34 Recently, Yan et al33 found miR-34a to be actively expressed in melanocytes but not in uveal melanoma cells. The transfection of miR-34a into melanoma cells led to a significant repression of their growth and migration by downregulating the expression of c-Met directly and the expression of phosphorylated Akt (p-Akt) and other cell-cycle–related proteins indirectly.
Other miRNAs in Melanoma
Kitago et al64 recently reported that miR-532-5p directly targets the runt-related transcription factor 3 (RUNX3) tumor suppressor during the progression from melanocyte and skin to metastatic melanoma. MiR-532-5p was shown to be significantly upregulated in melanoma cells compared to normal melanocytes and in metastatic melanoma tissue compared to primary melanoma tissue. Transfection of anti−miR-532-5p molecules into the melanoma cells rescued the expression of RUNX3. Methylation analysis of the RUNX3 promoter region showed that transcriptional regulation was not a major regulatory mechanism for the downregulation of RUNX3 expression in melanoma, suggesting that miR-532-5p–induced posttranscriptional regulation of RUNX3 may play an important role in melanoma progression.
Zhang et al65 demonstrated that the expression of miR-210, the most prominent miRNA upregulated by hypoxia and a direct transcriptional target of hypoxia-inducible factors, was elevated in multiple cancer types and correlated with breast cancer and melanoma metastases, respectively. MiR-210 overexpression in cancer cells bypassed hypoxia-induced cell-cycle arrest by directly targeting the expression of MNT, an Myc antagonist. The miR-210–mediated abolishment of hypoxia-induced cell-cycle arrest was restored by the loss of Myc. This finding indicated that miR-210 influenced the hypoxia response in tumor cells by triggering a Myc-like response by targeting MNT expression.
CLINICAL FOCUS
Several years ago, we and other groups separately demonstrated that miRNAs were relatively more stable and tolerant of RNAases than were mRNAs, both in archived tissue samples and in blood samples,66-68 which suggests that miRNAs have the potential to be valuable, practical, and reliable biomarkers for disease states. Recently, several groups employed high-throughput microarray techniques to discover miRNA biomarkers from formalin-fixed and paraffin-embedded (FFPE) melanoma samples.69-71 A number of miRNAs have shown the potential to become diagnostic markers for melanoma based on data from clinical samples and array analyses.69-71 Radhakrishnan et al72 examined the presence of oncomirs in uveal melanoma using FFPE specimens by comparing miRNA expression profiles between noninvasive tumor and melanoma metastatic to the liver. They revealed 19 miRNAs that were expressed in nonmetastatic melanoma and absent in metastatic melanoma and 11 miRNAs with the opposite expression pattern.
Satzger et al73 determined the expression level of 16 miRNAs in 6 melanocytes versus 10 melanoma cell lines and in 11 nevi versus 16 melanoma cell lines, respectively, finding that miR-15b and miR-210 were significantly upregulated in parallel with the downregulation of miR-34a in melanomas compared to nevi. These 3 miRNAs were then analyzed in 128 primary melanoma samples from patients with detailed clinical follow-up information. Only high expression of miR-15b was significantly correlated with poor recurrence-free survival and overall survival by univariate Kaplan-Meier and multivariate Cox analyses. Transfection of anti−miR-15b into melanoma cells led to reduced tumor cell proliferation and increased apoptosis. These results showed that miR-15b might be a novel melanoma biomarker contributing to poor prognosis and tumorigenesis.
Worley et al32 were the first to use a genome-wide, microarray-based approach to investigate the value of miRNA expression patterns in predicting metastatic risk in uveal melanoma. They found that the most significant discriminator for classifying low and high metastatic risk was let-7b and miR-199a expression. A classifier system that included the top 6 miRNA discriminators accurately distinguished melanoma patient tissues of high metastatic propensity with 100% sensitivity and specificity. Their work suggested for the first time that miRNA expression might represent a highly accurate biomarker for metastatic risk in melanoma.
Because miRNAs are critical for regulating many cellular events and are highly deregulated in various cancers, including melanoma, it is likely that miRNAs could be effective targets for treatment. Sun et al74 recently found that genistein, an isoflavone isolated from soybeans, inhibited human uveal melanoma cell growth in vitro and in vivo. Additionally, it altered the expression of miR-27a and its target gene ZBTB10 (zinc finger and BTB domain containing 10; BTB represents bric-a-brac, tramtrack, Broad-Complex), hinting at the contributions of miR-27a to the inhibitory effect of genistein on melanoma growth. Kota et al75 demonstrated that the expression of miR-26a in liver cancer cells in vitro induces cell-cycle arrest by targeting cyclins D2 and E2, while miR-26a inhibited cancer cell proliferation and induced tumor-specific apoptosis without toxicity in a mouse model. These findings highlight the possibility that the delivery of miRNAs that are highly expressed, and therefore typically well tolerated, in normal tissues but lost in diseased cells may provide a general strategy for miRNA replacement therapies.
CONCLUSION
MiRNA regulation represents a promising, multifaceted means for targeted therapy against melanoma. Indeed, miRNAs have been known to interact with many of the most important regulatory pathways in melanoma development and progression, including MAPK/ERK and PI3K/PTEN/Akt. Their influence over normal and tumorigenic processes, particularly in regard to their small number, will almost certainly allow for the discovery of many new prognostic markers and key melanoma regulators. Furthermore, archived FFPE tissue samples will continue to be a valuable source of stable miRNAs for future comprehensive analysis. Understanding miRNA expression trends from melanocyte to metastatic melanoma and their roles in many complex regulatory networks may lead to the development of better therapeutic management.
REFERENCES
- 1.Jemal A., Siegel R., Ward E., Hao Y., Xu J., Thun M. J. Cancer statistics, 2009 [published online ahead of print May 27, 2009]. CA Cancer J Clin. 2009;59((4)):225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Knecht A. K., Bronner-Fraser M. Induction of the neural crest: a multigene process. Nat Rev Genet. 2002;3((6)):453–461. doi: 10.1038/nrg819. [DOI] [PubMed] [Google Scholar]
- 3.Gupta P. B., Kuperwasser C., Brunet J. P., et al. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation [published online ahead of print September 4, 2005]. Nat Genet. 2005;37((10)):1047–1054. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kasemeier-Kulesa J. C., Teddy J. M., Postovit L. M., et al. Reprogramming multipotent tumor cells with the embryonic neural crest microenvironment. Dev Dyn. 2008;237((10)):2657–2666. doi: 10.1002/dvdy.21613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rigel D. S. Cutaneous ultraviolet exposure and its relationship to the development of skin cancer. J Am Acad Dermatol. 2008;58((5 suppl 2)):S129–S132. doi: 10.1016/j.jaad.2007.04.034. [DOI] [PubMed] [Google Scholar]
- 6.Bulliard J. L., Cox B., Semenciw R. Trends by anatomic site in the incidence of cutaneous malignant melanoma in Canada, 1969-93. Cancer Causes Contro. 1999;10((5)):407–416. doi: 10.1023/a:1008964621225. [DOI] [PubMed] [Google Scholar]
- 7.Rigel D. S. The effect of sunscreen on melanoma risk. Dermatol Clin. 2002;20((4)):601–606. doi: 10.1016/s0733-8635(02)00024-4. [DOI] [PubMed] [Google Scholar]
- 8.Haass N. K., Smalley K. S., Li L., Herlyn M. Adhesion, migration and communication in melanocytes and melanoma. Pigment Cell Res. 2005;18((3)):150–159. doi: 10.1111/j.1600-0749.2005.00235.x. [DOI] [PubMed] [Google Scholar]
- 9.Mitra D., Fisher D. E. Transcriptional regulation in melanoma. Hematol Oncol Clin North Am. 2009;23((3)):447–465. doi: 10.1016/j.hoc.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 10.Bedogni B., Warneke J. A., Nickoloff B. J., Giaccia A. J., Powell M. B. Notch1 is an effector of Akt and hypoxia in melanoma development [published online ahead of print October 16, 2008]. J Clin Invest. 2008;118((11)):3660–3670. doi: 10.1172/JCI36157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoek K., Rimm D. L., Williams K. R., et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res. 2004;64((15)):5270–5282. doi: 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- 12.Pinnix C. C., Lee J. T., Liu Z. J., et al. Active Notch1 confers a transformed phenotype to primary human melanocytes [published online ahead of print June 23, 2009]. Cancer Res. 2009;69((13)):5312–5320. doi: 10.1158/0008-5472.CAN-08-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egger G., Liang G., Aparicio A., Jones P. A. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429((6990)):457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 14.Howell P. M., Jr, Liu S., Ren S., Behlen C., Fodstad O., Riker A. I. Epigenetics in human melanoma. Cancer Control. 2009;16((3)):200–218. doi: 10.1177/107327480901600302. [DOI] [PubMed] [Google Scholar]
- 15.Friedman R. C., Farh K. K., Burge C. B., Bartel D. P. Most mammalian mRNAs are conserved targets of microRNAs [published online ahead of print October 27, 2008]. Genome Res. 2009;19((1)):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis B. P., Burge C. B., Bartel D. P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120((1)):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 17.Bentwich I., Avniel A., Karov Y., et al. Identification of hundreds of conserved and nonconserved human microRNAs [published online ahead of print June 19, 2005]. Nat Genet. 2005;37((7)):766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 18.Lau N. C., Lim L. P., Weinstein E. G., Bartel D. P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294((5543)):858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 19.Okamura K., Phillips M. D., Tyler D. M., Duan H., Chou Y. T., Lai E. C. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution [published online ahead of print March 30, 2008]. Nat Struct Mol Biol. 2008;15((4)):354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116((2)):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 21.Ambros V. The functions of animal microRNAs. Nature. 2004;431((7006)):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 22.Lee Y. S., Kim H. K., Chung S., Kim K. S., Dutta A. Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation [published online ahead of print February 18, 2005]. J Biol Chem. 2005;280((17)):16635–16641. doi: 10.1074/jbc.M412247200. [DOI] [PubMed] [Google Scholar]
- 23.Takamizawa J., Konishi H., Yanagisawa K., et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64((11)):3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 24.Carmell M. A., Xuan Z., Zhang M. Q., Hannon G. J. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16((21)):2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- 25.Karube Y., Tanaka H., Osada H., et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96((2)):111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calin G. A., Dumitru C. D., Shimizu M., et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia [published online ahead of print November 14, 2002]. Proc Natl Acad Sci U S A. 2002;99((24)):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calin G. A., Sevignani C., Dumitru C. D., et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers [published online ahead of print February 18, 2004]. Proc Natl Acad Sci U S A. 2004;101((9)):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L., Huang J., Yang N., et al. MicroRNAs exhibit high frequency genomic alterations in human cancer [published online ahead of print June 5, 2006]. Proc Natl Acad Sci U S A. 2006;103((24)):9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaur A., Jewell D. A., Liang Y., et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67((6)):2456–2468. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- 30.Bemis L. T., Chen R., Amato C. M., et al. MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Res. 2008;68((5)):1362–1368. doi: 10.1158/0008-5472.CAN-07-2912. [DOI] [PubMed] [Google Scholar]
- 31.Felicetti F., Errico M. C., Segnalini P., Mattia G., Carè A. MicroRNA-221 and -222 pathway controls melanoma progression. Expert Rev Anticancer Ther. 2008;8((11)):1759–1765. doi: 10.1586/14737140.8.11.1759. [DOI] [PubMed] [Google Scholar]
- 32.Worley L. A., Long M. D., Onken M. D., Harbour J. W. Micro-RNAs associated with metastasis in uveal melanoma identified by multiplexed microarray profiling. Melanoma Res. 2008;18((3)):184–190. doi: 10.1097/CMR.0b013e3282feeac6. [DOI] [PubMed] [Google Scholar]
- 33.Yan D., Zhou X., Chen X., et al. MicroRNA-34a inhibits uveal melanoma cell proliferation and migration through downregulation of c-Met [published online ahead of print November 21, 2008]. Invest Ophthalmol Vis Sci. 2009;50((4)):1559–1565. doi: 10.1167/iovs.08-2681. [DOI] [PubMed] [Google Scholar]
- 34.Migliore C., Petrelli A., Ghiso E., et al. MicroRNAs impair MET-mediated invasive growth. Cancer Res. 2008;68((24)):10128–10136. doi: 10.1158/0008-5472.CAN-08-2148. [DOI] [PubMed] [Google Scholar]
- 35.Segura M. F., Hanniford D., Menendez S., et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor [published online ahead of print February 2, 2009]. Proc Natl Acad Sci U S A. 2009;106((6)):1814–1819. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mueller D. W., Rehli M., Bosserhoff A. K. miRNA expression profiling in melanocytes and melanoma cell lines reveals miRNAs associated with formation and progression of malignant melanoma [published online ahead of print February 12, 2009]. J Invest Dermatol. 2009;129((7)):1740–1751. doi: 10.1038/jid.2008.452. [DOI] [PubMed] [Google Scholar]
- 37.Garraway L. A., Widlund H. R., Rubin M. A., et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436((7047)):117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 38.Steingrimsson E., Copeland N. G., Jenkins N. A. Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet. 2004:38365–38411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- 39.Dhomen N., Marais R. BRAF signaling and targeted therapies in melanoma. Hematol Oncol Clin North Am. 2009;23((3)):529–545. doi: 10.1016/j.hoc.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 40.Ozsolak F., Poling L. L., Wang Z., et al. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22((22)):3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roush S., Slack F. J. The let-7 family of microRNAs [published online ahead of print September 4, 2008]. Trends Cell Biol. 2008;18((10)):505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 42.Schultz J., Lorenz P., Gross G., Ibrahim S., Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18((5)):549–557. doi: 10.1038/cr.2008.45. [DOI] [PubMed] [Google Scholar]
- 43.Müller D. W., Bosserhoff A. K. Integrin beta 3 expression is regulated by let-7a miRNA in malignant melanoma [published online ahead of print August 4, 2008]. Oncogene. 2008;27((52)):6698–6706. doi: 10.1038/onc.2008.282. [DOI] [PubMed] [Google Scholar]
- 44.He L., Thomson J. M., Hemann M. T., et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435((7043)):828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Landais S., Landry S., Legault P., Rassart E. Oncogenic potential of the miR-106-363 cluster and its implication in human T-cell leukemia. Cancer Res. 2007;67((12)):5699–5707. doi: 10.1158/0008-5472.CAN-06-4478. [DOI] [PubMed] [Google Scholar]
- 46.Levati L., Alvino E., Pagani E., et al. Altered expression of selected microRNAs in melanoma: antiproliferative and proapoptotic activity of miRNA-155. Int J Oncol. 2009;35((2)):393–400. [PubMed] [Google Scholar]
- 47.Alexeev V., Yoon K. Distinctive role of the cKit receptor tyrosine kinase signaling in mammalian melanocytes. J Invest Dermatol. 2006;126((5)):1102–1110. doi: 10.1038/sj.jid.5700125. [DOI] [PubMed] [Google Scholar]
- 48.Curtin J. A., Busam K., Pinkel D., Bastian B. C. Somatic activation of KIT in distinct subtypes of melanoma [published online ahead of print August 14, 2006]. J Clin Oncol. 2006;24((26)):4340–4346. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 49.Smalley K. S., Contractor R., Nguyen T. K., et al. Identification of a novel subgroup of melanomas with KIT/cyclin-dependent kinase-4 overexpression. Cancer Res. 2008;68((14)):5743–5752. doi: 10.1158/0008-5472.CAN-08-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang S., Luca M., Gutman M., et al. Enforced c-KIT expression renders highly metastatic human melanoma cells susceptible to stem cell factor-induced apoptosis and inhibits their tumorigenic and metastatic potential. Oncogene. 1996;13((11)):2339–2347. [PubMed] [Google Scholar]
- 51.Igoucheva O., Alexeev V. MicroRNA-dependent regulation of cKit in cutaneous melanoma [published online ahead of print January 4, 2009]. Biochem Biophys Res Commun. 2009;379((3)):790–794. doi: 10.1016/j.bbrc.2008.12.152. [DOI] [PubMed] [Google Scholar]
- 52.Lopez-Bergami P., Fitchman B., Ronai Z. Understanding signaling cascades in melanoma [published online ahead of print December 15, 2007]. Photochem Photobiol. 2008;84((2)):289–306. doi: 10.1111/j.1751-1097.2007.00254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palmieri G., Capone M., Ascierto M. L., et al. Main roads to melanoma. J Transl Med. 2009;7:86. doi: 10.1186/1479-5876-7-86. Available at http://www.translational-medicine.com/content/7/1/86. Accessed 20 May 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smalley K. S. Understanding melanoma signaling networks as the basis for molecular targeted therapy. J Invest Dermatol. 2010;130((1)):28–37. doi: 10.1038/jid.2009.177. [DOI] [PubMed] [Google Scholar]
- 55.Zhang L., Deng T., Li X., et al. microRNA-141 is involved in a nasopharyngeal carcinoma-related genes network [published online ahead of print January 6, 2010]. Carcinogenesis. 2010;31((4)):559–566. doi: 10.1093/carcin/bgp335. [DOI] [PubMed] [Google Scholar]
- 56.Garofalo M., Di Leva G., Romano G., et al. miR-221 & 222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16((6)):498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Costoya J. A., Hobbs R. M., Pandolfi P. P. Cyclin-dependent kinase antagonizes promyelocytic leukemia zinc-finger through phosphorylation [published online ahead of print February 4, 2008]. Oncogene. 2008;27((27)):3789–3796. doi: 10.1038/onc.2008.7. [DOI] [PubMed] [Google Scholar]
- 58.Gottschalk A. R., Basila D., Wong M., et al. p27Kip1 is required for PTEN-induced G1 growth arrest. Cancer Res. 2001;61((5)):2105–2111. [PubMed] [Google Scholar]
- 59.Mamillapalli R., Gavrilova N., Mihaylova V. T., et al. PTEN regulates the ubiquitin-dependent degradation of the CDK inhibitor p27(KIP1) through the ubiquitin E3 ligase SCF(SKP2). Curr Biol. 2001;11((4)):263–267. doi: 10.1016/s0960-9822(01)00065-3. [DOI] [PubMed] [Google Scholar]
- 60.Lee S. G., Su Z. Z., Emdad L., Sarkar D., Fisher P. B. Astrocyte elevated gene-1 (AEG-1) is a target gene of oncogenic Ha-ras requiring phosphatidylinositol 3-kinase and c-Myc [published online ahead of print November 6, 2006]. Proc Natl Acad Sci U S A. 2006;103((46)):17390–17395. doi: 10.1073/pnas.0608386103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thirkettle H. J., Mills I. G., Whitaker H. C., Neal D. E. Nuclear LYRIC/AEG-1 interacts with PLZF and relieves PLZF-mediated repression [published online ahead of print August 3, 2009]. Oncogene. 2009;28((41)):3663–3670. doi: 10.1038/onc.2009.223. [DOI] [PubMed] [Google Scholar]
- 62.He L., He X., Lim L. P., et al. A microRNA component of the p53 tumour suppressor network [published online ahead of print June 6, 2007]. Nature. 2007;447((7148)):1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lodygin D., Tarasov V., Epanchintsev A., et al. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer [published online ahead of print August 1, 2008]. Cell Cycle. 2008;7((16)):2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 64.Kitago M., Martinez S. R., Nakamura T., Sim M. S., Hoon D. S. Regulation of RUNX3 tumor suppressor gene expression in cutaneous melanoma [published online ahead of print March 31, 2009]. Clin Cancer Res. 2009;15((9)):2988–2994. doi: 10.1158/1078-0432.CCR-08-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Z., Sun H., Dai H., et al. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT [published online ahead of print September 29, 2009]. Cell Cycle. 2009;8((17)):2756–2768. doi: 10.4161/cc.8.17.9387. [DOI] [PubMed] [Google Scholar]
- 66.Xi Y., Nakajima G., Gavin E., et al. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples [published online ahead of print August 13, 2007]. RNA. 2007;13((10)):1668–1674. doi: 10.1261/rna.642907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang X., Chen J., Radcliffe T., Lebrun D. P., Tron V. A., Feilotter H. An array-based analysis of microRNA expression comparing matched frozen and formalin-fixed paraffin-embedded human tissue samples [published online ahead of print October 2, 2008]. J Mol Diagn. 2008;10((6)):513–519. doi: 10.2353/jmoldx.2008.080077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cortez M. A., Calin G. A. MicroRNA identification in plasma and serum: a new tool to diagnose and monitor diseases. Expert Opin Biol Ther. 2009;9((6)):703–711. doi: 10.1517/14712590902932889. [DOI] [PubMed] [Google Scholar]
- 69.Glud M., Klausen M., Gniadecki R., et al. MicroRNA expression in melanocytic nevi: the usefulness of formalin-fixed, paraffin-embedded material for miRNA microarray profiling [published online ahead of print November 13, 2009]. J Invest Dermatol. 2009;129((5)):1219–1224. doi: 10.1038/jid.2008.347. [DOI] [PubMed] [Google Scholar]
- 70.Liu A., Tetzlaff M. T., Vanbelle P., et al. MicroRNA expression profiling outperforms mRNA expression profiling in formalin-fixed paraffin-embedded tissues. Int J Clin Exp Pathol. 2009;2((6)):519–527. [PMC free article] [PubMed] [Google Scholar]
- 71.Ma Z., Lui W. O., Fire A., Dadras S. S. Profiling and discovery of novel miRNAs from formalin-fixed, paraffin-embedded melanoma and nodal specimens. J Mol Diagn. 2009;11((5)):420–429. doi: 10.2353/jmoldx.2009.090041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Radhakrishnan A., Badhrinarayanan N., Biswas J., Krishnakumar S. Analysis of chromosomal aberration (1, 3, and 8) and association of microRNAs in uveal melanoma. Mol Vis. 2009:152146–152154. [PMC free article] [PubMed] [Google Scholar]
- 73.Satzger I., Mattern A., Kuettler U., et al. MicroRNA-15b represents an independent prognostic parameter and is correlated with tumor cell proliferation and apoptosis in malignant melanoma. Int J Cancer. 2010;126((11)):2553–2562. doi: 10.1002/ijc.24960. [DOI] [PubMed] [Google Scholar]
- 74.Sun Q., Cong R., Yan H., et al. Genistein inhibits growth of human uveal melanoma cells and affects microRNA-27a and target gene expression. Oncol Rep. 2009;22((3)):563–567. doi: 10.3892/or_00000472. [DOI] [PubMed] [Google Scholar]
- 75.Kota J., Chivukula R. R., O'Donnell K. A., et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137((6)):1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Molnar V., Tamasi V., Bakos B., Wiener Z., Falus A. Changes in miRNA expression in solid tumors: an miRNA profiling in melanomas [published online ahead of print January 15, 2008]. Semin Cancer Biol. 2008;18((2)):111–122. doi: 10.1016/j.semcancer.2008.01.001. [DOI] [PubMed] [Google Scholar]