Abstract
Aims
Growth factor-induced repression of smooth muscle (SM) cell marker genes is an integral part of vascular SM (VSM) cell proliferation. This is partly regulated via translocation of extracellular signal-regulated kinase 1/2 (ERK1/2) to the nucleus which activates the transcription factor Elk-1. The mediators involved in ERK1/2 nuclear translocation in VSM cells are unknown. The aim of this study is to examine the mechanisms which regulate growth factor-induced nuclear translocation of ERK1/2 and gene expression in VSM cells.
Methods and results
In cultured human VSM cells, phospholipase C (PLC)γ1 expression was required for platelet-derived growth factor (PDGF)-induced ERK1/2 nuclear translocation, Elk-1 phosphorylation, and subsequent repression of SM α-actin gene expression. The mechanisms of a role for PLCγ1 in ERK1/2 nuclear localization were further examined by investigating interacting proteins. The ERK1/2-binding phosphoprotein, protein enriched in astrocytes-15 (PEA-15), was phosphorylated by PDGF and this phosphorylation required activation of PLCγ1. In cells pre-treated with PEA-15 siRNA, ERK1/2 distribution significantly increased in the nucleus and resulted in decreased SM α-actin expression and increased VSM cell proliferation. Overexpression of PEA-15 increased ERK1/2 localization in the cytoplasm. The regulatory role of PEA-15 phosphorylation was assessed. In VSM cells overexpressing a non-phosphorylatable form of PEA-15, PDGF-induced ERK1/2 nuclear localization was inhibited.
Conclusion
These results suggest that PEA-15 phosphorylation by PLCγ1 is required for PDGF-induced ERK1/2 nuclear translocation. This represents an important level of phenotypic control by directly affecting Elk-1-dependent transcription and ultimately SM cell marker protein expression in VSM cells.
Keywords: Vascular smooth muscle, Phospholipase C, Transcription, Mitogen-activated protein kinase
1. Introduction
Alterations in vascular structure and responsiveness are a hallmark of cardiovascular disease. These alterations occur via a phenotypic modulation of the growth properties of vascular smooth muscle (VSM) cells from a differentiated contractile phenotype to a more proliferative phenotype.1,2 This modulation typically occurs via the action of mitogens, including growth factors such as platelet-derived growth factor (PDGF) BB.2 It has been established that mitogenic signals decrease the expression of SM cell differentiation marker proteins by repressing expression of the respective genes.3 This repression of SM cell marker genes is probably an important step in the change towards a proliferative phenotype during the development of vascular disease.
Central to growth factor-induced repression of SM cell marker genes are the intracellular signals which regulate transcription. When a mitogenic signal is received, this leads to activation of the mitogen-activated protein kinases, extracellular signal-regulated kinase 1/2 (ERK1/2). ERK1/2 is a key regulatory element in the control of transcription following initiation of VSM cell proliferation.3,4 Following its activation, ERK1/2 translocates to the nucleus and directly phosphorylates the Ets transcription factor, Elk-1.4–6 Activated Elk-1 binds to serum response factor and this complex results in repression of several SM cell marker genes5,6 while also inducing expression of genes that play roles in atherogenesis.6,7 As the ERK1/2 signalling cascade is necessary for mediating mitogenic effects during VSM cell phenotypic modulation, the spatial and temporal regulation of ERK1/2 is critical, particularly with regard to its nuclear localization.8 ERK1/2 has a multiplicity of substrates (upwards of 100) and can phosphorylate targets in the cytoplasm or translocate to the nucleus to alter gene expression via effects on transcription factor activity.9 Such a promiscuous signalling complex requires additional modulatory processes to ensure integration from a range of signals and to direct outputs dependent on the cellular context. In addition to feedback loops which regulate ERK1/2 activity,10 there are several ERK1/2 scaffolding proteins that modulate outcomes following activation of this signalling pathway.9 For example, in some cell lines, ERK1/2 nuclear translocation can be regulated by changes in the cytoskeleton via interaction with tropomyosin.11 As the regulation of ERK1/2 nuclear translocation directly relates to Elk-1 phosphorylation and repression of SM cell marker genes in VSM cell, it is important to understand the modulatory mechanisms that may alter ERK1/2 localization in VSM cells following growth factor stimulation.
In the present study, we now demonstrate a novel mechanism in cultured VSM cells and in ex vivo blood vessels which is required for PDGF-induced Elk-1 phosphorylation and the repression of the SM cell differentiation marker gene, SM α-actin. The intracellular pathway involves activation of phospholipase C (PLC)γ1,12 a prerequisite for ERK1/2 nuclear translocation. We further reveal that the downstream target of PLCγ1 is a phosphoprotein previously uncharacterized in VSM cells, protein enriched in astrocytes-15 (PEA-15). PEA-15 is an ERK1/2-binding protein which acts as a cytoplasmic anchor to prevent ERK1/2 nuclear translocation. The PDGF-induced increase in PLCγ1 activation results in phosphorylation of PEA-15. This phosphorylation releases activated ERK1/2 and allows ERK1/2 translocation to the nucleus leading to ERK1/2-dependent activation of Elk-1. This pathway involving PLCγ1 and PEA-15 represents a hitherto unknown level of regulation in PDGF-induced VSM cell phenotypic modulation.
2. Methods
2.1. Reagents
Human PDGF-BB was purchased from R&D Systems (Abingdon, UK). Antibodies against the following proteins were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA), c-fos, GAPDH, PLCγ1, RhoGDI, lamin A, SM α-actin, haemagglutinin, phospho-Ser116PEA-15. Antibodies against ERK1/2, phospho-ERK1/2, phospho-Elk-1, PEA-15, and phospho-Ser104PEA-15 were from Cell Signaling (Beverly, MA, USA). HRP- and fluorescent-conjugated secondary antibodies were from Dako Ltd (Cambridge, UK). Lipofectamine 2000, Fura-2 AM, and antibody against phospho-Ser116PEA-15 were obtained from Invitrogen (San Diego, CA, USA). All other chemicals were from Sigma (Poole, Dorset, UK), unless otherwise stated.
2.2. Cell culture
Human coronary artery SM (HCoASM) cells, purchased from Lonza (Wokingham, UK), were maintained in Clonetics SM growth medium-2 containing 5% FBS, 0.5 μg/L EGF, 5 mg/L insulin, 2 mg/L FGF, 50 mg/L gentamicin and 50 mg/L amphotericin. Routinely, cells were used between passages 4 and 12 from at least three independent donors.
2.3. Tissue preparation
Segments of human saphenous vein were obtained directly following removal of the vessel during coronary bypass surgery following a protocol approved by the North of Scotland Research Ethics Committee and informed written consent was obtained from each patient. This investigation conforms with the principles outlined in the Declaration of Helsinki. Adventitia was removed from the vessel segments and the lumen scraped to remove the endothelial layer. Segments were maintained at 37°C for 1h before stimulation with growth factor. Following incubations, segments were flash frozen and homogenized in homogenising buffer as previously described.13
2.4. Immunoblotting
Protein samples were prepared and subjected to SDS–PAGE as previously described.13
2.5. Transfection of siRNA
For knockdown of PLCγ1 expression, a pool of four synthetic siRNAs corresponding to the human PLCγ1 mRNA sequence was used (Santa Cruz Biotechnology). Multiple functional, non-targeting siRNA sequences were used to verify on-target effects. Targeted knockdown of PLCγ1 was further verified using a different siRNA duplex (Dharmacon, Lafayette, CO, USA). A different non-targeted control siRNA containing at least four mismatches to any human, mouse, or rat gene was used in this case. There was no difference in knockdown, regardless of the source or sequence of siRNA. PLCγ1 knockdown did not affect PLCγ2 or PLCβ1 isoforms as confirmed by immunoblots (data not shown). PEA-15 knockdown was obtained using a pool of three target-specific siRNAs (Santa Cruz Biotechnology). As above, multiple functional, non-targeting siRNA sequences were used to verify targeted effects. Specificity was also verified by using pre-designed siRNA. Primary HCoASM cells were transfected with 100 nM siRNA using Lipofectamine 2000 (Invitrogen), according to the manufacturers’ instructions. Cells were incubated at 37°C for 24 h, transferred to serum-free medium, and incubated for a further 48 h prior to treatment.
2.6. Transfection of cDNA constructs
HA-tagged PEA-15 was created and subcloned into pcDNA3.1 as described previously.14 The non-phosphorylatable HA-PEA-15 has both the Ser phosphorylation site (Ser104 and Ser116) switched to Ala, designated as HA-PEA-15(SSAA), as described previously.14 This was also subcloned into pcDNA3.1. HCoASM cells were transfected with 5 μg DNA using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. Cells were incubated at 37°C for 24 h, transferred to serum-free medium, and incubated for a further 48 h prior to treatment. In the case of HA-PEA-15(SSAA), the transfection efficiency was low (routinely 10%). Different transfection methods and alterations to the protocol were tested but did not improve efficiency. This was particular to HCoASM cells as other cell types, such as Cos-7 cells, had ∼90% transfection efficiency.
2.7. Imaging of [Ca2+]i
HCoASM cells were incubated in serum-free medium for 24 h and PDGF-induced Ca2+ increase was measured as described previously.13
2.8. Preparation of nuclear and membrane fractions
Membrane and cytosolic fraction were prepared as described previously.15 In all cases, cytosolic and nuclear fractions were checked against the expression of RhoGDI (a characterized cytoplasmic marker) and lamin A (a nuclear localized protein).
2.9. Confocal microscopy
HCoASM cells were fixed and immunolabelled as described previously.15 For double labelling of phospho-ERK1/2 and HA, a different species origin of the primary antibodies was used.
2.10. Quantitative PCR
Quantitative real-time PCR was performed using Taqman gene expression assays from Applied Biosystems (Foster City, CA, USA). Following treatments, RNA was extracted from HCoASM cells using TRIzol (Invitrogen) according to the manufacturer's instructions. cDNA was reverse transcribed using a DyNAmo probe 2-step qRT-PCR kit (New England Biolabs, Ipswich, MA, USA) according to the manufacturer's instructions. Changes in gene expression of SM α-actin (assay ID: Hs00909449), GAPDH (assay ID: Hs99999905), and β-actin (assay ID: Hs99999903) were assessed using verified FAM-labelled hydrolysis probes. Samples were analysed using a Lightcycler 480 system for real-time PCR (Roche Applied Biosystems, Indianapolis, IN, USA). Changes in gene expression for smooth α-actin were expressed as a ratio of the corresponding GAPDH. The same results were obtained when expressed as a ratio of β-actin.
2.11. Bromodeoxyuridine assay
Cultured VSM cells were grown to 80% confluence in 96-well plates and deprived of serum for 24 h prior to treatment. Following treatment, incorporation of bromodeoxyuridine (BrdU) label was determined according to the manufacturer's instructions (Calbiochem).
2.12. Statistics
Data are expressed as mean ± SEM. Significance was tested by means of Student's t-test or analysis of variance where appropriate. A value of P < 0.05 was considered significant.
3. Results
3.1. PLCγ1 is required for PDGF-induced Elk-1 phosphorylation but not ERK1/2 activation
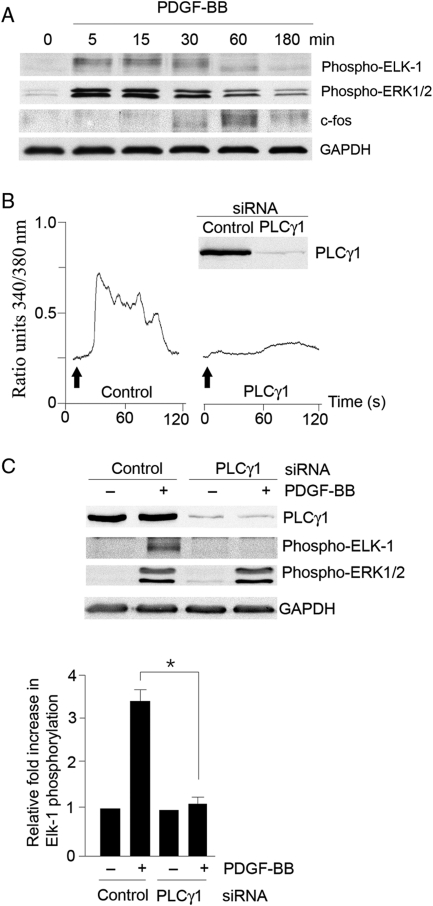
In HCoASM cells, PDGF (50 ng/mL) produced a maximal increase in ERK1/2 phosphorylation (6.2 ± 0.7-fold compared with control, n = 5) and Elk-1 phosphorylation (3.4 ± 0.6-fold compared with control, n = 3) at 15 min and decreased thereafter (Figure 1A). c-fos expression increased to maximum within 1 h (4.5 ± 1.1-fold increase, n = 3). As growth receptor tyrosine kinases activate PLCγ1, we determined whether this PLC isoform has a regulatory role in PDGF-induced Elk-1 phosphorylation. In HCoASM cells transfected with PLCγ1 siRNA, PDGF did not produce a rise in the intracellular Ca2+ concentration (Figure 1B). Despite a >95% knockdown of PLCγ1 expression, there was no effect on PDGF-induced ERK1/2 activation (Figure 1C). However, following PLCγ1 knockdown (94 ± 3% decrease in protein expression, n = 6), PDGF-induced Elk-1 activation was significantly inhibited.
Figure 1.
PDGF-induced activation of Elk-1 but not ERK1/2 in VSM cells requires the expression of PLCγ1. (A) HCoASM cells were stimulated with 50 ng/mL PDGF at 37°C over a time course of 3 h. Phosphorylated ERK1/2, phosphorylated Elk-1, and c-fos were assessed by immunoblotting using specific antibodies. GAPDH served as a loading control. Typical example of n = 3–5 for each blot. (B) HCoASM cells were transfected with either control or PLCγ1 siRNA (100 nmol/L) for 48 h at 37°C. Cell extracts were fractionated by SDS–PAGE and analysed for PLCγ1 expression by immunoblotting. PDGF-BB-induced intracellular Ca2+ release was assessed using single-cell fluorescence imaging of FURA-2-loaded cells (typical example of n = 6). (C) HCoASM cells were transfected with control or PLCγ siRNA (100 nmol/L) for 48 h at 37°C and transferred to serum-free medium for a further 24 h. Cells were treated with vehicle alone or with PDGF-BB (50 ng/mL) for 15 min at 37°C and analysed for the expression of PLCγ1, phospho-Elk-1, and phospho-ERK1/2 (typical blots shown of n = 4 for mean ± SEM data, *P < 0.05).
3.2. PLCγ1 is required for ERK1/2 nuclear localization
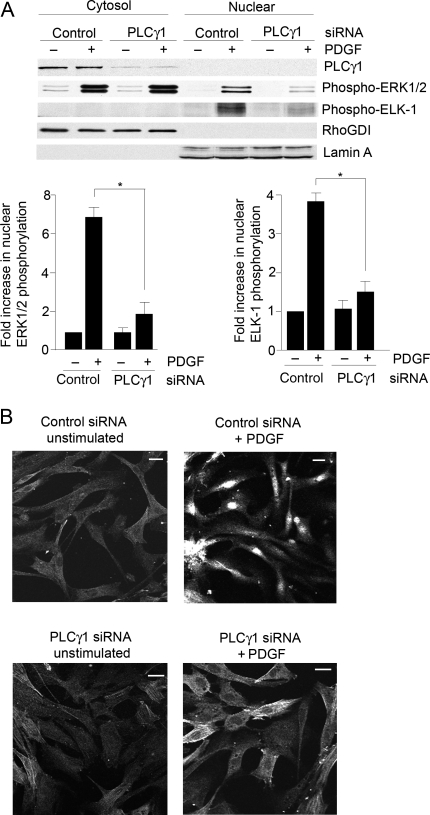
In HCoASM cells following PLCγ1 knockdown, the level of PDGF-induced ERK1/2 phosphorylation was significantly reduced in the nuclear fraction compared with the cytosol (Figure 2A). This decreased ERK1/2 nuclear translocation was also reflected in the decreased PDGF-induced Elk-1 phosphorylation. This was further studied using immunolocalization in HCoASM cells. Following 15 min stimulation, PDGF-induced nuclear translocation of activated ERK1/2 was inhibited in cells transfected with PLCγ1 siRNA (Figure 2B). In control non-stimulated cells, phospho-ERK1/2 had a cytoplasmic distribution, co-localizing partly with filamentous structures as previously observed in other cell types.11 In PDGF-treated cells, phospho-ERK1/2 had a nuclear localization.
Figure 2.
PLCγ1 knockdown reduces nuclear translocation of phospho-ERK1/2. (A) HCoASM cells were transfected with control or PLCγ1 siRNA (100 nmol/L) for 48 h, transferred to serum-free medium for a further 24 h, and treated with vehicle or PDGF-BB (50 ng/mL) for 15 min at 37°C. Cytosolic and nuclear extracts were fractionated by SDS–PAGE and analysed by immunoblotting for the expression of PLCγ1, phospho-Elk-1, and phospho-ERK1/2. RhoGDI and lamin A served as loading controls for the cytosolic and nuclear fractions, respectively. Typical blots and mean ± SEM data for n = 3 are shown; *P < 0.05. (B) HCoASM cells treated as above, stained with antibody against phospho-ERK1/2 and visualized by confocal immunofluorescence microscopy. Scale bar = 5 μm. Representative image of at least n = 3 individual experiments.
3.3. PLCγ1 is required for repression of SM α-actin
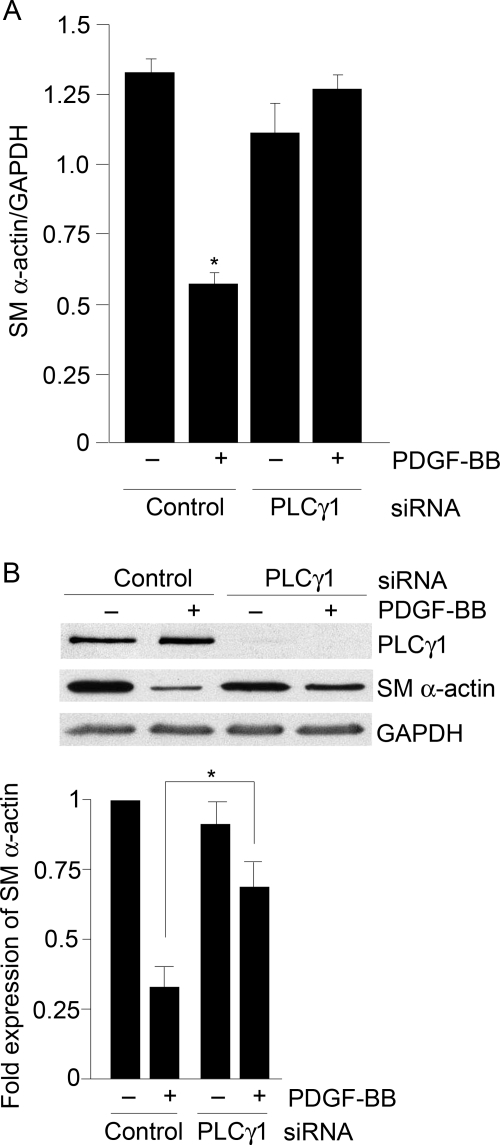
The role of PLCγ1 on SM α-actin expression was determined. In cells treated with control siRNA, the SM α-actin gene expression, as measured using qPCR, was significantly decreased (more than 50%) following 24 h PDGF stimulation. In HCoASM cells transfected with PLCγ1 siRNA, SM α-actin gene expression was maintained at control levels even after 24 h PDGF stimulation (Figure 3A). This was also reflected in SM α-actin protein expression. Following PLCγ1 knockdown, SM α-actin expression was maintained significantly closer to control levels following PDGF stimulation (Figure 3B).
Figure 3.
PLCγ1 knockdown inhibits PDGF-induced repression of SM α-actin gene and protein expression. (A) HCoASM cells were transfected with control or PLCγ1 siRNA (100 nmol/L) for 48 h, transferred to serum-free medium, and treated with PDGF-BB (50 ng/mL) for a further 24 h at 37°C. The expression of SM-α-actin was determined by qPCR using gene-specific probes and expressed as a ratio of the corresponding GAPDH gene expression. n = 3 for each treatment. *P < 0.05. (B) HCoASM cells were transfected with PLCγ1 siRNA as above, transferred to serum-free medium, and treated with PDGF-BB (50 ng/mL) for 72 h at 37°C. Cell extracts were analysed by immunoblotting for the expression of SM α-actin. Representative blots and mean ± SEM data of n = 3 experiments are shown. *P < 0.05.
3.4. The ERK1/2-binding protein PEA-15 sequesters ERK1/2 in the cytoplasm of VSM cells and is involved in PDGF-induced repression of SM α-actin
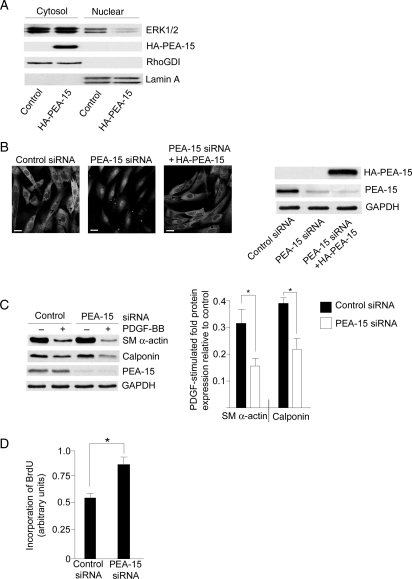
PEA-15, an ERK1/2-binding phosphoprotein, is expressed in HCoASM cells (Figure 4). In order to examine whether PEA-15 can regulate ERK1/2 localization in VSM cells, the expression of PEA-15 in HCoASM cells was manipulated using either transfection with HA-tagged PEA-15 or PEA-15 siRNA. In HCoASM cells overexpressing HA-PEA-15, a decrease in ERK1/2 localization to the nuclear fraction was observed compared with control cells (Figure 4A, 4.2 ± 0.3-fold decrease in HA-PEA-15-treated cells compared with control, n = 3). In HCoASM cells treated with PEA-15 siRNA, the localization of ERK1/2 was switched to a predominantly nuclear distribution as observed using immunolocalization (Figure 4B). In rescue experiments, cells were transfected with both PEA-15 siRNA and the HA-PEA-15 plasmid. In this case, ERK1/2 localization was restored to a cytoplasmic distribution.
Figure 4.
Changes in PEA-15 expression regulate ERK1/2 localization and proliferation. (A) HCoASM cells were transfected with control or HA-PEA-15 plasmid for 48 h at 37°C. Cytosolic and nuclear fractions were analysed for the distribution of ERK1/2 and HA-PEA-15. RhoGDI and lamin A served as loading controls for the cytosolic and nuclear fractions, respectively. Anti-HA antibody was used to demonstrate the expression of the HA-PEA-15. Typical blots of n = 3 independent samples. (B) HCoASM cells were transfected with control or PEA-15 siRNA for 48 h at 37°C. In some cases, cells were also simultaneously transfected with HA-PEA-15 plasmid. ERK1/2 localization was determined by confocal immunofluorescence microscopy. Scale bar = 5 μm. Corresponding immunoblots verify knockdown and/or overexpression of proteins. Typical images and blots of n = 3 independent samples. (C) HCoASM cells were transfected with control or PEA-15 siRNA for 48 h at 37°C followed by stimulation with PDGF-BB (50 ng/mL) for 15 min. Cells extracts were immunoblotted for proteins indicated. Typical blots and mean ± SEM data for n = 3 are shown; *P < 0.05. (D) Cells were transfected with PEA-15 siRNA as above and treated with 50 ng/mL PDGF-BB for 48 h. BrdU incorporation was measured using a spectrophotometer.
To assess whether PEA-15 was required for PDGF-induced repression of differentiated SM marker proteins, HCoASM cells were treated with PEA-15 siRNA followed by stimulation with PDGF for 48 h. In control cells, PDGF stimulation led to a decrease in SM α-actin expression. Cells treated with PEA-15 siRNA and stimulated with PDGF had a significantly greater decrease in SM α-actin expression compared with control siRNA-treated cells (Figure 4C). In addition, the expression of another marker of SM differentiation, calponin, was regulated similarly to SM α-actin following knockdown of PEA-15 (Figure 4C). This had a subsequent effect on proliferation. In HCoASM cells treated with PEA-15 siRNA and stimulated with 10% serum for 48 h, BrdU incorporation was significantly increased compared with control cells (Figure 4D).
3.5. PDGF induces phosphorylation of PEA-15 via PLCγ1
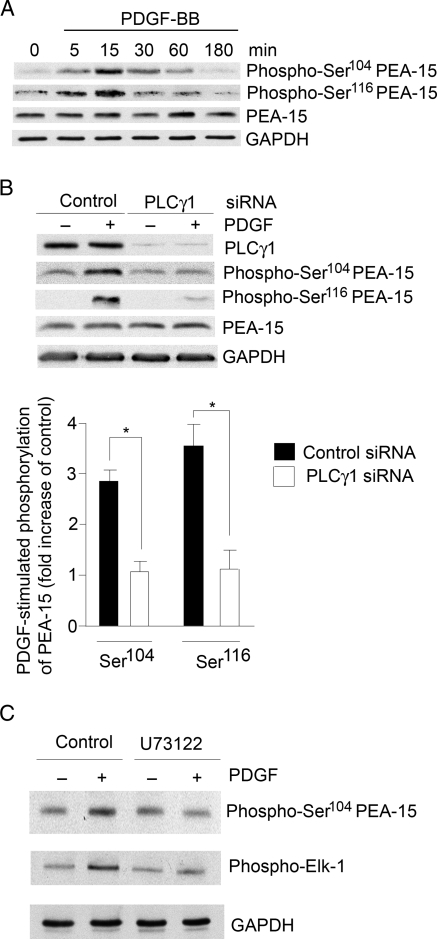
Recent evidence suggests that following PEA-15 phosphorylation, ERK1/2 constitutively bound to PEA-15 is released allowing ERK1/2 translocation to the nucleus.16,17 PEA-15 contains two serine phosphorylation sites (Ser104 and Ser116).18 PDGF stimulated an increased phosphorylation on both Ser104 and Ser116 of PEA-15 in a time-course compatible with a role in regulating ERK1/2 nuclear translocation (Figure 5A). In HCoASM cells treated with PLCγ1 siRNA, stimulation with PDGF for 15 min resulted in a significant inhibition of PEA-15 phosphorylation at both Ser104 and Ser116 (Figure 5B).
Figure 5.
PLCγ1 is required for phosphorylation of PEA-15 by PDGF. (A) HCoASM cells were treated with PDGF-BB (50 ng/mL) at 37°C for the times indicated. Cell extracts were analysed by immunoblotting for PEA-15, phospho-Ser104PEA-15, and phospho-Ser116PEA-15. GAPDH acted as a loading control. Typical blots of n = 3 independent experiments. (B) Cells were transfected with control or PLCγ1 siRNA (100 nmol/L) as previously and treated with vehicle or PDGF-BB (50 ng/mL) for 15 min at 37°C. Cell extracts were analysed by immunoblotting for expression of proteins indicated. Typical blots and mean ± SEM data for n = 3 experiments are shown; *P < 0.05. (C) Freshly isolated human saphenous vein segments were pre-incubated with the PLC inhibitor U73122 (4 mmol/L) for 30 min and stimulated with PDGF-BB (50 ng/mL) for 15 min at 37°C. Homogenates were subjected to SDS–PAGE and immunoblotting with antibodies as indicated. Blots are representative images of n = 3 experiments.
To determine whether PEA-15 phosphorylation was regulated similarly in human blood vessels, freshly isolated human saphenous veins were incubated with 50 ng/mL PDGF for 15 min. Some vein segments were pre-incubated with the PLC inhibitor U73122. Immunoblots demonstrated that PDGF can phosphorylate PEA-15 in human saphenous vein and this is dependent upon PLC activation (Figure 5C; control 2.4 ± 0.3-fold increase vs. U73122 0.9 ± 0.2-fold increase, n = 3, P < 0.05). Elk-1 phosphorylation was also significantly dependent on PLC activation (control 3.0 ± 0.5-fold increase vs. U73122 1.2 ± 0.2-fold increase, n = 3, P < 0.05).
3.6. Nuclear translocation of activated ERK1/2 is regulated by the phosphorylation of PEA-15
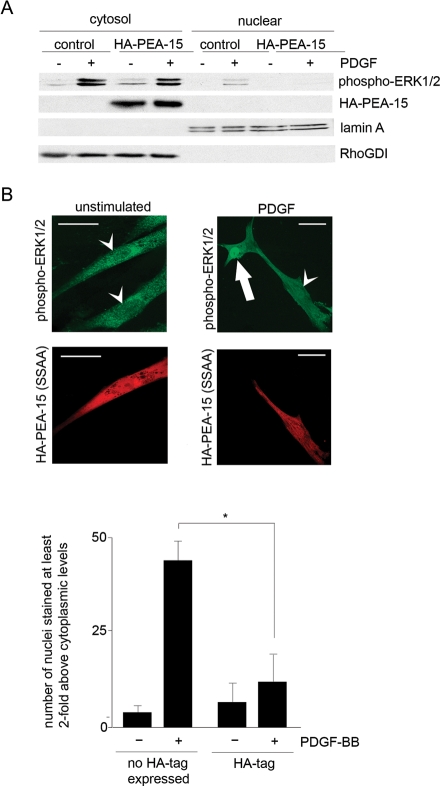
We next investigated whether PEA-15 phosphorylation regulates the nuclear translocation of PDGF-activated ERK1/2. In HCoASM cells, transfection with HA-PEA-15 prevented the increase in phosphorylated ERK1/2 in the nuclear fraction following PDGF stimulation for 15 min (Figure 6A; cytoplasm:nuclear % ratio of phosphorylated ERK1/2, control plasmid 59 ± 5% vs. HA-PEA-15 plasmid 14 ± 2%, n = 3, P < 0.05). In order to examine whether the phosphorylation of PEA-15 by PDGF was important for regulating the nuclear localization of activated ERK1/2, we transfected HCoASM cells with a non-phosphorylatable form of PEA-15 [HA-PEA-15(SSAA)]. Owing to the relatively high expression levels of endogenous PEA-15 in HCoASM cells and the low transfection rates obtained with this plasmid, cells were treated with both siRNA PEA-15 (to knockdown endogenous PEA-15 expression) and HA-PEA-15(SSAA). Following transfection, cells were stimulated with PDGF for 15 min. Preparations were also double-labelled for the HA-tag to identify cells which were expressing the HA-PEA-15(SSAA). In parallel experiments, cells were labelled with anti-PEA-15 antibody (data not shown). In all cases, labelling of PEA-15 was at the level of background fluorescence suggesting at least 95% knockdown of PEA-15 (as observed in immunoblots). In HCoASM cells under unstimulated conditions, phosphorylated ERK1/2 was at low levels and was present in both the cytoplasm and the nucleus, regardless of whether cells were expressing HA-PEA-15(SSAA) or not (Figure 6B). When cells were stimulated with PDGF, phosphorylated ERK1/2 translocated to the nucleus only if cells were not expressing HA-PEA-15(SSAA). In contrast, in cells expressing the non-phosphorylatable HA-PEA-15, the activated ERK1/2 did not translocate to the nucleus and remained predominantly cytoplasmic.
Figure 6.
PEA-15 phosphorylation regulates activated ERK1/2 nuclear translocation. (A) HCoASM cells were transfected with HA-PEA-15 for 24 h and transferred to serum-free medium for an additional 24 h. Cells were treated without or with PDGF-BB (50 ng/mL) for 15 min at 37°C, fractionated into cytosolic and nuclear extracts, and examined for the expression of HA-PEA-15 and phospho-ERK1/2. RhoGDI and Lamin A acted as loading controls for cytosolic and nuclear fractions, respectively. Anti-HA antibody was used to demonstrate the expression of the HA-PEA-15. (B) Cells were initially transfected with PEA-15 siRNA for 24 h followed by transfection with either control or non-phosphorylatable PEA-15 plasmid, HA-PEA-15(SSAA). Cells were treated with PDGF (50 ng/mL) for 15 min and double labelled for both phospho-ERK1/2 (top panels) and HA-tag (corresponding bottom panels). Localization was determined by confocal immunofluorescence microscopy. Bottom panels labelled with anti HA-antibody show cells transfected with HA-PEA-15(SSAA). Scale bar = 5 μm. Arrow denotes the nucleus of the cell showing PDGF-induced nuclear translocation of phospho-ERK1/2; arrowheads denote nuclei with no increase in phospho-ERK1/2 staining. Graph denotes quantitative data from three experiments, with 10 separate fields of view per experiment. Images were analysed using NIH Image. Nuclei with more than two-fold increase in staining compared with cytoplasm were counted as positive for phospho-ERK1/2 nuclear translocation. The presence of the HA-tag was used to demonstrate expression of the HA-PEA-15(SSAA) plasmid. *P < 0.05.
4. Discussion
The growth factor-induced repression of SM cell differentiation marker genes is a hallmark of vascular proliferation and occurs predominantly via Elk-1-dependent transcription.3–5 The regulation of Elk-1 activity is therefore of particular importance in controlling VSM cell phenotype. Elk-1 is regulated by direct phosphorylation via the mitogen-activated protein kinase family members, ERK1/2. As Elk-1 has a mostly nuclear distribution and ERK1/2 is predominantly activated in the cytoplasm, following growth factor stimulation there is a requirement for activated ERK1/2 to translocate to the nucleus. However, the mechanisms which regulate this translocation are largely unknown. In the current study, we reveal for the first time that PDGF-induced Elk-1 activation and subsequent repression of SM α-actin gene expression requires the sequential regulation of both PLCγ1 and ERK1/2. Our data demonstrate that PLCγ1 is necessary for the phosphorylation of the ERK1/2-sequestering protein PEA-15. PEA-15 acts as a cytoplasmic anchor for ERK1/2 and, following PEA-15 phosphorylation, ERK1/2 is released and translocates to the nucleus. This represents a hitherto unknown level of growth factor regulation for Elk-1-dependent transcription in VSM cells and may regulate proliferation in vivo.
PEA-15 was initially cloned from astrocytes where it is expressed in relative abundance.19 The protein sequence contains a N-terminal death effector domain (DED)20,21 and two phosphorylation sites; Ser104 (phosphorylated predominantly by protein kinase C) and Ser116 (phosphorylated by CaMKII or Akt).18,22 The DED domain regulates the anti-apoptotic effect of PEA-15 in astrocytes and cancer cell lines, via binding to FADD,16 and is also involved in regulating integrin activation.20 In addition, studies using yeast two-hybrid screens and cell lines have determined that PEA-15 binds and sequesters ERK1/2 in the cytoplasm by preventing nuclear entry.14,23,24 We now demonstrate in VSM cells that PEA-15 acts as a cytoplasmic anchor for ERK1/2. Interestingly, knockdown of PEA-15 expression does not decrease cytoplasmic ERK1/2 activation by PDGF in VSM cells but only alters ERK1/2 cytoplasmic distribution towards a predominantly nuclear localization. This suggests that PEA-15 does not affect upstream facilitators for ERK1/2 activation but acts as a downstream regulator of ERK1/2 functional effects. In some cell types, PEA-15 can also act as a cytoplasmic scaffolding protein which facilitates the activation of the ERK downstream target, p90 ribosomal S6 kinase 2.25 Importantly, variations in PEA-15 expression can regulate ERK1/2-dependent transcription in vivo. A recent study in T-cells from PEA-15−/− mice has shown that the proliferation rate is enhanced due to an increase in ERK1/2-dependent transcription.26 It is therefore possible that alterations in PEA-15 expression in VSM cells (possibly via pathological processes) may contribute to changes in VSM cell gene expression and ultimately phenotype through regulation of ERK1/2 signalling. Although the promoter regions of the PEA-15 gene have only recently been examined,27,28 the regulation of PEA-15 expression in vivo is not clear. Changes in PEA-15 expression during the development of pathological conditions, such as vascular disease, could alter ERK1/2 nuclear localization and may have important effects on VSM cell phenotype. This remains to be determined.
An important aspect regarding PEA-15 function is the role of PEA-15 phosphorylation.18,22 Studies suggest that the phosphorylation of Ser104 and Ser116 is involved in regulating PEA-15 interaction with other proteins.16,17 We now demonstrate in VSM cells that phosphorylation of PEA-15 is required for PDGF-induced ERK1/2 nuclear translocation and occurs in a time-course compatible with regulation of Elk-1 activation. We further demonstrate that PLCγ1 is a critical component in PDGF-induced Elk-1 activation and the subsequent repression of SM α-actin. Following growth factor receptor engagement, signalling pathways for ERK1/2 and PLCγ1 are both activated in VSM cells. Our results reveal that both these pathways are required for Elk-1 activation. Without activation of PLCγ1, PEA-15 is not phosphorylated. In this case, ERK1/2 can be activated but remains sequestered in the cytoplasm, bound to the unphosphorylated PEA-15. The possibility therefore exists that changes in the PLCγ1 pathway will directly alter growth factor-induced gene expression via regulation of PEA-15 phosphorylation. We have previously demonstrated that VSM cells from native blood vessels with a proliferating phenotype have different expression levels of PLCγ1 compared with the fully developed VSM cell phenotype.13 Although the importance of this phenotypic difference in regulating cell phenotype remains to be determined, it does provide a potential in vivo scenario whereby changes in PLCγ1 expression can alter PEA-15 signalling to have an important influence on growth factor-stimulated transcription.
In conclusion, the results of the present study indicate that PLCγ1 has an integral role in regulating ERK1/2 nuclear translocation which occurs via the ERK1/2-binding phosphoprotein PEA-15. PLCγ1-dependent phosphorylation is required for PDGF-induced repression of the SM α-actin gene expression in VSM cells. This growth factor-induced mechanism could be important in regulating VSM cell phenotype and may have implications for the development of vascular disease.
Conflict of interest: none declared.
Funding
G.F.N. was supported by The Wellcome Trust and KRF-2008-220-F00013 (Global Research Network) from the Research Foundation of Korea (NRF). J.W.R. was supported by National Institutes of Health, USA (RO1-CA93849).
References
- 1.Wamhoff BR, Bowles DK, Owens GK. Excitation-transcription coupling in arterial smooth muscle. Circ Res. 2006;98:868–878. doi: 10.1161/01.RES.0000216596.73005.3c. doi:10.1161/01.RES.0000216596.73005.3c. [DOI] [PubMed] [Google Scholar]
- 2.Kaplan-Albuquerque N, Bogaert YE, Van Putten V, Weiser-Evans MC, Nemenoff RA. Patterns of gene expression differentially regulated by PDGF and hypertrophic stimuli in vascular smooth muscle. J Biol Chem. 2005;280:19966–19976. doi: 10.1074/jbc.M500917200. doi:10.1074/jbc.M500917200. [DOI] [PubMed] [Google Scholar]
- 3.Kawai-Kowase K, Owens GK. Multiple repressor pathways contribute to phenotypic switching of vascular smooth muscle cells. Am J Physiol Cell Physiol. 2007;292:C59–C69. doi: 10.1152/ajpcell.00394.2006. doi:10.1152/ajpcell.00394.2006. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z, Whang D-Z, Hockmeyer D, McAnally J, Nordheim A, Olson EN. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–189. doi: 10.1038/nature02382. doi:10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- 5.Zhou J, Hu G, Herring BP. Smooth muscle-specific genes are differentially sensitive to inhibition by Elk-1. Mol Cell Biol. 2005;25:9874–9885. doi: 10.1128/MCB.25.22.9874-9885.2005. doi:10.1128/MCB.25.22.9874-9885.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miano JM. Serum response factor: toggling between disparate programs of gene expression. J Mol Cell Cardiol. 2003;35:577–593. doi: 10.1016/s0022-2828(03)00110-x. doi:10.1016/S0022-2828(03)00110-X. [DOI] [PubMed] [Google Scholar]
- 7.Khachigian LM. Early growth response-1 in cardiovascular pathobiology. Circ Res. 2006;98:186–191. doi: 10.1161/01.RES.0000200177.53882.c3. doi:10.1161/01.RES.0000200177.53882.c3. [DOI] [PubMed] [Google Scholar]
- 8.Pouysségur J, Volmat V, Lenormand P. Fidelity and spatio-temporal control in MAP kinase (ERKs) signalling. Biochem Pharmacol. 2002;64:755–763. doi: 10.1016/s0006-2952(02)01135-8. doi:10.1016/S0006-2952(02)01135-8. [DOI] [PubMed] [Google Scholar]
- 9.Ramos JW. The regulation of extracellular signal-regulated kinase in mammalian cells. Int J Biochem Cell Biol. 2008;40:2707–2719. doi: 10.1016/j.biocel.2008.04.009. doi:10.1016/j.biocel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 10.Morrison DK, Davis RJ. Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu Rev Cell Dev Biol. 2003;19:91–118. doi: 10.1146/annurev.cellbio.19.111401.091942. doi:10.1146/annurev.cellbio.19.111401.091942. [DOI] [PubMed] [Google Scholar]
- 11.Bharadwaj S, Vasanth G, Masuelli L, Thanawala R, Prasad GL. Inhibition of nuclear accumulation of phosphorylated ERK by tropomyosin-1-mediated cytoskeletal reorganization. J Cancer Mol. 2008;5:139–144. [Google Scholar]
- 12.Rhee SG. Regulation of phosphoinositide-specific phospholipase C. Annu Rev Biochem. 2001;70:281–312. doi: 10.1146/annurev.biochem.70.1.281. doi:10.1146/annurev.biochem.70.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egan CG, Wainwright CL, Wadsworth RM, Nixon GF. PDGF-induced signaling in proliferating and differentiated vascular smooth muscle: effects of altered Ca2+ regulation. Cardiovasc Res. 2005;67:308–316. doi: 10.1016/j.cardiores.2005.03.019. doi:10.1016/j.cardiores.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 14.Formstecher E, Ramos JW, Fauquet M, Calderwood DA, Hsieh JC, Canton B, et al. PEA-15 mediates cytoplasmic sequestration of ERK MAP kinase. Dev Cell. 2001;1:239–250. doi: 10.1016/s1534-5807(01)00035-1. doi:10.1016/S1534-5807(01)00035-1. [DOI] [PubMed] [Google Scholar]
- 15.Hunter I, Nixon GF. Spatial compartmentalization of tumor necrosis factor (TNF) receptor 1-dependent signaling pathways in human airway smooth muscle cells. Lipid rafts are essential for TNF-α-mediated activation of RhoA but dispensable for the activation of the NF-κB and MAPK pathways. J Biol Chem. 2006;281:34705–34715. doi: 10.1074/jbc.M605738200. doi:10.1074/jbc.M605738200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Renganathan H, Vaidyanathan H, Knapinska A, Ramos JW. Phosphorylation of PEA-15 switches its binding specificity from ERK/MAPK to FADD. Biochem J. 2005;390:729–735. doi: 10.1042/BJ20050378. doi:10.1042/BJ20050378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krueger J, Chou FL, Glading A, Schaefer E, Ginsberg MH. Phosphorylation of phosphoprotein enriched in astrocytes (PEA-15) regulates extracellular signal-regulated kinase-dependent transcription and cell proliferation. Mol Biol Cell. 2005;16:3552–3561. doi: 10.1091/mbc.E04-11-1007. doi:10.1091/mbc.E04-11-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubes M, Cordier J, Glowinski J, Girault JA, Chneiweiss H. Endothelin induces a calcium-dependent phosphorylation of PEA-15 in intact astrocytes: identification of Ser104 and Ser116 phosphorylated, respectively, by protein kinase C and calcium/calmodulin kinase II in vitro. J Neurochem. 1998;71:1307–1314. doi: 10.1046/j.1471-4159.1998.71031307.x. doi:10.1046/j.1471-4159.1998.71031307.x. [DOI] [PubMed] [Google Scholar]
- 19.Estellés A, Yokoyama M, Nothias F, Vincent JD, Glowinski J, Vernier P, et al. The major astrocytic phosphoprotein PEA-15 is encoded by two mRNAs conserved on their full length in mouse and human. J Biol Chem. 1996;271:14800–14806. doi: 10.1074/jbc.271.25.14800. doi:10.1074/jbc.271.25.14800. [DOI] [PubMed] [Google Scholar]
- 20.Ramos JW, Kojima TK, Hughes PE, Fenczik CA, Ginsberg MH. The death effector domain of PEA-15 is involved in its regulation of integrin activation. J Biol Chem. 1998;273:33897–33900. doi: 10.1074/jbc.273.51.33897. doi:10.1074/jbc.273.51.33897. [DOI] [PubMed] [Google Scholar]
- 21.Kitsberg D, Formstecher E, Fauquet M, Kubes M, Cordier J, Canton B, et al. Knock-out of the neural death effector domain protein PEA-15 demonstrates that its expression protects astrocytes from TNFalpha-induced apoptosis. J Neurosci. 1999;19:8244–8251. doi: 10.1523/JNEUROSCI.19-19-08244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trencia A, Perfetti A, Cassese A, Vigliotta G, Miele C, Oriente F, et al. Protein kinase B/Akt binds and phosphorylates PED/PEA-15, stabilizing its antiapoptotic action. Mol Cell Biol. 2003;23:4511–4521. doi: 10.1128/MCB.23.13.4511-4521.2003. doi:10.1128/MCB.23.13.4511-4521.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill JM, Vaidyanathan H, Ramos JW, Ginsberg MH, Werner MH. Recognition of ERK MAP kinase by PEA-15 reveals a common docking site within the death domain and death effector domain. EMBO J. 2002;21:6494–6504. doi: 10.1093/emboj/cdf641. doi:10.1093/emboj/cdf641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitehurst AW, Robinson FL, Moore MS, Cobb MH. The death effector domain protein PEA-15 prevents nuclear entry of ERK2 by inhibiting required interactions. J Biol Chem. 2004;279:12840–12847. doi: 10.1074/jbc.M310031200. doi:10.1074/jbc.M310031200. [DOI] [PubMed] [Google Scholar]
- 25.Vaidyanathan H, Opoku-Ansah J, Pastorino S, Renganathan H, Matter ML, Ramos JW. ERK MAP kinase is targeted to RSK2 by the phosphoprotein PEA-15. Proc Natl Acad Sci USA. 2007;104:19837–19842. doi: 10.1073/pnas.0704514104. doi:10.1073/pnas.0704514104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pastorino S, Renganathan H, Caliva MJ, Filbert EL, Opoku-Ansah J, Sulzmaier FJ, et al. The death effector domain protein PEA-15 negatively regulates T-cell receptor signaling. FASEB J. 2010;24:2818–2828. doi: 10.1096/fj.09-144295. doi:10.1096/fj.09-144295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ungaro P, Teperino R, Mirra P, Cassese A, Fiory F, Perruolo G, et al. Molecular cloning and characterization of the human PED/PEA-15 gene promoter reveal antagonistic regulation by hepatocyte nuclear factor 4alpha and chicken ovalbumin upstream promoter transcription factor II. J Biol Chem. 2008;283:30970–30979. doi: 10.1074/jbc.M803895200. doi:10.1074/jbc.M803895200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Condorelli G, Vigliotta G, Iavarone C, Caruso M, Tocchetti CG, Andreozzi F, et al. PED/PEA-15 gene controls glucose transport and is overexpressed in type 2 diabetes mellitus. EMBO J. 1998;17:3858–3866. doi: 10.1093/emboj/17.14.3858. doi:10.1093/emboj/17.14.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]