Abstract
Video tutorials are an effective way for researchers to quickly learn how to use online tools offered by biological databases. At MaizeGDB, we have developed a number of video tutorials that demonstrate how to use various tools and explicitly outline the caveats researchers should know to interpret the information available to them. One such popular video currently available is ‘Using the MaizeGDB Genome Browser’, which describes how the maize genome was sequenced and assembled as well as how the sequence can be visualized and interacted with via the MaizeGDB Genome Browser.
Database URL: http://www.maizegdb.org/
Introduction
Researchers who rely upon online data resources demand that these resources function intuitively. At the same time, researchers wish to expertly interact with the data available, but do not have much time to learn how to effectively and efficiently use some of the more complex tools available to them. In addition, not all researchers interacting with the data provided are aware of the caveats that should be known to properly interpret the data presented. Biological databases are required to satisfy different needs simultaneously in an integrated fashion. Database personnel must not only strive to continually create and recreate intuitive interfaces, but also train researchers to expertly use the more complex tools and datasets offered.
Towards this goal, we at MaizeGDB (1) strive to create intuitive interfaces, take steps to educate users and are determined to respond quickly to researchers’ stated needs and feedback requests. To accomplish this, we track questions posed over time and use researchers’ feedback to evaluate which tools within MaizeGDB seem least intuitive. Where possible, we update the interface and tools to increase ease of use. In cases where the complexity of the biological problem itself creates a complicated tool, we put effort into education and outreach. To educate our busy users, in this article we introduce our new online video tutorial web site and highlight the ‘Using the MaizeGDB Genome Browser’ video tutorial available at: http://tutorial.maizegdb.org/?p=365 to provide an example of the type of tutorials that can be developed by biological databases to improve the experience of their users.
Our outreach process
At MaizeGDB there are three curators (L.C.H., M.L.S. and J.M.G.) on staff, each with ≥20 years of bench research experience in maize genetics. Each curator works in a different location: L.C.H. in Albany and Berkeley, CA; M.L.S. in Columbia MO; J.M.G. in Tucson, AZ. The curators regularly interact with a diverse group of active maize researcher at their home location. While outreach is part of all MaizeGDB team members’ jobs, one curator (LH) specializes in outreach, and makes up to three visits per year to universities and other research centers to give live interactive demonstrations of tools, tutorials and one-on-one help. These direct interactions have two distinct outcomes for researchers: first, they receive clear explanations on how to use non-intuitive aspects of the web site or its features. Second, researchers learn about tools and functionalities offered by MaizeGDB that they otherwise might not discover. At the same time, these interactions allow the curators to identify needs expressed by many researchers and articulate those needs to the MaizeGDB Team so that a plan for meeting stated needs can be developed and implemented.
Researchers attending on-site outreach workshops fill out evaluations of the quality and usefulness of the visit. Although the evaluations are very positive, the number of people that can be reached through these local events is limited. More than 500 researchers attend the Annual Maize Genetics Conference each year, and the number of maize ‘Cooperators’ is over 1600 at present (where Cooperators are researchers who have attended at least one Annual Maize Genetics Conference or who have specifically requested to become a maize Cooperator). In order to reach a wider audience, we recently began to create video tutorials that are accessible directly from the MaizeGDB home page. To identify areas that may be universally more difficult for researchers and thus would warrant the creation of a video tutorial, we compiled web site feedback gathered using the Request Tracker software (2) and cataloged the questions asked via all types of interactions. Such documentation is maintained internally. The bulk of questions asked over the past 3 years concern how the genome of inbred line B73 was sequenced, how this sequence can be accessed and used, and caveats for interacting with the data. In addition, responses to the Maize Genetics Executive Committee’s 2010 survey of maize Cooperators documents (i) a need for increased support for training in maize genetics, genomics and bioinformatics and (ii) the continued development of an effective genome browser. These two items ranked among the top 10 resources, tools and suggestions needed to support current and future directions for maize research (http://maizemeeting.maizegdb.org/mgec-survey10/analyze_final.php).
To reach more maize researchers, we created a new MaizeGDB video tutorial web site, which can be accessed from our homepage at http://www.maizegdb.org. For B73, we have created a total of five different video tutorials on the topic of how best to access and make use of the genome sequence. One video entitled ‘Using the MaizeGDB Genome Browser’ (available at: http://tutorial.maizegdb.org/?p=365) is described in detail below to provide an example of how biological databases can create video tutorials to improve the experience of their users and to meet researchers’ stated education needs.
Developing a video tutorial
In the course of developing tutorial videos for the outreach web site, we have developed a set of guidelines for video creation:
The creators of the videos must have expert knowledge of the subject matter. In the case of the ‘Using the MaizeGDB Genome Browser’ video, although scientific literature contains background information needed to create the video, much of the useful information was not published. Thus, personal interactions with the data generators, i.e. the Maize Genome Sequencing Consortium (MGSC) (3), by a biologist who understands the complexities of sequencing was essential.
Feedback from researchers who use MaizeGDB must be obtained and implemented before deployment of any video to gauge whether the video addresses expressed needs. We have gone back to people that have provided feedback, and to additional maize researchers and asked them how we can improve the tutorials before its official deployment. This activity has resulted in improvements to the tutorials each and every time.
Background context and real-world examples of how to use the presented material must be included in all videos, especially early in the presentation, to fully engage the audience.
During outreach visits, it is clear that one has to provide scientific biological context when presenting new tools in order to emphasize their utility. For example, in demonstrating how to use MaizeGDB’s ‘Locus Lookup’ tool (4), it is necessary to outline the basic steps of how to clone a gene in maize by ‘walking’ (5), then to explain how the Locus Lookup tool can automate analyses to determine which genomic segments are likely to contain the researchers gene of interest based only on genetic maps. By providing context with specific examples, researchers are able to learn not only how to use a tool, but what to use it for and, more specifically, how the tool can help their research. We use the same approach in our video tutorials. For example, in our ‘Using the MaizeGDB Genome Browser’ tutorial, almost half of the tutorial is background information about how the maize genome sequence was generated. This information is essential to understanding the data served in the MaizeGDB Genome Browser. In the case of the B73 reference genome sequence, many maize researchers do not know the details as to how the sequence was generated, nor the caveats that they must be aware of before making inferences based on the sequence. Methods employed to assemble the maize B73 sequence (6) involved prior knowledge from both physical and genetic maps (7, 8), making the assembly process itself fairly complex.
Testing a video tutorial
The first draft of the tutorial ‘How to use the MaizeGDB Genome Browser’ was distributed to two groups: The scientists who generated the data (in this case the MGSC), and a group of willing beta testers drawn from researchers who have volunteered to help. We had two people from the MGSC (Sandra Clifton and Bob Fulton) to review the video, and received extremely helpful comments and improvements from them. After the factual content of the video was approved by the MGSC, we tested the ‘understandability’ of the video by showing it to approximately 35 maize researchers, asking for their suggestions, and incorporating those suggestions into the final product.
Content of the tutorial video: ‘Using the MaizeGDB Genome Browser’
This video describes how the maize genome was sequenced, assembled and how the sequence can be viewed and used via the MaizeGDB Genome Browser and is accessible online at http://tutorial.maizegdb.org/?p=365. Here, we outline the type and level of information we provided in the video, which is divided into six different sections. Note that the text presented here is tied to the schematic movies in the video.
Understanding the B73 genome assemblies
The Maize B73 genome was sequenced in a bacterial artificial chromosome (BAC)-by-BAC approach. There are currently three B73 genome assemblies, each released incrementally as an improvement on the former release. First, the Missouri Mapping Project (8) created a BAC library from the maize B73 inbred. In addition, the project created high-resolution intermated B73 × Mo17 (IBM) genetic maps (7), and used these maps to order the BACs relative to each other. The result was a series of overlapping BAC clones assembled in ‘contigs’. This series of overlapping BACs was called the ‘Fingerprint Contig’ (FPC) map or ‘the physical map’ (7). Next, the MGSC used the FPC map to select a minimum tiling path (MTP) of BAC clones for sequencing. The first assembly of sequences was called the ‘BAC-based Assembly’. This assembly is a series of overlapping BACs ordered by the FPC map with sequence elements mapped to those physical locations. The second version of the assembly was called ‘B73 RefGen_v1’. This was the first sequence-based pseudomolecule assembly for maize, and is the only assembly currently published or available via GenBank (3, 6). The current version is an improvement called ‘B73 RefGen_v2’. This assembly is a global reassembly of Bacterial Artificial Chromosome sequences integrating fosmid reads, B73 Optical Map data (9) and maize-sorghum synteny (F. Wei et al., manuscript in preparation). Subsequent versions are anticipated to be named incrementally: i.e. B73 RefGen_v3, would be next, etc.
How the Maize Genome Consortium Sequenced each BAC
A separate, more detailed video on this topic is available, but it is briefly covered in the ‘How to use the MaizeGDB Genome Browser’ movie, because how the BACs were sequenced is important for understanding what the sequence represents. Each BAC clone was shotgun sequenced and assembly was aided by scaffolding where possible (the scaffolding process is visually explained in our ‘How the Maize Genome Consortium Sequenced each BAC’ movie tutorial). Thus, each BAC sequence is as a series of about 5–12 segments with relative order and orientation noted, if known. One hundred ‘Ns’ are used to mark places in the pseudomolecule assembly where the sequence is not contiguous. This, of course, complicates the identification of genes, thus researchers benefit greatly from an awareness of the quality of the underlying sequence.
Creating a pseudomolecule
This part of the tutorial explains how the BACs and contigs were joined to make the pseudomolecule (again, a separate, more detailed video on this topic is available). Contigs and BACs are anchored to the Intermated B73 X Mo17 genetic map, which can usually provide order and orientation. Within a single contig, BAC sequences were merged based on strict criteria (≥99% identity, and other criteria outlined in the video). Once BACs within a contig were assembled, the contigs were strung together to make up a pseudomolecule for each chromosome. The gaps between contigs are represented by 1000 ‘Ns’ in the pseudomolecule, to distinguish this type of gap from the ones between pieces of sequence within each BAC (represented as 100 ‘Ns’).
How these assemblies look on the MaizeGDB Genome Browser
For the video, after each description of how the assembly was generated, the display fades to a view of how the pseudomolecule is represented within the context of the MaizeGDB Genome Browser. Most maize researchers have used the MaizeGDB Genome Browser, so this is an attempt to relate the visuals researchers are accustomed to with the methods that were used for sequencing and assembly.
Entering the MaizeGDB Genome Browser
There are several ways to enter the Genome Browser from the MaizeGDB home page, and each is shown in this section of the video. Researchers can click on ‘Genome Browser’ on the home page, or use the simple search box at the top of any header or footer, and select ‘genome browser’ from the pull down menu. Another way to enter is through the MaizeGDB implementation of BLAST (10). Researchers can BLAST a sequence against a genome assembly then upload hits to the browser as a separate, private track. We also have additional outreach videos that spend several minutes going through exactly how to do this.
General features of the genome browser
We use GBrowse (11), and many of the features available within the MaizeGDB implementation of GBrowse are similar to those offered at other databases [e.g. Flybase (12) and TAIR (13)]. Several features are shown in the videos, including ‘rubberbanding’ your region of choice (i.e. choosing a specific region on the genome browser by clicking the mouse button and holding it down while selecting a region), how to get more information on the data in each track, collapsing and expanding tracks, making customized views, and more.
In the next part of the tutorial, some lesser-known features of the MaizeGDB Genome Browser are discussed: these were included based upon specific questions we received from researchers. Examples include how to: download sequence files, bookmark a region, upload private tracks and share private tracks. At the end of the tutorial, viewers are invited again to submit questions anytime, and our contact information is given.
The tutorial views
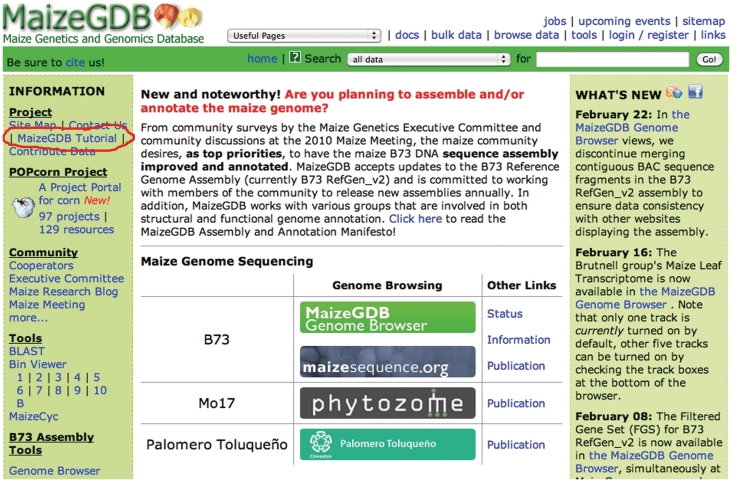
Tutorials can be accessed from our home page (Figure 1, red oval):
Figure 1.
Screen shot of the MaizeGDB home page.
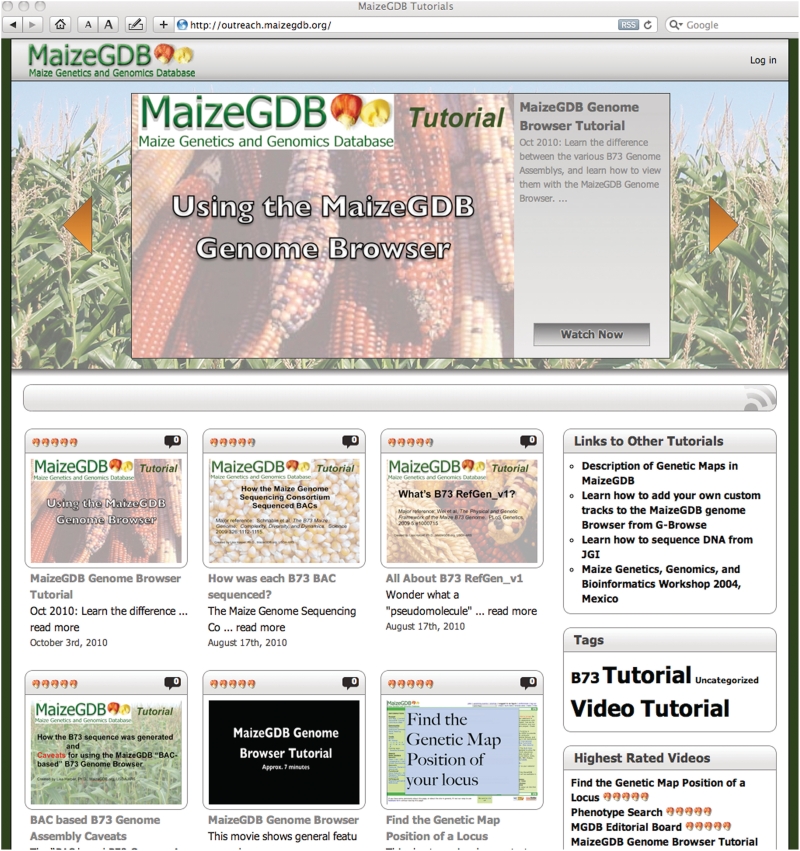
This directs the user to this page (http://tutorial.maizegdb.org) (Figure 2):
Figure 2.
MaizeGDB’s new tutorial page.
Clicking the ‘Using the MaizeGDB Genome Brower’ tutorial starts the video online.
Video tutorial web site usage
Our video tutorial web site contains videos aimed at all levels of researchers from undergraduates to experienced scientists. The idea is that if researchers have a question on a topic, they can view a video tutorial before asking for individual help. In Table 1, the usage statistics for the video tutorial site are listed. In Table 2, the usage statistics for the MaizeGDB web site proper are listed for comparison. In less than a year, the visits to the video site have increased 10-fold. The video tutorial web site has been advertised in two ways: (i) through email, verbal contacts and on-site demonstrations, as well as (ii) by adding links to the tutorials throughout the MaizeGDB web site. Recall that the estimated size of the maize research community is approximately 1600 people. From our usage statistics, it appears that users are looking at an average of 2–3 pages per visit, which means they are not just landing there and leaving.
Table 1.
2010 usage statisticsa for the MaizeGDB video tutorial web site reported by month
| Month | Visitsb | Pagesc | Filesd | Hitse |
|---|---|---|---|---|
| March | 115 | 142 | NA | NA |
| April | 103 | 124 | NA | NA |
| May | 127 | 169 | NA | NA |
| June | 76 | 100 | NA | NA |
| July | 90 | 123 | NA | NA |
| August | 889 | 2106 | 2835 | 5835 |
| September | 1208 | 1935 | 5774 | 8403 |
| October | 1283 | 3152 | 8302 | 11 229 |
| November | 1020 | 2713 | 7011 | 9175 |
| December | 1160 | 2936 | 5894 | 8981 |
| Totals | 6071 | 13 500 | 29 816 | 43 623 |
aWebalyzer Version 2.01’s log-based methods were used for gathering usage statistics.
bSeries of page requests from the same uniquely identified client with a time of no more than 30 min between each page request.
cA request for a file of type defined as a ‘page’.
dThe server sends something back to the client, such as a webpage or graphic image.
eA request for a file from the web server.
NA, Not available.
Table 2.
2010 usage statisticsa for the MaizeGDB web site reported by month
| Month | Visitsb | Pagesc | Filesd | Hitse |
|---|---|---|---|---|
| January | 114 770 | 895 226 | 1 655 973 | 1 830 566 |
| February | 75 132 | 1 231 974 | 1 383 189 | 1 612 616 |
| March | 101 377 | 1 017 406 | 1 288 128 | 1 637 521 |
| April | 87 050 | 839 195 | 1 171 477 | 1 342 145 |
| Mayf | 81 389 | 1 300 104 | 1 345 052 | 1 534 848 |
| Junef | 86 217 | 788 639 | 967 951 | 1 193 549 |
| Julyf | 82 353 | 851 508 | 955 118 | 1 148 732 |
| Augustf | 79 156 | 1 257 784 | 1 050 514 | 1 437 999 |
| Septemberf | 82 694 | 1 679 344 | 1 680 593 | 1 981 048 |
| October | 138 906 | 1 562 127 | 1 684 616 | 2 114 542 |
| November | 108 056 | 1 076 946 | 1 311 844 | 1 532 583 |
| December | 106 531 | 1 170 259 | 1 451 617 | 1 706 532 |
| Totals | 1 143 631 | 13 670 512 | 15 946 072 | 19 072 681 |
aWebalyzer Version 2.01’s log-based methods were used for gathering usage statistics.
bSeries of page requests from the same uniquely identified client with a time of no more than 30 min between each page request.
cA request for a file of type defined as a ‘page’.
dThe server sends something back to the client, such as a webpage or graphic image.
eA request for a file from the web server.
fUsage goes down in the summer months, when maize researchers are in the field. This trend is seen every year.
Updating content
It is very important that video tutorials be kept current. For example, the second sequence assembly version for B73 (i.e. B73 RefGen_v2) was released by the MGSC in March 2010 (F. Wei et al., unpublished data), with the third assembly scheduled for June 2011 release (D. Ware, personal communication). With each genome release some caveats are no longer relevant, and new issues emerge. However, because researchers might already have research outcomes generated based on the previous versions of the assembly, MaizeGDB keeps all versions of the assemblies, as well as all versions of the tutorials, available for the benefit of the maize researchers. As new assemblies come out, we will create and add new tutorials as needed.
Methods for creating video tutorials
Videos were created on a Macintosh computer with OS X 10.5. Movie screen shots of the browser were created using Snapz Pro (Ambrosia). Animations of the genome assembly process were created using Keynote ’09, version 5.0.4 (Apple). The movie was put together and sound equalized in iMovie 7 (in the iLife ’08 suite, Apple). Videos are uploaded to Vimeo, and converted to a format viewable on the web. The http://tutorial.maizegdb.org/ page was made in Wordpress (http://wordpress.org). We roughly estimate that it took about two full time months (∼340 person-hours) to create this video—including time to create content and learn how to use the software listed here. Less time is spent on every new video as personnel become more experienced in creating them.
Conclusion
From how to operate a smartphone to posting items for sale via online auctions, videos for learning are becoming more pervasive. Video tutorials for outreach represent a quick and easy way for researchers to learn new tools with less frustration. We have learned which items at MaizeGDB require further explanation by directly interacting with our user group and offer various tutorials to meet their training and outreach needs.
Funding
This work is supported by the United States Department of Agriculture-Agricultural Research Service and National Science Foundation grants #0743804 and #0703273. Additional support is provided to C.J.L. and T.Z.S. through; POPcorn, A Project Portal for Corn (C. Lawrence PI, NSF Award #0734804) as well as to C.J.L. via The Grass Regulome Initiative (E. Grotewold PI, NSF Award #0701405); The UniformMu Project (D. McCarty PI, NSF award #0703273), The Improving Water Acquisition in Maize Project (J. Lynch PI, NSF award #0965380); and The Center for Maize and Wheat Improvement (CIMMYT/USAID).
Conflict of interest. None declared.
Acknowledgements
MaizeGDB would like to acknowledge a diverse group of individuals, organizations and funding agencies whose enduring support makes the mission of MaizeGDB possible.
Core funding for MaizeGDB is provided by the USDA-ARS. Guidance is generously provided by maize researchers at large; the MaizeGDB Working Group (A. Barkin, O. Hoekenga, A. Lamblin, T. Lubberstedt, K. McGinnis, L. Mueller, M. Pop, M. Sachs, P. Schnable, A. Sylvester); the Maize Genetics Executive Committee (T. Brutnell, P. Schnable, M. Sachs, V. Walbot, W. Tracy, S. Wessler, J. Bennetzen, B. Boston, E. Buckler, C. Lawrence); and the National Corn Growers Association.
References
- 1.Sen TZ, Andorf C, Schaeffer L, et al. MaizeGDB becomes ‘sequence-centric’. Database. 2009;2009 doi: 10.1093/database/bap020. published online 7 December 2009, doi:10.1093/database/bap020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vincent J, Spier R, Rolsky R, et al. RT Essentials. O’Reilly Media: Sebastopol; 2008. [Google Scholar]
- 3.Schnable P, Ware D, Fulton R, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 4.Andorf C, Lawrence C, Harper L, et al. The Locus Lookup tool at MaizeGDB: identification of genomic regions in maize by integrating sequence information with physical and genetic maps. Bioinformatics. 2010;26:434–436. doi: 10.1093/bioinformatics/btp556. [DOI] [PubMed] [Google Scholar]
- 5.Bortiri E, Jackson D, Hake S. Advances in maize genomics: the emergence of positional cloning. Curr. Opin. Plant Biol. 2006;9:164–171. doi: 10.1016/j.pbi.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Wei F, Zhang J, Zhou S, et al. The physical and genetic framework of the Maize B73 genome. PLoS Genet. 2009;5:e1000715. doi: 10.1371/journal.pgen.1000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coe E, Cone K, McMullen M, et al. Access to the maize genome: an integrated physical and genetic map. Plant Physiol. 2002;128:9–12. doi: 10.1104/pp.128.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cone KC, McMullen MD, Bi IV, et al. Genetic, physical, and informatics resources for maize. On the road to an integrated map. Plant Physiol. 2002;130:1598–1605. doi: 10.1104/pp.012245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou J, Wei F, Nguyen J, et al. A single molecule scaffold for the maize genome. Plos Genet. 2009;5:e1000711. doi: 10.1371/journal.pgen.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altschul S, Gish W, Miller W, et al. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 11.Stein L, Mungall C, Shu S, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tweedie S, Ashburner MK, Falls K, et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swarbreck D, Wilks C, Lamesch P, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]