Abstract
The transcriptional repressor PLZF was identified by its translocation with retinoic acid receptor alpha in t(11;17) acute promyelocytic leukemia (APL). Ectopic expression of PLZF leads to cell cycle arrest and growth suppression, while disruption of normal PLZF function is implicated in the development of APL. To clarify the function of PLZF in cell growth and survival, we used an inducible PLZF cell line in a microarray analysis to identify the target genes repressed by PLZF. One prominent gene identified was c-myc. The array analysis demonstrated that repression of c-myc by PLZF led to a reduction in c-myc-activated transcripts and an increase in c-myc-repressed transcripts. Regulation of c-myc by PLZF was shown to be both direct and reversible. An interaction between PLZF and the c-myc promoter could be detected both in vitro and in vivo. PLZF repressed the wild-type c-myc promoter in a reporter assay, dependent on the integrity of the binding site identified in vitro. PLZF binding in vivo was coincident with a decrease in RNA polymerase occupation of the c-myc promoter, indicating that repression occurred via a reduction in the initiation of transcription. Finally, expression of c-myc reversed the cell cycle arrest induced by PLZF. These data suggest that PLZF expression maintains a cell in a quiescent state by repressing c-myc expression and preventing cell cycle progression. Loss of this repression through the translocation that occurs in t(11;17) would have serious consequences for cell growth control.
Acute promyelocytic leukemia (APL) is caused by translocation of nuclear receptor retinoic acid receptor alpha (RARα) to one of five partner genes (50). This leads to disruption of the normal activity of both genes targeted by the translocation and to the expression of two new chimeric proteins with additional effects on those regulatory pathways. The transcription factor PLZF is fused to RARα in t(11;17) APL. Notable features of PLZF are the BTB/POZ repression and multimerization domain, a second region (RD2) responsible for transcriptional repression, and the nine zinc fingers that create the DNA binding domain. The translocation observed in APL results in replacement of the activation domain of RARα with the repressive BTB/POZ domain of PLZF, creating the repressive nuclear receptor PLZF-RARα. The reciprocal fusion protein RARα-PLZF is formed by fusing seven of nine zinc fingers of PLZF to the RARα N-terminal activation domain (9). We previously showed that elucidation of the normal function of PLZF has led to a deeper understanding of how RARα fusion proteins function in leukemogenesis. The integrity of the BTB domain is responsible for dimerization of PLZF, correct nuclear localization, and part of the transcriptional function of PLZF (48). PLZF physically interacts with SMRT, mSin3a, and HDAC-1 through this domain, recruiting a repression complex to PLZF-bound promoters and shutting down the transcription of those genes (reference 76 and references therein). The exact nature of the DNA-protein complex that forms around PLZF, and the changes in chromatin configuration that occur during the silencing of transcription, is still under investigation. ETO, another corepressor, binds PLZF through the second repression domain and contributes further to the repression of transcription induced by PLZF expression (51).
PLZF-mediated transcriptional repression is associated with suppression of cellular proliferation. PLZF expression declines during differentiation of HL-60 and NB4 cells (9). In a variety of cell models, continued PLZF expression was associated with cell cycle arrest in G1 and eventual apoptosis (65, 71, 75). PLZF can also alter myeloid differentiation before induction of apoptosis. In 32D cells, constitutive PLZF blocked differentiation induced by granulocyte colony-stimulating factor and granulocyte-macrophage colony-stimulating factor, while in a U937 cell system, PLZF blocked vitamin D3-induced monocytic differentiation (71). A few target genes regulated by PLZF have been identified—the cell cycle protein cyclin A2 was directly reduced by expression of PLZF (75), and levels of the interleukin-3Rα (IL-3Rα) chain may also be repressed (J. D. Licht, unpublished data). To pursue the pathways and mechanisms by which PLZF alters cell differentiation and death, we used an inducible expression system and a series of high-density cDNA microarrays to identify both direct target genes and the downstream effectors of PLZF.
One of the most strikingly regulated genes identified in this study was the proto-oncogene c-myc. c-myc is central to the control of proliferation, apoptosis, and differentiation decisions in the cell. c-myc is commonly activated in human and animal tumors and has been shown to have multiple effects on cell behavior (12, 16, 17). Regulation of both the activity and the expression of c-myc is highly complex and tightly controlled. Many transcription factors have been shown to bind to the c-myc promoter directly and alter gene expression (5, 6, 10, 21, 24, 25, 28, 34, 39, 57, 61, 62, 70), and others, such as C/EBPα (35) and Smad-3 (74), regulate c-myc expression indirectly by interacting with and altering the action of direct transcription factors such as the E2F family. c-myc can also negatively autoregulate its own expression (20) by undefined mechanisms. We demonstrate here that PLZF directly suppresses the initiation of c-myc transcription and that growth suppression mediated by PLZF can be reversed by enforced expression of c-myc. Further, PLZF-regulated genes overlap with, and are regulated inversely to, c-myc target genes. Dysregulation of the normal repression of c-myc by the abnormal fusion proteins expressed in t(11;17) APL provides a possible mechanism for increased c-myc activity, which could contribute to the phenotype observed in leukemic cells.
MATERIALS AND METHODS
Cell lines and transfections.
The U937T:PLZF45 inducible PLZF system was previously described (71) and is based on the U937T autoregulatory tet-off system, in which withdrawal of tetracycline leads to gene expression (7a). A control cell line, U937T:Neo1, that does not express PLZF was also used. U937T cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, Calif.), 1 mg of G418 (Cellgro, Herndon, Va.) per ml, 0.5 μg of puromycin (Sigma, St. Louis, Mo.) per ml, and 0.1 μg of tetracycline (Roche, Indianapolis, Ind.) per ml and grown at 37°C with 5% CO2. PLZF expression was induced by washing the cells twice in phosphate-buffered saline (PBS; Invitrogen) and replating them in the medium described above, with the omission of tetracycline. 293T cells were maintained in Dulbecco modified Eagle medium with 10% fetal bovine serum and penicillin-streptomycin (Invitrogen). HEL cells were maintained in RPMI 1640 medium with 10% fetal calf serum and penicillin-streptomycin. U937T cells were transfected by electroporation as follows. Cells (107) were washed once in RPMI 1640 medium without additives, resuspended in 0.4 ml of additive-free RPMI 1640 medium with 20 μg of plasmid DNA in a cuvette with a 0.4-mm gap, and incubated at room temperature for 10 min. Electroporation was carried out at 72 W, 220 V, and 2,800 μF in a BTX 600 electroporator (Genetronics, San Diego, Calif.), and the cells were allowed to recover at room temperature for 10 min and plated into maintenance medium. 293T cells (3 × 105) were plated in 24-well dishes 16 h before transfection with Superfect (Qiagen, Valencia, Calif.). One-half microgram of plasmid DNA was combined with 2.5 μl of Superfect, mixed with 45 μl of additive-free Dulbecco modified Eagle medium (Invitrogen), and incubated at room temperature for 15 min. One-half milliliter of complete maintenance medium was added, and the DNA-Superfect-medium mixture was overlaid onto freshly washed 293T cells. This mixture was removed after 3 h and replaced with maintenance medium.
Plasmids.
The c-myc promoter reporter constructs cmyc2.5 and cmyc0.14 were described previously (35), as were the mutant minimal promoters (20). Expression vectors for PLZF and RARα-PLZF were described previously (42). The c-myc:ER/GFP plasmid was created by removal of the c-myc/ER coding region from c-myc:ER pBabepuro (45; gift of G. Evan, University of California San Francisco) by EcoRI digestion and insertion into MIGR1 cut with EcoRI.
Cell cycle analysis.
Cells were permeabilized and fixed by dropwise addition into ice-cold 70% ethanol, washed in PBS, and incubated in 5 μg of propidium iodide (PI) per ml in PBS with 250 μg of RNase A per ml for 30 min at 37°C. Cells were washed in PBS and analyzed for DNA content with CellQuest software on a FACScalibur system (Becton Dickinson, Franklin Lakes, N.J.).
Apoptosis detection.
One million cells were washed in PBS with 1% bovine serum albumin (BSA) and incubated with 5 μl of fluorescein isothiocyanate-labeled annexin V antibody (Roche) and 10 μl of PI (Sigma) for 30 min at room temperature. After being washed three times in PBS-1% BSA, the cells were analyzed by flow cytometry for annexin V and PI positivity with CellQuest software on a FACScalibur system (Becton Dickinson). Cells that were annexin V positive and PI negative were labeled early apoptosis. Cells that were PI positive were not included in the analysis.
RNA extraction and microarray analysis.
PLZF45 cells (108) and matched Neo1 control cells were collected at 0, 12, 24, and 48 h post tetracycline withdrawal. mRNA was harvested with a Fast-Track mRNA extraction kit (Invitrogen). For each experiment, fluorescent cDNA probes were prepared by reverse transcription of mRNAs with Cy3-dUTP and Cy5-dUTP (NEN Life Sciences, Beverly, Mass.) for PLZF and control samples, respectively. Labeled cDNAs were incubated overnight onto Lymphochip microarrays (2a).Fluorescent images of hybridized microarrays were obtained with a GenePix 4000A microarray scanner (Axon Instruments, Foster City, Calif.). Images were analyzed with GenePixPro3.0 (Axon Instruments), and single spots or areas of the array with obvious blemishes were flagged and excluded from subsequent analyses. Fluorescence ratios were stored in a custom database, and normalized data were extracted from this database for further analysis. Array data were filtered by selecting genes that presented data on at least 75% of the arrays and had a spot diameter of 25 μm and a signal of 200 in each channel or 1,000 in one channel and some baseline signal in the other channel. In many cases, the values shown are averages of several representations of one gene on the arrays. Genes are listed by the names most commonly used in the literature or, when they are ambiguous, based on Human Genome Organization-approved gene symbols. The cDNA clones on the Lymphochip microarray are available from Research Genetics. For the Affymetrix array, 107 cells were withdrawn from tetracycline and three independent samples were taken at 24 and 48 h. RNA was made with the Qiagen RNeasy Kit and biotinylated with an Ambion MessageAmp kit (Ambion, Austin, Tex.) in one round of amplification. Each biotinylated cRNA was hybridized to Affymetrix (Santa Clara, Calif.) TestArray3 chips to verify the quality of the labeled probe and then to a single Affymetrix HG_U95Av2 chip, resulting in a biological triplicate. The 0-h specimen was made into RNA and labeled once. It was then hybridized to three HG_U95Av2 chips as an experimental triplicate. We used the Silicon Genetics (Redwood City, Calif.) Cross-Gene error model based on replicates. The values for each time point were normalized such that values below 0 were set to 0. Each measurement was divided by the 50.0th percentile of all measurements in that sample. Specific samples were normalized to one another such that samples 1 to 9 were normalized against the median of control samples 1 to 3. Each measurement for each gene in those specific samples was divided by the median of that gene's measurements in the corresponding control samples.
Northern blot analysis.
mRNA was isolated from PLZF45 and Neo1 cells induced by withdrawal of tetracycline for the corresponding time points, and 20 μg of each mRNA was electrophoresed in a 1% denaturing formaldehyde gel. RNA was transferred to Hybond N membrane (Amersham, Piscataway, N.J.). After fixation and prehybridization, the membrane was incubated overnight with a 414-bp PstI fragment of the pCGN-myc plasmid (kindly provided by Z. Ronai, Mount Sinai School of Medicine) labeled with [α-32P]dCTP with the RediPrime II kit and protocol (Amersham). The blot was then washed three times for 15 min each time at 42°C in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) and once for 30 min at 65°C in 0.2× SSC-0.1% SDS. Signal was detected by phosphorimager (Amersham). The blot was stripped and reprobed with a human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) restriction fragment by the same protocol.
Western blotting.
Cells were lysed in 1% Triton X-100-140 mM NaCl-10 mM Tris (pH 8), and the lysates were denatured by being boiled in an equal volume of 2× SDS buffer (4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.25 M Tris [pH 6.9], 0.01% bromophenol blue). Proteins were separated by SDS-10% polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membrane (Millipore, Bedford, Mass.) in 25 mM Tris-192 mM glycine buffer. The membrane was blocked in PBS-5% skim milk powder overnight. Incubation of the membrane with the primary antibody was carried out at room temperature for 1 h in PBS-0.5% skim milk, membranes was washed three times for 5 min each time in PBS, and the appropriate horseradish peroxidase-conjugated secondary antibody was added to PBS at the concentration recommended by the manufacturer (Chemicon, Temecula, Calif.). The horseradish peroxidase conjugate was detected by chemiluminescence with an ECL kit (Amersham) and autofluorography.
Quantitative real-time PCR.
RNA was extracted either from 1 × 106 PLZF cells at different times after tetracycline withdrawal or from 1 × 105 to 5 × 105 green fluorescent protein (GFP)-positive cells purified by flow cytometry, and cDNA was produced with oligo(dT) primers and Superscript II reverse transcriptase (Invitrogen). PCR was carried out with either an Applied Biosystems 7700 Prism real-time PCR machine and the manufacturer's SYBR green kit and directions (Applied Biosystems, Foster City, Calif.) or with an Opticon DNA Engine (MJ Research) and a SYBR green master mix kit (Qiagen). The sequences of the primers used are listed in Table 1. The threshold cycle (Ct) value for the “+tet” sample was taken as baseline expression, and ΔCt, the difference between the +tet Ct and the Ct obtained after manipulation, was calculated for each PCR. A positive ΔCt value represented an increase, and a negative ΔCt value represented a decrease, over the baseline. The ΔCt for each transcript was expressed relative to the ΔCt for GAPDH in each induction.
TABLE 1.
Sequences of primers used in this study
| Primer | Target | Sequence (5′-3′) | Use |
|---|---|---|---|
| GAPDH ex5F | GAPDH coding region | CCAAAATCAAGTGGGGCGATG | Real-time PCR |
| GAPDH ex8R | GAPDH coding region | AAAGGTGGAGGAGTGGGTGTCG | Real-time PCR |
| PLZF F1 | PLZF coding region | CACTTACTGGCTCATTCAGCGG | Real-time PCR |
| PLZF R1 | PLZF coding region | CTTACACTCAAAGGGCTTCTCACC | Real-time PCR |
| hTERTF1 | hTERT coding region | ATCAGAGCCAGTCTCACCTTCAAC | Real-time PCR |
| hTERT | hTERT coding region | TTTCAGGATGGAGTAGCAGAGGG | Real-time PCR |
| c-mycF2 | c-myc coding region | TCGGATTCTCTGCTCTCCTCG | Real-time PCR |
| c-mycR2 | c-myc coding region | CTGCGTAGTTGTGCTGATGTGTG | Real-time PCR |
| Site1F | c-myc promoter | TTGGAAGCTACTATATTCACTTAACACTTG | EMSA |
| Site1R | c-myc promoter | TGCAGCTCAGCGTTCAAGTGTTAAGTGAATATA | EMSA |
| Site2F | c-myc promoter | ACGTTTGCGGGTTACATACAGTGCACTTTC | EMSA |
| Site2R | c-myc promoter | CTGAATACTAGTGAAAGTGCACTGTATGTA | EMSA |
| Site3F | c-myc promoter | TGGGGGTGGGACGGTGGGGTACAGACTGGCAGA | EMSA |
| Site3R | c-myc promoter | AGGTTGCCTGCTCTCTGCCAGTCTGTACCCCAC | EMSA |
| Site4F | c-myc promoter | ATGCGGTTTGTCAAACAGTACTGCTACGGA | EMSA |
| Site4R | c-myc promoter | CTCTGCTGCTCCTCCGTAGCAGTACTGTTT | EMSA |
| Site5F | c-myc promoter | CGGGGCTTTATCTAACTCGCTGTAGTAA | EMSA |
| Site5R | c-myc promoter | TCGCTGGAATTACTACAGCGAGTTAGATAA | EMSA |
| Site2mut4F | c-myc promoter | ACGTTTGCGGGTTACATATAGTGCACTTTC | EMSA |
| Site2mut4R | c-myc promoter | CTGAATACTAGTGAAAGTGCACTATATGTA | EMSA |
| c-myc5′2F | c-myc promoter | AATGCCTTTGGGTGAGGGAC | ChIP |
| c-myc5′2R | c-myc promoter | TCCGTGCCTTTTTTTGGGG | ChIP |
| bcl-6 F1 | Bcl-6 coding region | CGATGAGGAGTTTCGGGATGTC | ChIP |
| bcl-6 B1 | Bcl-6 coding region | TTTCTGGGGGCTCTGTGGACTAAC | ChIP |
| CycA F2 | Cyclin A2 promoter | GTGCCCCAGATTTTAGACC | ChIP |
| CycA B2 | Cyclin A2 promoter | CAAAGACGCCCAGAGATG | ChIP |
Electrophoretic mobility shift analysis.
Complementary oligonucleotide probes were annealed, the overhanging ends of the double-stranded oligonucleotides were extended with Klenow and [α-32P]dCTP, and then the oligonucleotide probes were purified by spin chromatography. Ten femtomoles of probe, corresponding to 50,000 cpm, was used in each binding reaction. Binding reactions were carried out in a volume of 10 μl. The probe was mixed with 3 μg of PLZF45 nuclear extract in 20 mM HEPES (pH 7.5)-1 mM MgCl2-10 μM ZnCl2-4% glycerol-100 μg of BSA per ml-50 ng sonicated salmon sperm DNA. Reaction mixtures were incubated on ice for 20 min, and then, depending on the experiment, either antibody or unlabeled double-stranded oligonucleotides were added and the mixture was incubated for a further 20 min. DNA loading dye was added, and samples were electrophoretically separated through a 0.5× Tris-borate-EDTA-nondenaturing polyacrylamide gel before autoradiography. The sequences of the probes used are listed in Table 1.
ChIP.
The antibodies used for chromatin immunoprecipitation (ChIP) were against PLZF (Calbiochem, San Diego, Calif.), FLAG M2 (Sigma), the large subunit of RNA polymerase II (Santa Cruz, Calif.), and normal rabbit immunoglobulin G (Zymed, San Francisco, Calif.). For each immunoprecipitation, 107 cells were fixed in 1% formaldehyde at 37°C for 10 min and quenched in 0.125 M glycine for 5 min at room temperature. Cells were washed twice in PBS containing Complete protease inhibitor (Roche) and lysed in 1.0 ml of lysis buffer (140 mM NaCl, 10 mM Tris [pH 8], 1% NP-40). Lysates were sonicated to break DNA into fragments of less than 1 kb and centrifuged at 14,000 rpm in an Eppendorf microcentrifuge for 10 min. Lysis buffer was added to a final volume of 1.5 ml, and supernatants were precleared for 45 to 60 min with protein A-agarose beads and 0.4 μg of salmon sperm DNA (Upstate Biotechnology, Lake Placid, N.Y.) per μl. After brief centrifugation, supernatant was removed and incubated with 1 μg of the precipitating antibody overnight at 4°C. Protein A-salmon sperm DNA was added, and the immune complex was collected for 1 h at 4°C. Complexes were washed for 5 min each in low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 150 mM NaCl), high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 500 mM NaCl), LiCl buffer (0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl [pH 8.1]) and then washed twice in Tris-EDTA. DNA was eluted twice for 15 min each time in 250 μl of 1% SDS-0.1 M NaHCO3, and the two eluates were combined. A 20-μl volume of 5 M NaCl was added, and eluates were incubated at 65°C overnight to reverse the cross-links. DNA was recovered by phenol-chloroform extraction and ethanol precipitation and then used in a PCR. The sequences of the primers used for the PCR are given in Table 1.
Reporter assays.
Reporter and effector plasmids were used in a 1:4 ratio, with 10 ng of Renilla luciferase included as an internal control for every microgram of plasmid DNA. 293T cells were transfected with Superfect (Qiagen) as described above. Transfected cells were harvested at 42 to 45 h posttransfection, and lysates were assayed for luciferase activity with a Dual Luciferase kit (Promega, Madison, Wis.) as recommended by the manufacturer. Raw values obtained from each experimental, performed in triplicate, were normalized to the Renilla value for each replicate. The standard error of the mean for each triplicate represents the error for each experiment.
BrdU labeling and GFP detection.
Electroporated cells were labeled by addition of 10 mM bromodeoxyuridine (BrdU) to the culture medium and incubation for 30 min. Cells were washed in PBS-1%BSA and fixed in 2% paraformaldehyde for 30 min at 4°C. After being washed in PBS-1% BSA, cells were permeabilized in 0.1% Triton X-100-PBS on ice for 2 min. After being washed in PBS-1% BSA, cells were resuspended in 1 ml of PBS (with CaCl2 and MgCl2) and 50 U of DNase I and incubated for 30 min at 37°C. Cells were resuspended in 100 μl of PBS, and the BrdU was labeled with 5 μl of Phoenix-Red-conjugated anti-BrdU monoclonal antibody (Phoenix Flow Systems, San Diego, Calif.). GFP positivity was measured by flow cytometry. GFP-positive cells were gated, and the proportion of BrdU-positive cells within the GFP-positive population was assessed.
RESULTS
PLZF expression induces cell cycle arrest and apoptosis.
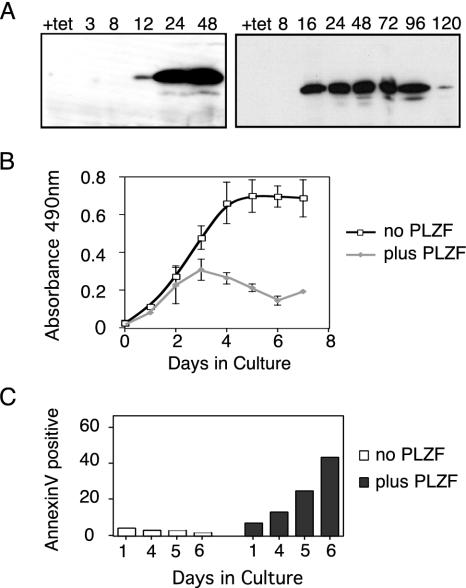
Constitutive expression of the transcriptional repressor PLZF results in rapid cell death, so we used a tetracycline-regulated U937 cell line to examine the effect of PLZF expression on cell growth and survival. Medium containing tetracycline prevented the expression of PLZF, while 12 h after withdrawal of tetracycline, PLZF protein could be detected by Western blotting (Fig. 1A). Expression was sustained until the point at which cellular proliferation (Fig. 1B) decreased significantly, approximately 96 to 120 h after tetracycline withdrawal. No distinguishable differences were observed in proliferation between parental U937 cells, a neomycin-resistant cell line that did not express PLZF (Neo1), and the PLZF45 line grown in the presence of tetracycline (data not shown). The effect of PLZF on cell cycle progression was also examined (Table 2). Up to 48 h after tetracycline withdrawal, there was no significant change in the proportion of cells in each phase of the cycle. However, at 48 h, there was a 50% decrease in the number of cells in G2/M and a simultaneous increase in the numbers of cells in both the G1 and sub-G1 populations. The accumulation of cells in G1 continued with further PLZF expression and was accompanied by an increasing sub-G1 apoptotic population. Five days after tetracycline withdrawal, more than 60% of the cells were sub-G1 and apoptotic, correlating with the block in proliferation. The apoptotic phenotype was confirmed by annexin V staining, by which early apoptosis was identified by annexin V positivity and exclusion of PI (Fig. 1C). PLZF45 cells grown in tetracycline had a basal level of apoptosis of approximately 5%, while expression of PLZF for 4 days induced annexin V positivity above the background. By day 6 postinduction, the percentage of cells in early apoptosis had reached 40%, consistent with a sub-G1 population of greater than 60%.
FIG. 1.
PLZF expression in U937T cells induces cell death. (A) At the indicated hour post tetracycline withdrawal, 106 U937T:PLZF45 cells were collected and blotted for PLZF expression with monoclonal antibody 2A9. (B) PLZF45 cells without or with PLZF expression were seeded at 5 × 104/ml, and 100 μl of each culture was assayed in triplicate with a CellTitre 96 Aq kit (Promega) at the time points indicated. The data shown are averages of three independent experiments, and the error bars represent standard deviations. (C) Induction of apoptosis was measured by annexin V positivity. Cells were grown in tetracycline-containing (no PLZF) or tetracycline-free (plus PLZF) medium for 6 days. At the time points indicated, 106 cells were withdrawn from each culture, incubated with fluorescein isothiocyanate-conjugated annexin V antibody and PI, and then analyzed by flow cytometry. The cells were separated by annexin V labeling, and the annexin V-positive, PI-negative population was quantified.
TABLE 2.
Effect of PLZF on cell cycle progression
| Condition | % of cells
|
|||
|---|---|---|---|---|
| G1 | S | G2/M | Sub-G1 | |
| No PLZF | 53 | 32 | 15 | 0 |
| 24 h of PLZF | 48 | 30 | 22 | 0 |
| 48 h of PLZF | 64 | 27 | 9 | 2 |
| 72 h of PLZF | 75 | 18 | 6 | 20 |
| 120 h of PLZF | 76 | 18 | 5 | 63 |
PLZF expression suppressed c-myc expression.
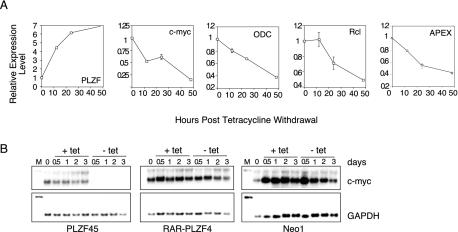
In order to understand what role PLZF played in G1 arrest and the subsequent apoptosis we observed, we used the inducible PLZF expression system to query Lymphochip, a collection of hematopoietic derived cDNA sequences arrayed on glass slides (2). This microarray was chosen to look primarily at the hematopoietic aspects of PLZF function. To this end, mRNA was collected from PLZF45 and Neo1 control cells at the time of induction and at 12, 24, and 48 h after induction. For samples at each time point, the PLZF45 mRNA was labeled with Cy3-dUTP, the Neo1 mRNA was labeled with Cy5-dUTP. Each matched mRNA set was hybridized to Lymphochip, and the relative expression profile was determined. We set a criterion of a 2.5-fold or greater change in gene expression, either induced or repressed, after PLZF induction. PLZF was represented on the array, and the increase in PLZF transcript at 12 h post tetracycline withdrawal was determined to be 4.5-fold, increasing to 7-fold at 48 h (Fig. 2A, part 1 [from the left]). Of the approximately 6,000 sequences represented on the array, there were 75 genes whose expression was significantly altered by PLZF expression, either positively or negatively. We further examined the subset with a significant change at 12 h, under the assumption that the direct changes would be the earliest to occur. Of this group, the most striking member was c-myc, whose expression had already decreased by 50% by 12 h after PLZF induction. By 48 h of PLZF expression, c-myc expression had decreased to 15% of the signal from the control cell line (Fig. 2A, part 2). Down-regulation of myc was accompanied by a decrease in several genes known to be up-regulated by myc, i.e., those for ODC, rcl, and APEX (parts 3 to 5).
FIG. 2.
PLZF suppresses expression of c-myc. PLZF45 and Neo1 cells were withdrawn from tetracycline, and mRNA was harvested at 12, 24, and 48 h, labeled with Cy3 or Cy5, and hybridized against Lymphochip. (A) Expression data for PLZF, c-myc, and c-myc target genes from the Lymphochip microarray analysis. Change in gene expression is taken from the average signal of at least three hybridization spots of the cDNA on the array and is expressed relative to the level at the zero time point for each message. (B) Total RNA was purified at the time points indicated either before or after tetracycline withdrawal and used in a Northern blot assay. c-myc and GAPDH expression in PLZF45 (left), RARα-PLZF (center), and Neo1 (right) cells was assessed in each case. M, marker. The values at the top are numbers of days in culture.
In order to examine this phenomenon more closely, we used the c-myc target gene database (http://www.myc-cancer-gene.org/site/mycTargetDB.asp) to query the PLZF expression profiles generated from the Lymphochip analysis. This analysis showed that a substantial proportion (20 [∼27%] of 75) of the genes significantly regulated in response to PLZF had been previously described as responsive to c-myc (Table 3). All of the genes repressed by PLZF were scored as activated by c-myc, while the genes activated in response to PLZF were repressed by c-myc, with only one exception. This reinforced the identification of c-myc as a PLZF target gene and suggested that regulation of c-myc may be a major axis of PLZF function.
TABLE 3.
c-myc target genesa versus the PLZF Lymphochip expression profile
| Gene | LocusLink IDc no. | PLZF action | c-myc actionb |
|---|---|---|---|
| NCL | 4691 | Down | Up |
| TERT | 7015 | Down | Up |
| AKAP1 | 8165 | Down | Up |
| FOSL1 | 8061 | Down | Up |
| C1qBP | 708 | Down | Up |
| ODC1 | 4953 | Down | Up |
| APEX | 328 | Down | Up |
| B4GALT7 | 11285 | Down | Up |
| FABP5 | 2171 | Down | Up |
| NME1 | 4830 | Down | Up |
| CDK4 | 1019 | Down | Up |
| PHB | 5245 | Down | Up |
| FBL | 2091 | Down | Up |
| HRMT1L2 | 3276 | Down | Up |
| PAICS | 10606 | Down | Up |
| DUSP6 | 1848 | Up | Down |
| KIAA0053 | 9938 | Up | Down |
| ITGAL | 3683 | Up | Down |
| CCND3 | 896 | Up | Up |
| CDKN1A | 1026 | Up | Down |
c-myc-responsive genes that were altered 2.5-fold or more by PLZF.
ID, identification.
PLZF is expressed throughout development and in adult nonhematopoietic tissues (4, 14, 59). To extend the correlation between PLZF and c-myc-responsive genes beyond hematopoiesis-related genes, we interrogated a different set of genes represented on the Affymetrix U95A_v2 chip. RNA was extracted in triplicate at 24 and 48 h and labeled with biotinylated UTP and CTP. PLZF transcript was induced 40-fold in this system. Confirming the Lymphochip analysis, we saw a profound reduction in c-myc expression, from 100% to 14%, at 24 h after PLZF induction. As described above, we used the c-myc target gene database to query the PLZF gene profiles. There were 244 genes altered by PLZF 2.5-fold or more in either direction. Of these, 52 (21%) were c-myc target genes. Of these 52 c-myc target genes, 45 (∼87%) were regulated concordantly with our hypothesis that PLZF results in reversal of c-myc action by reduction of c-myc transcription (Table 4). As in the Lymphochip analysis, genes repressed by PLZF almost completely correlated, with 29 (∼97%) of the 30 genes repressed by PLZF known to be activated by c-myc, while 16 (∼73%) of the 22 genes activated in response to PLZF were repressed by c-myc. Eight of the 20 c-myc target genes from the PLZF Lymphochip array were present in the c-myc target list from the PLZF Affymetrix array (those that encode FABP5, NME1, PHB, PAICS, DUSP6, KIAA0053, CCND3, and CDKN1A), and all of these were regulated in the same pattern in both systems.
TABLE 4.
c-myc target genesa versus the PLZF Affymetrix expression profile
| Gene name | LocusLink IDd no. | Change (fold) at:
|
PLZF action | c-myc actionc | |
|---|---|---|---|---|---|
| 24 h | 48 h | ||||
| PLZF | 7704 | 40.9 | 25.9 | NAb | NA |
| MYC | 4609 | 0.14 | 0.24 | NA | NA |
| HSPA9B | 3313 | 0.53 | 0.37 | Down | Up |
| RANBP1 | 5902 | 0.44 | 0.26 | Down | Up |
| ASS | 445 | 0.48 | 0.61 | Down | Up |
| DDX21 | 9188 | 0.57 | 0.45 | Down | Up |
| VARS1 | 7406 | 0.50 | 0.36 | Down | Up |
| FABP5 | 2171 | 0.61 | 0.39 | Down | Up |
| ABCE1 | 6059 | 0.49 | 0.34 | Down | Up |
| HSPE1 | 3336 | 0.55 | 0.37 | Down | Up |
| NME1 | 4830 | 0.61 | 0.31 | Down | Up |
| PAICS | 10606 | 0.57 | 0.41 | Down | Up |
| PTMA | 5757 | 0.49 | 0.48 | Down | Up |
| CHC1 | 1104 | 0.56 | 0.41 | Down | Up |
| METAP1 | 23173 | 0.64 | 0.45 | Down | Up |
| GCSH | 2653 | 0.32 | 0.25 | Down | Up |
| RRS1 | 23212 | 0.49 | 0.35 | Down | Up |
| NOLC1 | 9221 | 0.49 | 0.36 | Down | Up |
| RUVBL2 | 10856 | 0.51 | 0.48 | Down | Up |
| BOP1 | 23246 | 0.43 | 0.35 | Down | Up |
| NOL5A | 10528 | 0.54 | 0.34 | Down | Up |
| KIAA0664 | 23277 | 0.40 | 0.27 | Down | Up |
| HNRPH1 | 3187 | 0.69 | 0.33 | Down | Down/Up |
| LOC56902 | 56902 | 0.46 | 0.41 | Down | Up |
| RPL5 | 6125 | 0.41 | 0.21 | Down | Up |
| KIAA0116 | 23016 | 0.47 | 0.68 | Down | Up |
| MAC30 | 27346 | 0.49 | 0.54 | Down | Up |
| CTSB | 1508 | 0.76 | 0.49 | Down | Down |
| KIAA0179 | 23076 | 0.44 | 0.35 | Down | Up |
| PHB | 5245 | 0.52 | 0.34 | Down | Up |
| RAB40B | 10966 | 0.63 | 0.44 | Down | Up |
| TRAP1 | 16131 | 0.43 | 0.34 | Down | Up |
| IFIT1 | 3434 | 0.85 | 2.11 | Up | Down |
| E2-EPF | 27338 | 2.40 | 2.64 | Up | Up |
| IFI35 | 3430 | 1.96 | 2.76 | Up | Down |
| NCAM1 | 4684 | 1.71 | 2.38 | Up | Down |
| HLA-A | 3105 | 0.85 | 2.30 | Up | Down |
| DUSP6 | 1848 | 1.50 | 2.11 | Up | Down |
| NUCB1 | 4924 | 0.95 | 2.19 | Up | Down |
| CCND1 | 595 | 2.28 | 2.60 | Up | Down |
| OAS1 | 4938 | 3.30 | 4.14 | Up | Down |
| KIAA0053 | 9938 | 1.59 | 2.27 | Up | Down |
| CD48 | 962 | 1.15 | 2.52 | Up | Down |
| IRF7 | 3665 | 2.07 | 2.22 | Up | Down |
| LRMP | 4033 | 1.13 | 2.10 | Up | Down |
| GGTLA4 | 92086 | 1.82 | 2.33 | Up | Down |
| AIM1 | 202 | 1.78 | 3.17 | Up | Up |
| BAX | 581 | 1.53 | 2.54 | Up | Up |
| CDKN1A | 1026 | 3.68 | 5.51 | Up | Down |
| CCNG2 | 901 | 1.00 | 2.44 | Up | Down |
| CDC2 | 983 | 1.26 | 3.14 | Up | Up |
| CCND3 | 896 | 3.04 | 4.30 | Up | Up |
| IER3 | 8870 | 3.44 | 2.89 | Up | Down |
| YWHAE | 7531 | 1.75 | 2.29 | Up | Up |
c-myc-responsive genes that were altered 2.5-fold or more by PLZF.
NA, not applicable.
ID, identification.
The reduction in the c-myc transcript level upon PLZF expression was confirmed by Northern blot analysis (Fig. 2B). As seen in both microarray experiments, within 12 h of PLZF induction, c-myc expression was substantially reduced; by 24 h, it was below the level of detection by Northern blot assay. In contrast, the APL-associated fusion protein RARα-PLZF did not reduce c-myc transcript levels, consistent with the loss of the PLZF repression domain in this fusion protein. The decrease in c-myc transcript levels was also not seen in the control cell line Neo1.
PLZF physically interacts with the c-myc promoter.
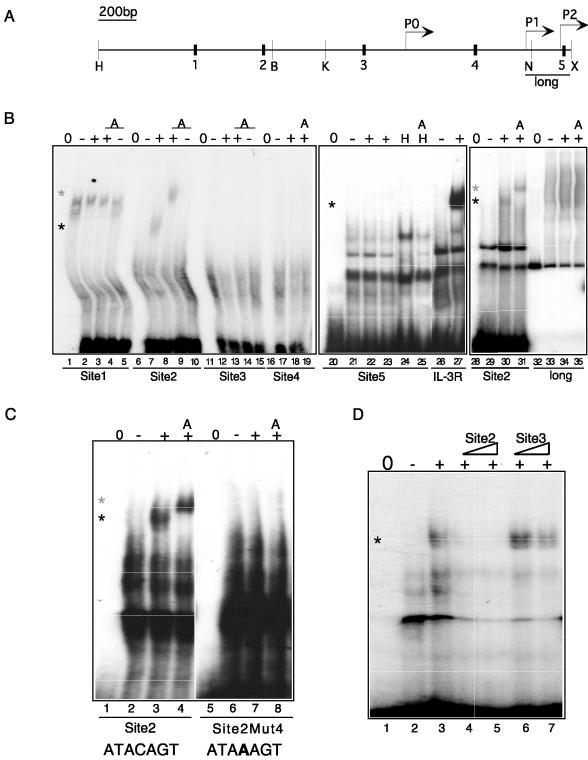
We next demonstrated direct regulation of the c-myc gene by PLZF. PLZF can recruit transcriptional repression machinery to specific DNA sequences (18, 43); therefore, we searched the c-myc promoter for putative PLZF binding sites. On the basis of information from our prior binding site selection experiments (3, 40), several potential PLZF DNA recognition sequences are located within a 2.5-kb region upstream of the major P2 promoter (Fig. 3A). Each one of these was examined for the ability to interact with PLZF in vitro. Electrophoretic mobility shift assays (EMSAs) were carried out with nuclear extracts made from PLZF45 cells grown in the presence of tetracycline (−PLZF) or after 24 h of tetracycline withdrawal (+PLZF). The same results were seen with PLZF from transfected 293T cells and HEL cells expressing endogenous PLZF (data not shown). The PLZF binding site identified in the IL-3Rα promoter (3) or Site2 (present study) was used as a positive control for PLZF binding. As shown in Fig. 3B, Site1 was bound by some factor in the PLZF45 extracts, forming a complex (lane 2) that changed upon induction of PLZF (lane 3). However, the complex itself did not contain PLZF, as demonstrated by the lack of supershifting upon addition of PLZF monoclonal antibody (lane 4). Site2, located 1.6 kb upstream from the P2 major transcript start site of c-myc, specifically bound PLZF, as demonstrated by a clear band shift (lane 8, black asterisk) and a supershift with PLZF monoclonal antibody 2A9 (lane 9, grey asterisk). Neither Site3, Site4, nor Site5 (lanes 11 through 25) bound PLZF. A larger probe spanning 240 bp of the proximal promoter region (including Site5) also showed no interaction with PLZF (Fig. 3B, lanes 32 through 35), nor did two smaller restriction fragments of these promoter sequences (data not shown). Thus, there appears to be a single site for direct interaction with PLZF in the c-myc upstream regulatory region. To confirm the specificity of the interaction between Site2 and PLZF, we carried out a mutational analysis. The fourth base in the PLZF response element in Site2 was changed from C to A, completely abolishing the PLZF interaction (Fig. 3C). The specificity of the PLZF interaction was further confirmed by competition assays with increasing doses of unlabeled Site2 (interacting) and Site3 (noninteracting) oligonucleotides (Fig. 3D).
FIG. 3.
PLZF interacts specifically with a sequence from the c-myc regulatory region. (A) Schematic of the c-myc promoter showing the location of EMSA probes Site1 (black box 1) through Site5 (black box 5) and the EMSA probes created by PCR and restriction digestion. H, HindIII site; B, BglI site; K, KpnI site; N, NotI site; X, XhoI site; long, full-length PCR probe. Locations of the P0, P1, and P2 promoters are indicated by arrows. Scale bar, 200 bases. (B) Nuclear extracts were used in an EMSA on Site1 through Site5 and the probes spanning the proximal promoter region, long. The extracts used are indicated as follows: 0, probe only; −, PLZF45 nuclear extract with no PLZF; +, PLZF45 nuclear extract with PLZF; H, HEL cell nuclear extract; A, anti-PLZF monoclonal antibody added. IL-3R, PLZF binding site from the IL-3Rα promoter. (C) The fourth base of the PLZF consensus site in Site2 was mutated from C to A (Site2Mut4) and used in EMSAs with nuclear extracts from PLZF45 cells with and without PLZF expression. 0, probe only; −, PLZF45 nuclear extract with no PLZF; +, PLZF45 nuclear extract with PLZF; A, anti-PLZF monoclonal antibody added. (D) The interaction between PLZF and Site2 (lane 3) was competed with increasing concentrations of unlabeled Site2 probe (lanes 4 and 5) but not by unlabeled noninteracting probe Site3 (lanes 6 and 7). Black asterisk, PLZF shift; grey asterisk, PLZF supershift.
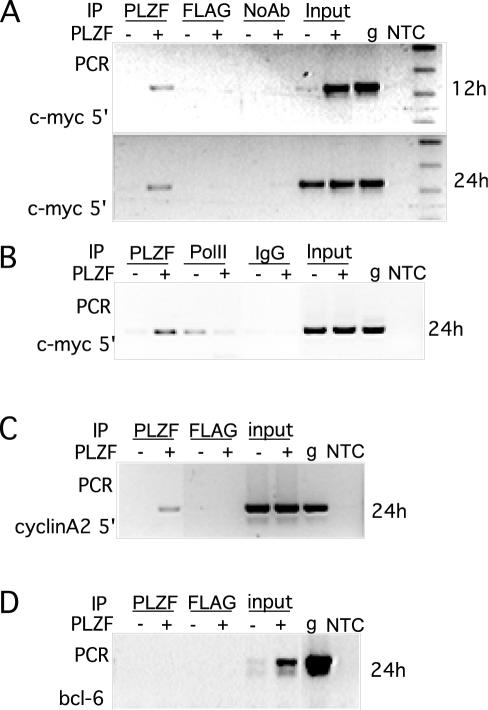
We next determined whether PLZF was recruited to the endogenous c-myc promoter coincident with the induction of transcriptional repression. Chromatin was immunoprecipitated from cells with no PLZF expression, or from cells at different time points after withdrawal of tetracycline, with either anti-PLZF monoclonal antibody 2A9 or the unrelated FLAG M2 monoclonal antibody. The precipitated chromatin was queried by PCR with primers to a region of the c-myc promoter. Directly correlating with the microarray and Northern data, PLZF could be found on the c-myc promoter as early as 12 h after withdrawal of tetracycline (Fig. 4A). The rest of the ChIP experiments were carried out at 24 h postinduction, when PLZF expression was high and the c-myc transcript level was clearly decreased. To determine the point at which PLZF interrupted c-myc expression, ChIP was performed with an antibody to the RNA polymerase II large subunit. Consistent with the transcript decrease associated with PLZF expression, the amount of RNA polymerase II associated with the c-myc promoter also decreased with PLZF expression, demonstrating that the effect of PLZF on the c-myc mRNA level was at the level of initiation of c-myc transcription (Fig. 4B). As a positive control, we examined the promoter of the other gene described as a direct target for PLZF, that which encodes cyclin A2 (75), and found that PLZF associated with this gene in vivo (Fig. 4C). We also examined the gene for Bcl6, which was not regulated in response to PLZF in the array analysis (the relative Bcl6 gene expression level at 24 h post tetracycline withdrawal was 1.04). PLZF antibody did not precipitate Bcl-6 chromatin (Fig. 4D), demonstrating the specificity of the interaction of PLZF with regulated sequences.
FIG. 4.
ChIP of PLZF on the c-myc promoter. (A) Lysates were made 12 and 24 h after tetracycline withdrawal as indicated, and chromatin was immunoprecipitated with antibodies to PLZF and FLAG M2 as a nonspecific control or no antibody. This was then purified and used as a template for PCR. PCR was carried out with primers to the c-myc 5′ regulatory region. −, no PLZF; +, PLZF expressing; PLZF, anti-PLZF monoclonal antibody; FLAG, anti-FLAG M2 epitope antibody; noAb, no antibody added; input, 10% of the immunoprecipitation input; g, genomic DNA; NTC, no-template control PCR. (B) Lysates from cells induced for 24 h were immunoprecipitated (IP) with either an anti-PLZF monoclonal antibody (PLZF), the RNA polymerase II large subunit (PolII), or preimmune rabbit immunoglobulin G (IgG), and the chromatin was used for PCR of the c-myc 5′ regulatory region. (C) Lysates from a 24-h induction were immunoprecipitated with either the anti-PLZF monoclonal antibody (PLZF) or the FLAG control, and the purified chromatin was queried with primers to the cyclin A2 promoter. (D) Lysates from a 24-h induction were immunoprecipitated with either anti-PLZF monoclonal antibody 2A9 or a FLAG control, and the purified chromatin was queried with primers to the bcl-6 promoter. The sequences of all of the primers used are listed in Table 1.
PLZF repressed the c-myc promoter in vitro.
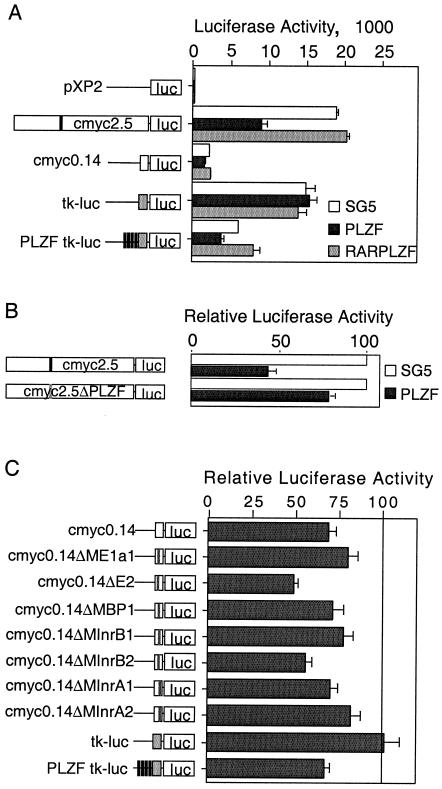
To extend these findings and to understand the molecular basis of PLZF-mediated repression of the c-myc promoter, we used a 2.5-kb fragment of the c-myc promoter, including the in vitro binding site identified in Fig. 3 (cmyc2.5), in a reporter assay. The data presented here are from 293T cells; similar results (not shown) were obtained with inducible PLZF45 cells. Consistent with changes to the endogenous transcript, expression of pSG5-PLZF suppressed the luciferase activity of cmyc2.5 by an average of 70% (Fig. 5A). RARα-PLZF, which contains the DNA binding domain of PLZF fused to the activation domain of RARα, did not suppress promoter activity. Instead, it activated it slightly (if not significantly), as has been previously observed for this fusion protein (40). This is consistent both with the effect of RARα-PLZF on the endogenous myc transcript (Fig. 2B) and with the lack of PLZF repression domains in this fusion protein. Together with the ChIP data from Fig. 4, this strongly supported the hypothesis that the c-myc upstream regulatory region contains sequences specifying PLZF recruitment, leading to transcriptional repression. We compared PLZF-mediated repression of cmyc2.5 to that of the P2 immediate proximal promoter (cmyc0.14), a region shown not to bind PLZF in vitro. This was only minimally repressed, consistent with the absence of PLZF binding. The same pattern was observed in the inducible PLZF45 cell line, where cmyc2.5 was repressed by PLZF greater than 50% and cmyc0.14 was repressed to a much lesser extent (data not shown).
FIG. 5.
PLZF represses a c-myc promoter reporter construct. 293T cells were seeded into 24-well dishes and transfected with 100 ng of reporter plasmid and 400 ng of effector plasmid. All luciferase activity is expressed relative to the expression of an internal Renilla luciferase promoter, and each bar represents the combined data from at least three experiments, each in triplicate. (A) The promoter constructs shown on the left, empty pXP2, two fragments of the c-myc promoter cloned into pXP2—full length (cmyc2.5) and 142 bp (cmyc0.14)—a minimal tk promoter (tkluc), or four PLZF binding sites upstream of the minimal tk promoter (PLZF-tkluc), were transfected with either empty expression vector SG5 (white bar), an SG5 vector expressing PLZF (black bar), or RARα-PLZF (grey bar). The white boxes represent the c-myc promoter fragments, the grey boxes represent the tk promoter, and the black bars represent PLZF binding sites. The effect of each plasmid on the activity of the promoter fragments is shown on the right. (B) A mutation in the PLZF binding site (black bar) was introduced into the 2.5-kb c-myc construct to create cmyc2.5ΔPLZF (grey bar). The effect of the mutation on PLZF-mediated repression is shown. Repression is expressed relative to the level of activity with the empty SG5 vector, which was normalized to 100%. (C) The effect of PLZF on the series of minimal promoter mutant constructs depicted was expressed relative to the activity of the construct in the presence of the empty vector SG5. White box, c-myc minimal promoter; grey bar, mutation. The grey box represents the tk promoter, and the black bars represent PLZF binding sites.
To assess the contribution of the binding site identified by EMSA to PLZF-mediated repression of c-myc, the site was mutated in a single position critical for in vitro binding (Fig. 3C). This reduced PLZF repression from 65% on wild-type cmyc2.5 to approximately 25% on cmyc2.5ΔPLZF (Fig. 5B). This demonstrated that the binding site identified by EMSA was responsible for most of the PLZF-induced repression. The residual 25% repression was the same as the PLZF repression of cmyc0.14, suggesting that PLZF might also interact with the immediate proximal region. C/EBPα represses cmyc0.14 indirectly via interaction with E2F (35), so we examined the eight transcription factor binding sites within the proximal region (20). One of these mutations (hMAZ) reduced the basal activity so much that repression could not be assessed. Of the remaining sites, none of the mutations completely blocked PLZF-mediated repression (Fig. 5C), indicating that PLZF, unlike C/EBPα, does not interact with any single factor to prevent activation of the c-myc promoter.
The data presented thus far indicate that PLZF mediates c-myc repression through direct interaction with a single binding site in the c-myc regulatory region, resulting in a decrease in the initiation of transcription.
PLZF repression of c-myc is rapidly reversible.
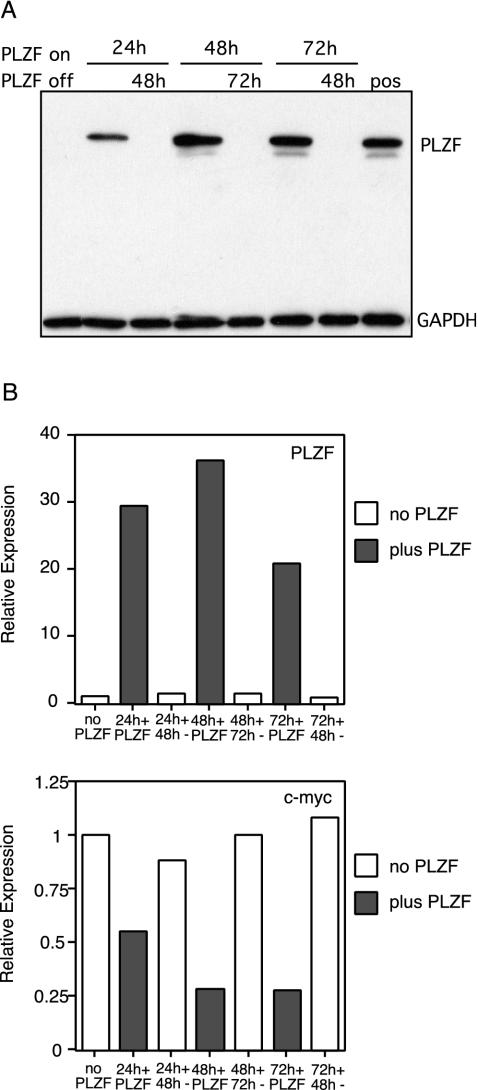
It was recently reported that expression of the APL fusion protein PML-RARα led to silencing of a target gene associated with hypermethylation of the promoter region (19). Given this, we asked if PLZF-mediated silencing would have a similar epigenetic effect on c-myc expression. The tet-VP16 activator was deactivated by addition of tetracycline back into the culture medium at various time-points, extinguishing PLZF expression. PLZF protein levels were completely undetectable 48 h after readdition of tetracycline, regardless of the length of the previous induction of PLZF expression (Fig. 6A). This was considered the point at which cells were negative for PLZF expression in subsequent experiments. PLZF, c-myc, and GAPDH transcript levels were analyzed by real-time PCR after PLZF induction and removal. Each PCR product was quantified by determination of the Ct value, and the change in PLZF and c-myc levels (ΔCt) was determined relative to the change in GAPDH (Fig. 6B). Amplification by each set of primers was close to 100% efficient (data not shown), so we used x = 2ΔCt to calculate the template concentration. The changes in PLZF mRNA mirrored the changes in the PLZF protein level. Withdrawal of tetracycline increased the PLZF transcript level approximately 30-fold, and replacement of tetracycline brought the message level back to the baseline. As observed by other methods, induction of PLZF led to a significant decrease in c-myc mRNA. Upon removal of PLZF, the c-myc transcript level rose back to the baseline regardless of how long PLZF had been expressed. Even after 72 h of PLZF expression, the point after which viability begins to decline and apoptosis becomes significant, c-myc was reexpressed by tetracycline addition, cells recovered from G1 arrest and resumed normal proliferation (data not shown). This demonstrated that the effect of PLZF on c-myc was both transient and tightly regulated and implies that no long-term silencing of c-myc occurs by PLZF, epigenetic or otherwise. Whether PLZF might still have an epigenetic role in the regulation of other genes remains to be determined, as does the extent of epigenetic regulation by APL-associated transcription factors.
FIG. 6.
PLZF-mediated c-myc repression is reversible. (A) Replacement of tetracycline into inducible cell medium turned off PLZF expression. PLZF45 cells were grown in tetracycline (lane 1) or withdrawn from tetracycline for 24 (lanes 2 and 3), 48 (lanes 4 and 5), or 72 (lanes 6 and 7) h. Tetracycline was added back to each culture for 48 (lanes 3 and 7) or 72 (lane 5) h, and then 106 cells were collected from each condition, lysed, and blotted with PLZF monoclonal antibody 2A9. The lower half of the blot was probed separately with a monoclonal antibody against GAPDH. Lane 8, control for PLZF expression. (B) mRNA was purified from the cultures in panel A and used in a real-time PCR. PLZF, c-myc, and GAPDH messages in each condition were quantified, and the Ct for each message in the absenceof PLZF was set as 1. For each message, the change in Ct (ΔCt) between samples was determined in triplicate and averaged and expressed relative to the change in GAPDH to convert into the relative change in expression.
c-myc overexpression reverses the cell cycle effect of PLZF.
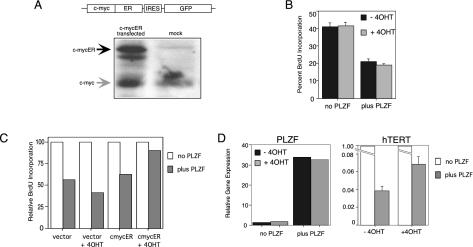
PLZF was clearly able to repress c-myc expression by transcriptional repression; therefore, we asked if this repression had any consequences in a cellular context. Continued expression of PLZF results in reduced proliferation, G1 arrest, an increase in adhesion marker expression, and eventually apoptosis. c-myc expression, in the presence of mitogenic stimulation, increases cell cycling and proliferation and acts to oppose differentiation. To assess the effect of c-myc on the cell cycle effect of PLZF, we created a bicistronic c-mycER:GFP vector with a mutant estrogen receptor (that is only activated in response to tamoxifen [4OHT]) fused to the c-myc coding region (45). Figure 7A shows a schematic of the construct and that a high level of expression of c-mycER, approximately equivalent to that of endogenous c-myc, was generated from this vector. PLZF45 cells grown in the presence or absence of tetracycline were pulsed with BrdU, and the number of cells in S phase was determined under each condition. Forty percent of PLZF45 cells with no PLZF expression were in S phase at any given time. Expression of PLZF for 4 days halved this to 20%, while addition of 4OHT had no effect on the S phase (Fig. 7B). We transfected PLZF45 cells with c-mycER:GFP and pulsed them with BrdU to determine the effect of c-myc on this PLZF-mediated decline in DNA synthesis (Fig. 7C). In the absence of PLZF, approximately 50% of the c-myc-expressing (GFP-positive) cells were in S phase, as expected, as observed with nontransfected cells (data not shown). PLZF expression in both vector-transfected and inactive c-mycER cells was associated with a block of the cell cycle, with only 25% of the cells entering S phase, a reduction of about 50%. Strikingly, activation of c-mycER blocked the ability of PLZF to inhibit cell cycle progression, as under these conditions there was only a 10% decrease in S-phase cells. Neither expression nor activation of c-mycER had any paracrine effect on the non-GFP-positive, nontransfected cells (data not shown).
FIG. 7.
c-myc expression rescued PLZF-mediated cell cycle arrest. (A) Schematic (top) of the bicistronic c-mycER:GFP vector that was transfected into 293T cells. The lysates were blotted with a monoclonal antibody to c-myc. The black arrow indicates the band resulting from the bicistronic c-myc-ER fusion construct, and the grey arrow indicates endogenous c-myc. (B) PLZF45 cells were split between −PLZF and +PLZF (by tetracycline withdrawal), pulsed with BrdU, and grown in the absence (black bar) or presence (grey bar) of 200 nM 4OHT. The proportion of BrdU-labeled cells in each condition was determined by fluorescence-activated cell sorter analysis with a Phoenix Red-conjugated anti-BrdU monoclonal antibody. (C) PLZF45 cells were electroporated with c-mycER:GFP or the GFP vector and then plated into +tet or −tet medium. Cultures were then split in two again and c-myc was activated by addition of 4OHT to one-half of each culture. After 4 days, cells were pulsed with BrdU and labeled with an anti-BrdU monoclonal antibody conjugated to Phoenix Red. Cells were analyzed by flow cytometry; those expressing GFP were gated, and the extent of BrdU incorporation was assessed in the GFP-positive cells. The percentage of BrdU-labeled cells for −PLZF (white bars) and +PLZF (grey bars) under each condition is expressed relative to the −PLZF value under each condition, which was designated 100. Vector, MIGR1 transfected; c-mycER, c-mycER:GFP transfected. The data presented are from a representative experiment, and the same result was observed in three repetitions. (D) Cells were electroporated and treated with 4OHT as in the experiment whose results are shown in panel C. Three days postelectroporation, GFP-positive cells were separated by flow cytometry and RNA was extracted and transcribed into cDNA. PLZF (left panel) and hTERT (right panel) transcript levels under each condition were assessed by quantitative reverse transcription-PCR, and each change was expressed relative to GAPDH. The experiment was performed in triplicate, and the error bars represent the standard error of the mean. The data shown are from a representative experiment; the same effect was observed in three repetitions.
In addition, c-mycER activation by 4OHT reversed the ability of PLZF to repress c-myc target genes. GFP-positive cells were isolated by flow cytometry under each of the conditions described above, and levels of the c-myc target gene hTERT were determined by quantitative real-time PCR. Expression of PLZF was not altered by addition of 4OHT (Fig. 7D, part 1). PLZF expression led to a 95% decrease in hTERT expression. Activation of c-mycER by 4OHT in the presence of PLZF approximately doubled the hTERT transcript level (Fig. 7D, part 2), which was not observed in the vector-transfected, non-mycER-expressingcells (data not shown). From this, we conclude that down-modulation of c-myc expression is a major mechanism by which PLZF controls cell growth.
DISCUSSION
The mechanism by which the transcriptional repressor PLZF alters gene expression has been extensively studied, and our group has focused previously on the function of the BTB/POZ domain and the role of nuclear corepressors (48, 49, 71). These studies showed that growth suppression by PLZF is critically linked to transcriptional repression. What has remained poorly understood is the identity of the genes regulated by PLZF relevant to growth control. To date, the only direct targets described have been cyclin A2 (75) and, very recently, HoxD11 (4). The work described here used a cDNA array approach to identify c-myc as a gene directly responsive to PLZF, a finding that was confirmed by both Northern blotting and real-time PCR and shown to occur via transcriptional repression. Modulation of c-myc expression may explain much of the effect of PLZF on the cell cycle and cell growth. Further, we confirmed the direct nature of the interaction between PLZF and the cyclin A2 promoter, another pro-proliferative factor whose repression by PLZF contributes to the cell cycle arrest mediated by PLZF expression.
The Lymphochip analysis showed a 7-fold induction of PLZF on tetracycline withdrawal, whereas both the Affymetrix chip and the quantitative PCR demonstrated 30-fold induction, implying that the cDNA glass slide arrays were less sensitive than the oligonucleotide arrays. However, the same patterns were observed with both array systems, including several of the same genes, suggesting that specificity is not reduced in cDNA arrays. In the Lymphochip analysis, approximately 27% of the B-lymphoid-related genes altered in response to PLZF are, in fact, c-myc target genes. In general, these genes were regulated by PLZF in a manner opposite to the action of c-myc. This suggested that PLZF-mediated reversal of c-myc action is a major mode of action of PLZF in hematopoiesis and growth control. The same pattern was observed with a much larger gene set derived from a nonbiased, predominantly nonoverlapping gene population represented on the Affymetrix U95A_v2 chip. Eighty-seven percent of the c-myc target genes were regulated by PLZF inversely to the action of c-myc, rising to 97% for PLZF-repressed, c-myc-activated genes. The lower correlation for PLZF-activated, c-myc-repressed genes is due to two factors. First, c-myc-activated genes are found more frequently than c-myc-repressed genes and were more highly represented on the array (13, 27, 52, 55, 63, 72), and second, the mechanism by which c-myc induces repression is not fully understood and may not be direct (12). Similarly, gene activation by PLZF is very unlikely to be a direct transcriptional effect.
c-myc drives proliferation through activation of growth regulators and repression of differentiative genes such as that which encodes the cyclin-dependent kinase inhibitor p21. The identification of these genes and the nature of the regulatory mechanism have been debated for many years, but the use of high-throughput cDNA screening protocols is bringing about consensus on the genes regulated in response to c-myc (13, 27, 52, 55, 63, 72). A large number of these genes are involved in the nucleic acid and protein metabolism necessary for cell cycle transition. The pro-differentiation genes downregulated by c-myc include those that encode p21, CDC2, and NCAM1, while the c-myc-activated pro-growth genes include those that encode prohibitin, cyclin D3, RAB40B, and TRAP1. We demonstrated that PLZF reduced the expression of these pro-growth genes and increased the expression of the cell cycle arrest and differentiation genes, directly impacting cell growth.
A direct interaction was shown between the endogenous c-myc promoter and PLZF, which was correlated to a decrease in RNA polymerase II occupancy of the c-myc promoter upon PLZF expression. This reflects a reduction in the initiation of transcription. This was again correlated to repression of a c-myc promoter construct, which was shown to be largely dependent on a PLZF binding site. Electrophoretic mobility shift analysis indicated the presence of only a single direct binding site for PLZF in the 2.5 kb of upstream c-myc sequences, and mutation of this site significantly reduced the ability of PLZF to repress the 2.5-kb c-myc promoter. The repression of the myc reporter by PLZF was less robust than that of the endogenous c-myc transcript, illustrating the limited ability of a reporter system to necessarily reflect actual promoter activity. However, the reporter assay did indicate the necessity for an intact PLZF binding site for PLZF-mediated repression.
Repression of c-myc by PLZF was rapidly reversible upon removal of PLZF. It was reported that the PML moiety of the PML-RARα fusion protein mediated interaction of the fusion protein with DNA methyltransferases. Expression of the fusion protein led to methylation and partial silencing of a target promoter that could only be reversed by a demethylating agent and not removal of PML-RARα (19). This clearly was not the case with PLZF repression of c-myc, which was rapidly reversible with the removal of PLZF from the cell. This may be because PLZF does not interact with the DNA-methylating machinery, a possibility that is under further investigation. Alternatively, c-myc may be too critical a regulator to be completely shut down by one factor. Regulation of c-myc occurs at many different levels, transcriptional, posttranscriptional, and translational, and by many different proteins. The multiple factors positively regulating c-myc promoter architecture and transcription (1, 11, 25, 30, 34, 41, 53, 57, 66, 70) are unlikely to all be overcome by PLZF. Whether PLZF can lead to long-term silencing of the other targets, cyclin A2 and HoxD11, remains to be determined.
A decrease in c-myc expression is absolutely necessary for terminal differentiation (reviewed in references 31 and 32). The PLZF-mediated decrease in c-myc expression adds to the body of evidence indicating that despite the induction of apoptosis resulting from overexpression, PLZF has a positive effect on some aspectsof blood cell differentiation. Previous work showed that induction of PLZF expression in U937 cells resulted in increased expression of cell surface markers CD11b and CD11c (71), which was confirmed by the array analysis in this study. c-myc expression decreases both the differentiation and the expression of adhesion markers (47), and the effective removal of c-myc by PLZF could conceivably result in the increased CD11b/CD18 expression observed upon PLZF induction. From the array experiments, we also observed that PLZF increased the levels of the CDK inhibitors p21 and p19, which presumably mediates, in part, the cell cycle arrest induced by PLZF. This could also occur through modulation of c-myc levels, as c-myc has been demonstrated to repress p21 expression (23, 54, 73). However, expression of p21 can be sufficient to mediate myeloid differentiation in the appropriate background (46), reflecting the importance of cell cycle withdrawal for differentiation. Another indication that PLZF has a positive role in differentiation comes from megakaryocytes, where PLZF expression is retained throughout differentiation of CD34+ cells into mature megakaryocytes (37).
Whether c-myc expression induces apoptosis or proliferation is dependent on the growth factor status of the cell. In this system, cells were grown in serum and the effect of c-myc expression was a very slight increase in the number of cells in S phase at a given time. In cells without mitogenic stimulation, c-myc expression leads to apoptosis. On the basis of the current assay system, it was impossible to draw any conclusions about the effect of PLZF on c-myc-mediated apoptosis. It is tempting to speculate, given that induction of high levels of apoptosis by PLZF takes several days of continued expression, an initial down-regulation of the proapoptotic function of c-myc would occur upon PLZF expression. This, in turn, suggests that the apoptosis that eventually results from PLZF expression is the result of another process, perhaps associated with an abortive attempt at differentiation.
Our working model of the action of PLZF is as follows. The PLZF target genes described by our group to date, c-myc and that which encodes cyclin A2, both have a positive function in cell cycle progression and proliferation. Repression by PLZF prevents further proliferation and leads to exit from the cell cycle, leaving the cell poised in G0. As differentiation occurs down erythroid and myeloid lineages, PLZF expression declines (15, 37). The cellular environment will also have an impact on cell fate—if lineage-specific factors such as C/EBPα and PU.1 are present, differentiation will occur along a particular lineage. PLZF may also alter cell fate by protein-protein interaction—expression in U937 cells blocks differentiation induced by 1,25-dihydroxyvitamin D3 by preventing the action of the vitamin D3 receptor (71).
In light of the fact that PLZF profoundly represses c-myc expression, loss of PLZF can be considered an oncogenic event. We believe that there are three molecular axes to the development of t(11;17) APL. The first is abnormal repression of RARα target genes by PLZF-RARα (22, 33, 36, 38, 44, 50, 56, 58, 60, 67, 68, 76, 77, 78), the second is loss of PLZF-mediated repression of PLZF target genes, and the third is activation of PLZF target genes by RARα-PLZF. As the use of murine models of APL showed, development of the complete phenotype associated with the t(11;17) translocation requires either the expression of both PLZF-RARα and RARα-PLZF or PLZF-RARα expression in the absence of PLZF (29). This implies that loss of normal PLZF repression of a given target, and possibly activation of that target by RARα-PLZF, is essential for the cell to escape normal growth controls and become leukemic. The importance of pro-proliferative genes such as c-myc and cyclin A2 as targets of PLZF is underscored by the double-transgenic PLZF-RARα/RARα-PLZF mice, which exhibit an enhanced rate of DNA synthesis and a decreased apoptosis rate (29). PLZF−/− mice have a similar phenotype, with some developing organ sites showing decreased cell death and increased cell division (4). These findings might be explained by deregulated c-myc and cyclin A2 expression resulting from loss of normal PLZF repression and possibly additional activation of such genes by RARα-PLZF. The status of c-myc and cyclin A2 levels in these animals has not yet been reported. However, there is evidence that the c-myc gene dosage is important in human myeloid disease. In t(15;17) APL with PML-RARα translocation, complete or partial trisomy 8 occurs in 50% of the patients who have a chromosomal change in addition to the primary translocation (26, 64). The pathogenic region has been localized to 8q22∼qter (69), which would increase the gene dose of c-myc located at 8q24. Other forms of AML commonly have high rates of trisomy 8 or c-myc amplification as either primary or secondary events (7, 8, 26). In the absence of 8q abnormalities in t(11;17) APL, one can speculate that loss of PLZF could increase the effective dose of c-myc, hence contributing to leukemogenesis.
Identification of PLZF target genes has been critical to understanding both the role PLZF plays in differentiation and the effect of translocation in APL. The description of c-myc as a target for PLZF repression has greatly enhanced our understanding of the normal function of PLZF and helps to explain why dysregulation of PLZF is so disastrous for the developing cell.
Acknowledgments
This work was supported by National Institutes of Health grant CA59936, the Chemotherapy Foundation, and American Cancer Society grant DHP160.
We thank Dan Tenen and Linda Penn for c-myc promoter constructs and helpful discussions, Chi Dang for assistance with the myc target gene database, Carol Bodian for statistical advice, and the Mount Sinai Flow Cytometry Shared Research Facility for assistance.
REFERENCES
- 1.Albert, T., J. Wells, J. O. Funk, A. Pullner, E. E. Raschke, G. Stelzer, M. Meisterernst, P. J. Farnham, and D. Eick. 2001. The chromatin structure of the dual c-myc promoter P1/P2 is regulated by separate elements. J. Biol. Chem. 276:20482-20490. [DOI] [PubMed] [Google Scholar]
- 2.Alizadeh, A., M. Eisen, R. E. Davis, C. Ma, H. Sabet, T. Tran, J. I. Powell, L. Yang, G. E. Marti, D. T. Moore, J. R. Hudson, Jr., W. C. Chan, T. Greiner, D. Weisenburger, J. O. Armitage, I. Lossos, R. Levy, D. Botstein, P. O. Brown, and L. M. Staudt. 1999. The lymphochip: a specialized cDNA microarray for the genomic-scale analysis of gene expression in normal and malignant lymphocytes. Cold Spring Harbor Symp. Quant. Biol. 64:71-78. [DOI] [PubMed] [Google Scholar]
- 2a.Alizadeh, A. A., M. B. Eisen, R. E. Davis, C. Ma, I. S. Lossos, A. Rosenwald, J. C. Boldrick, H. Sabet, T. Tran, X. Yu, J. I. Powell, L. Yang, G. E. Marti, T. Moore, J. Hudson, Jr., L. Lu, D. B. Lewis, R. Tibshirani, G. Sherlock, W. C. Chan, T. C. Greiner, D. D. Weisenburger, J. O. Armitage, R. Warnke, R. Levy, W. Wilson, M. R. Grever, J. C. Byrd, D. Botstein, P. O. Brown, and L. M. Staudt. 2000. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403:503-511. [DOI] [PubMed] [Google Scholar]
- 3.Ball, H. J., A. Melnick, R. Shaknovich, R. A. Kohanski, and J. D. Licht. The promyelocytic leukemia zinc finger (PLZF) protein binds DNA in a high molecular weight complex associated with cdc2 kinase. Nucleic Acids Res. 27:4106-4113. [DOI] [PMC free article] [PubMed]
- 4.Barna, M., N. Hawe, L. Niswander, and P. P. Pandolfi. 2000. Plzf regulates limb and axial skeletal patterning. Nat. Genet. 25:166-172. [DOI] [PubMed] [Google Scholar]
- 5.Barrett, T. J., N. P. Sandhu, A. J. Tomlinson, L. M. Benson, M. Subramaniam, S. Naylor, and T. C. Spelsberg. 2000. Interactions of the nuclear matrix-associated steroid receptor binding factor with its DNA binding element in the c-myc gene promoter. Biochemistry 39:753-762. [DOI] [PubMed] [Google Scholar]
- 6.Bartley, P. A., J. K. Lutwyche, and T. J. Gonda. 2001. Identification and validation of candidate Myb target genes. Blood Cells Mol. Dis. 27:409-415. [DOI] [PubMed] [Google Scholar]
- 7.Batanian, J. R., E. Ma, Y. Huang, and B. Gadre. 2001. Co-existence of alternative forms of 8q gain in cytogenetic clones of three patients with acute myeloid leukemia, pointing to 8q22 approximately 8qter as a region of biologic significance. Cancer Genet. Cytogenet. 126:20-25. [DOI] [PubMed] [Google Scholar]
- 7a.Boer, J., J. Bonten-Surtel, and G. Grosveld. 1998. Overexpression of the nucleoporin CAN/NUP214 induces growth arrest, nucleocytoplasmic transport defects, and apoptosis. Mol. Cell. Biol. 18:1236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruckert, P., R. Kappler, H. Scherthan, H. Link, F. Hagmann, and H. Zankl. 2000. Double minutes and c-MYC amplification in acute myelogenous leukemia: are they prognostic factors? Cancer Genet. Cytogenet. 120:73-79. [DOI] [PubMed] [Google Scholar]
- 9.Chen, Z., N. J. Brand, A. Chen, S. J. Chen, J. H. Tong, Z. Y. Wang, S. Waxman, and A. Zelent. 1993. Fusion between a novel Kruppel-like zinc finger gene and the retinoic acid receptor-alpha locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. EMBO J. 12:1161-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chernukhin, I. V., S. Shamsuddin, A. F. Robinson, A. F. Carne, A. Paul, A. I. El-Kady, V. V. Lobanenkov, and E. M. Klenova. 2000. Physical and functional interaction between two pluripotent proteins, the Y-box DNA/RNA-binding factor, YB-1, and the multivalent zinc finger factor, CTCF. J. Biol. Chem. 275:29915-29921. [DOI] [PubMed] [Google Scholar]
- 11.Cogswell, J. P., P. C. Cogswell, W. M. Kuehl, A. M. Cuddihy, T. M. Bender, U. Engelke, K. B. Marcu, and J. P.-Y. Ting. 1993. Mechanism of c-myc regulation by c-Myb in different cell lineages. Mol. Cell. Biol. 13:2858-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole, M. D., and S. B. McMahon. 1999. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene 18:2916-2924. [DOI] [PubMed] [Google Scholar]
- 13.Coller, H. A., C. Grandori, P. Tamayo, T. Colbert, E. S. Lander, R. N. Eisenman, and T. R. Golub. 2000. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc. Natl. Acad. Sci. USA 97:3260-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cook, M., A. Gould, N. Brand, J. Davies, P. Strutt, R. Shaknovich, J. Licht, S. Waxman, Z. Chen, S. Gluecksohn-Waelsch, et al. 1995. Expression of the zinc-finger gene PLZF at rhombomere boundaries in the vertebrate hindbrain. Proc. Natl. Acad. Sci. USA 92:2249-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai, M. S., N. Chevallier, S. Stone, M. C. Heinrich, M. McConnell, T. Reuter, H. E. Broxmeyer, J. D. Licht, L. Lu, and M. E. Hoatlin. 2002. The effects of the Fanconi anemia zinc finger (FAZF) on cell cycle, apoptosis, and proliferation are differentiation stage-specific. J. Biol. Chem. 277:26327-26334. [DOI] [PubMed] [Google Scholar]
- 16.Dang, C. V. 1999. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 19:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dang, C. V., L. M. Resar, E. Emison, S. Kim, Q. Li, J. E. Prescott, D. Wonsey, and K. Zeller. 1999. Function of the c-Myc oncogenic transcription factor. Exp. Cell Res. 253:63-77. [DOI] [PubMed] [Google Scholar]
- 18.David, G., L. Alland, S. H. Hong, C. W. Wong, R. A. DePinho, and A. Dejean. 1998. Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene 16:2549-2556. [DOI] [PubMed] [Google Scholar]
- 19.Di Croce, L., V. A. Raker, M. Corsaro, F. Fazi, M. Fanelli, M. Faretta, F. Fuks, F. Lo Coco, T. Kouzarides, C. Nervi, S. Minucci, and P. G. Pelicci. 2002. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science 295:1079-1082. [DOI] [PubMed] [Google Scholar]
- 20.Facchini, L. M., S. Chen, W. W. Marhin, J. N. Lear, and L. Z. Penn. 1997. The Myc negative autoregulation mechanism requires Myc-Max association and involves the c-myc P2 minimal promoter. Mol. Cell. Biol. 17:100-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feo, S., D. Arcuri, E. Piddini, R. Passantino, and A. Giallongo. 2000. ENO1 gene product binds to the c-myc promoter and acts as a transcriptional repressor: relationship with Myc promoter-binding protein 1 (MBP-1). FEBS Lett. 473:47-52. [DOI] [PubMed] [Google Scholar]
- 22.Freemantle, S. J., J. S. Kerley, S. L. Olsen, R. H. Gross, and M. J. Spinella. 2002. Developmentally-related candidate retinoic acid target genes regulated early during neuronal differentiation of human embryonal carcinoma. Oncogene 21:2880-2889. [DOI] [PubMed] [Google Scholar]
- 23.Gartel, A. L., X. Ye, E. Goufman, P. Shianov, N. Hay, F. Najmabadi, and A. L. Tyner. 2001. Myc represses the p21(WAF1/CIP1) promoter and interacts with Sp1/Sp3. Proc. Natl. Acad. Sci. USA 98:4510-4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh, A. K., I. Grigorieva, R. Steele, R. G. Hoover, and R. B. Ray. 1999. PTEN transcriptionally modulates c-myc gene expression in human breast carcinoma cells and is involved in cell growth regulation. Gene 235:85-91. [DOI] [PubMed] [Google Scholar]
- 25.Grigorieva, I., V. G. Grigoriev, M. K. Rowney, and R. G. Hoover. 2000. Regulation of c-myc transcription by interleukin-2 (IL-2): identification of a novel IL-2 response element interacting with STAT-4. J. Biol. Chem. 275:7343-7350. [DOI] [PubMed] [Google Scholar]
- 26.Grimwade, D., H. Walker, F. Oliver, K. Wheatley, C. Harrison, G. Harrison, J. Rees, I. Hann, R. Stevens, A. Burnett, and A. Goldstone. 1998. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. Blood 92:2322-2333. [PubMed] [Google Scholar]
- 27.Guo, Q. M., R. L. Malek, S. Kim, C. Chiao, M. He, M. Ruffy, K. Sanka, N. H. Lee, C. V. Dang, and E. T. Liu. 2000. Identification of c-myc responsive genes using rat cDNA microarray. Cancer Res. 60:5922-5928. [PubMed] [Google Scholar]
- 28.He, L., J. Liu, I. Collins, S. Sanford, B. O'Connell, C. J. Benham, and D. Levens. 2000. Loss of FBP function arrests cellular proliferation and extinguishes c-myc expression. EMBO J. 19:1034-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He, L. Z., M. Bhaumik, C. Tribioli, E. M. Rego, S. Ivins, A. Zelent, and P. P. Pandolfi. 2000. Two critical hits for promyelocytic leukemia. Mol. Cell 6:1131-1141. [DOI] [PubMed] [Google Scholar]
- 30.He, T. C., A. B. Sparks, C. Rago, H. Hermeking, L. Zawel, L. T. da Costa, P. J. Morin, B. Vogelstein, and K. W. Kinzler. 1998. Identification of c-MYC as a target of the APC pathway. Science 281:1509-1512. [DOI] [PubMed] [Google Scholar]
- 31.Hoffman, B., A. Amanullah, M. Shafarenko, and D. A. Liebermann. 2002. The proto-oncogene c-myc in hematopoietic development and leukemogenesis. Oncogene 21:3414-3421. [DOI] [PubMed] [Google Scholar]
- 32.Hoffman, B., and D. A. Liebermann. 1998. The proto-oncogene c-myc and apoptosis. Oncogene 17:3351-3357. [DOI] [PubMed] [Google Scholar]
- 33.Hummel, J. L., T. Zhang, R. A. Wells, and S. Kamel-Reid. 2002. The retinoic acid receptor alpha (RARα) chimeric proteins PML-, PLZF-, NPM-, and NuMA-RARα have distinct intracellular localization patterns. Cell Growth Differ. 13:173-183. [PubMed] [Google Scholar]
- 34.Itkes, A., C. J. Allegra, and M. Zajac-Kaye. 2000. Multiprotein complexes present at the MIF motifs flanking the promoter of the human c-myc gene. FEBS Lett. 474:23-28. [DOI] [PubMed] [Google Scholar]
- 35.Johansen, L. M., A. Iwama, T. A. Lodie, K. Sasaki, D. W. Felsher, T. R. Golub, and D. G. Tenen. 2001. c-Myc is a critical target for C/EBPα in granulopoiesis. Mol. Cell. Biol. 21:3789-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kerley, J. S., S. L. Olsen, S. J. Freemantle, and M. J. Spinella. 2001. Transcriptional activation of the nuclear receptor corepressor RIP140 by retinoic acid: a potential negative-feedback regulatory mechanism. Biochem. Biophys. Res. Commun. 285:969-975. [DOI] [PubMed] [Google Scholar]
- 37.Labbaye, C., M. T. Quaranta, A. Pagliuca, S. Militi, J. D. Licht, U. Testa, and C. Peschle. 2002. PLZF induces megakaryocytic development, activates Tpo receptor expression and interacts with GATA1 protein. Oncogene 21:6669-6679. [DOI] [PubMed] [Google Scholar]
- 38.Lee, K.-H., M.-Y. Chang, J.-I. Ahn, D.-H. Yu, S.-S. Jung, J.-H. Choi, Y.-H. Noh, Y.-S. Lee, and M.-J. Ahn. 2002. Differential gene expression in retinoic acid-induced differentiation of acute promyelocytic leukemia cells, NB4 and HL-60 cells. Biochem. Biophys. Res. Commun. 296:1125-1133. [DOI] [PubMed] [Google Scholar]
- 39.Lee, T. C., and E. B. Ziff. 1999. Mxi1 is a repressor of the c-Myc promoter and reverses activation by USF. J. Biol. Chem. 274:595-606. [DOI] [PubMed] [Google Scholar]
- 40.Li, J. Y., M. A. English, H. J. Ball, P. L. Yeyati, S. Waxman, and J. D. Licht. 1997. Sequence-specific DNA binding and transcriptional regulation by the promyelocytic leukemia zinc finger protein. J. Biol. Chem. 272:22447-22455. [DOI] [PubMed] [Google Scholar]
- 41.Li, L. H., C. Nerlov, G. Prendergast, D. MacGregor, and E. B. Ziff. 1994. c-Myc represses transcription in vivo by a novel mechanism dependent on the initiator element and Myc box II. EMBO J. 13:4070-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Licht, J. D., R. Shaknovich, M. A. English, A. Melnick, J. Y. Li, J. C. Reddy, S. Dong, S. J. Chen, A. Zelent, and S. Waxman. 1996. Reduced and altered DNA-binding and transcriptional properties of the PLZF-retinoic acid receptor-alpha chimera generated in t(11;17)-associated acute promyelocytic leukemia. Oncogene 12:323-336. [PubMed] [Google Scholar]
- 43.Lin, R. J., L. Nagy, S. Inoue, W. Shao, W. H. Miller, Jr., and R. M. Evans. 1998. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature 391:811-814. [DOI] [PubMed] [Google Scholar]
- 44.Lin, R. J., T. Sternsdorf, M. Tini, and R. M. Evans. 2001. Transcriptional regulation in acute promyelocytic leukemia. Oncogene 20:7204-7215. [DOI] [PubMed] [Google Scholar]
- 45.Littlewood, T. D., D. C. Hancock, P. S. Danielian, M. G. Parker, and G. I. Evan. 1995. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 23:1686-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu, M., M. H. Lee, M. Cohen, M. Bommakanti, and L. P. Freedman. 1996. Transcriptional activation of the Cdk inhibitor p21 by vitamin D3 leads to the induced differentiation of the myelomonocytic cell line U937. Genes Dev. 10:142-153. [DOI] [PubMed] [Google Scholar]
- 47.Lopez-Rodriguez, C., M. D. Delgado, A. Puig-Kroger, A. Nueda, E. Munoz, J. Leon, C. Bernabeu, and A. L. Corbi. 2000. c-Myc inhibits CD11a and CD11c leukocyte integrin promoters. Eur. J. Immunol. 30:2465-2471. [DOI] [PubMed] [Google Scholar]
- 48.Melnick, A., K. F. Ahmad, S. Arai, A. Polinger, H. Ball, K. L. Borden, G. W. Carlile, G. G. Prive, and J. D. Licht. 2000. In-depth mutational analysis of the promyelocytic leukemia zinc finger BTB/POZ domain reveals motifs and residues required for biological and transcriptional functions. Mol. Cell. Biol. 20:6550-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Melnick, A., G. Carlile, K. F. Ahmad, C. L. Kiang, C. Corcoran, V. Bardwell, G. G. Prive, and J. D. Licht. 2002. Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors. Mol. Cell. Biol. 22:1804-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Melnick, A., and J. D. Licht. 1999. Deconstructing a disease: RARα, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood 93:3167-3215. [PubMed] [Google Scholar]
- 51.Melnick, A. M., J. J. Westendorf, A. Polinger, G. W. Carlile, S. Arai, H. J. Ball, B. Lutterbach, S. W. Hiebert, and J. D. Licht. 2000. The ETO protein disrupted in t(8;21)-associated acute myeloid leukemia is a corepressor for the promyelocytic leukemia zinc finger protein. Mol. Cell. Biol. 20:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Menssen, A., and H. Hermeking. 2002. Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc. Natl. Acad. Sci. USA 99:6274-6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller, T. L., Y. Jin, J. M. Sun, A. S. Coutts, L. C. Murphy, and J. R. Davie. 1996. Analysis of human breast cancer nuclear proteins binding to the promoter elements of the c-myc gene. J. Cell. Biochem. 60:560-571. [DOI] [PubMed] [Google Scholar]
- 54.Mitchell, K. O., and W. S. El-Deiry. 1999. Overexpression of c-Myc inhibits p21WAF1/CIP1 expression and induces S-phase entry in 12-O-tetradecanoylphorbol-13-acetate (TPA)-sensitive human cancer cells. Cell Growth Differ. 10:223-230. [PubMed] [Google Scholar]
- 55.Neiman, P. E., A. Ruddell, C. Jasoni, G. Loring, S. J. Thomas, K. A. Brandvold, R. Lee, J. Burnside, and J. Delrow. 2001. Analysis of gene expression during myc oncogene-induced lymphomagenesis in the bursa of Fabricius. Proc. Natl. Acad. Sci. USA 98:6378-6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pandolfi, P. P. 2001. In vivo analysis of the molecular genetics of acute promyelocytic leukemia. Oncogene 20:5726-5735. [DOI] [PubMed] [Google Scholar]
- 57.Ramana, C. V., N. Grammatikakis, M. Chernov, H. Nguyen, K. C. Goh, B. R. Williams, and G. R. Stark. 2000. Regulation of c-myc expression by IFN-γ through Stat1-dependent and -independent pathways. EMBO J. 19:263-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Redner, R. L. 2002. Variations on a theme: the alternate translocations in APL. Leukemia 16:1927-1932. [DOI] [PubMed] [Google Scholar]
- 59.Reid, A., A. Gould, N. Brand, M. Cook, P. Strutt, J. Li, J. Licht, S. Waxman, R. Krumlauf, and A. Zelent. 1995. Leukemia translocation gene, PLZF, is expressed with a speckled nuclear pattern in early hematopoietic progenitors. Blood 86:4544-4552. [PubMed] [Google Scholar]
- 60.Rusiniak, M. E., M. Yu, D. T. Ross, E. C. Tolhurst, and J. L. Slack. 2000. Identification of B94 (TNFAIP2) as a potential retinoic acid target gene in acute promyelocytic leukemia. Cancer Res. 60:1824-1829. [PubMed] [Google Scholar]
- 61.Sakatsume, O., H. Tsutsui, Y. Wang, H. Gao, X. Tang, T. Yamauchi, T. Murata, K. Itakura, and K. K. Yokoyama. 1996. Binding of THZif-1, a MAZ-like zinc finger protein, to the nuclease-hypersensitive element in the promoter region of the c-MYC protooncogene. J. Biol. Chem. 271:31322-31333. [DOI] [PubMed] [Google Scholar]
- 62.Schmidt, M., V. Nazarov, L. Stevens, R. Watson, and L. Wolff. 2000. Regulation of the resident chromosomal copy of c-myc by c-Myb is involved in myeloid leukemogenesis. Mol. Cell. Biol. 20:1970-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schuldiner, O., and N. Benvenisty. 2001. A DNA microarray screen for genes involved in c-MYC and N-MYC oncogenesis in human tumors. Oncogene 20:4984-4994. [DOI] [PubMed] [Google Scholar]
- 64.Sessarego, M., G. Fugazza, E. Balleari, R. Bruzzone, A. Ballestrero, and F. Patrone. 1997. High frequency of trisomy 8 in acute promyelocytic leukemia: a fluorescence in situ hybridization study. Cancer Genet. Cytogenet. 97:161-164. [DOI] [PubMed] [Google Scholar]
- 65.Shaknovich, R., P. L. Yeyati, S. Ivins, A. Melnick, C. Lempert, S. Waxman, A. Zelent, and J. D. Licht. 1998. The promyelocytic leukemia zinc finger protein affects myeloid cell growth, differentiation, and apoptosis. Mol. Cell. Biol. 18:5533-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simonsson, T., M. Pribylova, and M. Vorlickova. 2000. A nuclease hypersensitive element in the human c-myc promoter adopts several distinct i-tetraplex structures. Biochem. Biophys. Res. Commun. 278:158-166. [DOI] [PubMed] [Google Scholar]
- 67.Spinella, M. J., J. S. Kerley, K. A. White, and J. C. Curtin. 2003. Retinoid target gene activation during induced tumor cell differentiation: human embryonal carcinoma as a model. J. Nutr. 133:273S-276S. [DOI] [PubMed] [Google Scholar]
- 68.Truckenmiller, M. E., M. P. Vawter, C. Cheadle, M. Coggiano, D. M. Donovan, W. J. Freed, and K. G. Becker. 2001.Gene expression profile in early stage of retinoic acid-induced differentiation of human SH-SY5Y neuroblastoma cells. Restor. Neurol. Neurosci. 18:67-80. [PubMed] [Google Scholar]
- 69.Vial, J. P., F. X. Mahon, A. Pigneux, A. Notz, F. Lacombe, J. Reiffers, and C. Bilhou-Nabera. 2003. Derivative (7)t(7;8)(q34;q21), a new additional cytogenetic abnormality in acute promyelocytic leukemia. Cancer Genet Cytogenet. 140:78-81. [DOI] [PubMed] [Google Scholar]
- 70.Vindigni, A., A. Ochem, G. Triolo, and A. Falaschi. 2001. Identification of human DNA helicase V with the far upstream element-binding protein. Nucleic Acids Res. 29:1061-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ward, J. O., M. J. McConnell, G. W. Carlile, P. P. Pandolfi, J. D. Licht, and L. P. Freedman. 2001. The acute promyelocytic leukemia-associated protein, promyelocytic leukemia zinc finger, regulates 1,25-dihydroxyvitamin D3-induced monocytic differentiation of U937 cells through a physical interaction with vitamin D3 receptor. Blood 98:3290-3300. [DOI] [PubMed] [Google Scholar]
- 72.Watson, J. D., S. K. Oster, M. Shago, F. Khosravi, and L. Z. Penn. 2002. Identifying genes regulated in a Myc-dependent manner. J. Biol. Chem. 277:36921-36930. [DOI] [PubMed] [Google Scholar]
- 73.Wu, S., C. Cetinkaya, M. J. Munoz-Alonso, N. Von Der Lehr, F. Bahram, V. Beuger, M. Eilers, J. Leon, and L. G. Larsson. 2003. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene 22:351-360. [DOI] [PubMed] [Google Scholar]
- 74.Yagi, K., M. Furuhashi, H. Aoki, D. Goto, H. Kuwano, K. Sugamura, K. Miyazono, and M. Kato. 2002. c-myc is a downstream target of the Smad pathway. J. Biol. Chem. 277:854-861. [DOI] [PubMed] [Google Scholar]
- 75.Yeyati, P. L., R. Shaknovich, S. Boterashvili, J. Li, H. J. Ball, S. Waxman, K. Nason-Burchenal, E. Dmitrovsky, A. Zelent, and J. D. Licht. 1999. Leukemia translocation protein PLZF inhibits cell growth and expression of cyclin A. Oncogene 18:925-934. [DOI] [PubMed] [Google Scholar]
- 76.Zelent, A., F. Guidez, A. Melnick, S. Waxman, and J. D. Licht. 2001. Translocations of the RARα gene in acute promyelocytic leukemia. Oncogene 20:7186-7203. [DOI] [PubMed] [Google Scholar]
- 77.Zhang, D., S. Vuocolo, V. Masciullo, T. Sava, A. Giordano, D. R. Soprano, and K. J. Soprano. 2001. Cell cycle genes as targets of retinoid induced ovarian tumor cell growth suppression. Oncogene 20:7935-7944. [DOI] [PubMed] [Google Scholar]
- 78.Zhao, X., and R. A. Spanjaard. 2003. The apoptotic action of the retinoid CD437/AHPN: diverse effects, common basis. J. Biomed. Sci. 10:44-49. [DOI] [PubMed] [Google Scholar]