Abstract
Cycloheximide acts at the large subunit of the ribosome to inhibit translation. Here we report that ubiquitin levels are critical for the survival of Saccharomyces cerevisiae cells in the presence of cycloheximide: ubiquitin overexpression confers resistance to cycloheximide, while a reduced ubiquitin level confers sensitivity. Consistent with these findings, ubiquitin is unstable in yeast (t1/2 = 2 h) and is rapidly depleted upon cycloheximide treatment. Cycloheximide does not noticeably enhance ubiquitin turnover, but serves principally to block ubiquitin synthesis. Cycloheximide also induces UBI4, the polyubiquitin gene. The cycloheximide-resistant phenotype of ubiquitin overexpressors is also characteristic of partial-loss-of-function proteasome mutants. Ubiquitin is stabilized in these mutants, which may account for their cycloheximide resistance. Previous studies have reported that ubiquitin is destabilized in the absence of Ubp6, a proteasome-associated deubiquitinating enzyme, and that ubp6 mutants are hypersensitive to cycloheximide. Consistent with the model that cycloheximide-treated cells are ubiquitin deficient, the cycloheximide sensitivity of ubp6 mutants can be rescued either by ubiquitin overexpression or by mutations in proteasome subunit genes. These results also show that ubiquitin wasting in ubp6 mutants is proteasome mediated. Ubiquitin overexpression rescued cells from additional translational inhibitors such as anisomycin and hygromycin B, suggesting that ubiquitin depletion may constitute a widespread mechanism for the toxicity of translational inhibitors.
Cells maintain their viability under adverse conditions through elaborate regulatory mechanisms known as stress responses. Processes under the control of such pathways include transcription, translation, DNA repair, and protein degradation. In particular, the ubiquitin-proteasome pathway has been implicated in the response of cells to stresses such as high temperature, starvation, heavy metal exposure, UV light exposure, alkylation damage, and oxidative damage (16). Among the most-well-studied stresses is heat shock, which generates misfolded proteins that are preferred substrates for the ubiquitin pathway. The burst of substrates for ubiquitination that results from heat shock can lead to rapid depletion of free ubiquitin (10, 23, 35). The deleterious effects of ubiquitin depletion are counteracted by induction of the polyubiquitin gene and suppression of the translation of constitutive mRNAs (3), possibly because newly synthesized proteins are highly susceptible to misfolding and thus are preferred substrates for ubiquitination.
The induction of the heat shock response can be effectively mimicked by exposing cells to amino acid analogs, which give rise to misfolded proteins and consequently impose an increased load on the ubiquitin-proteasome pathway. Even in the absence of amino acid analogs, a large fraction of newly synthesized proteins is rapidly degraded, and this may constitute a major load on the ubiquitin-proteasome pathway (38). Ubiquitin pathway mutants are highly sensitive to amino acid analogs (5, 10, 37, 39), yet the effects of drugs that simply inhibit translation are also profoundly altered in many ubiquitin pathway mutants (12, 18). The reason for this has been unclear and is addressed in this study.
Keeven et al. (18) recently characterized gain-of-function mutations in the PDR2 gene of Saccharomyces cerevisiae. PDR2 encodes a transcription factor that regulates multidrug resistance (6). The PDR2-2 mutation confers resistance to cycloheximide, a bacterially produced inhibitor of protein synthesis that interferes with tRNA translocation when bound to the 60S subunit of the ribosome (29, 34). PDR2-2 appears to confer drug resistance by increasing the synthesis of drug efflux pumps that transport cycloheximide. However, the cycloheximide resistance conferred by PDR2-2 required the wild-type product of the UBP6 gene (18), which encodes one of over 15 deubiquitinating enzymes expressed in S. cerevisiae (50). ubp6Δ cells were found to be hypersensitive to cycloheximide in the absence of PDR2-2 as well (18). To account for these results, it was suggested that although cycloheximide has no known effect on the fidelity of translation, it might, as an inhibitor of the ribosome, nonetheless promote the formation of truncated and hence abnormal and potentially toxic proteins (18).
Mutations in ubiquitin-proteasome pathway genes other than UBP6 have been found to confer cycloheximide resistance rather than sensitivity. Indeed, all known crl mutations—temperature-sensitive lethal mutations that confer cycloheximide resistance—have been mapped to genes encoding proteasome subunits (12, 21). Moreover, only mutations that produce a deficit in the protein-degrading capacity of the proteasome resulted in cycloheximide resistance. It is implicit in the model described above, in which cycloheximide promotes the formation of misfolded and toxic translation products, that loss-of-function mutations in the proteasome would, like ubp6Δ, confer cycloheximide sensitivity. Since these mutations instead confer resistance, some other mechanism must account for the effects of cycloheximide on proteasome subunit mutants. Interestingly, Ubp6 is a proteasome component as well, and several lines of evidence suggest that Ubp6 function is obligatorily coupled to the proteasome. First, free Ubp6 shows only low levels of activity, whereas proteasome-bound Ubp6 is stimulated approximately 300-fold (20). Second, a large fraction of Ubp6 in whole-cell extracts is proteasome-associated (unpublished observations), and Ubp6 is an abundant component of affinity-purified proteasomes (20, 47). Finally, deletion of the proteasome-binding motif in Ubp6 produces an apparent null phenotype (20). In summary, the ubiquitin pathway mutants known to affect survival in the presence of cycloheximide are all components of the proteasome. However, some confer sensitivity, such as ubp6Δ, while others confer resistance.
A model that could potentially explain these divergent observations was suggested by the finding that cells lacking UBP6 exhibit rapid degradation of ubiquitin (20). Thus, when present in conjugated species, ubiquitin is apparently susceptible to degradation by the proteasome, unless it is released from these species in a timely fashion through the action of Ubp6 (and presumably other proteasomal deubiquitinating enzymes). In this model, ubp6Δ mutants are sensitive to cycloheximide because they degrade ubiquitin at a rapid rate and thus cannot maintain ubiquitin pools when protein synthesis is reduced. On the other hand, proteasome subunit mutants would exhibit cycloheximide resistance because their attenuated proteasomes would degrade ubiquitin at a reduced rate and thus allow cells to survive when protein synthesis is compromised. A necessary assumption of the model (which we verify below) is that ubiquitin is degraded at a significant rate even in the presence of wild-type Ubp6.
In the present work, we test this model and find that overexpression of ubiquitin can rescue cells from the toxic effects of cycloheximide as well as a number of other translational inhibitors. ubp6Δ cells could also be rescued by overexpression of ubiquitin. Furthermore, cycloheximide was shown to induce a depletion of total cellular ubiquitin. Also consistent with the model, cycloheximide-resistant proteasome hypomorphs showed higher levels of ubiquitin than did wild-type cells after cycloheximide treatment. We therefore propose that the depletion of ubiquitin contributes critically to the toxicity of translational inhibitors and that this depletion of ubiquitin occurs largely through the proteasome.
MATERIALS AND METHODS
Yeast strains, methods, and media.
The strains employed in this work are listed in Table 1. YEp96 is a TRP1-marked plasmid that expresses ubiquitin in a copper-inducible manner (7). YEp46Δ is the control plasmid. For all plasmid-bearing strains, at least two independent transformants were tested. Cultures were grown at 30°C in YPD, YPRafGal, or synthetic media as indicated. YPD medium consisted of 1% yeast extract, 2% Bacto Peptone, and 2% dextrose. YPRafGal medium consisted of 1% yeast extract, 2% Bacto Peptone, 2% raffinose, and 2% galactose. Synthetic media consisted of 0.7% Difco yeast nitrogen base supplemented with amino acids, adenine, uracil, and 2% dextrose as described previously (36). All drugs were dissolved in H2O, except for hygromycin B, nocodazole, and rapamycin, which were dissolved in phosphate-buffered saline (hygromycin B) or dimethyl sulfoxide (nocodazole and rapamycin).
TABLE 1.
Characteristic of the yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| SUB62 | MATalys2-801 leu2-3, 2-112 ura 3-52 his3-Δ200 trp1-1 | 10 |
| SJH30 | MATalys2-801 leu2-3, 2-112 ura3-52 his3-Δ200 trp-1-1 [YEp46Δ] | This work |
| SJH34 | MATalys2-801 leu2-3, 2-112 ura3-52 his3-Δ200 trp1-1 [YEp96] | This work |
| SJH31 | MATalys2-801 leu2-3, 2-112 ura3-52 his3-Δ200 trp1-1 ubp6::URA3 [YEp46Δ] | This work |
| SJH35 | MATalys2-801 leu2-3, 2-112 ura3-52 his3-Δ200 trp1-1 ubp6::URA3 [YEp96] | This work |
| SUB328 | MATalys2-801 leu2-3, 2-112 ura3-52 his3-Δ200 trp1-1 ubil-Δ1::TRP1 ubi2-Δ2::URA3 ubi3Δub-2 ubi4-Δ2::LEU2 [pUB146] [pUB100] | 41 |
| SUB246 | MATalys2-801 leu2-3, 2-112 ura3-52 his3-Δ200 trp1-1ubi3Δub-2 | 9 |
| SUB254 | MATalys2-801 leu 2-3, 2-112 ura 3-52 his3-Δ200 trp 1-1 ubil-Δ1::TRP1 ubi2-Δ2::URA3 [pUB100] | 9 |
| SUB276 | MATalys2-801 leu 2-3, 2-112 ura 3-52 his3-Δ200 trp 1-1 ubil-Δ1::TRP1 ubi2-Δ2::URA3 ubi3Δub-2 [pUB100] | This work |
| YHI29W | MATaura3 leu2-3, 112 his3-11,15 | 12 |
| YRG11a | MATapre3-1 ura3 leu2-3, 112 his3-11,15 | 12 |
| YRG16a | MATapre3-6 (crl21) ura3 leu2-3,112 his3-11,15 | 12 |
| SJH42a | MATaura3 leu2-3,112 his3-11,1 ubp6::URA3 | This work |
| SJH44a | MATapre3-6 (crl21) ura3 leu2-3,112 his3-11,15 ubp6::URA3 | This work |
These strains are isogenic to YH129W. All other strains are isogenic to SUB62.
Assays of drug sensitivity.
Cultures were grown in synthetic media lacking tryptophan or YPD as indicated. Cultures were normalized to an optical density at 600 nm (OD600) of 0.2 and spotted in successive threefold serial dilutions onto plates consisting of either synthetic media or YPD supplemented with copper sulfate (100 μM) and the appropriate drug as indicated. Plates were incubated at 30°C for 2 to 7 days. pH sensitivity experiments were carried out at pH 3.5 as described previously (33).
Protein synthesis assay.
Wild-type (SUB62) cells were grown in exponential phase at 30°C in media lacking methionine. Cultures were treated with cycloheximide at the indicated concentrations. After 1 h of treatment, an equivalent number of cells from each sample, as determined by measurement of OD600, were incubated with 50 μCi of 35S-labeled methionine at 30°C for 15 min. Ten microliters of each sample was spotted onto filters pretreated with 50% trichloroacetic acid (TCA). Dried filters were boiled for 5 min in 10% TCA, washed twice in 10% TCA, washed once in 100% ethanol, dried, and subjected to scintillation counting. Error bars represent the standard deviations of an experiment carried out in triplicate.
Northern blot analysis.
Total yeast RNA was prepared and transferred to Hybond-N membrane (Amersham) by Northern transfer under standard conditions (2). After transfer, RNA was cross-linked to the membrane by UV irradiation (1,200 J/m2) and probed with a 2.4-kb, 32P-labeled fragment from the ubiquitin-coding sequence of the UBI4 gene generated by endonuclease cleavage with EcoRI (31). Blots were stripped and then reprobed with a 250-bp 32P-labeled fragment of the ACT1 actin gene as a control. Duplicate samples were stained with ethidium bromide to visualize rRNA.
Analysis of ubiquitin turnover.
Overnight cultures of the indicated strains were adjusted to an equal OD600 and allowed to grow in the exponential phase for 4 h at 30°C. Cultures were again normalized by OD600, and cycloheximide was added. To examine the effect of cycloheximide on cellular ubiquitin levels (see Fig. 2), a concentration of 200 μg/ml was employed. For experiments examining turnover of ubiquitin in ubp6Δ strains (see Fig. 5), a concentration of 50 μg/ml was employed to minimize ubiquitin depletion by cycloheximide. Aliquots were taken at the indicated time points after cycloheximide treatment, and measurements of OD600 were taken to ensure that an equal number of cells were collected at each time point. The cycloheximide-free cultures were diluted out with fresh YPD over the course of the experiment to maintain the cells in exponential-phase growth. Cell aliquots were pelleted, resuspended in 300 μl of 1× Laemmli loading buffer, boiled for 5 min, and stored at −80°C.
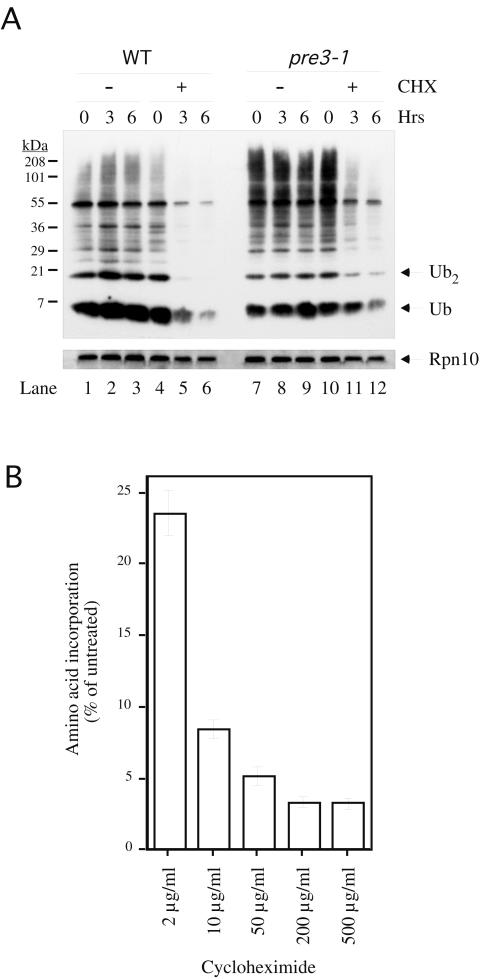
FIG. 2.
Depletion of ubiquitin by cycloheximide in wild-type and pre3-1 cells. (A) Wild-type (WT) (YHI29W) or pre3-1 cells (YRG11) were grown in YPD at 30°C in the exponential phase with or without 200-μg/ml cycloheximide (CHX). Aliquots containing equivalent amounts of cells were taken at the indicated time points and analyzed by immunoblotting with antiubiquitin antibody (top panel) or anti-Rpn10 control antibody (bottom panel). Essentially identical results were obtained for the wild-type SUB62 strain (data not shown). (B) Equivalent numbers of wild-type cells pretreated for 1 h with cycloheximide at the indicated concentrations were incubated with 35S-labeled methionine for 15 min. TCA-insoluble material was subjected to scintillation counting. Error bars represent the standard deviation of an experiment carried out in triplicate.
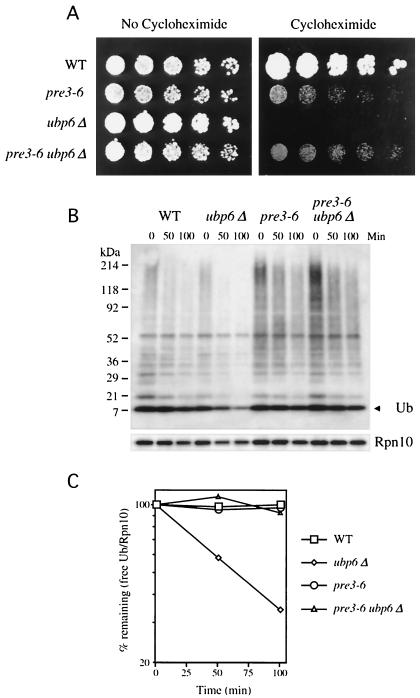
FIG. 5.
Ubiquitin wasting in ubp6 mutants is mediated by the proteasome. (A)Threefold serial dilutions of wild-type (WT) (YHI29W), pre3-6 (YRG16), ubp6Δ (SJH42), and pre3-6 ubp6Δ (SJH44) were applied as spots to YPD plates with or without 0.5-μg/ml cycloheximide and grown at 30°C for 2 (left) or 7 days (right). Note that pre3-6 is not more resistant to cycloheximide than is the wild type at this concentration of drug, but only at higher cycloheximide levels. (B) Wild-type (YHI29W), ubp6Δ (SJH42), pre3-6 (YRG16), and pre3-6 ubp6Δ (SJH44) cells were grown in YPD at 30°C in exponential phase. Cycloheximide was added to a final concentration of 50 μg/ml, and aliquots containing equivalent amounts of cells were taken at the indicated time points and analyzed by immunoblotting with an antiubiquitin antibody (upper panel) or anti-Rpn10 control antibody (lower panel). Ub, ubiquitin. (C) Quantitation of the data in panel B. The immunoblot from panel B was stripped and reprobed with the same antiubiquitin antibody or anti-Rpn10 antibody followed by 125I-labeled protein A. The data shown have been normalized against Rpn10 levels, which is necessary to correct for total protein loads. It should be noted that free ubiquitin levels appear to be slightly decreased over the 100-min time course in the wild-type, pre3-6, and pre3-6 ubp6Δ samples (see panel B), but show no decrease when normalized to Rpn10. It is therefore possible that the decay curves of panel C slightly underestimate the rate of disappearance of free ubiquitin.
Thirty microliters of each aliquot was analyzed on an 18% sodium dodecyl sulfate (SDS) gel (200:1 acrylamide-bisacrylamide) (Fig. 2) or on a 4 to 20% SDS-polyacrylamide gel (Invitrogen) (Fig. 5), and proteins were transferred to Hybond-P membrane (Amersham). The membrane was boiled in water for 30 min and then probed with an antiubiquitin antibody (Affiniti). The blot was visualized with the ECL enhanced chemiluminescence system (Amersham). For quantification, blots were stripped and reprobed with an antiubiquitin antibody followed by 125I-labeled protein A and then analyzed with a PhosphorImager. Rpn10 controls were processed similarly.
Determination of ubiquitin half-life.
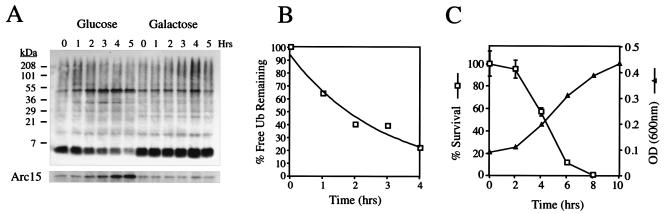
The strain SUB328 bears a single, galactose-inducible ubiquitin gene and has been described elsewhere (41). Cultures of SUB328 were grown in YPRafGal in the exponential phase at 30°C. Cells were then harvested, washed in H2O, and transferred to YPD or back to YPRafGal, and aliquots were taken at the indicated time points. For cultures growing in glucose, an equal culture volume was taken at each time point to prevent dilution of the total ubiquitin pool by cell division; for cultures growing on galactose, an equal number of cells were taken at each time point, as determined by OD600. Samples were analyzed on an 18% SDS-polyacrylamide gel as described above.
For quantitation, the immunoblot was stripped and reanalyzed with an antiubiquitin antibody followed by 125I-labeled protein A. This blot was analyzed on a PhosphorImager and quantitated with NIH Image. There was a lag in ubiquitin degradation of approximately 1 h, presumably due to the time required for mRNA depletion. Therefore, the half-life was calculated from the 1-h time point onwards. The curve was fit to the equation y = 94.28e−0.352x, resulting in a calculated half-life of 2.0 h. Calculation of ubiquitin half-life from time zero resulted in a slightly longer half-life of 2.5 h.
Growth curves of SUB328 growing in glucose or galactose were determined during exponential-phase growth by measurement of culture density at OD600 at the indicated time points. To determine cell viability as a function of ubiquitin depletion, cultures of SUB328 growing in galactose or glucose in the exponential phase were sampled at the indicated time points. An equivalent number of cells was plated on YPRafGal and incubated at 30°C, and colonies were counted 2 days later. The death rate is expressed as the rate of survival on glucose divided by the rate of survival on galactose. The experiment was performed in triplicate.
The level of ubiquitin required for viability was calculated by determination of the fold-reduction in free ubiquitin levels (see Fig. 6B) that correlated with a 50% decrease in viability in SUB328 cells (Fig. 6C) and adjusting that figure to reflect free ubiquitin levels in SUB328 under inducing conditions, which are approximately 50% that of the wild type (data not shown).
FIG. 6.
Determination of the half-life of ubiquitin. (A) SUB328 cells expressing a single galactose-inducible ubiquitin gene were grown in galactose in exponential phase. At time zero, cells were either transferred to glucose-containing media or maintained on galactose, and aliquots were taken at the indicated time points. For cells growing on glucose, an equal culture volume was taken at each time point to prevent dilution of the total ubiquitin pool by cell division; for cells growing on galactose, an equivalent number of cells were taken. Cells were analyzed by immunoblotting for ubiquitin (upper panel), followed by enhanced chemiluminescence. Loads were normalized by using an antibody to Arc15 as described previously (lower panel). (B) The immunoblot from panel A was stripped and reprobed with the same antiubiquitin antibody, followed by 125I-labeled protein A, and quantitated on a PhosphorImager with NIH Image. There was a lag time of approximately 1 h in ubiquitin (Ub) shutoff by glucose, and therefore the half-life was determined from 1 to 5 h postshift. (C) Cultures of SUB328 growing in glucose or galactose in exponential phase (as in panel A) were sampled at the indicated time point, and an equivalent amount of OD600 units were plated on YPRafGal plates. Colonies were counted 2 days later. The survival curve is expressed as the percent plating efficiency of the glucose-grown culture divided by that of the galactose culture (□). Error bars represent the standard deviation from an experiment carried out in triplicate. Cultures of SUB328 shifted to glucose media were normalized by OD and allowed to grow at 30°C in the exponential phase. ODs were determined at the indicated time points (▵).
RESULTS
Rescue of cycloheximide toxicity by ubiquitin overexpression.
To test the hypothesis that ubiquitin depletion contributes to cycloheximide toxicity in yeast, we transformed wild-type cells with a plasmid that expresses ubiquitin from the copper-inducible CUP1 promoter (7) or with a control plasmid lacking the ubiquitin gene. At a cycloheximide concentration of 2 μg/ml, colony formation by wild-type yeast was completely inhibited (Fig. 1). Overexpression of ubiquitin by the addition of cupric sulfate to the medium restored plating efficiency to levels approximating those seen when no drug was present.
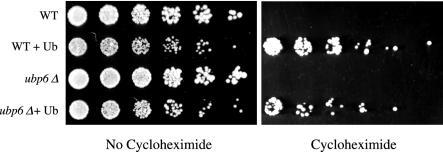
FIG. 1.
Cycloheximide resistance of wild-type and ubp6Δ cells due to ubiquitin overexpression. Threefold serial dilutions of wild-type (WT) cells (SJH30), wild-type cells overexpressing ubiquitin (Ub) (SJH34), ubp6Δ cells (SJH31), and ubp6Δ cells overexpressing ubiquitin (SJH35) were applied as spots to selective plates containing copper sulfate (100 μM) as described with or without 2-μg/ml cycloheximide and grown at 30°C for 3 (left panel) or 7 (right panel) days. Cycloheximide-containing plates were incubated longer to correct for reduced growth rates that result from translational inhibition.
There are two possible explanations for the ubiquitin rescue effect: cycloheximide could deplete ubiquitin pools, leading to a failure of cellular proliferation, or alternatively, high-level ubiquitin overexpression could artificially perturb some aspect of cellular physiology in a way that indirectly confers cycloheximide resistance. In support of the ubiquitin depletion model, the cycloheximide tolerance conferred by the ubiquitin-expressing plasmid did not require the addition of copper to the medium (data not shown). Basal expression from the CUP1 promoter is significant and results in a ubiquitin expression level comparable to that of wild-type cells (7, 41). Thus, in the absence of copper, only modest overexpression can be achieved. In addition, where copper was used as an inducer, cells were not exposed to copper prior to cycloheximide addition (see Materials and Methods). The modest amounts of ubiquitin that can be produced through leaky protein synthesis in the presence of cycloheximide would not be expected to grossly alter cellular physiology. Additional evidence in support of the ubiquitin depletion model is presented below.
Deletion of the UBP6 gene results in a ubiquitin wasting phenotype (20). In particular, ubiquitin is rapidly turned over in Δubp6 cells, most likely reflecting a failure to regenerate ubiquitin at the proteasome. ubp6 mutants are more sensitive than the wild type to cycloheximide (18). The cycloheximide sensitivity of ubp6Δ cells, like that of the wild type, is rescued by ubiquitin overexpression (Fig. 1). Rescue of cycloheximide toxicity by ubiquitin in ubp6Δ cells is less efficient than in wild-type cells, probably reflecting a balance between ubiquitin supplementation via the overexpression plasmid and ubiquitin wasting due to the lack of UBP6. This differential rescue effect was observed for a number of other drugs (described below and data not shown). Ubiquitin overexpression did not enhance growth in the absence of cycloheximide treatment. Under these conditions, both wild-type and ubp6Δ cells showed lower levels of growth when overexpressing ubiquitin (Fig. 1), regardless of copper induction (data not shown).
Cycloheximide-treated cells are ubiquitin deficient.
The rescue of cycloheximide toxicity by ubiquitin suggested that a critical effect of cycloheximide is to deplete cellular levels of ubiquitin. We therefore examined ubiquitin levels in wild-type cells treated with cycloheximide. In untreated cells, ubiquitin levels remained relatively constant over time (Fig. 2A, lanes 1 to 3). However, cells displayed significantly reduced levels of ubiquitin after cycloheximide treatment (Fig. 2A; compare lanes 1 to 3 with lanes 4 to 6). Both free ubiquitin and high-molecular-weight ubiquitin conjugates were progressively depleted. The rapid loss of immunoreactive material appears specific to ubiquitin, because a control protein, Rpn10, showed no significant depletion phenotype in the presence of cycloheximide (Fig. 2A, bottom panel). Rapid cycloheximide-dependent ubiquitin depletion as seen in Fig. 2A has not been observed previously in yeast (20, 43), most likely because previous experiments employed shorter time courses and lower levels of cycloheximide. Indeed, maximal inhibition of translation in vivo was achieved only at a concentration of 200 μg/ml (Fig. 2B) and not at 50 μg/ml, the concentration employed in previous studies. Therefore, although low concentrations of cycloheximide are effective at achieving strong partial inhibition of translation, maximal inhibition can be achieved only at significantly higher concentrations. We monitored translation rates up to 6 h after cycloheximide addition and observed no adaptation or recovery of translation during later time points (data not shown). The dose dependence of translational inhibition in vivo in yeast cells (Fig. 2B) is consistent with previous findings obtained with partially purified mammalian ribosomes (49). Interestingly, in both this work and the study by Wettstein et al. (49), a residual amount of label incorporation occurred even in the presence of maximal inhibition by cycloheximide.
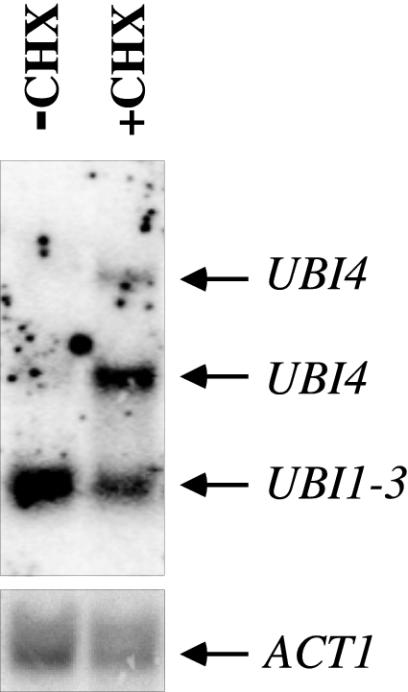
UBI4 is induced by treatment with cycloheximide.
The model that cycloheximide-treated cells are in a critical state of ubiquitin deficiency predicts that cycloheximide treatment should result in a compensatory induction of ubiquitin gene transcription. In yeast, there are four ubiquitin genes. UBI4 is the major stress-inducible ubiquitin gene, while UBI1-3 provides the bulk of ubiquitin under favorable growth conditions (10, 31). In untreated wild-type cells, low levels of UBI4 mRNA were detected (Fig. 3), as previously reported (10). Treatment with cycloheximide resulted in a strong induction of UBI4 (Fig. 3), as previously observed in Dictystelium discoideum, Aspergillus nidulans, and cyclic AMP-deficient yeast mutants (25, 28, 46). This response might reflect a mechanism designed to restore ubiquitin levels. However, in the presence of strong translational inhibition, this response is insufficient to maintain ubiquitin levels or cell viability (Fig. 1 and 2).
FIG. 3.
Induction of the UBI4 gene by cycloheximide. Wild-type (SUB62) cells were grown in the exponential phase for 2 h in the presence (+CHX) or absence (−CHX) of 200-μg/ml cycloheximide. mRNA was isolated and analyzed as described in Materials and Methods. The bands correspond to the UBI1-3 and UBI4 (1.5 and 2.6 kb) genes as indicated (top panel) or ACT1 (bottom panel).
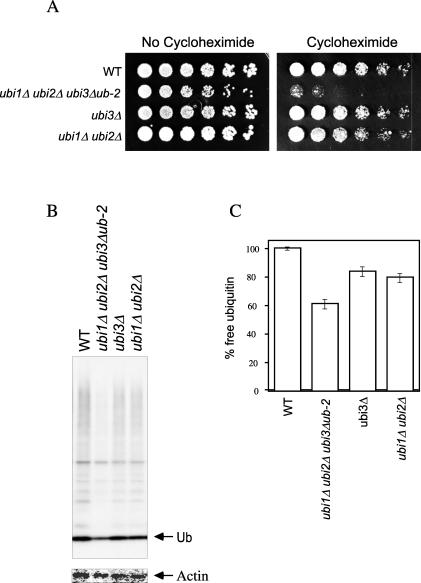
A mutant with reduced ubiquitin levels is cycloheximide sensitive.
The data described above suggest that the cycloheximide resistance of ubiquitin overexpressors reflects depletion of cellular ubiquitin pools by translational inhibition in wild-type cells. This interpretation predicts that mutants with reduced ubiquitin levels should be sensitive to cycloheximide. Because there are four ubiquitin genes in yeast, graded decreases in ubiquitin levels can be achieved by deleting one or more of these genes. The UBI1, UBI2, and UBI3 genes each express a 5′ ubiquitin-coding element that is cotranslated with a downstream sequence encoding a ribosomal protein. Ubiquitin is cleaved from the ribosomal protein during or shortly after translation. To test for the effect of reduced ubiquitin levels on cycloheximide sensitivity, we used strains in which the ribosomal protein components of each gene product were expressed independently of ubiquitin (see the legend to Fig. 4 and reference 9 for details). A triple deletion of UBI1, UBI2, and UBI3, which reduced ubiquitin levels by 40%, resulted in a marked sensitivity to cycloheximide (Fig. 4). A more modest reduction in ubiquitin levels was achieved by deleting either the ubiquitin element from UBI3 (the ubi3-Δub2 allele) or by deleting both UBI1 and UBI2. In these cases, no sensitivity to cycloheximide was observed, at least at the concentration tested (1 μg/ml). Thus, elevated ubiquitin levels confer cycloheximide resistance, whereas reduced ubiquitin levels confer cycloheximide sensitivity. For this and other reasons given above, the resistance phenotype is unlikely to result from an indirect, nonphysiological effect of high-level ubiquitin overexpression.
FIG. 4.
Cycloheximide sensitivity of a ubi1Δ ubi2Δ ubi3Δub-2 mutant. (A) Threefold serial dilutions of wild-type (WT) (SUB62), ubi1Δ ubi2Δ ubi3Δub-2 (SUB276), ubi3Δ (SUB246), and ubi1Δ ubi2Δ (SUB254) cells were applied as spots to YPD plates with or without 1.0-μg/ml cycloheximide and grown at 30°C for 2 (left panel) or 4 days (right panel). Note that assays of cycloheximide sensitivity are carried out at 0.5 to 1 μg/ml, whereas assays of resistance are carried out at 2 μg/ml, because control cells are sensitive to 2 μg/ml but resistant to 0.5 to 1 μg/ml. (B) Wild-type and mutant cells as described above were grown in YPD at 30°C in the exponential phase. An equivalent number of cells were analyzed for free ubiquitin (Ub) levels by immunoblotting with antiubiquitin antibody followed by 125I-labeled protein A (upper panel). The same immunoblot was stripped and reprobed with antiactin antibody followed by 125I-labeled protein A (lower panel). (C) Quantitation of free ubiquitin levels from the data in panel B. The data shown have been normalized against actin levels to correct for total protein loads. Error bars represent the standard deviations of the experiment carried out in duplicate.
Proteasome mutants stabilize ubiquitin and confer cycloheximide resistance.
Strains bearing partial-loss-of-function mutations in the proteasome are generally cycloheximide resistant, as described above. The hypothesis that ubiquitin depletion contributes to cycloheximide toxicity predicts that such strains should show reduced depletion of ubiquitin. We therefore examined the levels of cellular ubiquitin in a cycloheximide-resistant strain bearing a mutation in the gene PRE3, which encodes a subunit of the catalytic core of the proteasome (25). In the absence of drug, the pre3-1 strain showed enhanced levels of high-molecular-weight ubiquitin conjugates compared to the wild type (Fig. 2, compare lanes 7 to 9 with lanes 1 to 3), consistent with previously reported results (12). Although some depletion of ubiquitin pools in the pre3-1 strain was evident upon cycloheximide treatment, there were higher levels of both free ubiquitin and high-molecular-weight conjugates after cycloheximide treatment compared to the wild type (Fig. 2, lanes 4 to 6 and 10 to 12). Similar results were obtained with a second crl mutant, pre3-6 (data not shown). These data suggest that the cycloheximide resistance phenotype of proteasome loss-of-function mutations is due to ubiquitin sparing and provide additional evidence that ubiquitin wasting underlies cycloheximide toxicity. The stabilization of ubiquitin observed in proteasome mutants also suggests that the major agent of cycloheximide-induced ubiquitin wasting is the proteasome.
Proteasome mutants suppress the cycloheximide sensitivity of ubp6 mutants.
To further test whether ubiquitin stabilization by proteasome loss-of-function mutations is relevant to their cycloheximide resistance, we combined the pre3-6 hypomorphic mutation with the ubiquitin-destabilizing null mutation ubp6Δ. The pre3-6 mutation was able to substitute for ubiquitin overexpression in conferring cycloheximide tolerance to ubp6Δ (Fig. 5A). We tested whether the ability of pre3-6 to rescue ubp6Δ was mediated by an increase in free ubiquitin levels by immunoblot analysis. The ubiquitin-wasting phenotype of the ubp6Δ mutant was corrected in the double mutant (Fig. 5B and C). In summary, these data support the model that the opposing effects of pre3-6 and ubp6 mutations on cycloheximide tolerance result from perturbations of ubiquitin pools. In addition, these data confirm that ubiquitin wasting in ubp6 mutants is mediated by the proteasome.
The half-life of ubiquitin in yeast.
In the presence of cycloheximide (200 μg/ml), free ubiquitin disappears from yeast with a half-life of several hours (Fig. 2) (data not shown). To determine whether this represents the steady-state turnover rate of ubiquitin or a cycloheximide-induced destabilization, we measured ubiquitin turnover in the absence of drug treatment. The abundance of ubiquitin that had been expressed from a galactose-dependent promoter was measured during a chase incubation after a shift to glucose. By this method, the half-life of ubiquitin was estimated at 2 h (Fig. 6B). Thus, ubiquitin turnover is rapid even in the absence of cycloheximide. In the galactose-shutoff experiment, cells rapidly lose viability, as measured by colony-forming ability; after being switched to glucose-based media, the survival rate falls to approximately 50% within 4 h (Fig. 6C). At this point, continued growth of the culture is evident, although depletion of ubiquitin has dramatically reduced colony-forming ability (Fig. 6C). A comparison of the immunoblotting and viability data indicates that the steady-state level of ubiquitin in wild-type cells is approximately fivefold in excess over the critical level for viability (50% survival; see Materials and Methods for details).
Ubiquitin overexpression confers resistance to multiple translational inhibitors.
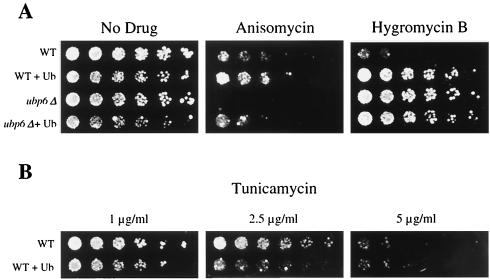
To test whether ubiquitin wasting may be a general mechanism for the toxicity of translational inhibitors, we examined the capacity of ubiquitin to rescue cells from a number of other drugs. Anisomycin is, like cycloheximide, an inhibitor of the peptidyltransferase activity of the ribosome (19). ubp6Δ cells have also been shown to be hypersensitive to anisomycin (18). As shown in Fig. 7A, ubiquitin supplementation was able to rescue both wild-type and ubp6Δ cells from anisomycin toxicity.
FIG. 7.
Resistance to multiple translational inhibitors mediated by ubiquitin overexpression. Threefold serial dilutions of wild-type (WT) cells (SJH30), wild-type cells overexpressing ubiquitin (Ub) (SJH34), ubp6Δ (SJH31) cells, and ubp6Δ cells overexpressing ubiquitin (SJH35) were applied as spots to selective plates containing copper sulfate (100 μM) and no drug, anisomycin (50 μg/ml), or hygromycin B (100 μg/ml) (A) or tunicamycin (B) (concentrations as indicated) and grown at 30°C for 2 to 7 days.
We next sought to determine whether ubiquitin depletion mediates the toxicity of only peptidyltransferase inhibitors or of translational inhibitors in general. The peptidyltransferase inhibitors work largely by decreasing the overall rate of protein synthesis (1), and they are thought to have little or no effect on the fidelity of translation. A second group of translational inhibitors induces misreading and thus generates aberrant protein products. An inhibitor of this class, hygromycin B, blocked the growth of wild-type cells at a concentration of 100 μg/ml (Fig. 7A). In contrast, supplementation with ubiquitin enhanced plating efficiency to levels observed in the absence of drug.
Whereas ubp6Δ cells were found to be generally hypersensitive to peptidyltransferase inhibitors, they were resistant to hygromycin B (Fig. 7A). Interestingly, hygromycin resistance is the first ubp6 phenotype that is not corrected by ubiquitin overexpression and thus not linked to ubiquitin deficiency. Hygromycin resistance is a rare phenotype, typically indicating reduced function of the plasma membrane H+-ATPase, Pma1, which results in a hygromycin uptake defect (27). However, ubp6 mutants are unlikely to have reduced Pma1 function, because they grow normally at low pH (data not shown). We are currently examining the hygromycin-resistant phenotype of ubp6 mutants in more detail. A similar resistance of ubp6 mutants was observed for neomycin (data not shown), another translational inhibitor that induces misreading, suggesting that ubp6Δ cells may be generally resistant to this class of drug. In contrast, the proteasome-defective crl mutants are sensitive to hygromycin (22). Thus, as with cycloheximide, ubp6 and crl mutants show opposing phenotypes.
Our data indicate that ubiquitin overexpression can rescue cells from the toxic effects of a diverse set of translational inhibitors. Thus, ubiquitin wasting may be a general feature of the mechanism of toxicity of these antibiotics. Supplementation of ubiquitin conferred no growth advantage to wild-type cells in the presence of rapamycin, tunicamycin, nocodazole, and fluconazole (Fig. 7B) (data not shown; see Materials and Methods for details). Thus, the capacity of ubiquitin to rescue drug-treated cells appears to show specificity for translational inhibitors.
DISCUSSION
Translational inhibitors and the ubiquitin-proteasome pathway.
A large and well-studied class of natural antibiotics function by binding to the ribosome and inhibiting translation. Here we show that the growth-suppressing activity of several translation inhibitors is critically linked to the depletion of ubiquitin. The role of ubiquitin in mediating the cellular effects of translational inhibition appears to be highly specific and has to our knowledge not been predicted. The notion that ubiquitin is a key target of translational inhibition stands in contrast to an implicit model that translational inhibitors stop growth simply by virtue of an across-the-board cessation of new protein synthesis (as they must do at very high levels). In the wild, where translational inhibitors mediate competition among microbes, intermediate levels of inhibitor may be prevalent.
In yeast, ubiquitin is not known to be turned over at a significant rate (20, 43). However, we show here that the ability of ubiquitin overexpression to rescue cell growth in the presence of translational inhibitors is a reflection of ubiquitin turnover, which depletes ubiquitin soon after drug treatment. Our results may explain several previous findings concerning the ubiquitin-proteasome pathway and translational inhibitors. First, a screen for mutants that are both temperature-sensitive and resistant to cycloheximide, although initially assumed to yield mutations in translational machinery, eventually proved to identify only loss-of function mutations in proteasome subunits (12, 21). This result was problematic because, if one assumes that cycloheximide treatment results in the accumulation of aberrant and truncated translation products that must be degraded by proteasomes (see reference 18), the loss-of-function mutation should have instead resulted in sensitivity to cycloheximide rather than resistance. The resistance of proteasome hypomorphs to cycloheximide can potentially be explained on the basis of our finding of a critical role of ubiquitin levels in the cycloheximide resistance of wild-type cells. The ubiquitin-sparing effect of proteasome hypomorphic mutations can be observed directly by monitoring ubiquitin levels and genetically by the ability of these mutations to suppress the cycloheximide-sensitive ubp6Δ mutation, which causes enhanced ubiquitin turnover.
The finding of Keeven and coworkers (18) that the resistance of PDR2-2 mutants to cycloheximide is dependent on the wild-type UBP6 gene, may be explained by our results as well. The requirement for Ubp6 reflects a critical role of ubiquitin levels in cycloheximide resistance as well as the requirement for Ubp6 to maintain ubiquitin levels. The instability of ubiquitin shown in the present work and the dependence of its turnover on the proteasome indicate that ubiquitin recycling from proteasomal substrates is not fully efficient, even when the proteasome is associated with a wild-type complement of deubiquitinating enzymes. This is probably also the case in mammalian cells, where ubiquitin turnover has been shown to be nonlysosomal and ATP dependent (13, 15). Ubiquitin may be translocated into the core particle in parallel with its covalently bound substrate or as a part of a proteasome-bound remnant generated prior to substrate translocation into the core particle. The deubiquitinating enzymes Ubp6 (20) and Rpn11 (48, 52) function to reduce the frequency of ubiquitin translocation into the core particle, but together do not fully eliminate ubiquitin turnover. Presumably, the ubiquitin pool would be depleted very rapidly if the proteasome did not discriminate between ubiquitin and the substrates that it targets for degradation. Thus, degradation of ubiquitin by the proteasome is generally regarded as an error in proteasome function. It cannot be excluded, however, that ubiquitin turnover by the proteasome serves in the regulation of ubiquitin pools.
Why is the effect of translational inhibition specific for ubiquitin?
It is surprising that, of the thousands of proteins expressed in yeast, ubiquitin in particular should be so critical for the survival of protein synthesis inhibition. In principle, this could reflect either an unusually short half-life of ubiquitin or, if the amount of ubiquitin required for normal cellular function is close enough to the steady-state level, that modest perturbations in its abundance would not be tolerated. However, a half-life of 2 h, while short, does not place ubiquitin among the most unstable proteins, whose half-lives have been estimated at 5 min or less—a group that includes cyclins, CDK inhibitors, and some transcriptional regulators. Effects on viability appear to require a substantial (∼5-fold) reduction in abundance when ubiquitin is depleted over a short term of several hours (Fig. 6C). The levels of free ubiquitin required for viability in long-term assays such as in Fig. 1 may be higher, however. In addition to its half-life, more specific functional properties of ubiquitin may underlie its importance under conditions of reduced protein synthesis. For example, since the turnover of many other proteins depends on ubiquitin, a decline in the level of ubiquitin may function as a brake on the degradation of other unstable proteins.
Our data may alternatively point to a coupling between protein degradation and synthesis that has yet to be fully understood. Ubiquitin is cotranslated with several ribosomal proteins (9) and is a major covalent modifier of the ribosome (42). A ubiquitin mutation that abrogates formation of Lys-63-linked multiubiquitin chains confers marked sensitivity to certain translational inhibitors (42), but the overall specificity of those effects is significantly different from that observed in the ubiquitin overexpression experiments reported in this work, and it also differs from that of the ubp6-null mutant. For example, ubiK63R mutants are slightly resistant to cycloheximide, although highly sensitive to anisomycin, and comparable to the wild type in their resistance to hygromycin. Given these phenotypic properties, it is unlikely that ubiquitin depletion confers sensitivity to translational inhibitors simply because it results in deficient formation of Lys-63-linked multiubiquitin chains on the ribosome. In addition, the UbK63R mutation does not result in reduced ubiquitin levels (41).
Functional significance of ubiquitin pools.
Although the regulation of ubiquitin gene expression remains poorly understood, various observations point to the importance of ubiquitin levels when cells are challenged by drug treatment, stress, mutation, or disease (10, 11, 23, 35, 38, 43-45). Indeed, free ubiquitin levels may be partially rate limiting for many ubiquitin-dependent processes. For example, overexpression of ubiquitin led to decreased abundance of postsynaptic glutamate receptors in Caenorhabditis elegans, while mutations that decrease glutamate receptor ubiquitination increase their abundance (4). Thus, free ubiquitin levels may be partially rate limiting for glutamate receptor turnover in the wild-type organism.
Another example of the significance of ubiquitin levels is in retrovirus budding. Patnaik et al. (32) have shown that the inhibition of virus budding by proteasome inhibitors is an indirect effect mediated by depletion of free ubiquitin. Indeed, proteasome inhibitors may reduce free ubiquitin levels by as much as 95% within less than 2 h (23, 32). Thus, some of the functions attributed to proteasomes on the basis of proteasome inhibitor experiments may instead reflect exhaustion of free ubiquitin pools.
It is paradoxical that proteasome inhibitors deplete free ubiquitin, while proteasome hypomorphic mutations in proteasome subunit genes are ubiquitin sparing. We suggest that this can be understood on the basis of two opposing effects: depletion of free ubiquitin by driving it into high-molecular-weight forms and an inherently slower effect of enhancing free ubiquitin levels by reducing the rate of ubiquitin degradation. In the case of proteasome inhibitor treatment as well as proteasome hypomorphs, both effects operate at the same time; however, the former effect predominates for proteasome inhibitors, whereas the latter effect predominates for the hypomorphs. One explanation for this is that experiments on proteasome inhibitors examine an acute response and are thus more sensitive to the rapid effect of driving conjugate formation, while the effect of the hypomorphic mutations is chronic. Under acute conditions, induced expression of ubiquitin is likely too slow to maintain adequate free pools. Perhaps more importantly, proteasome hypomorphs such as the crl mutants almost certainly impair proteasome function to a lesser degree than do proteasome inhibitors under conditions shown to result in ubiquitin depletion. Consequently, in the hypomorphs, the fraction of total ubiquitin driven into conjugated forms (Fig. 2) is much less than that seen in the presence of proteasome inhibitors (see for example, reference 23).
Ubiquitin pools in neurons.
Exhaustion of free ubiquitin pools may also explain the neurodegenerative phenotype observed in mice with ataxia. Although these mutants display a complex phenotype, including tremor, paralysis, and early death, their most prominent defects appear to reflect synaptic dysfunction (51). The affected gene in ataxic mice encodes Usp14, the ortholog of Ubp6 (51). Thus, assuming that this deubiquitinating enzyme functions similarly in yeast and in mammals, ubiquitin depletion may underlie the synaptic defects of the ataxic mice. We are currently testing this possibility. It is interesting that the effect shows specificity for synapses. The ubiquitin pathway is highly active in synapses (8), which may therefore be sites of elevated ubiquitin use (4, 14, 26, 51). Due to the distance between synapses and cell bodies, a local ubiquitin deficiency in the synapse may not be sensed efficiently in the nucleus of the neuron. Recent studies suggest that ubiquitin depletion may not be restricted to neurons that lack Usp14. While this article was under review, a neuronal ubiquitin-wasting phenotype was described for null mutants in the deubiquitinating enzyme UCH-L1 (30). Neurons have been suggested to be susceptible to ubiquitin depletion when injured, and such ubiquitin deficiency may lead to cell death and neurodegeneration (44, 45). Reduced levels of free ubiquitin have also been associated with hippocampal damage by surgical lesions and by sublethal ischemia (17, 24).
Free ubiquitin exists in a rapid and complex equilibrium with multiple conjugated forms, including free chains (50). Thus, free ubiquitin pools may be depleted through a number of mechanisms. Cycloheximide depletes free ubiquitin by preventing synthesis of new protein; ubp6 mutations deplete ubiquitin by enhancing its rate of degradation; and proteasome inhibitors, by allowing the accumulation of ubiquitination substrates, drive ubiquitin into conjugated forms. Misfolded and aggregated proteins, which have been implicated in various neurodegenerative diseases, may be an additional and most physiologically relevant inducer of ubiquitin depletion. Misfolded proteins are preferentially ubiquitinated (40). If not efficiently degraded, they may, like chemical proteasome inhibitors, interfere with protein degradation both by inhibiting the proteasome and by depleting free ubiquitin pools through enhanced formation of conjugates.
Acknowledgments
We thank R. Li for antibodies to Arc15, D. Wolf and W. Hilt for crl strains, Z. Moqtaderi for ACT1 primers, and R. Vierstra for antibodies to Rpn10. We also thank C. Walsh for helpful discussions and S. Elsasser for critical reading of the manuscript.
This work was supported by NIH grant GM43601 (D.F.). J.H. was supported by the Medical Scientist Training Program through grant T32 GM07753 to Harvard Medical School.
REFERENCES
- 1.Abraham, A. K., and A. Pihl. 1983. Effect of protein synthesis inhibitors on the fidelity of translation in eukaryotic systems. Biochim. Biophys. Acta 741:197-203. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1994. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 3.Brostrom, C. O., and M. A. Brostrom. 1998. Regulation of translational initiation during cellular responses to stress. Prog. Nucleic Acid Res. Mol. Biol. 58:79-125. [DOI] [PubMed] [Google Scholar]
- 4.Burbea, M., L. Dreier, J. S. Dittman, M. E. Grunwald, and J. Kaplan. 2002. Ubiquitin and AP180 regulate the abundance of GLR-1 glutamate receptors at postsynaptic elements in C. elegans. Neuron 35:107-120. [DOI] [PubMed] [Google Scholar]
- 5.Chen, Y., and P. W. Piper. 1995. Consequences of overexpression of ubiquitin in yeast: elevated tolerances of osmostress, ethanol and canavanine, yet reduced tolerances of cadmium, arsenite and paromomycin. Biochim. Biophys. Acta 1268:59-64. [DOI] [PubMed] [Google Scholar]
- 6.Cui, Z., T. Shiraki, D. Hirata, and T. Miyakawa. 1998. Yeast gene YRR1, which is required for resistance to 4-nitroquinoline N-oxide, mediates transcriptional activation of the multidrug resistance transporter gene SNQ2. Mol. Microbiol. 29:1307-1315. [DOI] [PubMed] [Google Scholar]
- 7.Ecker, D. J., M. I. Khan, J. Marsh, T. R. Butt, and S. T. Cooke. 1987. Chemical synthesis and expression of a cassette adapted ubiquitin gene. J. Biol. Chem. 262:3524-3527. [PubMed] [Google Scholar]
- 8.Ehlers, M. D. 2003. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat. Neurosci. 6:231-242. [DOI] [PubMed] [Google Scholar]
- 9.Finley, D., B. Bartel, and A. Varshavsky. 1989. The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature 338:394-401. [DOI] [PubMed] [Google Scholar]
- 10.Finley, D., E. Ozkaynak, and A. Varshavsky. 1987. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell 48:1035-1046. [DOI] [PubMed] [Google Scholar]
- 11.Galan, J.-M., and R. Haguenauer-Tsapis. 1997. Ubiquitin Lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 16:5847-5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerlinger, U. M., R. Guckel, M. Hoffmann, D. H. Wolf, and W. Hilt. 1997. Yeast cycloheximide-resistant crl mutants are proteasome mutants defective in protein degradation. Mol. Biol. Cell 8:2487-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haas, A. L., and P. M. Bright. 1987. The dynamics of ubiquitin pools within cultured human lung fibroblasts. J. Biol. Chem. 262:345-351. [PubMed] [Google Scholar]
- 14.Hegde, A. N., and A. DiAntonio. 2002. Ubiquitin and the synapse. Nat. Rev. Neurosci. 3:854-861. [DOI] [PubMed] [Google Scholar]
- 15.Hiroi, Y., and M. Rechsteiner. 1992. Ubiquitin metabolism in HeLa cells starved of amino acids. J. Biol. Chem. 307:156-161. [DOI] [PubMed] [Google Scholar]
- 16.Jentsch, S. 1992. The ubiquitin-conjugation system. Annu. Rev. Genet. 26:179-207. [DOI] [PubMed] [Google Scholar]
- 17.Kato, H., T. Chen, X. H. Liu, N. Nakata, and K. Kogure. 1993. Immunohistochemical localization of ubiquitin in gerbil hippocampus with induced tolerance to ischemia. Brain Res. 619:339-343. [DOI] [PubMed] [Google Scholar]
- 18.Keeven, J., D. Ko, J. Shallom, B. Uccelini, and J. Golin. 2002. PDR2 gain-of-function mutations eliminate the need for Pdr1 and require the UBP6 product for resistance to translational inhibitors. Curr. Genet. 41:11-19. [DOI] [PubMed] [Google Scholar]
- 19.Kirillov, S., B. T. Porse, B. Vester, P. Wooley, and R. A. Garrett. 1997. Movement of the 3′-end of tRNA through the peptidyl transferase center and its inhibition by antibiotics. FEBS Lett. 406:223-233. [DOI] [PubMed] [Google Scholar]
- 20.Leggett, D. S., J. Hanna, A. Borodovsky, B. Crosas, M. Schmidt, R. T. Baker, T. Walz, H. Ploegh, and D. Finley. 2002. Multiple associated proteins regulate proteasome structure and function. Mol. Cell 10:495-507. [DOI] [PubMed] [Google Scholar]
- 21.McCusker, J. H., and J. E. Haber. 1988. Cycloheximide-resistant temperature-sensitive lethal mutations of Saccharomyces cerevisiae. Genetics 119:303-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCusker, J. H., and J. E. Haber. 1988b. Crl mutants of Saccharomyces cerevisiae resemble both mutants affecting general control of amino acid biosynthesis and omnipotent translational suppressor mutants. Genetics 119:317-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mimnaugh, E. G., H. Y. Chen, J. R. Davie, J. E. Celis, and L. Neckers. 1997. Rapid deubiquitination of nucleosomal histones in human tumor cells caused by proteasome inhibitors and stress response inducers: effects on replication, transcription, and the cellular stress response. Biochemistry 36:14418-14429. [DOI] [PubMed] [Google Scholar]
- 24.Mizukami, K., M. Ishikawa, M. Iwakiri, S. Hidaka, N. Kato, and T. Asada. 2002. Alterations of ubiquitin immunoreactivity in the hippocampal formation after perforant pathway lesion. Acta Neuropathol. 103:453-457. [DOI] [PubMed] [Google Scholar]
- 25.Müller-Taubenberger, A., J. Hagmann, A. Noegel, and G. Gerisch. 1988. Ubiquitin gene expression in Dictystelium is induced by heat and cold shock, cadmium, and inhibitors of protein synthesis. J. Cell Sci. 90:51-58. [DOI] [PubMed] [Google Scholar]
- 26.Murphey, R. K., and T. A. Godenschwege. 2002. New roles for ubiquitin in the assembly and function of neuronal circuits. Neuron 36:5-8. [DOI] [PubMed] [Google Scholar]
- 27.Na, S., D. S. Perlin, D. Seto-Young, G-F. Wang, and J. Haber. 1993. Characterization of yeast plasma membrane H+-ATPase mutant pma1-A135 and its revertants. J Biol. Chem. 268:11792-11797. [PubMed] [Google Scholar]
- 28.Noventa-Jordao, M. A., A. M. do Nacimento, M. H. S. Goldman, H. F. Terenzi, and G. H. Goldman. 2000. Molecular characterization of ubiquitin genes from Aspergillus nidulans: mRNA expression on different stress and growth conditions. Biochim. Biophys. Acta 1490:237-244. [DOI] [PubMed] [Google Scholar]
- 29.Obrig, T. G., W. J. Culp, W. L. McKeehan, and B. Hardesty. 1971. The mechanism by which cycloheximide and related glutarimide antibiotics inhibit peptide synthesis on reticulocyte ribosomes. J. Biol. Chem. 246:174-181. [PubMed] [Google Scholar]
- 30.Osaka, H., Y. Wang, K. Takada, S. Takizawa, R. Setsuie, H. Li, Y. Sato, K. Nishikawa, Y. Sun, M. Sakurai, T. Harada, Y. Hara, I. Kimura, S. Chiba, K. Namikawa, H. Kiyama, M. Noda, S. Aoki, and K. Wada. 2003. Ubiquitin carboxy-terminal hydrolase L1 binds to and stabilizes monoubiquitin in neurons. Hum. Mol. Gen. 12:1945-1958. [DOI] [PubMed] [Google Scholar]
- 31.Özkaynak, E., D. Finley, and A. Varshavsky. 1984. The yeast ubiquitin gene: head-to-tail repeats encoding a polyubiquitin precursor protein. Nature 312:663-666. [DOI] [PubMed] [Google Scholar]
- 32.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perlin, D. S., C. L. Brown, and J. E. Haber. 1988. Membrane potential defect in hygromycin B-resistant pma1 mutants of Saccharomyces cerevisiae. J. Biol. Chem. 34:18118-18122. [PubMed] [Google Scholar]
- 34.Rao, S. S., and A. P. Grollman. 1967. Cycloheximide resistance in yeast: a property of the 60S ribosomal subunit. Biochem. Biophys. Res. Commun. 29:696-704. [DOI] [PubMed] [Google Scholar]
- 35.Rose, I. A., and J. V. B. Warms. 1987. A specific endpoint assay for ubiquitin. Proc. Natl. Acad. Sci. USA 84:1477-1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Rubin, D. M., M. H. Glickman, C. N. Larsen, S. Dhruvakumar, and D. Finley. 1998. Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J. 17:4909-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schubert, U., L. C. Anton, J. Gibbs, C. C. Norbury, J. W. Yewdell, and J. R. Bennink. 2000. Rapid degradation of a large fraction of newly-synthesized proteins by proteasomes. Nature 404:770-774. [DOI] [PubMed] [Google Scholar]
- 39.Seufert, W., and S. Jentsch. 1990. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 9:543-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sherman, M. Y., and A. L. Goldberg. 2001. Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron 29:15-32. [DOI] [PubMed] [Google Scholar]
- 41.Spence, J., S. Sadis, A. L. Haas, and D. Finley. 1995. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol. Cell. Biol. 15:1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spence, J., R. R. Gali, G. Dittmar, F. Sherman, and D. Finley. 2000. Cell cycle regulated modification of the ribosome by a variant multiubiquitin chain. Cell 102:67-76. [DOI] [PubMed] [Google Scholar]
- 43.Swaminathan, S., A. Y. Amerik, and M. Hochstrasser. 1999. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol. Biol. Cell 10:2583-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan, Z., W. Tu, W. Liu, M. Baudry, and S. S. Scheiber. 2000. p53 accumulation due to down-regulation of ubiquitin: relevance for neuronal apoptosis. Cell Death Differ. 7:675-681. [DOI] [PubMed] [Google Scholar]
- 45.Tan, Z., W. Tu, and S. S. Scheiber. 2001. Downregulation of free ubiquitin: a novel mechanism of p53 stabilization and neuronal cell death. Mol. Brain Res. 91:179-188. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka, K., K. Matsumoto, and A. Toh-e. 1988. Dual regulation of the polyubiquitin gene by cyclic AMP and heat shock in yeast. EMBO J. 7:495-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verma, R., S. Chen, R. Feldman, D. Schieltz, J. Yates, J. Dohmen, and R. J. Deshaies. 2000. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell 11:3425-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verma, R., R. Oania, W. H. McDonald, J. R. Yates III, E. V. Koonin, and R. J. Deshaies. 2002. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298:611-615. [DOI] [PubMed] [Google Scholar]
- 49.Wettstein, F. O., H. Noll, and S. Penman. 1964. Effect of cycloheximide on ribosomal aggregates engaged in protein synthesis in vitro. Biochim. Biophys. Acta 87:525-527. [DOI] [PubMed] [Google Scholar]
- 50.Wilkinson, K. D. 2000. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin. Cell Dev. Biol. 11:141-148. [DOI] [PubMed] [Google Scholar]
- 51.Wilson, S. M., B. Battacharyya, R. A. Rachel, V. Coppola, L. Tessarollo, D. B. Householder, C. F. Fletcher, R. J. Miller, N. G. Copeland, and N. A. Jenkins. 2002. Synaptic defects in ataxia mice result from a mutation in Usp14, encoding a ubiquitin-specific protease. Nat. Genet. 32:420-425. [DOI] [PubMed] [Google Scholar]
- 52.Yao, T., and R. E. Cohen. 2002. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419:403-407. [DOI] [PubMed] [Google Scholar]