Abstract
Genomic imprinting at the Igf2/H19 locus originates from allele-specific DNA methylation, which modifies the affinity of some proteins for their target sequences. Here, we show that AT-rich DNA sequences located in the vicinity of previously characterized differentially methylated regions (DMRs) of the imprinted Igf2 gene are conserved between mouse and human. These sequences have all the characteristics of matrix attachment regions (MARs), which are known as versatile regulatory elements involved in chromatin structure and gene expression. Combining allele-specific nuclear matrix binding assays and real-time PCR quantification, we show that retention of two of these Igf2 MARs (MAR0 and MAR2) in the nuclear matrix fraction depends on the tissue and is specific to the paternal allele. Furthermore, on this allele, the Igf2 MAR2 is functionally linked to the neighboring DMR2 while, on the maternal allele, it is controlled by the imprinting-control region. Our work clearly demonstrates that genomic imprinting controls matrix attachment regions in the Igf2 gene.
On the distal part of mouse chromosome 7, the Igf2 and H19 genes are separated by 72 kbp. At this locus, imprinting originates from paternal germ line-specific methylation that impairs the binding of a protein factor (CTCF) to the intergenic imprinting-control region (ICR). On the maternal allele, binding of CTCF results in activation of an insulator, which prevents Igf2 expression by blocking promoter access to a set of enhancers located downstream of H19 (2, 14). Thus, Igf2 gene expression is restricted to the paternal allele, whereas H19 is maternally expressed. However, Igf2 imprinting also involves additional differentially methylated regions (DMR0, DMR1, and DMR2) that control regional epigenetic modifications in a hierarchical and tissue-specific fashion (19). DMR1, located upstream of the fetal Igf2 promoters, is preferentially methylated on the paternal allele and acts as a methylation-sensitive silencer in several mesodermal tissues (5). DMR2 maps to exons 5 to 6 and augments transcription on the methylated paternal allele (24). Finally, DMR0 is maternally methylated in the placenta and overlaps the placenta-specific P0 promoter (23). All these DMRs display tissue-specific methylation patterns, and we have recently proposed that their methylation profiles are regionally coordinated through long-range chromatin interactions (19).
Here, we identify intragenic AT-rich DNA sequences in the Igf2 gene that are contiguous to previously identified DMRs. These sequences have all the characteristics of matrix attachment regions (MARs) and are conserved in humans. MARs are operationally defined in MAR assays by their ability to associate to a nuclear matrix or scaffold (17) and have been implicated in the regulation of chromatin structure and gene expression. They are frequently associated with enhancers (4, 10, 11) and promote chromatin accessibility and histone acetylation (7, 9, 15, 20, 26). MARs may also sometimes act as boundaries, preventing the spreading of potential repressive effects from the adjacent chromatin (25). In this work, using allele-specific genomic MAR assays, we show that genomic imprinting controls nuclear matrix association in the Igf2 gene, most likely reflecting the presence of allele-specific MAR binding proteins in vivo.
MATERIALS AND METHODS
Genomic MAR assays.
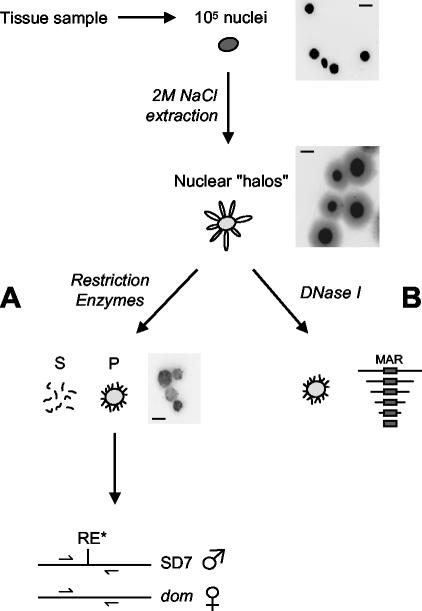
For each assay, samples were obtained from several mice, and nuclei were isolated as previously described (21). For nuclear halo preparation, we adapted a procedure from the NaCl method (22) (see below). Three major modifications of the standard procedure proved crucial for improving the reliability of the MAR assays. First, nuclei are kept very diluted to prevent aggregation. Second, each assay is performed in a single filtration unit to avoid centrifugation. Finally, matrix-associated DNA is carefully quantified by real-time PCR.
We incubated 105 nuclei on ice for 15 min in 1 ml of permeabilization buffer (10 mM Tris-HCl [pH 7.4], 3 mM MgCl2, 100 mM NaCl, 0.3 M sucrose, 0.5% Triton X-100) and stabilized at 37°C for 20 min. Stabilized nuclei were transferred in a 2-ml Ultrafree-CL filter (low-binding Durapore polyvinylidene difluoride membrane 0.22 μm) and filtered by applying a gentle pressure on the top of the tube using a rubber piston. Nuclei were then extracted with 1 ml of high-salt buffer (20 mM Tris-HCl pH 7.4, 2 M NaCl, 10 mM EDTA, 0.125 mM spermidine) to obtain nuclear halos. Each nuclear halo arises from a single nucleus and consists of a nuclear matrix surrounded by histone-depleted DNA loops that remain bound to the nuclear matrix by matrix attachment regions (MARs). Nuclear halos were washed four times on the filter with 1 ml of restriction buffer and treated with 500 U of appropriate restriction enzymes per ml for 3 h (see Fig. 2A). Digestion with XbaI, HindIII, and BamHI was performed in 6 mM Tris-HCl (pH 7.4)-6 mM MgCl2-50 mM NaCl-0.2 mM spermidine and digestion with HindIII and PstI was performed in 50 mM Tris-HCl (pH 8.0)-10 mM MgCl2-50 mM NaCl-0.2 mM spermidine.
FIG. 2.
MAR assays. We extracted 105 nuclei from a tissue sample with 2 M NaCl. The resulting nuclear halos were digested with restriction enzymes (A). Matrix-bound DNA (pellet, P) was isolated from loop DNA (supernatant, S) by ultrafiltration (see Materials and Methods). The relative enrichment of target sequences in the pellet fraction was determined by real-time quantitative PCR. For allele-specific MAR assays, primers were designed on either side of polymorphic restriction sites and quantifications were performed on DNA from the pellet fraction undigested or digested with the polymorphic enzyme (see Materials and Methods). Alternatively, for precise mapping of nuclear matrix attachments, nuclear halos were digested with DNase I before performing real-time PCR quantifications on the pellet fraction (B). Pictures show nuclei, nuclear halos, and restriction enzyme-treated halos stained with Hoechst. Scale bars, 10 μm.
Loop DNA (supernatant fraction) was isolated from nuclear matrices (pellet fraction) by ultrafiltration (Ultrafree-CL). Matrices remaining on the filter were washed in 1 ml of digestion buffer. DNA from both fractions was purified by proteinase K treatment and phenol-chloroform extraction and recovered by ethanol precipitation. Half of the DNA was loaded on agarose gel to control each assay; no more than 5% of total DNA should be retained in the matrix fraction. For precise mapping of nuclear matrix-attached sequences, the nuclear halos were digested with DNase I (Fig. 2B). Nuclear halos were washed four times on the filter in 1 ml of digestion buffer (10 mM Tris-HCl, pH 7.4, 100 mM NaCl, 25 mM KCl, 1 mM CaCl2, 0.25 mM spermidine) and treated with 50 U of DNase I (Sigma) for 50 min at 4°C. Digested matrices were washed three times in 1 ml of digestion buffer, and matrix-associated DNA was purified by proteinase K treatment and phenol-chloroform extraction and recovered by ethanol precipitation. DNA was loaded on agarose gel to check the size of matrix-associated fragments.
Quantitative analysis of MAR attachment.
For each MAR assay, target DNA sequences were quantified by real-time PCR with a SYBR Green mix and a LightCycler apparatus (Roche Molecular Biochemicals, software version 3.5). Melting curves were systematically performed to check for proper amplifications. For the restriction enzyme method, 1/50 of the DNA from the supernatant and pellet fractions was amplified with indicated primers, and we calculated the ratio of the quantifications in the pellet fraction versus the supernatant fraction. To standardize each assay, ratio values were normalized against the ratio obtained for a negative control, which is a sequence located upstream of Igf2 between the Ins and RpL13 genes with no predicted MAR. The well-characterized MAR of the Igκ gene (4) was used as a positive control, and gapdh was used to assess the basal enrichment of highly transcribed genes in the matrix fraction (see Fig. 3A, right panel).
FIG. 3.
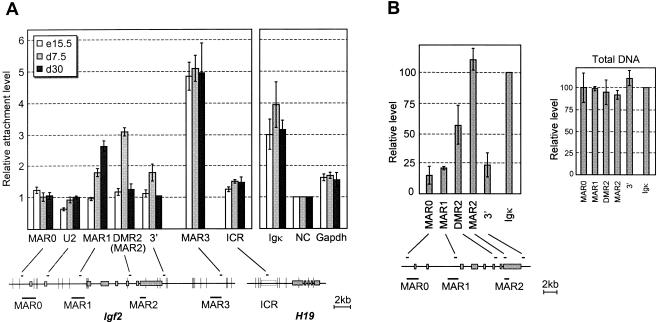
Matrix association patterns of Igf2 MARs during liver development. Below the bar graphs, the positions of restriction sites (vertical bars), real-time PCR-amplified sequences (small horizontal bars), potential MARs, and exons (solid boxes) are indicated. (A) Nuclear halos prepared from liver nuclei at the indicated developmental stages were digested with XbaI, HindIII, and BamHI. The graph shows the relative matrix attachment levels, expressed as the ratio between the amount of target sequence in the loop (S) and MAR fractions (P) after normalization to a negative control (NC), which was given the value of 1 (see Materials and Methods). In our assays, highly expressed genes such as gapdh and H19 (not shown) are weakly retained in the matrix fraction relative to the negative control. Error bars show the standard deviation for four (embryonic days 15.5 and 7.5) and two (embryonic day 30) independent experiments. In this experiment, MAR2 enrichment was investigated with the DMR2 primers located in the same restriction fragment. (B) Nuclear halos prepared from 7.5-day-old mouse liver were treated with DNase I. The left panel shows PCR quantifications on matrix-associated DNA, normalized to the Igκ MAR. A control performed on total genomic DNA is also shown (right panel). Error bars show the standard deviation for two independent experiments.
For the DNase I method, we carefully selected matrix-associated DNA fragments of size ranging between 600 and 2500 bp on agarose gels to avoid PCR bias. 1/50 of the DNA was amplified with indicated primers. To standardize each assay, values were normalized to the Igκ MAR. A control was performed on 600- to 2,500-bp DNA fragments from total genomic DNA treated with DNase I to check the lack of PCR bias (Fig. 3B, left panel).
The primer sequences were as follows: MAR0, 5′-GACTGCCTCACTCTGATAAGC-3′ and 5′-CCCACTAGTCTCTACCCTTTGCAC-3′; DMR0, 5′-TGGGTAGGTGGCTGGGGACTT-3′ and 5′-CAGCTGTGTACTCCTGCTGGGAAG-3′; U2, 5′-CTCTAGCACAGGAGCATCAGAGCTC-3′ and 5′-CTGCCTCGTGTCTGCTTGGC-3′; MAR1, 5′-CCCGTGTTGCTCATGCCTGG-3′ and 5′-GGGTTCCTAAATTATGGGGCCTG-3′; DMR2, 5′-GGAGCTTGTTGACACGCTTCAG-3′ and 5′-GGATGGCCAAGGTCCAGCTCTC-3′; MAR2, 5′-CCTGCTGACTAGCACCTCCTCTC-3′ and 5′-GTGATGGAACTGTCCCTGCTC-3′; Igf2 3′, 5′-CTCTGAGGAACCCAGAGGGTAG-3′ and 5′-GGGACAGACTCTAGCATAGC-3′; MAR3, 5′-GGTTCAAGTGAGAACTGTCCTCC-3′ and 5′-GTAGAGGCATGGCAGCAACC-3′; ICR, 5′-GCACATCTATGAGGACACCTGAC-3′ and 5′-GACAGTGCAAAACAGGTGAACCC-3′; Igκ, 5′-CTCCTAGGCAGGTGGCCCAG-3′ and 5′-GGACAGGCCTTAAGCCAGGG-3′; negative control, 5′-CCTGCCACACCAAGACTATC-3′ and 5′-TCTAGGCCCTCTTCCATCTC-3′; and Gapdh, 5′-ACAGTCCATGCCATCACTGCC-3′ and 5′-GCCTGCTTCACCACCTTCTTG-3′.
Quantification of relative allele abundance.
To assess the allelic specificity of MAR attachment, we performed MAR assays on nuclei from hybrid mice obtained by mating C57BL/6J × CBA F1 Mus musculus domesticus with SD7 mice. SD7 is a congenic C57BL/6J × CBA strain harboring the distal part of Mus spretus chromosome 7. For quantification of relative allele abundance, primers for real-time PCR were designed on either side of a polymorphic restriction site (Fig. 2). The relative proportion of both alleles was deduced from quantifications on undigested DNA and DNA digested with the polymorphic enzyme. One-fifth of the DNA from the pellet fraction was treated or not with 30 U of polymorphic enzyme for 3 h. For each primer pair, the reliability of the method was assessed with total genomic DNA to verify that quantification gives a 1:1 allelic ratio (not shown).
For MAR2 and MAR3, amplification biases for the domesticus versus spretus alleles were detected and carefully corrected with a mathematical model (31) adapted from reference 30. The bias values are 0.47 (MAR2) and 2.18 (MAR3). Primer sequences were as follows: DMR0 primers around the BamHI*SD7 site for MAR0; DMR2 primers around the EcoNI*SD7 site for MAR2; 5′-CCTGGAAGCTGATGATCCTTGG-3′ and 5′-GACACCAAACCCAATCATGGTAGC-3′ primers around the DraI*SD7 site for MAR3; 5′-CAGGACTCCAAAATCGGGACTC-3′ and 5′-CCAGCTAGGAAGACCGGTATGG-3′ primers around the DraI*SD7 site for MAR1.
RESULTS
Potential matrix attachment regions at the Igf2/H19 locus.
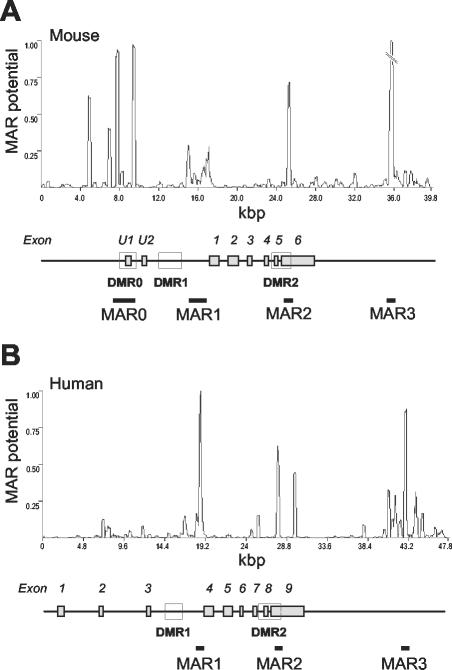
Bioinformatics analysis of the entire mouse Igf2/H19 locus predicted only four MARs located in and around the Igf2 gene (Fig. 1A). Remarkably, the first three (MAR0, MAR1, and MAR2) map next to the three Igf2 DMRs, which are important for Igf2 gene expression and/or imprinting (5, 24). The last one (MAR3) lies 8 kbp downstream of Igf2. Potential MAR0, MAR1, and MAR3 have previously been described to bind in vitro to purified nuclear matrices (13). Potential MAR1, MAR2, and MAR3 are conserved in human, where they map at equivalent positions relative to IGF2 DMRs and exons (Fig. 1B), whereas potential MAR0, which is located in a divergent region upstream of the mouse Igf2 gene (23), is not found in the human gene.
FIG. 1.
Conserved potential MARs in the murine and human Igf2 loci. Murine (A) and human (B) Igf2 sequences were analyzed with the MAR-Wiz software (version 1.5 [www.futuresoft.org]). A window size of 300 bp stepped at 50-bp intervals was used. The line graph shows the MAR potential score versus nucleotide position. For the murine gene (A), the MAR3 potential peak was clipped to 50% of the maximum. The gene structures are shown below the graphs (solid boxes, exons; open boxes, DMRs).
Tissue-specific patterns of matrix association in the Igf2 gene during mouse development.
Combining an improved nuclear matrix binding assay and real-time PCR quantification (Fig. 2 and Materials and Methods), we first analyzed nuclear matrix attachment in the mouse liver at three perinatal stages: embryonic day 15.5 and day 7.5 or 30 after birth. We have previously shown by run-on experiments that the Igf2 gene is actively transcribed in embryonic and early postnatal stages, but completely repressed after postnatal day 18 (21). Interestingly, each potential MAR at the Igf2 locus showed a specific attachment pattern (left panel of Fig. 3A).
The extragenic MAR3 maintained high nuclear matrix attachment levels throughout the period, while the predicted MAR0 and a restriction fragment containing the U2 exon of Igf2 remained unbound. Conversely, MAR1 and MAR2 attachments are modulated during the perinatal period in mouse liver. At embryonic day 15.5, no nuclear matrix association of MAR1 or MAR2 was detected. In contrast, in a 7.5-day-old mouse liver, MAR2 was attached to the nuclear matrix at a relative level threefold higher than the negative control. However, the 3′ part of the gene is also weakly retained in the nuclear matrix fraction, suggesting that a carryover may occur for DNA fragments around MAR2.
To define more precisely the intragenic attachment region, a genomic MAR assay was performed on 7.5-day-old mouse liver, and genomic DNA retained in the nuclear matrix was treated with DNase I (see Fig. 2B and Materials and Methods). With an additional PCR primer pair within the predicted MAR2 sequence, we confirmed that intragenic attachment is indeed located at MAR2, immediately adjacent to the differentially methylated region 2 of Igf2 (Fig. 3B). This attachment is lost in 30-day-old mouse liver, where Igf2 is repressed, when, surprisingly, MAR1 becomes attached to the nuclear matrix (Fig. 3A, left panel).
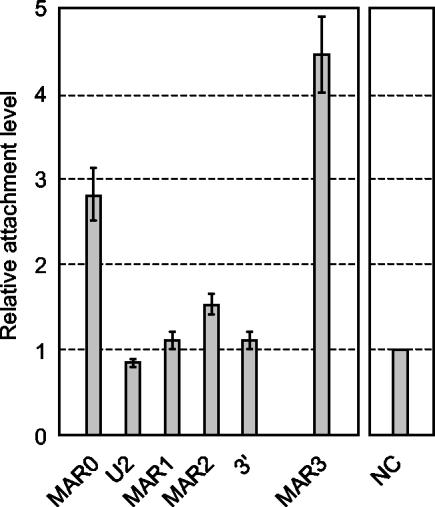
Since the predicted MAR0 was not found attached in liver and because the adjacent promoter (P0) and DMR (DMR0) are placenta specific (23), we analyzed the pattern of Igf2 MAR attachment in the placenta at embryonic day 18.5. As expected, MAR0 was highly associated with the nuclear matrix in the placenta (Fig. 4), while MAR1 and MAR2 were not or very weakly associated. Again, as in the liver, MAR3 showed a high attachment level in the placenta.
FIG. 4.
Matrix association patterns of Igf2 MARs in placenta. Nuclear halos were prepared from placenta at embryonic day 18.5 and analyzed as described for Fig. 3A. Error bars show the standard deviation for two independent experiments.
In some instances, MARs are known to have insulator activities (33). Thus, as previously suggested (6), we tested if the insulator upstream of H19 was bound to the nuclear matrix, even though no MAR was predicted in this region. We found that the ICR is very weakly associated with the nuclear matrix in mouse liver (Fig. 3A, left panel), and thus its insulator function is unlikely to result from MAR activity.
Genomic imprinting controls nuclear matrix association of Igf2 MARs.
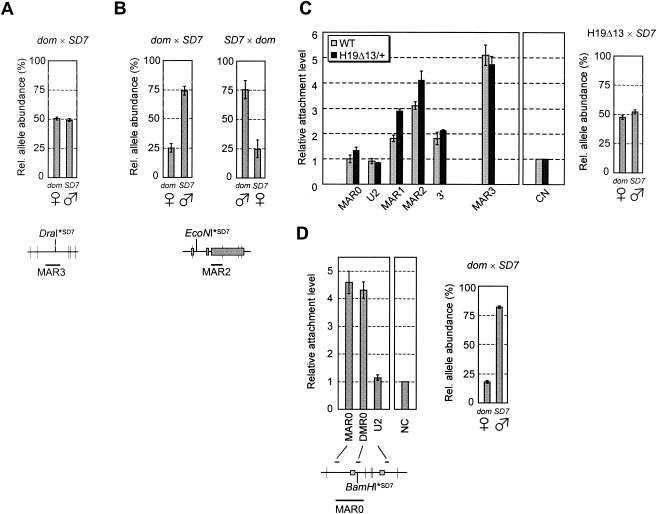
We then investigated the possibility that nuclear matrix attachments could be restricted to one of the parental alleles. MAR assays were performed on liver nuclei from 7.5-day-old hybrid mice obtained by mating a Mus musculus domesticus female with an SD7 male (Fig. 2A). SD7 is a congenic C57BL/6J × CBA strain harboring the distal part of Mus spretus chromosome 7. We calculated the proportion of each parental allele in the matrix-associated DNA fraction by using polymorphic restriction sites and real-time PCR quantification (see Materials and Methods). In the case of MAR3, both alleles were found to be equally retained in the nuclear matrix (Fig. 5A). In contrast, MAR2 clearly showed a preferential attachment of the paternally expressed Igf2 allele in 7.5-day-old mouse liver (Fig. 5B, left panel). Given the background level of our assay (see negative control in Fig. 3A, right panel), we conclude that MAR2 is not significantly attached on the maternal allele.
FIG. 5.
Allelic specificity of matrix association at the Igf2 locus. The allelic ratio of the XbaI, HindIII, and BamHI (vertical bars) restriction fragments containing MAR3 (A) or MAR2 (B and C) were assessed by real-time PCR quantification with a polymorphic DraI (MAR3) or EcoNI (MAR2) site specific for SD7 mice (see Materials and Methods). (A) Relative allele abundance of MAR3 in matrix-associated DNA (pellet fraction) from 7.5-day-old hybrid mouse liver. (B and C) Relative allele abundance of MAR2 in matrix-associated DNA prepared from liver nuclei of wild-type 7.5-day-old hybrid mice (B) or 7.5-day-old hybrid mice carrying a maternal deletion of the H19 region (H19Δ13) (18) (C, right panel). (C, left panel) Matrix association pattern of Igf2 MARs in the liver of H19Δ13 mutant mice. The pattern of Igf2 MAR attachment in wild-type mice (wt) is shown for comparison (data from Fig. 3A). (D) To assess the allelic specificity of MAR0 attachment in placenta, nuclear halos were digested with HindIII and PstI (vertical bars) (left panel). The allelic ratio of the MAR0 restriction fragment in the matrix-associated DNA was calculated with a polymorphic BamHI site specific for SD7 mice (right panel). Error bars show the standard deviation for two or three independent experiments. dom, Mus domesticus. Symbols for female and male denote maternal and paternal alleles, respectively.
Hybrid mice obtained from the reverse cross (SD7 female × Mus musculus domesticus male) gave identical results (Fig. 5B, right panel), confirming that this allelic specificity is due to parental imprinting and not to a genotype-specific effect. The influence of genomic imprinting on MAR2 attachment was then directly tested with liver samples from hybrid mice carrying a maternal transmission of the H19Δ13 deletion (18), where Igf2 has lost imprinting and shows biallelic expression due to the removal of the imprinting-control region (28, 29). In these samples, MAR2 attachment level is increased (Fig. 5C, left panel), as both parental alleles were recovered from the nuclear matrix fraction (Fig. 5C, right panel).
To determine on which allele MAR0 is attached in the placenta, we performed MAR assays on nuclei from the placenta of hybrid mice. Attachment of a new restriction fragment, containing the two major predicted peaks for MAR0 (see Fig. 1A) and a polymorphic restriction site, was quantified. We first saw that MAR0 had a higher attachment level in this assay than in the first one (compare Fig. 4 and Fig. 5D, left panel), suggesting that MAR0 may extend further from the previously described fragment (13), into the surrounding predicted peaks. Importantly, MAR0 was shown to be almost exclusively attached on the expressed paternal allele (Fig. 5D, right panel). Noteworthy, the neighboring DMR0 is known to be mostly unmethylated on the paternal allele in the placenta (23), whereas in liver, DMR2 is highly methylated on that allele (8, 32). Thus, like MAR2 in liver, MAR0 attachment occurs on the paternal Igf2 allele, although the adjacent DMR shows the converse allelic methylation pattern.
Finally, MAR1 attachment occurs equally on both parental alleles in wild-type 30-day-old mouse liver (data not shown). As the global attachment level of this MAR is low (see Fig. 3A), each allele is actually weakly associated compared to the matrix-associated alleles in MAR2 and MAR3.
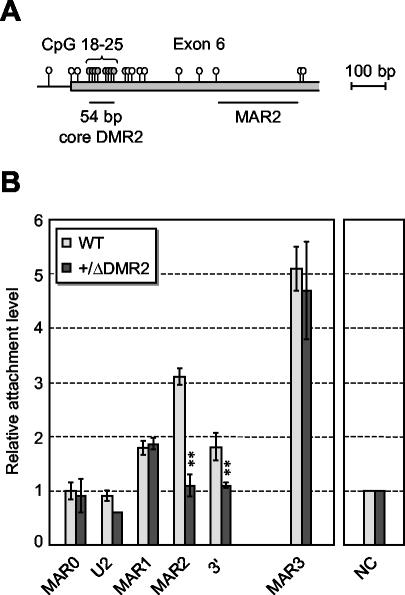
To gain further insight into the mechanisms by which MARs are linked to Igf2 imprinting, we investigated potential relationships between Igf2 MAR2 and the adjacent DMR2. To this end, MAR assays were performed on mice carrying a 54-bp deletion of a core region of DMR2 (ΔDMR2) (24). This deletion leaves MAR2 intact, as it removes only a small sequence located about 300 bp upstream the MAR (Fig. 6A). Strikingly, we found that in the livers of 7.5-day-old mice with paternal transmission of the ΔDMR2 deletion, MAR2 attachment remained at background levels (Fig. 6B). No significant effect was detected on matrix association of other sequences in the locus. This demonstrates that, on the paternal allele, the core DMR2 is required for MAR2 attachment. Finally, one also confirms here that, in liver, the contribution of the wild-type maternal allele to MAR2 attachment is insignificant.
FIG. 6.
Matrix association patterns of Igf2 MARs in liver of ΔDMR2 mice. (A) Map of the Igf2 intron 5/exon 6 region, showing the position of CpG dinucleotides (lollipops) and the location of MAR2 relative to the core DMR2 region (CpGs 18 to 25) which is deleted in ΔDMR2 mutant mice (24). (B) Nuclear halos prepared from liver nuclei of 7.5-day-old mutant mice with paternal transmission of the ΔDMR2 deletion (+/ΔDMR2) were analyzed as described for Fig. 3A. The pattern of Igf2 MAR attachment in wild-type mice (wt) is shown for comparison (data from Fig. 3A). Double stars indicate significant variations between mutant and wild-type mice. Error bars show the standard deviation for two independent experiments.
DISCUSSION
In this work, we used allele-specific MAR assays and quantitative real-time PCR to perform a detailed analysis of the Igf2 MARs during mouse development. We show that MAR3, which is located 8 kbp downstream of Igf2, is constitutively attached on both parental alleles in all tissues investigated, whereas nuclear matrix association of the intragenic MAR0 and MAR2 is tissue specific and highly modulated during development. This tissue specificity is remarkably consistent with the known tissue specificity of the neighboring DMRs: MAR0 and DMR0 are placenta-specific (23) while MAR2 and DMR2 are active in liver (24).
Importantly, both MAR0 and MAR2 attachments are restricted to the paternal allele, while the neighboring DMRs display opposite allelic methylation patterns. In placenta, DMR0 is preferentially methylated on the maternal allele (23) while in liver, DMR2 is highly methylated on the paternal allele (8, 32). Moreover, MAR2 becomes detached on the paternal allele in 30-day-old mouse liver, while DMR2 remains highly methylated (32), and, therefore, DNA methylation on its own is clearly not sufficient to explain nuclear matrix attachment. In mice carrying an H19 ICR deletion, where Igf2 imprinting is lost (18), MAR2 is attached on both alleles (Fig. 5C, right panel). This experiment indicates that, on the maternal allele, genomic imprinting impairs the activity of MARs through the active ICR/CTCF insulator. As CTCF has been described to have an AT-hook motif distinct from the zinc finger DNA binding motifs (1), it could directly contact MAR2 and prevent its attachment to the nuclear matrix. However, we cannot exclude that the influence of the ICR may be more indirect. This result reveals a functional link between MARs and imprinting which is also reinforced by the finding that MAR2 nuclear matrix association depends on the adjacent DMR2 (Fig. 6).
Two previous studies have addressed the relationship between MARs and imprinting (12, 16). Using fluorescent in situ hybridization techniques, both studies reported a preferential association of one parental allele of the human SNRPN gene to the nuclear matrix, the identity of which remains controversial. In the case of the Igf2 locus, we show that each MAR has a specific attachment pattern, even though they are separated by only a few kilobase pairs. Furthermore, the patterns depend on the tissue and the developmental stage. Such complexity may explain the controversial data obtained for matrix attachments at the SNRPN locus, which was analyzed by techniques of lower resolution.
In the Igf2 gene, intragenic MAR activity appears closely linked to expression. However, several lines of evidence clearly demonstrate that, in our assays, MAR activity cannot be considered a mere consequence of gene activity. First, highly expressed genes such as gapdh (Fig. 3A) and H19 (data not shown) are weakly retained in our quantitative assays compared to Igf2 MARs. Furthermore, in placenta (Fig. 4) and myoblastic cells (data not shown), the Igf2 gene body (including MAR2) is not significantly retained in the MAR fraction despite high expression levels. Finally, in the liver of embryonic day 15 embryos and ΔDMR2 mutant mice, MAR2 is inactive despite significant Igf2 transcription levels (21, 24), demonstrating that there is no direct correlation between MAR2 activity and Igf2 transcription.
We have previously postulated that a methylation-sensitive factor may bind the methylated core DMR2 on the paternal Igf2 allele, thus favoring transcription (24). This putative factor may help to regulate the association of MAR2 with the nuclear matrix. We propose that intragenic MARs and DMRs are part of a common tissue-specific regulatory mechanism controlled by genomic imprinting and required for high Igf2 expression levels on the paternal allele. One attractive model is that MARs induce a specific chromatin-loop organization that would favor long-range interactions between the Igf2 gene and distal enhancers downstream of the H19 gene. This model is strengthened by preliminary results indicating that the endodermic enhancers, located 8 kb downstream of the H19 promoter, are also recovered from the nuclear matrix fraction during liver development despite there being no predicted MAR in this region (data not shown). It would also be consistent with recent results demonstrating that, in the mouse, promoters and distant enhancers physically interact in vivo (3). Alternatively, MARs could act locally to stabilize the gene in the transcription complex or serve as entry sites for chromatin remodeling factors (27, 34) or components of the transcriptional machinery.
Altogether, our work confirms that MAR attachment reflects in vivo epigenetic characteristics of MAR sequences, most likely the presence of MAR-binding factors, and reveals that the association of such factors is a new epigenetic feature controlled by genomic imprinting at the Igf2 locus.
Acknowledgments
We thank Y. Vassetzky for discussions and advice about nuclear halo preparation, S. Tilghman for providing the H19Δ13 mouse strain, S. Heeson for technical assistance, and L. Milligan, T. Kohwi-Shigematsu, and colleagues from IGMM for critical reading of the manuscript. We also thank the staff from the animal units at IGMM and the Babraham Institute for help with mouse breeding.
This work was supported by the BBSRC, the CRUK, grants from the Association pour la Recherche contre le Cancer (ARC no. 4274) and from the Fond National de la Science (ACI jeune chercheur) given to T. Forné and by laboratory funds from the Centre National de la Recherche Scientifique and Université Montpellier II given to C. Brunel. M. Weber and H. Hagège were supported by Ph.D. fellowships from Allocations de Moniteur Normalien (Ministère de l'Education Nationale, de la Recherche et de la Technologie).
REFERENCES
- 1.Aravind, L., and D. Landsman. 1998. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 26:4413-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405:482-485. [DOI] [PubMed] [Google Scholar]
- 3.Carter, D., L. Chakalova, C. S. Osborne, Y. F. Dai, and P. Fraser. 2002. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 32:623-626. [DOI] [PubMed] [Google Scholar]
- 4.Cockerill, P. N., and W. T. Garrard. 1986. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell 44:273-282. [DOI] [PubMed] [Google Scholar]
- 5.ConstÂncia, M., W. Dean, S. Lopes, T. Moore, G. Kelsey, and W. Reik. 2000. Deletion of a silencer element in Igf2 results in loss of imprinting independent of H19. Nat. Genet. 26:203-206. [DOI] [PubMed] [Google Scholar]
- 6.Efstratiadis, A. 1994. Parental imprinting of autosomal mammalian genes. Curr. Opin. Genet. Dev. 4:265-280. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez, L. A., M. Winkler, and R. Grosschedl. 2001. Matrix attachment region-dependent function of the immunoglobulin mu enhancer involves histone acetylation at a distance without changes in enhancer occupancy. Mol. Cell. Biol. 21:196-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forné, T., J. Oswald, W. Dean, J. R. Saam, B. Bailleul, L. Dandolo, S. M. Tilghman, J. Walter, and W. Reik. 1997. Loss of the maternal H19 gene induces changes in Igf2 methylation in both cis and trans. Proc. Natl. Acad. Sci. 94:10243-10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forrester, W. C., L. A. Fernandez, and R. Grosschedl. 1999. Nuclear matrix attachment regions antagonize methylation-dependent repression of long-range enhancer-promoter interactions. Genes Dev. 13:3003-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forrester, W. C., C. van Genderen, T. Jenuwein, and R. Grosschedl. 1994. Dependence of enhancer-mediated transcription of the immunoglobulin mu gene on nuclear matrix attachment regions. Science 265:1221-1225. [DOI] [PubMed] [Google Scholar]
- 11.Gasser, S. M., and U. K. Laemmli. 1986. Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell 46:521-530. [DOI] [PubMed] [Google Scholar]
- 12.Greally, J. M., T. A. Gray, J. M. Gabriel, L. Song, S. Zemel, and R. D. Nicholls. 1999. Conserved characteristics of heterochromatin-forming DNA at the 15q11-q13 imprinting center. Proc. Natl. Acad. Sci. 96:14430-14435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greally, J. M., M. E. Guinness, J. McGrath, and S. Zemel. 1997. Matrix-attachment regions in the mouse chromosome 7F imprinted domain. Mamm, Genome 8:805-810. [DOI] [PubMed] [Google Scholar]
- 14.Hark, A. T., C. J. Schoenherr, D. J. Katz, R. S. Ingram, J. M. Levorse, and S. M. Tilghman. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405:486-489. [DOI] [PubMed] [Google Scholar]
- 15.Jenuwein, T., W. C. Forrester, L. A. Fernandez-Herrero, G. Laible, M. Dull, and R. Grosschedl. 1997. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature 385:269-272. [DOI] [PubMed] [Google Scholar]
- 16.Kagotani, K., H. Nabeshima, A. Kohda, M. Nakao, H. Taguchi, and K. Okumura. 2002. Visualization of transcription-dependent association of imprinted genes with the nuclear matrix. Exp. Cell Res. 274:189-196. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli, U. K., E. Kas, L. Poljak, and Y. Adachi. 1992. Scaffold-associated regions: cis-acting determinants of chromatin structural loops and functional domains. Curr. Opin. Genet. Dev. 2:275-285. [DOI] [PubMed] [Google Scholar]
- 18.Leighton, P. A., R. S. Ingram, J. Eggenschwiler, A. Efstratiadis, and S. M. Tilghman. 1995. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 375:34-39. [DOI] [PubMed] [Google Scholar]
- 19.Lopes, S., A. Lewis, P. Hajkova, W. Dean, J. Oswald, T. Forné, A. Murrell, M. ConstÂncia, M. Bartolomei, J. Walter, and W. Reik. 2003. Epigenetic modifications in an imprinting cluster are controlled by a hierarchy of DMRs suggesting long-range chromatin interactions. Hum. Mol. Genet. 12:295-305. [DOI] [PubMed] [Google Scholar]
- 20.Martens, J. H., M. Verlaan, E. Kalkhoven, J. C. Dorsman, and A. Zantema. 2002. Scaffold/matrix attachment region elements interact with a p300-scaffold attachment factor A complex and are bound by acetylated nucleosomes. Mol. Cell. Biol. 22:2598-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milligan, L., E. Antoine, C. Bisbal, M. Weber, C. Brunel, T. Forné, and G. Cathala. 2000. H19 gene expression is up-regulated exclusively by stabilization of the RNA during muscle cell differentiation. Oncogene 19:5810-5816. [DOI] [PubMed] [Google Scholar]
- 22.Mirkovitch, J., M. E. Mirault, and U. K. Laemmli. 1984. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell 39:223-232. [DOI] [PubMed] [Google Scholar]
- 23.Moore, T., M. ConstÂncia, M. Zubair, B. Bailleul, R. Feil, H. Sasaki, and W. Reik. 1997. Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse Igf2. Proc. Natl. Acad. Sci. 94:12509-12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murrell, A., S. Heeson, L. Bowden, M. ConstÂncia, W. Dean, G. Kelsey, and W. Reik. 2001. An intragenic methylated region in the imprinted Igf2 gene augments transcription. EMBO Rep. 2:1101-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Namciu, S. J., K. B. Blochlinger, and R. E. Fournier. 1998. Human matrix attachment regions insulate transgene expression from chromosomal position effects in Drosophila melanogaster. Mol. Cell. Biol. 18:2382-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pemov, A., S. Bavykin, and J. L. Hamlin. 1998. Attachment to the nuclear matrix mediates specific alterations in chromatin structure. Proc. Natl. Acad. Sci. 95:14757-14762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reyes, J. C., C. Muchardt, and M. Yaniv. 1997. Components of the human SWI/SNF complex are enriched in active chromatin and are associated with the nuclear matrix. J. Cell Biol. 137:263-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ripoche, M. A., C. Kress, F. Poirier, and L. Dandolo. 1997. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev. 11:1596-1604. [DOI] [PubMed] [Google Scholar]
- 29.Thorvaldsen, J. L., K. L. Duran, and M. S. Bartolomei. 1998. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 12:3693-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warnecke, P. M., C. Stirzaker, J. R. Melki, D. S. Millar, C. L. Paul, and S. J. Clark. 1997. Detection and measurement of PCR bias in quantitative methylation analysis of bisulphite-treated DNA. Nucleic Acids Res. 25:4422-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber, M., H. Hagège, G. Lutfalla, L. Dandolo, C. Brunel, G. Cathala, and T. Forné. 2003. A real-time polymerase chain reaction assay for quantification of allele ratios and correction of amplification bias. Anal. Biochem. 320:252-258. [DOI] [PubMed] [Google Scholar]
- 32.Weber, M., L. Milligan, A. Delalbre, E. Antoine, C. Brunel, G. Cathala, and T. Forné. 2001. Extensive tissue-specific variation of allelic methylation in the Igf2 gene during mouse fetal development: relation to expression and imprinting. Mech. Dev. 101:133-141. [DOI] [PubMed] [Google Scholar]
- 33.West, A. G., M. Gaszner, and G. Felsenfeld. 2002. Insulators: many functions, many mechanisms. Genes Dev. 16:271-288. [DOI] [PubMed] [Google Scholar]
- 34.Yasui, D., M. Miyano, S. Cai, P. Varga-Weisz, and T. Kohwi-Shigematsu. 2002. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature 419:641-645. [DOI] [PubMed] [Google Scholar]