Abstract
The stability of the tumor suppressor protein p53 is regulated via the ubiquitin-proteasome-dependent proteolytic pathway. Like most substrates of this pathway, p53 is modified by the attachment of polyubiquitin chains prior to proteasome-mediated degradation. However, the mechanism(s) involved in the delivery of polyubiquitylated p53 molecules to the proteasome are currently unclear. Here, we show that the human DNA repair protein hHR23 binds to polyubiquitylated p53 via its carboxyl-terminal ubiquitin-associated (Uba) domain shielding p53 from deubiquitylation in vitro and in vivo. In addition, downregulation of hHR23 expression within cells by RNA interference results in accumulation of p53. Since the Ubl domain of hHR23 has been shown to interact with the 26S proteasome, we propose that hHR23 is intrinsically involved in the delivery of polyubiquitylated p53 molecules to the proteasome. In this model, the Uba domain of hHR23 binds to polyubiquitin chains formed on p53 and protects them from deubiquitylation, while the Ubl domain delivers the polyubiquitylated p53 molecules to the proteasome.
Most substrate proteins of the 26S proteasome are covalently modified by the attachment of so-called polyubiquitin chains that serve as recognition signals for the proteasome. Conjugation of ubiquitin (ubiquitylation) to proteins requires the concerted action of several enzymes, including the ubiquitin-activating enzyme E1, ubiquitin-conjugating enzymes E2, ubiquitin-protein ligases E3, and ubiquitin assembly factors E4. E3 proteins are considered to specifically interact with individual substrate proteins, thus providing the specificity of the ubiquitin conjugation reaction (19, 39, 53).
Over the past few years, it has become clear that modification of proteins with ubiquitin is not only involved in protein degradation but also serves additional functions. For example, ubiquitylation has been implicated in inducing endocytosis of certain membrane proteins or in regulating the biochemical activity of proteins in a reversible manner, similar to phosphorylation events (14, 19, 23, 40, 53). The eventual fate of a ubiquitylated protein appears to be determined by the mode of ubiquitylation. Proteins destined for proteasome-mediated degradation seem to be modified by a polyubiquitin chain(s), in which the individual ubiquitin moieties are linked via lysine 48 or 29 of ubiquitin. In contrast, proteins not destined for degradation are modified by single ubiquitin moieties or by polyubiquitin chains, in which the individual ubiquitin moieties are linked via lysine residues other than lysines 48 and 29 (19, 38).
Covalent attachment of a lysine 48-linked tetraubiquitin chain is necessary and sufficient for a model substrate to be recognized and degraded by the 26S proteasome in vitro (48). Furthermore, at least two subunits of the 19S regulatory particle of the 26S proteasome have the capacity to physically interact with ubiquitin, suggesting that proteins in addition to the 26S proteasome are not required for the degradation of polyubiquitylated proteins (13, 33). However, there is accumulating evidence that additional proteins can modulate the transfer of polyubiquitylated proteins from the site of ubiquitylation to the proteasome. One group of such proteins is represented by the Saccharomyces cerevisiae proteins Rad23 and Dsk2 and their respective human homologs (hHR23A and hHR23B, hPlic-1, and Chap1/hPlic-2) (11, 18, 24, 28, 43, 44, 52). The architectures of these proteins are similar in that they bear an N-terminal ubiquitin-like (Ubl) domain and one (Dsk2, whose human homologue is hPlic) or two (Rad23 and hHR23) more C-terminally located ubiquitin-associated (Uba) domains. The Ubl domain of Rad23 and the hHR23 proteins has been shown to interact with subunits of the 19S complex of the 26S proteasome, while Uba domains in general have the capacity to bind to polyubiquitin chains (5, 11, 12, 16, 18, 24, 28, 43, 44, 52). Together with genetic analyses, these biochemical data indicate that at least some Ubl/Uba-containing proteins can act as bridging proteins between polyubiquitylated proteins and the 26S proteasome (12, 43). Furthermore, Rad23 has been reported to interfere with the ubiquitylation of proteins, suggesting an additional role for Rad23 in the regulation of ubiquitin-dependent proteolysis or in the regulation of ubiquitin conjugation in general (11, 36).
Rad23 and the hHR23 proteins are involved in the nucleotide excision repair pathway (34, 41). Moreover, hHR23 interacts with E6-AP, an E3 of the Hect domain family (27, 46), and its half-life has been reported to be regulated in a cell cycle-dependent manner (31). Interestingly with respect to nucleotide excision repair, degradation of hHR23 seems to be blocked upon UV-induced DNA damage, providing a possible mechanism by which the activity of hHR23 can be regulated (31). E6-AP was originally isolated as a protein that interacts with the E6 oncoprotein of cancer-associated human papillomaviruses (HPVs) and mediates the E6-induced ubiquitylation and degradation of the tumor suppressor protein p53 in HPV-positive cancer cells (22, 26, 45). In normal (i.e., HPV-negative) cells, however, ubiquitylation and subsequent degradation of p53 is mediated by the RING finger-type E3 Hdm2 (or Mdm2 in murine cells) (1, 3, 17, 25).
p53 is a key protein of the DNA damage response. Upon DNA damage, p53 levels increase and the activated p53 protein induces cell cycle arrest and/or apoptosis (1, 3). This indicates that under normal growth conditions, the antiproliferative activities of p53 need to be tightly controlled, a task that is in part achieved by Hdm2-induced degradation of p53. The Hdm2 proto-oncoprotein possesses four evolutionarily conserved domains, including an amino-terminal p53 binding domain, a carboxyl-terminal RING finger with E3 ligase activity, and a central acidic domain with an as-yet-unknown function. However, alanine substitutions of putative phosphorylation sites within this central acidic domain interfere with p53 degradation, while they still allow ubiquitylation to occur (7). This indicates that ubiquitylation of p53 is not sufficient for its degradation and that additional steps are required.
To gain insight into the mechanism(s) involved in linking p53 ubiquitylation and degradation, we investigated whether the hHR23 proteins affect p53 ubiquitylation and/or p53 degradation. Here we report that in vitro, hHR23 protects polyubiquitylated p53 from deubiquitylation as well as from proteasome-mediated degradation and that the Uba2 domain of hHR23 is necessary and sufficient for this effect. In addition, siRNA-mediated downregulation of hHR23 expression within cells results in the accumulation of p53. Taken together, our results indicate that hHR23 is intrinsically involved in the degradation of p53 and are consistent with the notion that hHR23 acts as a bridging factor between polyubiquitylated forms of p53 and the proteasome.
MATERIALS AND METHODS
Plasmids, siRNAs, and protein expression.
For in vitro translation and transient expression, the constructs pcDNA3-Flag-hHR23A, pcDNA3-Flag-hHR23B, pcDNA3-HA-hHR23A, and pcDNA3-HA (with cDNAs encoding the various deletion mutants of hHR23A [ΔUbl, ΔUba2, ΔUbl/ΔUba2, Uba2] [see Fig. 1A ]), and pcDNA3-p53, pcDNA3-Hrs, and pcDNA3-Hdm2 (human Mdm2) were generated by PCR-based methods (further details will be provided upon request). pcDNA3-Mdm2 was obtained by digesting pcocmdm2X2 (4) with EcoRI and ligating the resulting fragment into the EcoRI site of the pcDNA3 vector. The plasmid expressing His-tagged ubiquitin was a gift of Sibylle Mittnacht, the plasmid expressing CD44 was a gift of Veronique Orian-Rousseau, and the plasmid expressing green fluorescent protein (GFP) was provided by Sakari Hietanen.
FIG. 1.
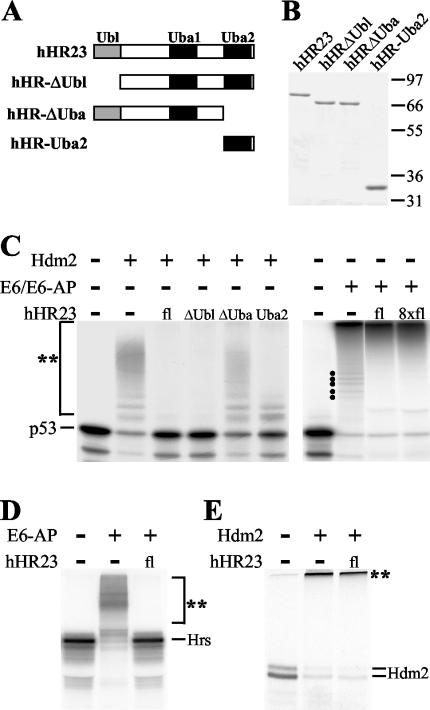
hHR23 interferes with Hdm2-mediated ubiquitylation of p53 but not with E6-mediated ubiquitylation in vitro. (A) The structure of hHR23 and the deletion mutants used in this study is schematically represented with respect to the location of the Ubl domain and the two Uba domains. (B) The indicated forms of hHR23 were expressed as GST fusion proteins in E. coli and purified by affinity chromatography. Similar amounts of the various proteins were used in the subsequent characterization of their biochemical properties. The running positions of molecular mass markers (in kilodaltons) are indicated. (C) In vitro-translated 35S-labeled p53 was ubiquitylated by Hdm2 or by the HPV-16 E6 oncoprotein and E6-AP under standard ubiquitylation conditions in the presence (+) or absence (−) of the various hHR23 forms in increasing amounts, as indicated. After 2 h at 30°C, the reactions were stopped and the complete reaction mixtures were electrophoresed on SDS-10% polyacrylamide gels. Radiolabeled p53 was detected by fluorography. (D) Ubiquitylation of in vitro-translated Hrs by baculovirus-expressed E6-AP in the absence and in the presence of hHR23. (E) Ubiquitylation of in vitro-translated Hdm2 by bacterially expressed GST-Hdm2 in the absence and in the presence of hHR23. Ubiquitylated forms of the respective radiolabeled proteins are indicated by a double asterisk. Running positions of ubiquitylated forms of p53 that are generated by E6/E6-AP but not observed in the additional presence of hHR23 are indicated by dots. Lanes fl, full-length protein.
For in vitro experiments, p53, Hrs, or Hdm2 was generated in the rabbit reticulocyte lysate (RRL) system (Promega) in the presence of [35S]methionine. Furthermore, Hdm2, HPV-16 E6, the Uba domain of Chap1/hPlic-2, and hHR23A and its various deletion mutants were expressed as glutathione S-transferase (GST) fusion proteins in Escherichia coli BL21/DE3. The ubiquitin-activating enzyme E1, the ubiquitin-conjugating enzyme UbcH5, S. cerevisiae UBP1 (provided by R. T. Baker) (2), and ubiquitin mutants (R48, K48, and K0) (provided by C. M. Pickart) were expressed in E. coli BL21/DE3 as described previously (22, 55). As a source of recombinant E6-AP, E6-AP was expressed in the baculovirus system and purified by anion-exchange chromatography (45).
siRNAs against mRNAs encoding hHR23A and hHR23B were obtained from Dharmacon Research. The respective target sequences were as follows: for hHR23AI, 5′CTTCCTCCTGAGTCAGAAC; for hHR23AII, 5′GGGTCGTGATGCCTTCCCC; for hHR23BI, 5′CTGACAGTACATCGGGTGA; and for hHR23BII, 5′TTGCAGCCCTGAGAGCCAG. For a control, the target sequence was 5′CCCCUUUUAAAAAGGGGG.
Cell lines, transfection, and microinjection.
Primary human fibroblasts (GM1604; National Institute of General Medical Sciences, Coriell Institute for Medical Research, Human Genetic Cell Repository, Camden, N.J.), H1299 lung carcinoma cells (p53 negative; American Type Culture Collection), and U2OS osteosarcoma cells (wild-type [wt] p53) (8) were cultured in Dulbecco's modified Eagle medium (Invitrogen) supplemented with 15% (for GM1604) or 10% (for H1299 and U2OS) fetal calf serum (Invitrogen). Transfections were carried out by calcium phosphate coprecipitation (9) or by lipofection (with DOTAP) according to the manufacturer's instructions (Roche Molecular Biochemicals). Where indicated, the proteasome inhibitor MG132 (Calbiochem) was added to a final concentration of 10 μM for 5 h.
Primary human fibroblasts grown on glass coverslips were injected with a vector expressing GFP (0.25 μg/ml), with a vector expressing GFP (0.2 μg/ml) and control siRNA (16 μM), or with a vector expressing GFP (0.2 μg/ml) and a mixture of siRNAs for hHR23A (I and II) and hHR23B (I and II), each at a final concentration of 4 μM, by using the Eppendorf 5242 microinjector and 5170 micromanipulator.
Antibodies and immunofluorescence.
For Western blot analyses, the following antibodies were used: DO-1, monoclonal anti-p53 antibody (50); 4B2, monoclonal anti-Mdm2 antibody (10); M2, monoclonal anti-Flag antibody (Sigma); and PC-10, monoclonal anti-proliferating-cell nuclear antigen antibody (51). Horseradish peroxidase-coupled goat anti-mouse immunoglobulin G (IgG) (DAKO) was used as a secondary antibody.
Immunofluorescence staining was performed as previously described (6) with the polyclonal anti-p53 antibody CM-1 (35) or the monoclonal anti-p53 antibody DO-1, the anti c-Jun antibody sc-45 (Santa Cruz), and monoclonal antibodies against the hemagglutinin (HA) tag (provided by C. Englert) and CD44 (provided by J. Sleeman). Cy2-conjugated anti-mouse IgG and Cy3-conjugated anti-rabbit IgG (Dianova) were used as secondary antibodies.
In vitro assays.
HPV E6-mediated and Hdm2-mediated ubiquitylation of p53 was assayed as described previously (22) by using in vitro-translated p53 and bacterially expressed GST-E6 and GST-Hdm2 fusion proteins, respectively. For Hdm2-mediated ubiquitylation of Hdm2 (“auto-ubiquitylation”), 2 μl of in vitro-translated Hdm2 was incubated in the absence or in the presence of 50 ng of bacterially expressed GST-Hdm2 under standard ubiquitylation conditions (22). Similarly, for E6-AP-mediated ubiquitylation of Hrs, 2 μl of in vitro-translated Hrs was incubated in the absence or in the presence of 200 ng of baculovirus-expressed E6-AP. All ubiquitylation reaction mixtures contained 8 μg of commercially available ubiquitin (Sigma) or the same amount of bacterially expressed ubiquitin, where indicated.
For coprecipitation analyses, p53 was ubiquitylated in the presence of Hdm2 or HPV-E6. After 1 h, the ubiquitylation reaction was stopped by the addition of 20 mM EDTA (final concentration). The capacity of p53 and its ubiquitylated forms to bind to GST-hHR23 or its various deletion mutants was then determined in GST pulldown experiments as described previously (26).
For UBP1-mediated deubiquitylation, p53, Hdm2, or Hrs were ubiquitylated as described above in the presence of wt ubiquitin or mutated forms of ubiquitin as indicated. After 1 h, the ubiquitylation reaction was stopped by the addition of 20 mM EDTA, and the ability of UBP1 to deubiquitylate the respective ubiquitylated forms of p53, Hdm2, or Hrs determined in the absence and in the presence of wt GST-hHR23 or its mutant forms.
For in vitro degradation of p53 and Hdm2, p53 was ubiquitylated in the presence of HPV E6 and Hdm2 was auto-ubiquitylated. After 1 h, degradation was initiated by the addition of 5 μl of untreated RRL (Promega) as a source of the 26S proteasome in the presence or in the absence of GST-hHR23 (or its various mutant forms) as indicated below. After incubation at 30°C for 1 h, total reaction mixtures were electrophoresed in sodium dodecyl sulfate (SDS)-10% polyacrylamide gels, and 35S-labeled p53 was detected by fluorography.
In vivo ubiquitylation assay.
H1299 cells were transfected overnight with plasmids encoding p53, His-tagged ubiquitin, hHR23, Mdm2, and vector DNA (to adjust the amount of transfected DNA). Cells were harvested 24 h after transfection. After being washed twice in ice-cold phosphate-buffered saline, 7.5 × 106 cells were lysed in 6 ml of guanidinium lysis buffer (6 M guanidinium-HCl, 0.1 M phosphate [pH 8], 0.01 M Tris [pH 8], 5 mM imidazole, 10 mM β-mercaptoethanol). Ni2+-nitrilotriacetic acid-agarose beads (75 μl) were added to the lysate, and the mixture was incubated at room temperature overnight. The next morning, beads were successively washed with the following buffers: guanidinium lysis buffer, urea buffer (8 M urea, 0.1 M phosphate [pH 8], 0.01 M Tris [pH 8], 10 mM β-mercaptoethanol), buffer A (8 M urea, 0.1 M phosphate [pH 6.3], 0.01 M Tris [pH 6.3], β-mercaptoethanol), buffer A plus 0.2% Triton X-100, and buffer A plus 0.1% Triton X-100. Ubiquitylated proteins were eluted with 200 mM imidazole in 5% SDS-0.15 M Tris [pH 6.7]-30% glycerol-0.72 M β-mercaptoethanol. The eluate was subjected to SDS-polyacrylamide gel electrophoresis. After transfer onto polyvinylidene difluoride-membrane (Millipore), the proteins were probed with the anti-p53 antibody DO-1.
RESULTS
hHR23 interferes with Hdm2-mediated but not with E6-mediated ubiquitylation of p53 in vitro.
Yeast Rad23 has recently been reported to interfere with the ubiquitylation of proteins (11, 36). To determine if hHR23A and hHR23B, the human homologs of Rad23, have similar properties, the effect of these proteins on the ubiquitylation of p53 mediated either by Hdm2 or by the HPV E6 oncoprotein was studied in vitro. (From now on, we will not distinguish between hHR23A and hHR23B, since similar results were obtained for these proteins.) As shown in Fig. 1C, addition of hHR23 resulted in a complete inhibition of Hdm2-mediated ubiquitylation of p53. Furthermore, the Uba2 domain of hHR23 was necessary and sufficient for this inhibitory effect. In contrast, hHR23 did not interfere with the E6-dependent reaction. In fact, in the presence of hHR23 only the highly ubiquitylated forms of p53 migrating at the top of the gel were observed, while the ladder of ubiquitylated forms of p53 with a lower molecular mass (indicated by dots in the right panel of Fig. 1C) disappeared. This result may suggest that hHR23 promotes E6-mediated polyubiquitylation of p53 or stabilizes polyubiquitylated forms of p53.
E6-facilitated ubiquitylation of p53 is mediated by E6-AP, which interacts with hHR23 (31). Thus, it seemed possible that the inability of hHR23 to interfere with E6-mediated ubiquitylation of p53 is a result of the interaction between hHR23 and E6-AP. To address this possibility, we studied the effect of hHR23 on E6-AP-mediated ubiquitylation of Hrs (29), another in vitro substrate for E6-AP (S. Glockzin and M. Scheffner, unpublished data). As shown in Fig. 1D, addition of hHR23 efficiently interfered with E6-AP-mediated ubiquitylation of Hrs, indicating that binding to E6-AP does not result in a general inactivation of the ability of hHR23 to inhibit ubiquitylation processes. Furthermore, Hdm2-mediated auto-ubiquitylation was not affected by hHR23 (Fig. 1E).
Our results indicate that the actual effect of hHR23 on ubiquitylation depends on the actual E3-substrate pair studied. However, the ability of hHR23 to interfere with ubiquitylation processes does not require a direct interaction between hHR23 and the respective E3 or substrate. This conclusion is based on the observation that the Uba2 domain of hHR23, which does not bind to E6-AP (31), Hdm2, Hrs, and p53 (data not shown), is necessary and sufficient to block both Hdm2-mediated ubiquitylation of p53 and E6-AP-mediated ubiquitylation of Hrs (Fig. 1C and data not shown).
hHR23 preferentially binds to K48-ubiquitylated p53.
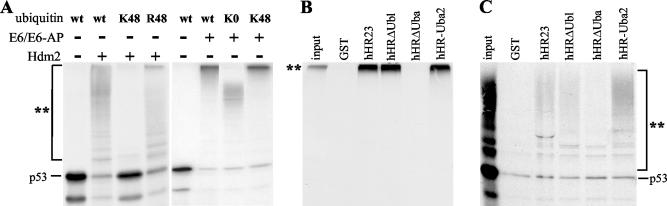
Based on published data (32), Hdm2-mediated ubiquitylation of p53 in vitro results predominantly in the attachment of single ubiquitin moieties to a number of different lysine residues of the p53 protein rather than in the attachment (or formation) of polyubiquitin chains. To further characterize Hdm2-mediated ubiquitylation of p53 in our system, three ubiquitin mutants were employed, a ubiquitin mutant in which lysine 48 was replaced by an arginine (R48-ubiquitin), a mutant in which all lysine residues were replaced by arginine except for lysine 48 (K48-ubiquitin), and a mutant in which all lysine residues were replaced by arginine (K0-ubiquitin). In the presence of R48-ubiquitin, the pattern of ubiquitylated forms of p53 generated in the presence of Hdm2 was similar to the pattern observed in the presence of wt ubiquitin (Fig. 2A). In contrast, p53 was not ubiquitylated by Hdm2 in the presence of the K48 mutant or the K0 mutant (not shown). In addition, similar results were obtained for E6-AP-mediated ubiquitylation of Hrs (i.e., similar ubiquitylation patterns with wt ubiquitin and R48-ubiquitin, while ubiquitylation was not observed in the presence of the K48 and K0 mutant; data not shown). The reason for why the K48 mutant and the K0 mutant are not used by Hdm2 or E6-AP in the ubiquitylation of p53 and Hrs, respectively, is currently unclear. However, the possibility that these mutants are generally defective in protein ubiquitylation can be excluded, since p53 was ubiquitylated by E6/E6-AP in the presence of the K48 mutant or the K0 mutant (see below). Thus, although it was not possible to determine whether, under the conditions used, p53 and/or Hrs is modified at several lysines by single ubiquitin molecules or by ubiquitin chains, it appears that, if ubiquitin chains are formed, the individual ubiquitin moieties are linked mainly via lysine residues other than K48. This assumption is supported by the fact that degradation of Hdm2-ubiquitylated p53 and of E6-AP-ubiquitylated Hrs, respectively, was not observed (data not shown) (note that K48-linked ubiquitin chains are commonly assumed to function as recognition signals for the 26S proteasome). In contrast, E6/E6-AP-mediated ubiquitylation results in the attachment of one or more K48-linked ubiquitin chains on p53 (Fig. 2A), targeting p53 for degradation by the 26S proteasome (see Fig. 5). Similarly, auto-ubiquitylation of Hdm2 results in highly ubiquitylated Hdm2 forms and the individual ubiquitin moieties within these chains are at least in part linked via K48 (L. K. Linares and M. Scheffner, unpublished observation). Taken together, the results presented above indicate that in vitro hHR23 does not interfere with modification of proteins with K48-linked ubiquitin chains (“K48-ubiquitylation”), whereas it inhibits modification of proteins with ubiquitin chains linked via lysine residues other than K48 (“non-K48-ubiquitylation”).
FIG. 2.
hHR23 binds to ubiquitylated p53. (A) In vitro-translated 35S-labeled p53 was ubiquitylated by Hdm2 or the E6/E6-AP complex in the presence of wt ubiquitin or ubiquitin mutants as indicated. After 2 h at 30°C, the reactions were analyzed as described in the legend to Fig. 1. K48 is a ubiquitin mutant in which all lysines were replaced by arginine except for lysine at position 48, R48 is a ubiquitin mutant in which the lysine residue at position 48 was replaced by arginine, and K0 is a ubiquitin mutant in which all lysine residues were replaced by arginine. (B and C) Radiolabeled p53 was ubiquitylated in the presence of the HPV-16 E6 oncoprotein (B) or in the presence of Hdm2 (C). After 1 h, the reactions were stopped and the ability of the respective reaction products to interact with GST or the various forms of hHR23 was determined by coprecipitation analysis (Materials and Methods). Ubiquitylated forms of the respective radiolabeled proteins are indicated by a double asterisk.
FIG. 5.
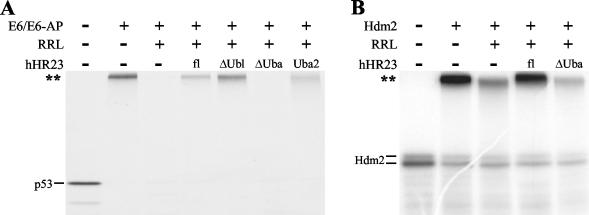
hHR23 inhibits degradation of ubiquitylated p53 and of ubiquitylated Hdm2. (A) Radiolabeled p53 was ubiquitylated by the E6/E6-AP complex. After 1 h, untreated RRL was added as a source of the 26S proteasome and degradation of ubiquitylated p53 studied in the absence (−) or in the presence (+) of the hHR23 forms indicated. (B) Radiolabeled Hdm2 was ubiquitylated in the presence of GST-Hdm2. Degradation of ubiquitylated Hdm2 was tested as described in panel A for p53. The running positions of the ubiquitylated forms of p53 and Hdm2 are indicated by a double asterisk.
Uba domains have been reported to bind to ubiquitin chains with higher affinity than to monomeric ubiquitin (5, 43, 54). To test if hHR23 also binds to K48-ubiquitylated and/or non-K48-ubiquitylated p53, p53 was ubiquitylated by E6 or Hdm2 in a standard ubiquitylation reaction (22). After 60 min, the reactions were stopped and the ability of p53 and its modified forms to interact with hHR23 was determined in coprecipitation experiments using GST fusion proteins of full-length hHR23 or the various hHR23 deletion mutants (Fig. 1A) as indicated (Fig. 2B and C). All forms of hHR23 that contain the Uba2 domain bound to both K48-ubiquitylated and non-K48-ubiquitylated p53, while binding to nonmodified p53 was not above background levels (as defined by the amount of nonmodified p53 bound to GST in the experiment shown in Fig. 2C). However, binding to K48-ubiquitylated p53 appeared to be significantly more efficient than binding to non-K48-ubiquitylated p53. Thus, the inability of hHR23 to interfere with K48-ubiquitylation of p53 is not due to the possibility that hHR23 does not efficiently interact with K48-ubiquitylated p53.
hHR23 interferes with deubiquitylation of K48-ubiquitylated p53 in vitro.
To obtain insight into the possible functional consequence(s) of the interaction of hHR23 with ubiquitylated p53, the effect of hHR23 on deubiquitylation of p53 was examined. As deubiquitylating enzyme, the yeast ubiquitin-specific protease UBP1 (49) was used. Although there is no evidence at present that the human ortholog of UBP1 is involved in deubiquitylation of p53 in vivo, recombinant UBP1 very efficiently deubiquitylates p53 in vitro under the conditions used (Fig. 3A).
FIG. 3.
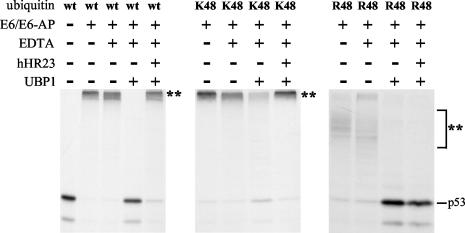
hHR23 protects E6-ubiquitylated p53 from deubiquitylation. (A) Radiolabeled p53 was ubiquitylated by Hdm2 or by E6/E6-AP as indicated. After 1 h, the reactions were stopped by the addition of EDTA, and the deubiquitylation reaction was started by the addition of the ubiquitin-specific protease UBP1 in the absence (−) or in the presence (+) of hHR23 or its deletion mutants or the Uba domain of Chap1/hPlic-2 as indicated. Similarly, the effect of UBP1 on auto-ubiquitylated Hdm2 (B) and on E6-AP-ubiquitylated Hrs (C) was tested. Ubiquitylated forms of the respective radiolabeled proteins are indicated by a double asterisk.
After ubiquitylation of p53 by Hdm2 or E6/E6-AP, the ubiquitylation reaction was stopped by the addition of EDTA. Then, UBP1 was added and deubiquitylation of p53 was measured in the absence or presence of hHR23 (Fig. 3A). In the absence of hHR23, both non-K48-ubiquitylated p53 (generated by Hdm2) and K48-ubiquitylated p53 (generated by E6) were efficiently deubiquitylated, as indicated by the disappearance of the ubiquitylated forms of p53 and the concomitant appearance of the nonmodified form of p53. However, in the presence of hHR23, deubiquitylation of K48-ubiquitylated p53 was significantly blocked, while deubiquitylation of non-K48-ubiquitylated p53 was not affected. Similarly, addition of hHR23 interfered with the deubiquitylation of auto-ubiquitylated Hdm2 (Fig. 3B), while deubiquitylation of E6-AP-ubiquitylated Hrs was not inhibited (Fig. 3C).
As observed for the binding to ubiquitylated proteins (Fig. 2), the isolated Uba2 domain of hHR23 was necessary and sufficient for this K48-ubiquitin chain-stabilizing activity of hHR23 (Fig. 3A, right panel). However, this does not appear to be a general property of Uba domains. Although the Uba domain of Chap1/hPlic-2, a protein that was previously shown to interfere with proteasome-mediated degradation of proteins upon overexpression in vivo (28), efficiently binds to K48-ubiquitylated p53 (data not shown), it did not interfere with UBP1-mediated deubiquitylation of p53 under the conditions used (Fig. 3A). Thus, although Uba domains in general seem to bind to ubiquitin chains, the functional consequence(s) may differ between individual Uba domains.
The above results indicate that hHR23 protects K48-ubiquitylated proteins from deubiquitylation but not non-K48-ubiquitylated proteins. To obtain further evidence for this hypothesis, E6/E6-AP-mediated ubiquitylation of p53 with subsequent deubiquitylation was assayed using the bacterially expressed ubiquitin mutants K48-ubiquitin and R48-ubiquitin (Fig. 4). The pattern of p53 ubiquitylation observed in the presence of R48 is similar, or identical, to the pattern observed with K0-ubiquitin (compare the respective lanes in Fig. 2A with the ones in Fig. 4). This clearly indicates that E6/E6-AP-mediated ubiquitylation of p53 results mainly, or exclusively, in the attachment of ubiquitin chains linked via K48. Moreover, hHR23 (or the isolated Uba2 domain; data not shown) efficiently inhibited UBP1-mediated deubiquitylation of K48-ubiquitin-modified p53 (note that for unknown reasons, K48-ubiquitin chains are less well recognized by UBP1 as substrates for deubiquitylation than wt chains). In contrast, hHR23 had no effect on deubiquitylation of the R48-ubiquitin-modified p53. Thus, the ability of hHR23 to interfere with p53 deubiquitylation seems to be specific for K48-ubiquitylated forms.
FIG. 4.
hHR23 protects K48-ubiquitylated p53 from deubiquitylation. Radiolabeled p53 was partially purified by anion-exchange chromatography to remove any ubiquitin from the RRL used for in vitro translation. The partially purified p53 was then ubiquitylated by E6/E6-AP in the presence of bacterially expressed wt ubiquitin or the indicated mutants (for further details on K48 and R48, see the legend to Fig. 2A). After 1 h, reactions were stopped by the addition of EDTA, and UBP1-mediated deubiquitylation of p53 was examined in the absence (−) or presence (+) of hHR23. The running positions of the ubiquitylated forms of p53 are indicated by a double asterisk.
hHR23 inhibits degradation of p53 and of Hdm2 in vitro.
The notion that E6-facilitated ubiquitylation results mainly in p53 molecules attached to K48-linked ubiquitin chains is strongly supported by the fact that E6-induced ubiquitylation of p53 is known to result in its efficient degradation by the proteasome (47). To test if hHR23 has an effect on proteasome-mediated degradation, E6-ubiquitylated p53 was generated, and its degradation was initiated by the addition of RRL as a source for the 26S proteasome. (Note that the degradation reaction requires the addition of RRL as the RRL used for in vitro translation of p53 was treated with a proteasome inhibitor.) As shown in Fig. 5A, E6-ubiquitylated p53 was efficiently degraded under the conditions used. However, when ubiquitylated p53 was incubated with full-length hHR23 prior to the addition of RRL, its degradation was efficiently blocked. Furthermore, the Uba2 domain was necessary and sufficient for this protective effect of hHr23. Similarly, degradation of auto-ubiquitylated Hdm2 was inhibited in the presence of hHR23 (Fig. 5B). Similar to the results obtained for deubiquitylation, the isolated Uba domain of Chap1/hPlic-2 did not interfere with proteasome-mediated degradation of p53 in this in vitro system (data not shown).
hHR23 stabilizes ubiquitylated forms of p53 in vivo.
Since in vitro hHR23 inhibits both deubiquitylation and proteasome-mediated degradation of p53, we wondered whether hHR23 has similar effects in living cells. Therefore, we first tested the effect of overexpressed hHR23 on Mdm2-mediated degradation of ectopically expressed p53 in cell culture experiments. As expected (21, 30), coexpression of Mdm2 and p53 resulted in the efficient degradation of p53, which was inhibited by addition of the proteasome inhibitor MG132 (Fig. 6A). While coexpressed hHR23 had no influence on the expression levels of p53 in the absence of additional Mdm2, Mdm2-induced degradation of p53 was severely attenuated in the presence of hHR23. Most importantly, in the presence of hHR23 distinct p53 forms with an apparent higher molecular mass than that of unmodified p53, indicative of ubiquitylated p53 species, were detectable by ordinary Western blotting of whole-cell lysate (Fig. 6A, upper panel). Indeed, when we affinity purified ubiquitylated proteins and probed them with a p53-specific antibody, we detected a significant increase in ubiquitylated p53 (Fig. 6A, lower panel). This result shows that regardless of the presence or absence of MG132, ubiquitylated p53 species accumulated in the presence of both hHR23 and Mdm2. Thus, overexpression of hHR23 interferes with both deubiquitylation and degradation of p53 within a living cell. The fact that Mdm2 targets p53 for degradation in vivo is obviously different from the situation in vitro (where Hdm2 does not induce p53 degradation [see above]), indicating that in contrast to the in vitro system, p53 is modified by K48 ubiquitin chains in the presence of Hdm2 in vivo. A likely explanation for this difference is that Hdm2 requires the activity of an E4 for K48-ubiquitylation of p53 and this E4 is missing in the in vitro system. This hypothesis is supported by a recent study indicating that p300 acts as an E4 in Hdm2-induced ubiquitylation of p53 (20).
FIG. 6.
Overexpression of hHR23 in the presence of Mdm2 results in the accumulation of ubiquitylated forms of p53. (A) H1299 cells (p53 null cells) were transfected with expression constructs for p53, Mdm2, Flag-tagged hHR23, and His-tagged ubiquitin (for further details, see Materials and Methods). Twenty-four hours after transfection, cells were harvested and whole-cell lysates were prepared from an aliquot of the cells. Using the whole-cell lysate, expression levels of p53, Mdm2, and Flag-tagged hHR23 were determined by Western blot analysis with antibodies against the respective proteins. Endogenous proliferating-cell nuclear antigen levels were determined for loading control. From the remaining cells, ubiquitylated proteins were isolated by affinity chromatography and ubiquitylated forms of p53 were detected by Western blot analysis using a p53-specific antibody. The running positions of ubiquitylated forms of p53 are indicated by a double asterisk. The running positions of putative ubiquitylated forms of p53 detected in a whole-cell extract with a p53-specific antibody (most upper panel) are indicated by dots. Where indicated, transfected cells were treated with the proteasome inhibitor MG132 for 5 h prior to the preparation of cell extracts. (B) The effect of hHR23 overexpression on E6-mediated degradation of p53 was determined in cotransfection experiments as described in panel A for Mdm2-induced p53 degradation in the absence of an expression vector for His-tagged ubiquitin.
Similar to its effect on Hdm2-mediated p53 degradation, overexpression of hHR23 also interfered with E6-induced degradation of p53 in transient-transfection experiments (Fig. 6B). However, E6-induced degradation of p53 was reproducibly less affected by hHR23 overexpression than Mdm2-induced p53 degradation. The reason for this difference is currently unknown.
siRNA-mediated downregulation of hHR23 expression results in the accumulation of endogenous p53.
To provide evidence that hHR23 is involved in the degradation of endogenous p53, we overexpressed hHR23 in the U2OS cell line, which possesses endogenous wt p53. As shown in Fig. 7A, overexpression of hHR23 resulted in the accumulation of endogenous p53. The effect of hHR23 overexpression was specific for p53 insofar that the stability of another known substrate of the proteasome, c-Jun (37), was not affected. Note that it is still a matter of debate if the transfection procedure per se may result in elevated p53 levels. The finding, however, that transfection of a CD44 expression construct did not cause an increase in endogenous p53 levels excludes this possibility.
FIG. 7.
Downregulation of hHR23 results in accumulation of endogenous p53. (A) U2OS cells (which express endogenous wt p53) were transfected with an expression construct for HA-tagged hHR23 or with an expression construct for CD44 for control. Twenty-four hours h after transfection, levels of p53, c-Jun, hHR23, or CD44 were determined by immunofluorescence using the anti-p53 antibody CM-1, the anti-c-Jun antibody sc-45, a monoclonal anti-HA antibody, and a monoclonal antibody against CD44. p53 and c-Jun are stained in red. hHR23 and CD44 are stained in green. (B) Primary human fibroblasts were microinjected with an expression vector encoding GFP either alone, in combination with control siRNA, or in combination with siRNAs directed against mRNAs encoding hHR23A and hHR23B. Three days after microinjection, p53 expression was determined by immunofluorescence using the p53-specific antibody DO-1. GFP expression is shown in green, and the p53 protein is stained in red.
While results obtained in overexpression studies often serve as first indication if a protein is involved in the degradation of another protein, they can be misleading. For example, overexpression of hHR23 may titrate out factors involved in degradation of p53 and, thus, may indirectly result in p53 stabilization. Therefore, to obtain evidence that endogenous hHR23 is directly involved in p53 degradation, siRNAs (15) directed against mRNAs encoding hHR23A and hHR23B were introduced by microinjection into human primary fibroblasts expressing wt p53 (Fig. 7B). While cells microinjected with control siRNA and an expression vector for GFP (as microinjection control) did not show any signs of p53 accumulation, more than 90% of the cells injected with a mix of siRNAs directed against hHR23 mRNAs showed significant accumulation of p53. Although we were not able to determine the effect of the siRNAs used on the expression level of endogenous hHR23 proteins (endogenous hHR23 proteins were not detectable by the commercially available hHR23 antibodies), the siRNAs directed against the hHR23A and hHR23B mRNA efficiently downregulated the expression of ectopically expressed hHR23 proteins in cotransfection experiments (data not shown). Thus, the siRNA experiments indicate that hHR23 is involved in p53 degradation in vivo.
DISCUSSION
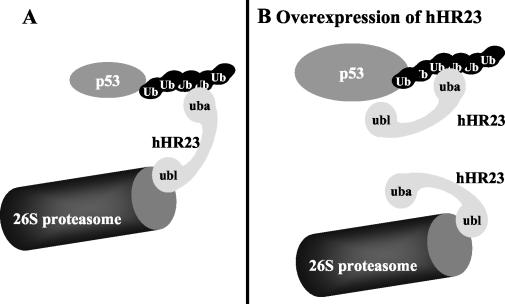
The Rad23 protein has been shown to play a role in protein degradation mediated by the ubiquitin-proteasome system (12, 43, 44). Rad23 contains an N-terminal Ubl domain and two Uba domains. Uba domains bind to ubiquitin chains (5, 18, 43, 54), and it was recently reported that Rad23 preferentially binds to ubiquitin chains that perform proteolytic functions (i.e., K48-linked polyubiquitin chains) (43). Since the Ubl domain of Rad23 interacts with the 19S regulatory complex of the 26S proteasome (16, 44), a current model is that Rad23 and structurally related proteins (i.e., proteins containing both Ubl and Uba domains) are involved in the transport of ubiquitylated proteins to the proteasome or, alternatively, facilitate the formation of ubiquitylated proteins in close proximity to the proteasome (12, 18, 43). In agreement with this model, we found that the human homologs of Rad23, hHR23A and hHR23B, bind to K48-ubiquitylated proteins, including p53, shielding them from deubiquitylation, and that overexpression of hHR23 interferes with p53 degradation in vitro and in vivo. Moreover, siRNA directed against hHR23 mRNAs result in the accumulation of endogenous p53. Taken together, these observations (both downregulation and upregulation of hHR23 levels result in enhanced p53 levels) indicate that hHR23 is intrinsically involved in p53 degradation, acting as a bridging factor between ubiquitylated p53 and the 26S proteasome (Fig. 8).
FIG. 8.
Model on the role of hHR23, in p53 degradation. hHR23 can interact with the 26S proteasome via its N-terminal Ubl domain and with polyubiquitylated p53 via its C-terminal Uba domain. At appropriate levels, a given hHR23 molecule can thus simultaneously interact with the 26S proteasome and a polyubiquitylated p53 molecule, thereby facilitating degradation of p53 (A). After overexpression of hHR23 (B), formation of a ternary complex between polyubiquitylated p53, hHR23, and 26S proteasomes is disturbed, since the respective interaction sites for hHR23 on polyubiquitylated p53 and the 26S proteasome are occupied by two different hHR23 molecules. In consequence, p53 protein levels accumulate.
Similar to the results presented here, it was recently reported that hHR23 interferes with deubiquitylation and degradation of artificial substrate proteins (42). A likely possibility is that binding of hHR23 to ubiquitin chains via the Uba2 domain sterically hinders the interaction of ubiquitylated proteins, including p53 with the proteasome. However, since overexpression of hHR23 does not interfere with proteasome-mediated degradation in general, other yet unknown properties of hHR23 may contribute to its inhibitory effect. Moreover, the different effect of hHR23 on different substrates of the proteasome suggests that different Ubl/Uba-containing proteins are involved in the degradation of specific subsets of proteins.
In addition to the proposed function as an adaptor between K48-ubiquitylated proteins and the proteasome, Rad23 was reported to impede “multiubiquitylation” of proteins (11, 36, 42). We also observed that hHR23 interferes with protein ubiquitylation, but only in those cases in which the respective proteins were modified by the attachment of single ubiquitin molecules to several lysine residues of the substrate protein or by the attachment of ubiquitin chains that are mainly linked via lysine residues other than K48 (termed non-K48-ubiquitylation within the frame of this paper). In contrast, modification of proteins with K48-linked ubiquitin chains that are considered to target the modified proteins for proteasomal degradation was not affected by hHR23. According to these results, we speculate that the ubiquitylation-inhibitory activity of hHR23 is linked not to its role in protein degradation but rather to other yet-unknown processes.
The hHR23 proteins contain two Uba domains, one of which is located close to the C-terminal end (Uba2), while the other is located in a more central region of hHR23 (Uba1). Interestingly, the presence of the Uba2 domain was necessary and sufficient for hHR23 to bind to ubiquitylated proteins in vitro. A possible explanation for this finding is that within the context of the full-length hHR23 protein, the Uba1 domain may not, or may only weakly, interact with ubiquitin and may rather serve as an interaction site for other proteins. Furthermore, although both the isolated Uba2 domain of hHR23 and the Uba domain of Chap1/hPlic-2 bind to ubiquitylated p53, only the hHR23 Uba2 domain protected K48-ubiquitylated p53 from deubiquitylation in vitro. This finding suggests that the functional consequence of the interaction with ubiquitylated proteins differs between different Uba domains or that the actual effect of a given Uba domain depends on the individual substrate or the nature of the ubiquitin chain.
Similarly to the effect on deubiquitylation, the Uba domain of Chap1/hPlic-2 did not interfere with proteasome-mediated degradation of p53 in vitro. This result seems to be in contrast to the previously reported finding that overexpression of Chap1/hPlic-2 leads to the accumulation of p53 in vivo (28). Since we also observed an inhibitory effect of Chap1/hPlic-2 on p53 degradation in transient-transfection experiments (F.-X. Ogi and C. Blattner, unpublished data), a possible explanation for this discrepancy is that the Uba domain of Chap1/hPlic-2 does not function properly under the conditions used in our in vitro systems.
Finally, the observation that both overexpression and downregulation of hHR23 interfere with p53 degradation suggests that in general, the expression levels of Ubl/Uba-containing proteins need to be finely tuned within cells. Both a decrease as well as an increase in expression levels can be expected to result in decreased degradation rates of the respective target proteins. Whether this model is correct or not and whether hHR23 and, possibly, Chap1/hPlic-2 are the only Ubl/Uba-containing proteins involved in p53 degradation remain to be determined.
Acknowledgments
M.S. and C.B. share senior authorship of this work.
We are grateful to Helen Morrison for her comments on the manuscript; to David Lane, Jonathan Sleeman, and Christoph Englert for providing antibodies; and to Sibylle Mittnacht, Veronique Orian-Rousseau, Sakari Hietanen, Cecile M. Pickart, and Rohan T. Baker for providing plasmids.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (C.B. and M.S.) and from Köln Fortune (M.S.) and by the Fonds der Chemischen Industrie (M.S.).
REFERENCES
- 1.Alarcon-Vargas, D., and Z. Ronai. 2002. p53-Mdm2—the affair that never ends. Carcinogenesis 23:541-547. [DOI] [PubMed] [Google Scholar]
- 2.Baker, R. T., J. W. Tobias, and A. Varshavsky. 1992. Ubiquitin-specific proteases of Saccharomyces cerevisiae. Cloning of UBP2 and UBP3, and functional analysis of the UBP gene family. J. Biol. Chem. 267:23364-23375. [PubMed] [Google Scholar]
- 3.Balint, E. E., and K. H. Vousden. 2001. Activation and activities of the p53 tumour suppressor protein. Br. J. Cancer 85:1813-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barak, Y., E. Gottlieb, G. T. Juven, and M. Oren. 1994. Regulation of mdm2 expression by p53: alternative promoters produce transcripts with non-identical translation potential. Genes Dev. 8:1739-1749. [DOI] [PubMed] [Google Scholar]
- 5.Bertolaet B. L., D. J. Clarke, M. Wolff, M. H. Watson, M. Henze, G. Divita, and S. I. Reed SI. 2001. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat. Struct. Biol. 8:417-422. [DOI] [PubMed] [Google Scholar]
- 6.Blattner, C., A. Sparks, and D. P. Lane. 1999. Transcription factor E2F-1 is upregulated in response to DNA damage in a manner analogous to that of p53. Mol. Cell. Biol. 19:3704-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blattner, C., T. Hay, D. W. Meek, and D. P. Lane. 2002. Hypophosphorylation of Mdm2 augments p53 stability. Mol. Cell. Biol. 22:6170-6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bottger, A., V. Bottger, A. Sparks, W.-L. Liu, S. F. Howard, and D. P. Lane. 1997. Design of a synthetic Mdm2-binding mini protein that activates the p53 response in vivo. Curr. Biol. 7:860-869. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C., and H. Okayama. 1987. High efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, J. D., V. Marechal, and A. J. Levine. 1993. Mapping of the p53 and Mdm2 interaction domains. Mol. Cell. Biol. 13:4107-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, L., U. Shinde, T. G. Ortolan, and K. Madura. 2001. Ubiquitin-associated (UBA) domains in Rad23 bind ubiquitin and promote inhibition of multi-ubiquitin chain assembly. EMBO Rep. 2:933-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, L., and K. Madura. 2002. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol. Cell. Biol. 22:4902-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deveraux, Q., V. Ustrell, C. Pickart, and M. Rechsteiner. 1994. A 26 S protease subunit that binds ubiquitin conjugates. J. Biol. Chem. 269:7059-7061. [PubMed] [Google Scholar]
- 14.Dupre, S., C. Volland, and R. Haguenauer-Tsapis. 2001. Membrane transport: ubiquitylation in endosomal sorting. Curr. Biol. 11:R932-R934. [DOI] [PubMed] [Google Scholar]
- 15.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 16.Elsasser, S., R. R. Gali, M. Schwickart, C. N. Larsen, D. S. Leggett, B. Muller, M. T. Feng, F. Tubing, G. A. Dittmar, and D. Finley. 2002. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol. 4:725-730. [DOI] [PubMed] [Google Scholar]
- 17.Fang, S., J. P. Jensen, R. L. Ludwig, K. H. Vousden, and A. M. Weissman. 2000. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 275:8945-8951. [DOI] [PubMed] [Google Scholar]
- 18.Funakoshi, M., T. Sasaki, T. Nishimoto, and H. Kobayashi. 2002. Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proc. Natl. Acad. Sci. USA 99:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glickman, M. H., and A. Ciechanover. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82:373-428. [DOI] [PubMed] [Google Scholar]
- 20.Grossman, S. R., M. E. Deato, C. Brignone, H. M. Chan, A. L. Kung, H. Tagami, Y. Nakatani, and D. M. Livingston. 2003. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science 300:342-344. [DOI] [PubMed] [Google Scholar]
- 21.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296-299. [DOI] [PubMed] [Google Scholar]
- 22.Hengstermann, A., L. K. Linares, A. Ciechanover, N. J. Whitaker, and M. Scheffner. 2001. Complete switch from Mdm2 to human papillomavirus E6-mediated degradation of p53 in cervical cancer cells. Proc. Natl. Acad. Sci. USA 98:1218-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hicke, L. 2001. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell. Biol. 2:195-201. [DOI] [PubMed] [Google Scholar]
- 24.Hiyama, H., M. Yokoi, C. Masutani, K. Sugasawa, T. Maekawa, K. Tanaka, J. H. Hoeijmakers, and F. Hanaoka. 1999. Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J. Biol. Chem. 274:28019-28025. [DOI] [PubMed] [Google Scholar]
- 25.Honda, R., and H. Yasuda. 2000. Activity of MDM2, a ubiquitin ligase, toward p53 or itself is dependent on the RING finger domain of the ligase. Oncogene 19:1473-1476. [DOI] [PubMed] [Google Scholar]
- 26.Huibregtse, J. M., M. Scheffner, and P. M. Howley. 1993. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol. Cell. Biol. 13:775-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huibregtse, J. M., M. Scheffner, S. Beaudenon, and P. M. Howley. 1995. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. USA 92:2563-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleijnen, M. F., A. H. Shih, P. Zhou, S. Kumar, R. E. Soccio, N. L. Kedersha, G. Gill, and P. M. Howley. 2000. The hPLIC proteins may provide a link between the ubiquitylation machinery and the proteasome. Mol. Cell 6:409-419. [DOI] [PubMed] [Google Scholar]
- 29.Komada, M., and N. Kitamura. 1995. Growth factor-induced tyrosine phosphorylation of Hrs, a novel 115-kilodalton protein with a structurally conserved putative zinc finger domain. Mol. Cell. Biol. 15:6213-6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kubbutat, M. H., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 387:299-303. [DOI] [PubMed] [Google Scholar]
- 31.Kumar, S., A. L. Talis, and P. M. Howley. 1999. Identification of HHR23A as a substrate for E6-associated protein-mediated ubiquitylation. J. Biol. Chem. 274:18785-18792. [DOI] [PubMed] [Google Scholar]
- 32.Lai, Z., K. V. Ferry, M. A. Diamond, K. E. Wee, Y. B. Kim, J. Ma, T. Yang, P. A. Benfield, R. A. Copeland, and K. R. Auger. 2001. Human mdm2 mediates multiple mono-ubiquitylation of p53 by a mechanism requiring enzyme isomerization. J. Biol. Chem. 276:31357-31367. [DOI] [PubMed] [Google Scholar]
- 33.Lam, Y. A., T. G. Lawson, M. Velayutham, J. L. Zweier, and C. M. Pickart. 2002. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature 416:763-767. [DOI] [PubMed] [Google Scholar]
- 34.Masutani, C., K. Sugasawa, J. Yanagisawa, T. Sonoyama, M. Ui, T. Enomoto, K. Takio, K. Tanaka, P. J. van der Spek, D. Bootsma, et al. 1994. Purification and cloning of a nucleotide excision repair complex involving the xeroderma pigmentosum group C protein and a human homologue of yeast RAD23. EMBO J. 13:1831-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Midgley, C. A., C. J. Fisher, J. Bartek, B. Vojtesek, D. P. Lane, and D. M. Barnes. 1992. Analysis of p53 expression in human tumours: an antibody raised against human p53 expressed in Escherichia coli. J. Cell Sci. 101:183-189. [DOI] [PubMed] [Google Scholar]
- 36.Ortolan, T. G., P. Tongaonkar, D. Lambertson, L. Chen, C. Schauber, and K. Madura. 2000. The DNA repair protein rad23 is a negative regulator of multi-ubiquitin chain assembly. Nat. Cell Biol. 2:601-608. [DOI] [PubMed] [Google Scholar]
- 37.Pahl, H. L., and P. A. Baeuerle. 1996. Control of gene expression by proteolysis. Curr. Opin. Cell Biol. 8:340-347. [DOI] [PubMed] [Google Scholar]
- 38.Pickart, C. M. 2000. Ubiquitin in chains. Trends Biochem. Sci. 25:544-548. [DOI] [PubMed] [Google Scholar]
- 39.Pickart, C. M. 2001. Mechanisms underlying ubiquitylation. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 40.Pickart, C. M. 2001. Ubiquitin enters the new millennium. Mol. Cell 8:499-504. [DOI] [PubMed] [Google Scholar]
- 41.Prakash, S., and L. Prakash. 2000. Nucleotide excision repair in yeast. Mutat. Res. 51:13-24. [DOI] [PubMed] [Google Scholar]
- 42.Raasi, S., and C. M. Pickart. 2003. Rad23 ubiquitin-associated domains (UBA) inhibit 26 S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J. Biol. Chem. 78:8951-8959. [DOI] [PubMed] [Google Scholar]
- 43.Rao, H., and A. Sastry. 2002. Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. J. Biol. Chem. 77:11691-11695. [DOI] [PubMed] [Google Scholar]
- 44.Schauber, C., L. Chen, P. Tongaonkar, I. Vega, D. Lambertson, W. Potts, and K. Madura. 1998. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature 91:715-718. [DOI] [PubMed] [Google Scholar]
- 45.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitylation of p53. Cell 75:495-505. [DOI] [PubMed] [Google Scholar]
- 46.Scheffner, M., U. Nuber, and J. M. Huibregtse. 1995. Protein ubiquitylation involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature 373:81-83. [DOI] [PubMed] [Google Scholar]
- 47.Shkedy, D., H. Gonen, B. Bercovich, and A. Ciechanover. 1994. Complete reconstitution of conjugation and subsequent degradation of the tumor suppressor protein p53 by purified components of the ubiquitin proteolytic system. FEBS Lett. 348:126-130. [DOI] [PubMed] [Google Scholar]
- 48.Thrower, J. S., L. Hoffman, M. Rechsteiner, and C. M. Pickart. 2000. Recognition of the polyubiquitin proteolytic signal. EMBO J. 19:94-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tobias, J. W., and A. Varshavsky. 1991. Cloning and functional analysis of the ubiquitin-specific protease gene UBP1 of Saccharomyces cerevisiae. J. Biol. Chem. 266:12021-12028. [PubMed] [Google Scholar]
- 50.Vojtesek, B., H. Dolezalova, L. Lauerova, M. Svitakiva, P. Havlis, J. Kovarik, C. A. Midgley, and D. P. Lane. 1995. Conformational changes in p53 analyzed using new antibodies to the core DNA binding domain of the protein. Oncogene 10:389-393. [PubMed] [Google Scholar]
- 51.Waasem, N. H., and D. P. Lane. 1990. Monoclonal antibody analysis of the proliferating cell nuclear antigen (PCNA)—structural conservation and the detection of a nucleolar form. J. Cell Sci. 96:121-129. [DOI] [PubMed] [Google Scholar]
- 52.Walters, K. J., M. F. Kleijnen, A. M. Goh, G. Wagner, and P. M. Howley. 2002. Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry 41:1767-1777. [DOI] [PubMed] [Google Scholar]
- 53.Weissman, A. M. 2001. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell. Biol. 2:169-178. [DOI] [PubMed] [Google Scholar]
- 54.Wilkinson, C. R., M. Seeger, R. Hartmann-Petersen, M. Stone, M. Wallace, C. Semple, and C. Gordon. 2001. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat. Cell Biol. 3:939-943. [DOI] [PubMed] [Google Scholar]
- 55.You, J., R. E. Cohen, and C. M. Pickart. 1999. Construct for high-level expression and low misincorporation of lysine for arginine during expression of pET-encoded eukaryotic proteins in Escherichia coli. BioTechniques 27:950-954. [DOI] [PubMed] [Google Scholar]