SUMMARY
Metastasis of tumor cells to distant organs is the leading cause of death in melanoma. Yet, the mechanisms of metastasis remain poorly understood. One key question is whether all cells in a primary tumor are equally likely to metastasize or whether subpopulations of cells preferentially give rise to metastases. Here, we identified a subpopulation of uveal melanoma cells expressing the multidrug resistance transporter ABCB1 that are highly metastatic compared to ABCB1− bulk tumor cells. ABCB1+ cells also exhibited enhanced clonogenicity, anchorage independent growth, tumorigenicity and mitochondrial activity compared to ABCB1− cells. A375 cutaneous melanoma cells contained a similar subpopulation of highly metastatic ABCB1+ cells. These findings suggest that some uveal melanoma cells have greater potential for metastasis than others, and that a better understanding of such cells may be necessary for more successful therapies for metastatic melanoma.
Keywords: ABCB1, metastasis, uveal melanoma, mitochondrial respiration, immunodeficient mouse model
INTRODUCTION
Metastasis of tumor cells to distant organs is the leading cause of death in most forms of cancer. Yet, the mechanisms of metastasis remain poorly understood. One key question is whether all cells in a primary tumor are equally likely to metastasize or whether there are subpopulations of cells that preferentially give rise to metastases. Several lines of evidence suggest that many forms of cancer contain subpopulations of cells with preferential capacity for metastasis. These cells can exhibit properties reminiscent of stem cells, including the capacity for self-renewal and differentiation into multiple cell types, slow cycling rate, formation of a side population on flow cytometry based on the ability to efflux intracellular dyes, in vitro spheroid formation, and other characteristics (Visvader and Lindeman, 2008).
In cutaneous melanoma, several markers have been identified in cell lines and patient samples that label subsets of tumor cells that exhibit some or all of these properties, including ABCB5 (Fukunaga-Kalabis et al., 2010; Schatton et al., 2008), ALDH (Boonyaratanakornkit et al., 2010), CD133 (Monzani et al., 2007; Rappa et al., 2008), CD271/NGFR/p75 (Boiko et al., 2010), and JARID1 (Roesch et al., 2010). ABCB5, CD133 and CD271/NGFR/p75 were specifically shown to mark cells with an increased capacity for metastasis. However, the presence of such cells has not been explored in non-cutaneous forms of melanoma, such as uveal melanoma, which is the most common cancer of the eye and the second most common form of melanoma.
In this study, we identified a subpopulation of uveal melanoma cells in fresh patient samples and in cultured cells that express the multidrug resistance protein encoded by ABCB1 (also known as MDR1 and P-glycoprotein). ABCB1+ cells were highly metastatic in vivo and exhibited the capacity for multipotent differentiation, enhanced clonogenicity, anchorage independence, and tumorigenicity. Further, these cells showed preferential up-regulation of the mitochondrial respiration transcriptional program and enhancement of mitochondrial activity. A similar subpopulation of ABCB1+ cells was found in cutaneous melanoma cells, indicating that this finding may not be unique to uveal melanoma. These studies provide biological insights that may guide future therapies for metastatic disease.
RESULTS
Uveal melanomas contain a side population of dye-effluxing cells
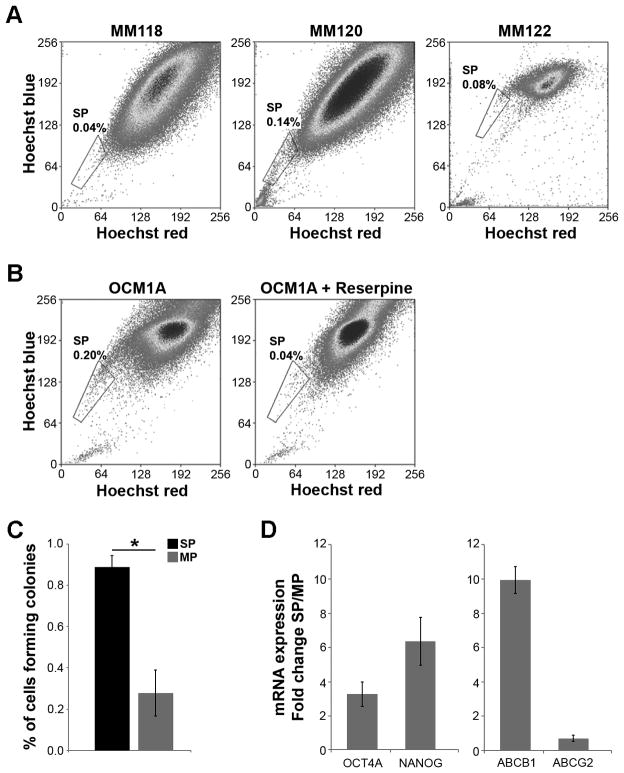
In primary tumor samples from three different patients, a Hoechst 33342 dye-effluxing side population was present, ranging from 0.04–0.14% of the total tumor cell population (Figure 1A). Similarly, OCM1A uveal melanoma cells, which are frequently used in studies of tumorigenicity and metastasis, displayed a dye-effluxing side population of 0.2%, which could be blocked by the addition of reserpine (Figure 1B). In soft agar clonogenic assays, a measure of anchorage independent proliferation, sorted OCM1A cells from the side population cells formed colonies much more efficiently than cells from the main population (P=0.02) (Figure 1C). Further, the side population cells exhibited enhanced mRNA expression of the stem cell markers OCT4A and NANOG compared to cells in the main population (Figure 1D).
Figure 1.
Uveal melanomas contain a side population that excludes Hoechst 33342 dye. (A) Hoechst dye efflux assay in primary uveal melanoma patient samples. The size of the side population (gate) as a percentage of total number of melanoma cells is indicated. The blue Hoechst fluorescence is plotted as a function of the red Hoechst fluorescence. (B) Hoechst dye efflux assay in OCM1A uveal melanoma cells. Control sample was treated with reserpine to verify the side population dependence on ABC transporter activity (right panel). (C) Soft agar colony forming assays in cells from the side population (SP) and main population (MP). (D) mRNA levels measured by real-time qPCR of embryonic stem cell markers OCT4A and NANOG, and of ABC transporters ABCB1 and ABCG2, expressed as fold change SP/MP in OCM1A cells. Data are representative of three independent experiments. *P<0.05.
Surprisingly, mRNA expression of ABCG2, which encodes a related membrane transporter that has also been associated with dye-effluxing side populations (Zhou et al., 2001), was not increased in the side population cells. In contrast, mRNA expression of the ABCB1 transporter was increased ten-fold in the side population compared to the main population (Figure 1D). These findings suggested that a subpopulation of ABCB1+ cells was being enriched in uveal melanoma side population.
Uveal melanomas contain a subpopulation of ABCB1+ cells
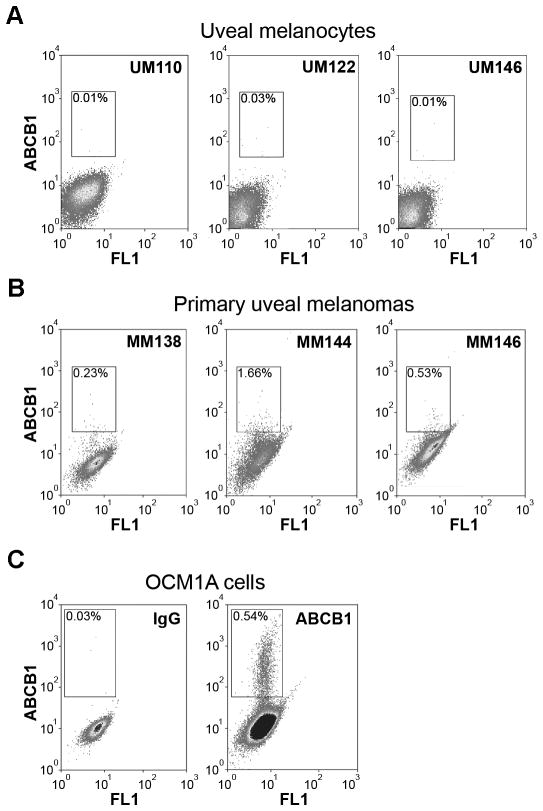
An ABCB1 shift assay was used to identify ABCB1+ cells in untreated cells from clinical specimens. Three different human samples of normal uveal melanocytes were found to be devoid of ABCB1+ cells, comparable to IgG isotype control background (Figure 2A). In contrast, primary uveal melanoma cells from three different patients contained an ABCB1+ subpopulation ranging from 0.23–1.66% of the total population (Figure 2B). OCM1A cells also contained an ABCB1+ subpopulation (Figure 2C). About 30% of OCM1A cells expressed detectable ABCB1 protein, and there was a smaller subpopulation of 0.54% cells that expressed high levels of functional ABCB1, defined as PE fluorescence above ~102 (Figure 2C, gate). This high expressing subpopulation was used for further experiments.
Figure 2.
Uveal melanomas contain a subpopulation of ABCB1+ cells. (A) ABCB1 shift assay in normal uveal melanocytes from three different patients. (B) ABCB1 shift assay in primary uveal melanoma tumors from three different patients. (C) ABCB1 shift assay in OCM1A uveal melanoma cells. Left panel depicts control assay with non-specific IgG antibody. Right panel depicts results with ABCB1 antibody. The gate delineates the ABCB1+ subpopulation. X-axes indicate autofluorescence, and Y-axes ABCB1 labeling.
ABCB1+ cells exhibit enhanced clonogenic and migratory capacity
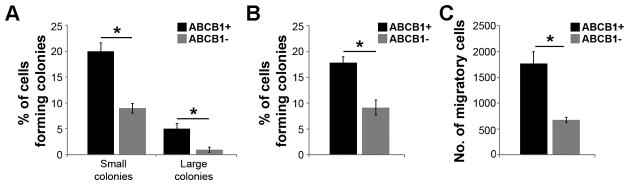
We tested the ability of ABCB1+ and ABCB1− cells to form colonies in single cell dilution assays. OCM1A cells were sorted for ABCB1expression using the ABCB1 shift assay, single cells were seeded onto 96-well plates, and multi-cellular colonies were counted after seven days. The percentage of cells able to form small colonies (<64 cells) and large colonies (≥64 cells) was significantly greater for ABCB1+ cells compared to ABCB1− cells (P=0.008 and P=0.04, respectively) (Figure 3A). When colonies from both groups were re-plated as single cells in 96-well plates, the ABCB1+ cells again gave rise to larger colonies with greater efficiency than ABCB1− cells (Supplementary Figure S1). In soft agar assays, ABCB1+ cells exhibited greater capacity for anchorage independent colony formation than ABCB1− cells (P=0.02) (Figure 3B). Additionally, ABCB1+ cells showed greater migratory behavior than ABCB1− cells in transwell migration assays (P=0.007) (Figure 3C).
Figure 3.
ABCB1+ cells exhibit increased clonogenicity and migration. (A) Single-cell clonogenic assays in ABCB1+ and ABCB1− cells. Small colonies, < 64 cells; large colonies ≥ 64 cells. (B) Soft agar colony forming assays in ABCB1+ and ABCB1− cells. (C) Cell migration assays in ABCB1+ and ABCB1− cells. *P<0.05.
ABCB1+ cells are tumorigenic in vivo
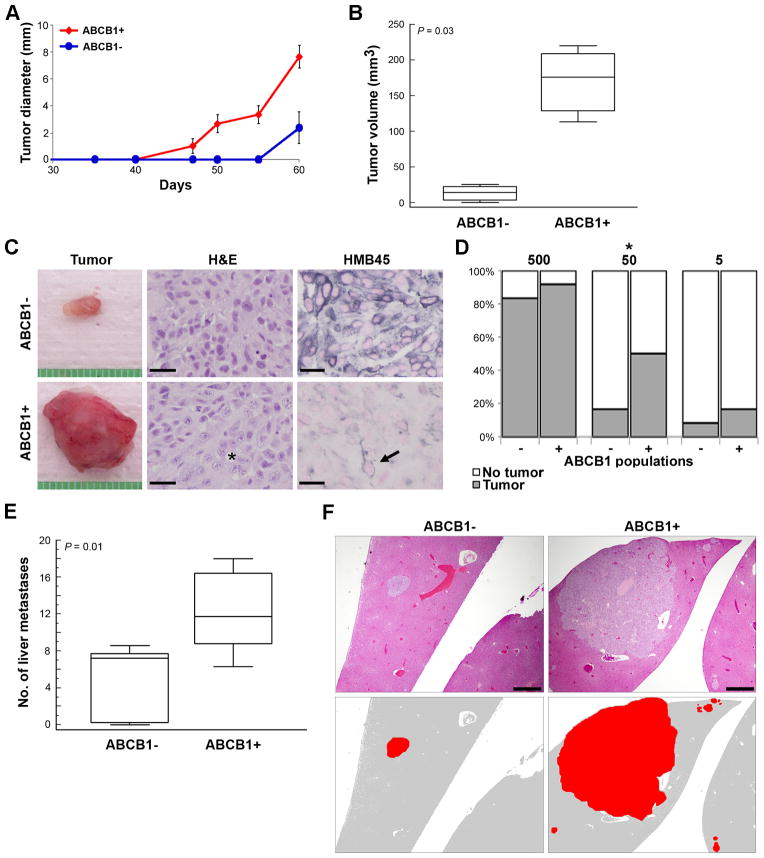
To study in vivo tumorigenicity, both ABCB1+ and ABCB1− sorted OCM1A cells were injected subcutaneously into the flanks of SCID mice (500 cells per injection). Mice were monitored closely for the development of palpable tumors. At day 40, ABCB1+ cells had formed palpable tumors in 100% of animals, compared to 0% for ABCB1− cells (Figure 4A). No tumors were detected in ABCB1− tumors until day 55. When final tumor volumes were measured at day 60, all tumors derived from ABCB1+ cells were ≥ 110 mm3, whereas all ABCB1− tumors were < 25 mm3 (P=0.03) (Figure 4B). At day 60, all mice were euthanized, and tumors were obtained for analysis (Figure 4C). ABCB1− tumors were composed mostly of spindle-shaped cells that are typical of well differentiated, low grade melanomas (Figure 4C). In contrast, ABCB1+ tumors exhibited a heterogeneous mixture of cells ranging from differentiated spindle cells to epithelioid, undifferentiated and multinucleated cells (Figure 4C). Consistent with these findings, the melanocyte differentiation marker HMB45 was strongly expressed in ABCB1− tumor cells, but this marker was expressed only in minority of cells in ABCB1+ tumors (Figure 4C). Titration experiments were performed in which subcutaneous flank injections of 500, 50 or 5 ABCB1− or ABCB1+ sorted cells were performed in NOD SCID gamma mice. ABCB1+ cells formed tumors more efficiently than ABCB1− cells. Significant difference in tumor initiation rate was observed in the 50 cell group (P=0.045) (Figure 4D).
Figure 4.
ABCB1+ uveal melanoma cells are highly tumorigenic and metastatic. (A) Graph showing the growth (in diameter) of tumors arising from injection of 500 ABCB1+ versus ABCB1− OCM1A uveal melanoma cells into the flanks of SCID mice (n=7 for each condition). (B) Volumes of ABCB1− and ABCB1+ flank tumors at the time of necropsy (day 60). Middle line represents median; box, 25th to 75th percentiles; outer bars, minimum and maximum values. (C) Representative gross appearance of ABCB1− and ABCB1+ flank tumors. stained with H&E or immunostained with HMB45 antibody (positive staining appears in blue). Scale bar, 25 μm. Asterisk indicates area of undifferentiated melanoma cells, and it is immediately next to an anaplastic multinucleated tumor cell. Arrow indicates a rare differentiated tumor cell in the ABCB1+ tumor. (D) Absolute tumor initiation rate in ABCB1+ and ABCB1− populations calculated by titrated xenotransplantation assays using 500, 50 and 5 cells (n=12 for each condition). *P<0.05. (E) Liver metastases counted at 7 weeks following tail vein injection of 105 ABCB1+ versus ABCB1− OCM1A cells (n=7 for each condition). Middle line represents median; box, 25th to 75th percentiles; outer bars, minimum and maximum values. (F) Representative photomicrographs of liver metastases in mice injected with ABCB1+ versus ABCB1− cells. Top panels show sections stained with H&E. Bottom panels are cartoons depicting tumors (red) in the corresponding top panels. Scale bar, 500 μm.
ABCB1+ cells exhibits enhanced propensity for metastasis
To assess metastatic capacity, both ABCB1+ and ABCB1− sorted OCM1A cells were injected into the tail vein of NOD SCID gamma mice (105 cells per animal). Mice were euthanized at 7 weeks. At necropsy, occasional metastatic lesions were identified in lung, lymph nodes, ovaries, kidneys and pancreas (data not shown), but the most common site of metastasis was liver, consistent with the pattern of uveal melanoma metastasis in humans (Harbour, 2003). The number of liver metastases were significantly greater in mice injected with ABCB1+ cells compared to ABCB1− cells (P=0.01) (Figure 4E). Further, the ABCB1+ metastatic liver tumors tended to be larger and more invasive than ABCB1− tumors (Figure 4F). Cells from both ABCB1− and ABCB1+ metastatic liver tumors were collected, re-plated in methylcellulose, and analyzed for their ability to form colonies. The ABCB1+ cells exhibited much greater colony forming ability than the ABCB1− cells (Supplementary Figure S2).
Cutaneous melanoma cells also contain an ABCB1+ subpopulation with enhanced metastatic capacity
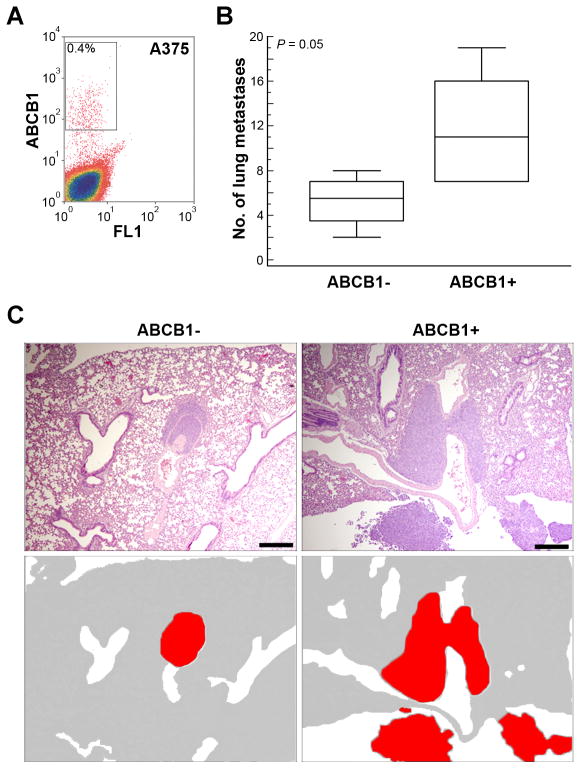
To determine whether an ABCB1+ subpopulation may also be present in cutaneous melanomas, A375 cells were analyzed using the ABCB1 shift assay. This cell line contained a 0.4% subpopulation of ABCB1+ cells (Figure 5A). ABCB1+ and ABCB1− sorted A375 cells were injected into the tail vein of NOD SCID gamma mice (5×103 cells per animal), which were euthanized at 6 weeks and subjected to necropsy. Unlike the OCM1A uveal melanoma cells, only occasional metastatic lesions were identified in the liver, and some lesions were found in the mediastinum (data not shown), but the most consistent site of metastasis for A375 cutaneous melanoma cells was lung. Even so, the number of lung metastases were significantly greater in mice injected with ABCB1+ cells compared to ABCB1− cells (P=0.05) (Figure 5B–C).
Figure 5.
Cutaneous melanoma cells contain an ABCB1+ subpopulation with enhanced metastatic capacity. (A) ABCB1 shift assay in A375 cutaneous melanoma cells, showing population of ABCB1+ cells (gate). (B) Lung metastases counted at 6 weeks following tail vein injection of 5×103 ABCB1+ versus ABCB1− A375 cells (n= 5 for each condition). Middle line represents median; box, 25th to 75th percentiles; outer bars, minimum and maximum values. (C) Representative photomicrographs of ABCB1− and ABCB1+ metastases in lung sections stained with H&E. Bottom panels are cartoons depicting tumors (red) in the corresponding top panels. Scale bar, 500 μm.
ABCB1+ cells exhibit enhancement of mitochondrial respiration
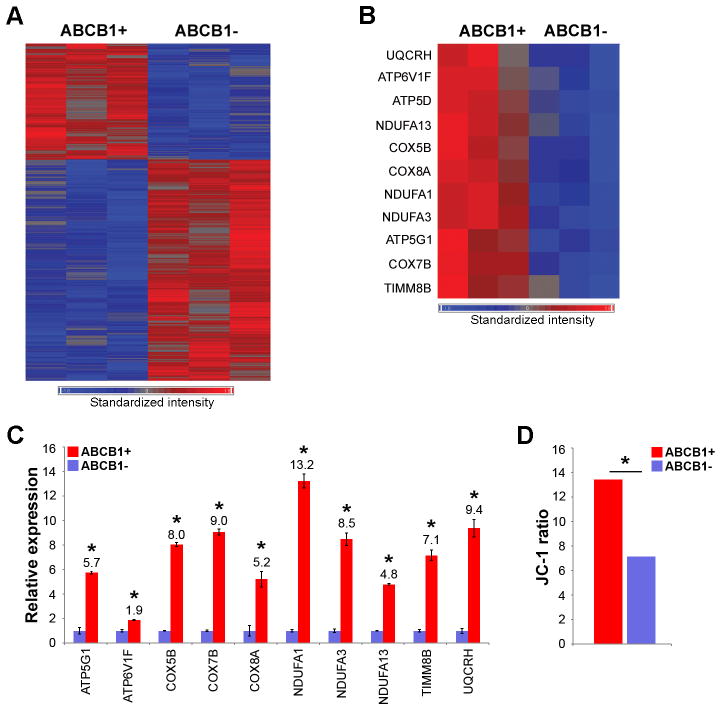
Next, we used gene expression profiling to gain functional insights into the ABCB1+ cells. ABCB1+ and ABCB1− OCM1A cells were collected from three independent ABCB1 shift assays. RNA was isolated, and gene expression profiling was performed using Illumina HumanRef-8 Expression BeadChip arrays. Using ANOVA and a significance threshold of P<0.01, 718 genes were identified to be differentially expressed between ABCB1+ and ABCB1− cells (Figure 6A and Supplementary Table S3). To confirm the microarray results, eight of the top discriminating genes were validated by real-time qPCR, including ABCB1 (Supplementary Figure S3). Using Gene Set Enrichment Analysis to identify significant functional patterns in the gene expression data, we found a striking up-regulation of genes involved in mitochondrial respiration in ABCB1+ cells (e.g., ATP5G1, ATP6V1F, COX5B, COX7B, COX8A, NDUFA1, NDUFA3, NDUFA13, PPARGC1, TIMM8B and UQCRH) (Figure 6B–C). Consistent with this finding, ABCB1+ cells exhibited significantly increased mitochondrial activity, as measured by mitochondrial membrane potential, compared to ABCB1− cells (P=0.004) (Figure 6D).
Figure 6.
Gene expression profiling shows up-regulation of mitochondrial respiration transcriptional program in ABCB1+ cells. (A) Heat map of 718 discriminating genes that were highly differentially expressed between ABCB1+ and ABCB1− OCM1A cells. P<0.01 was used as a significance threshold. (B) Heat map of a subset of discriminating genes involved in mitochondrial respiration. (C) Validation of microarray results using real-time qPCR for a subset of genes involved in mitochondrial respiration. (D) JC-1 assay comparing the mitochondrial membrane potential of ABCB1+ versus ABCB1− OCM1A cells. The JC-1 ratio was measured as red J-aggregates/green monomers (see Materials and Methods for details). Data are representative of three independent experiments. *P<0.05.
DISCUSSION
In this study, we showed that clinical samples from primary uveal melanomas, but not normal uveal melanocytes, contained a subpopulation of cells that express the multidrug resistance marker encoded by ABCB1. The ABCB1+ subpopulation of cells was highly tumorigenic and metastatic, and they showed up-regulated mitochondrial activity, compared to ABCB1− cells. These findings suggest that uveal melanomas contain subpopulations of cells that are preferentially responsible for metastasis, and they provide insights that may be important in the development of successful therapies for metastatic uveal melanoma.
ABCB1 encodes the P-glycoprotein (P-gp), which was initially identified as a drug-effluxing transporter protein that conveyed multidrug resistance in cancer cells (Kartner et al., 1983). ABCB1 has been implicated as a marker for aggressive subpopulations in cutaneous melanoma, lung cancer and gastrointestinal malignancies (Keshet et al., 2008; Gutova et al., 2007; Haraguchi et al., 2006), and expression of the ABCB1 protein has been linked to poor survival in uveal melanoma (Dunne et al., 1998).
Uveal melanomas are highly metastatic, giving rise to fatal hematogeneous metastasis in up to half of afflicted patients (Harbour, 2003). Several uveal melanoma biomarkers are highly predictive of metastasis, including a well-characterized gene expression profile (GEP) that has now been incorporated as a clinical prognostic test into the AJCC TNM classification (Finger, 2009). Tumors with the class 1 signature rarely give rise to metastasis, whereas those with the class 2 signature have a very high rate of metastasis (Onken et al., 2004). Interestingly, the tumors formed by ABCB1− and ABCB1+ cells closely resembled human class 1 and class 2 tumors, respectively. ABCB1− tumors, like class 1 tumors, had a low capacity for metastasis and were composed mainly of differentiated spindle cells (Onken et al., 2006). ABCB1+ tumors, like class 2 tumors, were highly metastatic and contained heterogeneous populations of tumor cells ranging from differentiated spindle cells to undifferentiated epithelioid cells (Onken et al., 2006). Also, several class 2 discriminating genes were differentially expressed in ABCB1+ cells, including PLXNB1, MTUS1, IL1RAP, TIMP3, ECM1 and RGS1. ABCB1+ cells exhibited a marked up-regulation of genes involved in mitochondrial respiration, and they showed significantly increased mitochondrial activity, compared to ABCB1− cells. We recently found that the vast majority of metastasizing class 2 tumors harbor inactivating mutations in the tumor suppressor gene BAP1 (Harbour et al., 2010). Interestingly, depletion of BAP1 in class 1 uveal melanoma cells caused a phenotypic switch to class 2-like cells, and this was accompanied by a highly significant up-regulation of genes involved in ATP synthesis and mitochondrial respiration, similar to our findings in ABCB1+ cells (J.W.H., unpublished data). These findings may shed light on the highly aggressive nature of this subpopulation of cells. We speculate that the increased mitochondrial activity of the ABCB1+ cells, and the BAP1-deficient cells, may reflect a selective adaptation to hypoxia, which is known to promote dedifferentiation and malignant progression in cancer cells (Lofstedt et al., 2004; Helczynska et al., 2003). Consistent with this possibility, class 2 uveal melanomas show differential up-regulation of hypoxia inducible factor 1-alpha compared to class 1 tumors (Chang et al., 2008). Further experimental work is needed to explore the relationship between hypoxia and metastasis in uveal melanoma.
ABCB1+ cells in uveal melanoma exhibited several stem-like features, including enhanced clonogenicity, anchorage independent growth, and the ability to generate daughter cells with multiple differentiated phenotypes (Visvader and Lindeman, 2008). Further, metastasizing class 2 tumors display significant transcriptomic similarities to neural and ectodermal stem cells (Chang et al., 2008), and aggressive uveal melanoma cells exhibit primitive, embryonic-like features such as vasculogenic mimicry (Hendrix et al., 2003). The existence of stem cells in cuteneous melanoma was postulated by a few independent groups (Refaeli et al., 2009). However some recent studies indicate that melanomas may not follow the classic cancer stem cell model (Quintana et al., 2010), but rather, they may exhibit “dynamic stemness” in which tumor cells gain and lose aggressive stem-like properties over time, depending on microenvironmental influences (Roesch et al., 2010). So, it remains unclear which of the models is more representative of human melanomas (Refaeli et al., 2009). Nevertheless, we do not argue here that ABCB1+ is a cancer stem cell marker, since the purpose of this study was to identify cells with enhanced ability to metastasize, not necessarily ones with cancer stem cell properties, as they are customarily defined (Visvader and Lindeman, 2008). We believe that the most important finding in this study is the link between ABCB1 and metastatic behavior.
A limitation of this study is the absence of in vivo studies using primary uncultured uveal melanoma cells. Because of the rarity of uveal melanoma and the paucity of tumor cells obtained from these relatively small eye tumors, such studies are highly impractical. Nevertheless, the findings reported here will provide direction and testable hypotheses for future work in this area.
METHODS
Tumor samples
This study was approved by the Institutional Review Board of Washington University and adhered to the tenets of the Declaration of Helsinki. Primary uveal melanomas and normal uveal melanocytes were collected at the time of enucleation (Supplementary Table S1). Written informed consent was obtained. Tumor samples were collected in HAM’S F-12 medium, incubated in trypsin and collagenase, and grown at 4% oxygen on collagen-covered tissue culture plates in HAM’s F-12 supplemented with 10% BSA, SITE supplement (Sigma), B27 supplement (Invitrogen), bFGF (PeproTech), L-glutamine, gentamicin and fungizon (MDMF medium). Normal uveal melanocytes were handled in the same manner, except that they were maintained in FICmedia. OCM1A uveal melanoma cells were generously provided by Dr. June Kan-Mitchell. A375 cutaneous melanoma cells were obtained from the ATCC (#CRL-1619). Both cell lines were grown in RPMI-1640 supplemented with 10% FBS and L-glutamine/antibiotics.
Flow cytometry
For Hoechst dye efflux assay, 5×107 cells were incubated with 5 μg/ml Hoechst 33342 fluorescent dye (Sigma) for 90 min at 37°C. Reserpine (Sigma) was added (5 μM) during the Hoechst incubation to verify dependence of the side population on ABC transporter activity. The Hoechst 33342 fluorescent dye was excited with an UV laser at 351 nm, and fluorescence emission was measured with a 460/20 BP filter (Hoechst Blue) and a 680 LP optical filter (Hoechst Red). A 610 DRSP was used to separate the emission wavelengths.
The ABCB1 shift assay (Millipore) was performed according to the manufacturer’s protocol. 108 cells were used, and ABCB1+ and ABCB1− cells were collected in RPMI-1640 for experiments or in TRIzol for RNA isolation. The assay is performed by exposing cells to a low dose of vinblastine (22.5 μM) for a short period of time (10 min) to induce a conformational change in the cell membrane-bound ABCB1 protein, which allows it to be recognized by the UIC2 antibody
Cell analysis and sorting were conducted using a MoFlo high speed flow cytometer (Beckman Coulter). Viable cells for both assays were selected with scatter gate or PI staining. Gates for sorting were drawn using reserpine treated cells (side population) or IgG isotype control (ABCB1). Data were analyzed using the Summit software (Dako). Before doing further assays, the amount and viability of the sorted cells were evaluated by staining with Trypan blue. No difference in cell viability was detected in MP versus SP populations or ABCB1− versus ABCB1+ populations after cell sorting.
Cell culture experiments
For anchorage-independent colony formation assays, sorted cells were plated in 0.3% soft agar or 1.0% methylcellulose (R&D Systems) (diluted in MDMF medium) on non-treated 35 mm culture plates. After 14–18 days of growth, colonies were counted under dissecting microscope after staining with MTT (Sigma).
For single-cell dilution assays, ABCB1− and ABCB1+ cells were seeded one viable cell per well into ultra-low attachment 96-well plates containing MDMF medium using flow cytometer. Wells containing one live cell were marked using a phase contrast microscope and cell number for each well containing a colony bigger than 4 cells was assessed after 7 days.
For cell migration assays, ABCB1− and ABCB1+ sorted cells (1×104 cells in 100 μL of MDM medium) were plated in upper chambers of 12-well transwell plates (Corning) and incubated 24h at 37°C. Medium with 10% FBS was added to the lower wells of the chambers. The non-migrated cells were removed from the upper surface of the filters by rubbing gently with a cotton-tipped applicator. Migrated cells were fixed, stained with crystal violet and counted under a phase contrast microscope.
The mitochondrial membrane potential was evaluated using the JC-1 assay (Cayman Chemical), following the manufacturer’s 96-well fluorescence plate reader protocol (Spectra MAX Gemini EM; Molecular Devices). The lipophilic cationic probe JC-1 reversibly changes colors from green to red as the mitochondrial membrane potential increases. The mitochondrial membrane potential was presented as the red/green JC-1 ratio.
Real-time qPCR
Total RNA extracted with TRIzol (Invitrogen) was DNase-treated and reverse transcription was performed using the iScript cDNA Synthesis kit (Bio-Rad Laboratories). Primers were designed using Primer Express 3.0 (Applied Biosystems) and synthesized by Integrated DNA Technologies (Supplementary Table S2). At least one primer of each pair crossed exon-exonboundaries to prevent amplification of genomic DNA, and primer quality was confirmed by melting curve analysis. Real-time qPCR was performed using the IQ SYBR Green Supermix (Bio-Rad Laboratories). Quantification of target gene mRNA was performed using a standard curve and reference gene RNA (UBC).
Gene expression profiling
Total RNA from ABCB1− and ABCB1+ cells from three independent ABCB1 shift assays was collected in TRIzol, RNA was isolated according to manufacturer’s protocol and purified by ammonium acetate precipitation. RNA quality was assessed on the Bioanalyzer 2100 (Agilent Technologies). Samples were subjected to gene expression profiling using 24,528 probe HumanRef-8 v3 Expression BeadChip arrays (Illumina), with the assistance of the Washington University Microarray Core Facility. Raw expression data were subjected to rank invariant normalization and log transformation. ANOVA and hierarchical clustering were performed with Partek Genomics Suite (version 6.4) using a significance of P<0.01 as a threshold. Gene Set Enrichment Analysis (GSEA)version 2.0.4 was performed using the Wilcoxon statistic and a false discovery rate of Q=0.001. Gene expression data have been deposited in NCBI Gene Expression Omnibus and are accessible through GEO Series accession number GSE21425 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21425).
Animal studies
Animal experiments were approved by the Washington University Animal Studies Committee. For tumorigenic assays, 500 ABCB1− or ABCB1+ sorted cells resuspended in matrigel (Trevigen) were injected subcutaneously into the flanks of SCID mice (Jackson Laboratory). Mice were monitored twice a week and euthanized after 60 days. Tumors were collected and measured using the ellipsoid volume formula (π xyz/6 mm3). Titration experiments were performed in which 5, 50 or 500 ABCB1− or ABCB1+ sorted cells resuspended in matrigel were injected subcutaneously into the flanks of NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ JAX® (NOD SCID gamma) mice (Jackson Laboratory). For metastasis assays, ABCB1− and ABCB1+ sorted cells (105 OCM1A uveal melanoma cells or 5×103 A375 cutaneous melanoma cells) suspended in 100 μl PBS and injected into the tail vein of NOD SCID gamma mice. Mice were euthanized at week 6 or 7, or when signs of sickness appeared. Cells isolated from ABCB1− and ABCB1+ liver metastases were re-plated in 1.0% methylcellulose for colony formation assays. Organs were harvested, fixed in 10% formalin and embedded in paraffin. Four micron sections were stained with H&E. Whole livers and lungs were serially sectioned, and five sections per organ were examined.
For immunostaining, sections were deparaffinized, rehydrated with ethanol, treated with 0.3% hydrogen peroxide and methanol, and subjected to heat-induced epitope retrieval in citrate buffer. Immunostaining was performed for HMB45 (1:500 dilution; Dako) using the streptavidin–biotin method with the Vector ABC Elite kit (Vector Laboratories). Blue stain was used and slides were counterstained with nuclear fast red.
Statistical analysis
Except where otherwise noted, data were analyzed for statistical significance using MedCalc software, version 9.5.1.0 (http://medcalcsoftware.com/medcalc.php).
Supplementary Material
Re-plating of OCM1A cells from ABCB1+ and ABCB1− colonies shows persistent increased colony forming ability of ABCB1+ cells.
Re-plating of OCM1A cells from ABCB1+ and ABCB1− metastatic liver tumors shows persistent increased colony forming ability of ABCB1+ cells.
Microarray gene expression profiling was validated by qPCR of top discriminating genes, as identified by ANOVA. *P<0.05.
Acknowledgments
The authors would like to thank Dr. Michael Onken for help with microarray data analyses, Belinda McMahan in the Immunomorphology Core Lab for preparation of histopathologic sections and William Eades, Jon Christopher Holley, and Jacqueline Hughes in the Siteman Cancer Center High Speed Sorter Core Facility for performing cell sorting (NCI Cancer Center Support Grant #P30 CA91842). This work was supported by Fonds de la Recherche en Santé du Québec Postdoctoral Training Award (SL), NIH/NIDCDT32 Research Training Program for Otolaryngology (ZTK), Natural Sciences and Engineering Research Council of Canada (CS), R01 CA125970 (JWH), R01 EY13169 (JWH), Barnes-Jewish Hospital Foundation (JWH), Kling Family Foundation (JWH), Tumori Foundation (JWH), Horncrest Foundation (JWH) and Research to Prevent Blindness (JWH), and by an unrestricted grant to the Department of Ophthalmology and Visual Sciences from a Research to Prevent Blindness, Inc. and the NIH Vision Core Grant P30 EY02687. JWH and Washington University may receive income based on a license of related technology by the University to Castle Biosciences, Inc. This work was not supported by Castle Biosciences, Inc.
References
- Boiko AD, Razorenova OV, van de Rijn M, et al. Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature. 2010;466:133–137. doi: 10.1038/nature09161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonyaratanakornkit JB, Yue L, Strachan LR, Scalapino KJ, LeBoit PE, Lu Y, Leong SP, Smith JE, Ghadially R. Selection of tumorigenic melanoma cells using ALDH. J Invest Dermatol. 2010;130:2799–2808. doi: 10.1038/jid.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SH, Worley LA, Onken MD, Harbour JW. Prognostic biomarkers in uveal melanoma: evidence for a stem cell-like phenotype associated with metastasis. Melanoma Res. 2008;18:191–200. doi: 10.1097/CMR.0b013e3283005270. [DOI] [PubMed] [Google Scholar]
- Dunne BM, McNamara M, Clynes M, Shering SG, Larkin AM, Moran E, Barnes C, Kennedy SM. MDR1 expression is associated with adverse survival in melanoma of the uveal tract. Hum Pathol. 1998;29:594–598. doi: 10.1016/s0046-8177(98)80008-7. [DOI] [PubMed] [Google Scholar]
- Finger PT. The 7th edition AJCC staging system for eye cancer: an international language for ophthalmic oncology. Arch Pathol Lab Med. 2009;133:1197–1198. doi: 10.5858/133.8.1197. [DOI] [PubMed] [Google Scholar]
- Fukunaga-Kalabis M, Martinez G, Nguyen TK, Kim D, Santiago-Walker A, Roesch A, Herlyn M. Tenascin-C promotes melanoma progression by maintaining the ABCB5-positive side population. Oncogene. 2010;29:6115–6124. doi: 10.1038/onc.2010.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutova M, Najbauer J, Gevorgyan A, Metz MZ, Weng Y, Shih CC, Aboody KS. Identification of uPAR-positive chemoresistant cells in small cell lung cancer. PLoS ONE. 2007;2:e243. doi: 10.1371/journal.pone.0000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi N, Utsunomiya T, Inoue H, Tanaka F, Mimori K, Barnard GF, Mori M. Characterization of a side population of cancer cells from human gastrointestinal system. Stem Cells. 2006;24:506–513. doi: 10.1634/stemcells.2005-0282. [DOI] [PubMed] [Google Scholar]
- Harbour JW. Clinical overview of uveal melanoma: introduction to tumors of the eye. In: Albert DM, Polans A, editors. Ocular Oncology. New York: Marcel Dekker; 2003. pp. 1–18. [Google Scholar]
- Harbour JW, Onken MD, Roberson ED, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helczynska K, Kronblad A, Jogi A, Nilsson E, Beckman S, Landberg G, Pahlman S. Hypoxia promotes a dedifferentiated phenotype in ductal breast carcinoma in situ. Cancer Res. 2003;63:1441–1444. [PubMed] [Google Scholar]
- Hendrix MJ, Seftor EA, Hess AR, Seftor RE. Vasculogenic mimicry and tumour-cell plasticity: lessons from melanoma. Nat Rev Cancer. 2003;3:411–421. doi: 10.1038/nrc1092. [DOI] [PubMed] [Google Scholar]
- Kartner N, Riordan JR, Ling V. Cell surface P-glycoprotein associated with multidrug resistance in mammalian cell lines. Science. 1983;221:1285–1288. doi: 10.1126/science.6137059. [DOI] [PubMed] [Google Scholar]
- Keshet GI, Goldstein I, Itzhaki O, et al. MDR1 expression identifies human melanoma stem cells. Biochem Biophys Res Commun. 2008;368:930–936. doi: 10.1016/j.bbrc.2008.02.022. [DOI] [PubMed] [Google Scholar]
- Lofstedt T, Jogi A, Sigvardsson M, Gradin K, Poellinger L, Pahlman S, Axelson H. Induction of ID2 expression by hypoxia-inducible factor-1: A role in dedifferentiation of hypoxic neuroblastoma cells. J Biol Chem. 2004;279:39223–39231. doi: 10.1074/jbc.M402904200. [DOI] [PubMed] [Google Scholar]
- Monzani E, Facchetti F, Galmozzi E, et al. Melanoma contains CD133 and ABCG2 positive cells with enhanced tumourigenic potential. Eur J Cancer. 2007;43:935–946. doi: 10.1016/j.ejca.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Onken MD, Ehlers JP, Worley LA, Makita J, Yokota Y, Harbour JW. Functional gene expression analysis uncovers phenotypic switch in aggressive uveal melanomas. Cancer Res. 2006;66:4602–4609. doi: 10.1158/0008-5472.CAN-05-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004;64:7205–7209. doi: 10.1158/0008-5472.CAN-04-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana E, Shackleton M, Foster HR, Fullen DR, Sabel MS, Johnson TM, Morrison SJ. Phenotypic heterogeneity among tumorigenic melanoma cells from patients that is reversible and not hierarchically organized. Cancer Cell. 2010;18:510–523. doi: 10.1016/j.ccr.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappa G, Fodstad O, Lorico A. The stem cell-associated antigen CD133 (Prominin-1) is a molecular therapeutic target for metastatic melanoma. Stem Cells. 2008;26:3008–3017. doi: 10.1634/stemcells.2008-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refaeli Y, Bhoumik A, Roop DR, Ronai ZA. Melanoma-initiating cells: a compass needed. EMBO Rep. 2009;10:965–972. doi: 10.1038/embor.2009.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch A, Fukunaga-Kalabis M, Schmidt EC, et al. A Temporarily Distinct Subpopulation of Slow-Cycling Melanoma Cells Is Required for Continuous Tumor Growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatton T, Murphy GF, Frank NY, et al. Identification of cells initiating human melanomas. Nature. 2008;451:345–349. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- Zhou S, Schuetz JD, Bunting KD, et al. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat Med. 2001;7:1028–1034. doi: 10.1038/nm0901-1028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Re-plating of OCM1A cells from ABCB1+ and ABCB1− colonies shows persistent increased colony forming ability of ABCB1+ cells.
Re-plating of OCM1A cells from ABCB1+ and ABCB1− metastatic liver tumors shows persistent increased colony forming ability of ABCB1+ cells.
Microarray gene expression profiling was validated by qPCR of top discriminating genes, as identified by ANOVA. *P<0.05.