Abstract
Nuclear and mitochondrial genomes are under continuous assault by a combination of environmentally and endogenously derived reactive oxygen species, inducing the formation and accumulation of mutagenic, toxic, and/or genome-destabilizing DNA lesions. Failure to resolve these lesions through one or more DNA-repair processes is associated with genome instability, mitochondrial dysfunction, neurodegeneration, inflammation, aging, and cancer, emphasizing the importance of characterizing the pathways and proteins involved in the repair of oxidative DNA damage. This review focuses on the repair of oxidative damage–induced lesions in nuclear and mitochondrial DNA mediated by the base excision repair (BER) pathway in mammalian cells. We discuss the multiple BER subpathways that are initiated by one of 11 different DNA glycosylases of three subtypes: (a) bifunctional with an associated β-lyase activity; (b) monofunctional; and (c) bifunctional with an associated β,δ-lyase activity. These three subtypes of DNA glycosylases all initiate BER but yield different chemical intermediates and hence different BER complexes to complete repair. Additionally, we briefly summarize alternate repair events mediated by BER proteins and the role of BER in the repair of mitochondrial DNA damage induced by ROS. Finally, we discuss the relation of BER and oxidative DNA damage in the onset of human disease. Antioxid. Redox Signal. 14, 2491–2507.
Introduction
The cellular condition in which reactive oxygen species (ROS) exceed the antioxidant scavenging ability of a cell or when cellular antioxidant systems are unable to neutralize the effects of prooxidants such as free radicals or reactive oxygen and nitrogen species is termed oxidative stress. These ROS molecules include superoxide anions, hydroxyl radicals, and hydrogen peroxide, among others. Such molecules stem from endogenous sources through cellular metabolism and exogenous sources mediated by environmental exposure. A large proportion of cellular ROS are generated in the mitochondria as a result of oxidative phosphorylation (100), although not all ROS are derived from the mitochondrial respiratory chain. Additional endogenous sources include microsomes, cytosolic enzymes such as NADPH oxidases, and phagocytes (2). Exogenous sources of ROS include asbestos, crystalline silica, coal, metal ions, chemotherapeutic and chemopreventive agents, cigarette smoke, ultraviolet light, psoralen and ultraviolet A (PUVA), ionizing radiation, pesticides, and related neurotoxins such as paraquat and 1-methyl-4-phenylpyridinium [MPP(+)] (46, 66, 88). Depending on the extent and type of ROS exposure, all macromolecules in the cell are subject to ROS-mediated damage, including proteins, lipids, RNA, and DNA.
The chemical makeup of the many DNA base and genome modifications triggered by ROS has been extensively reviewed over the past 25-year period, covering the chemistry of the modifications, lesion formation and repair, the utility of lesion characterization, and the identification of DNA lesions as biomarkers for human disease (9, 23, 62, 89). In most cases, DNA damage from ROS-generating agents is mediated by Fenton chemistry, giving rise to the formation of chronic and persistent DNA damage, including nucleotide base modifications, apurinic/apyrimidinic (AP) sites, single- and double-strand DNA breaks, and DNA crosslinks (30). Both cellular genomes (nuclear and mitochondrial) are damaged upon ROS exposure or oxidative stress. However, it has been demonstrated that mitochondrial DNA accumulates a greater level of damage as compared with nuclear DNA after oxidative stress (95).
Several DNA-repair pathways are engaged in response to oxidative damage (3, 20, 30, 94). Base excision repair (BER) and nucleotide excision repair (NER) are two pathways that are responsible for the repair of a majority of the DNA lesions induced by ROS (20, 30). If the cell is unable to repair replication-blocking lesions before the advancing DNA replication fork, tolerance of such lesions is possible through the action of lesion-bypass polymerases (30). These DNA polymerases provide a level of lesion tolerance but also induce an increase in replication-based DNA mutations. Repair of ROS-induced base lesions, AP sites, and single-strand DNA breaks is primarily the responsibility of the BER pathway (3, 20, 30). However, ROS gives rise to DNA double-strand breaks requiring repair by homologous recombination (HR) and nonhomologous end joining (NHEJ) (3, 30, 94). In addition, ROS can induce the formation of base modifications that can be repaired by the mismatch DNA repair (MMR) pathway or bulky DNA lesions and modifications from lipid peroxidation (LPO) (32) that are repaired by BER, NER, or direct reversal-repair proteins (3, 30, 94). Further, ROS and LPO induce formation of DNA intra- and interstrand crosslinks that may be repaired by the Fanconi anemia pathway (3, 30, 94).
This review focuses on the repair of oxidative damage–induced lesions in nuclear and mitochondrial DNA mediated by the BER pathway in mammalian cells. In the course of this review, we discuss the multiple BER subpathways that are initiated by one of 11 different DNA glycosylases of three subtypes: (a) bifunctional with an associated β-lyase activity; (b) monofunctional; and (c) bifunctional with an associated β,δ-lyase activity. These three subtypes of DNA glycosylases all initiate BER, dependent on the base lesion, but yield different chemical intermediates and hence different BER complexes to complete repair (3). Additionally, we briefly summarize alternate repair events mediated by BER proteins and the role of BER in the repair of mitochondrial DNA damage induced by ROS. Further, we discuss the relation of BER to the onset of genome-destabilizing events that can lead to oncogenesis.
Oxidative Base Damage Repaired by BER
The chemical makeup of spontaneous and exposure-induced DNA base modifications has been identified in purified DNA as well as nuclear and mitochondrial DNA from multiple organisms, including mammalian cells and tissues (9, 23, 62). Slight variations in the nomenclature are apparent in the literature with regard to DNA bases modified by ROS. To clarify, we have taken the suggestion recently proposed that the damage under discussion is referred to as oxidatively damaged DNA (15). In addition, where possible, we use nomenclature referring to the 2′-deoxyribonucleoside based on the major biologically relevant structure at pH 7.0, as suggested by the International Union of Pure and Applied Chemistry (IUPAC) (15). All four DNA bases or deoxynucleosides (deoxyadenosine, deoxyguanosine, thymidine, and deoxycytidine) plus the methylated form of deoxycytidine (5-methyl-deoxycytidine; 5dmC) can be oxidatively damaged. We do not intend to detail the structure of every modified base or deoxynucleoside here, but we focus on the major modifications, with an emphasis on those repaired by BER. Recognition and repair of oxidative base lesions by DNA glycosylases, BER, and other DNA-repair pathways are discussed in subsequent sections.
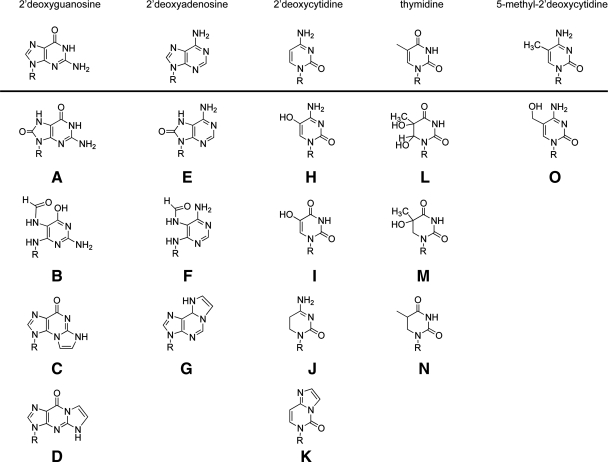
It has been more than 25 years since the identification of the oxidatively modified form of deoxyguanosine in DNA (47). This lesion is identified as 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG). The structures of 2′-deoxyguanosine (Fig. 1, top row) and 8-oxodG (Fig. 1A), as well as the other major oxidative lesion of guanine, 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapydG) (Fig. 1B), are shown. The 8-oxodG lesion, the most extensively studied DNA lesion resulting from oxidative damage, does not cause a significant block to DNA replication in mammalian cells, with the exception of DNA polymerase ι (97). In general, the 8-oxodG DNA lesion is minimally cytotoxic, but incorporation or formation of 8-oxodG is mutagenic. The increase in mutations, mostly G⇒T substitutions, arises from efficient DNA synthesis past the lesion and insertion of dAMP opposite the 8-oxodG DNA lesion. This is commonly referred to as the GO pathway and is discussed later. Replicative and repair DNA polymerases efficiently insert dAMP and dCMP opposite 8-oxodG, including DNA polymerase δ, κ, β, λ, and γ (36, 48, 54, 98). The 8-oxodG DNA lesion is more susceptible to oxidation and can be further oxidized to yield several mutagenic base lesions, including guanidinohydantoin and spiroiminodihydantoin (35, 69).
FIG. 1.
Oxidative damage of 2′-deoxynucleotides. Structures of 2′-deoxyguanosine and the oxidatively modified DNA lesions 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG) (A); 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapydG) (B); the LPO products N2,3-etheno-2′-deoxyguanosine (C); and 1,N2-etheno-2′-deoxyguanosine (D) (15, 19, 74). Structures of 2′-deoxyadenosine and the oxidatively modified DNA lesions 8-oxo-7,8-dihydro-2′-deoxyadenosine (8-oxodA) (E); 4,6-diamino-5-formamidopyrimidine (FapydA) (F); and the LPO product 1,N6-etheno 2′-deoxyadenosine (G) (15, 19, 74). Structures of 2′-deoxycytidine and the oxidatively modified DNA lesions 5-hydroxy-2′-deoxycytidine (OH5dC) (H); 5-hydroxy-2′-deoxyuridine (OH5dU) (I); 5,6-dihydro-2′-deoxyuridine (dHU) (J); and the LPO product 3,N4-ethenocytidine (K) (15, 19, 74). Structures of thymidine and the oxidatively modified DNA lesions thymine glycol (Tg) (L); 5-hydroxy-5,6-dihydrothymine (Th5) (M); and 5,6-dihydrothymine (dHT) (N) (15, 19). Structures of 5-methyl-2′-deoxycytidine and the oxidatively modified DNA lesion 5-(hydroxymethyl)-2′-deoxycytidine (hmdC) (O) (15). Where indicated, the R-group is 2′-deoxyribose.
Oxidative damage to 2′-deoxyadenosine (Fig. 1, top row) yields two major products: 8-oxo-7,8-dihydro-2′-deoxyadenosine (8-oxodA) and 4,6-diamino-5-formamidopyrimidine (FapydA) (Fig. 1E and F) (9). The FapydA lesion is present in both normal and cancerous tissues (71) and is the most abundant of the adenine lesions induced by γ-radiation (14). Both lesions are weakly mutagenic (45, 85), and tandem 8-oxodA lesions can be induced by hydroxyl radicals, precluding repair by BER.
A major oxidative product of 2′-deoxycytidine (Fig. 1, top row) is 5-hydroxy-2′-deoxycytidine (OH5dC), found in DNA both spontaneously and after exposure to H2O2 and related ROS-inducing agents (9). OH5dC can also arise by dehydration of 2′-deoxycytidine glycol (not shown) (9). In addition, OH5dC can deaminate to yield 5-hydroxy-2′-deoxyuridine (OH5dU) (9). Interestingly, OH5dC is efficiently incorporated into DNA but is not accurately replicated, resulting in C⇒T transition mutations (27). Structures for these oxidatively damaged nucleosides are shown in Fig. 1H–J.
One of the most highly studied oxidation products of thymine (Fig. 1, top row) is thymine glycol (Tg) (Fig. 1L). The Tg lesion is formed by hydroxyl radical attack on the double bond of thymine at C5 or C6 or by hydrogen abstraction from the methyl group (44). Tg was originally identified in purified DNA after oxidation with ionizing radiation (29). The Tg lesion is a block to human replicative DNA polymerases, but the lesion is tolerated because several translesion DNA polymerases readily bypass the lesion, including DNA polymerase η, κ, ν, β, and λ (6, 28, 50, 84), with a minimal increase in mutations. Additional oxidized thymine analogues include 5,6-dihydro-thymine (dHT) and 5-hydroxy-5,6-dihydro-thymine (Th5) (Fig. 1M and N).
A recently identified oxidatively modified DNA base lesion in mouse cells and human tissue is 5-hydroxymethyl-cytosine (hmdC), an oxidatively damaged form of 5dmC (49). Originally observed as a substitute for cytosine in T2, T4, and T6 bacteriophage genomes (30), hmdC is found in Purkinje neurons in the brain (49) and in mouse embryonic stem cells (83), derived from 5dmC after hydroxylation by the MLL partner protein TET1 (83). Interestingly, it was earlier predicted that human cells would accumulate hmdC at a rate of 20 per cell per day (87). The structure of 5dmC and the modified lesion hmdC are shown in Figure 1 (top row and O). Although it has been suggested that hmdC may be removed by a mammalian DNA glycosylase (12), the identity of this glycosylase is unknown. However, deamination of hmdC yields 5-hydroxymethyl-uracil (hmdU), a substrate for SMUG1, TDG, and MBD4 (64), suggesting that the hmdC lesion may be cleared from genomic DNA by deamination followed by BER.
In addition to lesions formed by direct oxidative damage to DNA from ROS, chronic induction of ROS promulgates a host of cellular alterations, including significantly elevated levels of LPO (5). LPO is a free radical chain reaction of polyunsaturated fatty acids that results in an accumulation of lipid hydroperoxides. These initial by-products of LPO decompose to generate a variety of breakdown products. Some of the most abundant breakdown products of lipid peroxidation include malondialdehyde, trans-4-hydroxy-2-nonenal (HNE), and trans,trans-2,4-decadienal (DDE) (61). Each of these lipid peroxidation products reacts directly with DNA, inducing mutagenic and genotoxic lesions. Reaction between the LPO products HNE or DDE and DNA yield predominantly the exocyclic etheno-base DNA adducts 1,N6-ethenoadenine (Fig. 1G); 3,N4-ethenocytosine (Fig. 1K); N2,3-ethenoguanine (Fig. 1C); and 1,N2-ethenoguanine (Fig. 1D). All four lesions are thought to be substrates for human DNA glycosylases (26). The guanine adduct 1,N2-ethenoguanine is repaired by BER, and, if not, is highly mutagenic (25, 53, 78). The adenine lesion 1,N6-ethenoadenine is formed in DNA after exposure to the vinyl chloride metabolite chloroacetaldehyde and is the preferred substrate for the mammalian methylpurine DNA glycosylase (MPG or AAG) (25, 26, 77, 79). The cytosine lesion 3,N4-ethenocytosine is also a substrate for BER (25). However, the glycosylase MPG will bind to the 3,N4-ethenocytosine lesion in DNA but not excise the lesion, effectively “hijacking” the repair protein (33).
The BER pathway appears to have evolved for repair of oxidatively damaged DNA. Of the 20 or more proteins defined as part of the BER machinery (3), almost all have a role in the repair of one or more lesions formed from oxidation of DNA. In the ensuing sections, we discuss the role of BER in the repair of these lesions and the specificity of each BER protein in the repair of oxidatively damaged DNA.
Multiple Base Excision Repair Subpathways That Depend on the Initiating Lesion and the Mechanism of Base Removal
Previously, we proposed a BER-pathway model that accommodates the multiple BER subpathways and variable BER protein complexes that are engaged for the repair of a wide range of BER-initiating lesions (3). As with most DNA-repair processes, it has been suggested by us and many others that BER functions through a series of protein complexes that assemble at the site of the DNA lesion. These protein complexes vary depending on the posttranslational modifications (PTMs) of the proteins involved (4), but most importantly, vary as a function of the chemical makeup of the initiating lesion as well as the chemistry of the repair intermediates. In this review, we describe and summarize the BER proteins and the subpathways of BER for the repair of oxidatively damaged DNA. Where appropriate, we discuss the role of PTMs in regulating these repair processes; however, a more complete update of BER protein PTMs (3) is forthcoming.
As we have discussed throughout this review, BER plays an essential role in removing DNA lesions that arise from oxidative stress that would otherwise result in either a block to the progression of the replicative DNA polymerases (and cell death) and/or an increase in the formation of DNA mutations as a prelude to oncogenesis (for mutations in the nuclear genome) or mitochondrial dysfunction (for mutations in the mitochondrial genome). Consistent with our overall BER model and regardless of the initiating lesion that is ultimately repaired (base lesions, abasic sites, DNA single-strand breaks), there are three functional steps that compose the repair of oxidatively damaged DNA (3): lesion recognition/strand scission, gap tailoring, and DNA synthesis/ligation. In line with this model (3), many similarities are found among the different complexes or assemblages of BER proteins brought to bear for removal of the many and varied DNA lesions that arise from oxidative damage (see Fig. 1). However, as expected, the protein complexes vary, depending on the initiating lesion, as well as the chemistry of the repair intermediates.
For a closer look at the precise mechanisms of base lesion removal by the human (and mammalian) DNA glycosylases and repair by the proteins of the BER pathway, we present three variations of this model. Each encompasses the three functional steps essential for BER: (a) lesion recognition/strand scission; (b) gap tailoring; and (c) DNA synthesis/ligation; yet each has both unique and common transient protein–protein interactions and protein complexes that facilitate repair. Further, we define the uniqueness of each subpathway by the chemical makeup of the repair intermediates that arise after glycosylase-mediated base lesion excision.
Repair initiated by bifunctional DNA glycosylases with associated β elimination
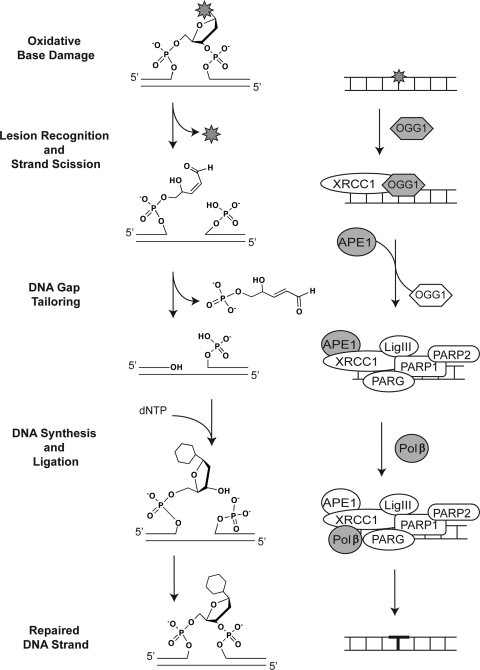
The most common or predominant BER mechanism for removal of oxidatively damaged DNA is initiated through bifunctional DNA glycosylases that excise the modified base and subsequently hydrolyze the DNA backbone through a β-elimination step, as detailed in Fig. 2. In this example, the lesion-recognition and strand-scission step is conducted by OGG1, one of the most highly studied DNA glycosylases for the repair of oxidatively damaged DNA (20). As shown (Fig. 2), the DNA backbone is hydrolyzed, 3′ to the incised base, leaving a 3′ unsaturated aldehyde (after β-elimination) and a 5′ phosphate at the termini of the repair gap. In addition to OGG1, both NTHL1 and NEIL3 are bifunctional DNA glycosylases with associated β-elimination activity (Table 1). Initial studies suggest that the expression of NEIL3 is limited to the nucleus (Table 1). Conversely, both OGG1 and NTHL1 are expressed in the nucleus and the mitochondria, supporting a role for these glycosylases in the repair of oxidatively damaged DNA in both subcellular compartments. As we discussed previously, many BER proteins are modified by PTMs, in some cases altering function or protein complex formation (3). OGG1 has two acetylation sites and four reported phosphorylation sites, one of which (S326) is mutated in the human population, giving rise to a Ser326Cys polymorphism (3). Although this mutant has a slight defect in activity, it has yet to be definitively determined if the Ser326Cys polymorphism is a risk factor for cancer, as numerous studies have been evaluated with differing results. Likewise, both NTHL1 and NEIL3 are modified by either phosphorylation or acetylation or both. Detailed information on human OGG1, NTHL1, and NEIL3 and additional orthologues can also be found on a new web-based DNA Repair database (24). In addition to the classic oxidative lesion 8-oxodG, these three glycosylases initiate BER by removal of a long list of lesions induced by oxidative damage, including the Fapy lesions (FapydG and FapydA) as well as those that arise from further oxidation of 8-oxodG. We have summarized a list of base lesions recognized and removed by the bi-functional (β-elimination) DNA glycosylases in Table 1. Only those lesions reported as glycosylase substrates, as determined by biochemical analysis of the human (or mouse) enzyme, where possible, are listed. A more complete list of substrates for all species may be found elsewhere (9).
FIG. 2.
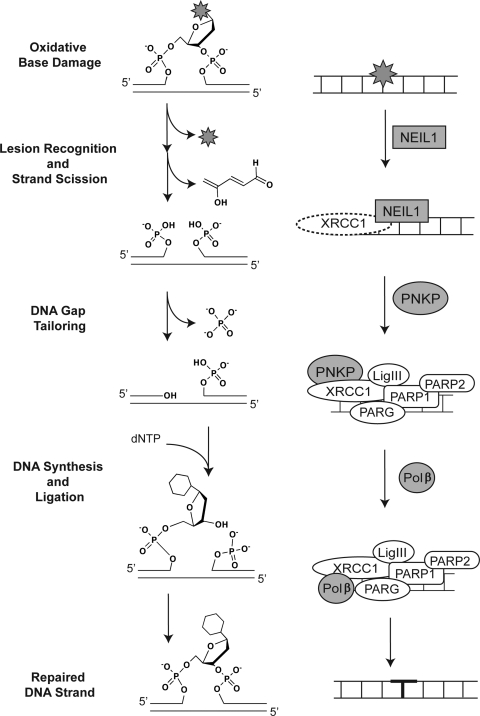
Bifunctional DNA glycosylase (via β-elimination)–initiated BER pathway. Repair of oxidative base damage (e.g., 8-oxodG) initiated by the bifunctional DNA glycosylase OGG1. The chemistry of the lesion and the repair intermediates throughout the repair process are shown (center), highlighting the three essential steps for BER: lesion recognition/strand scission, gap tailoring, and DNA synthesis/ligation (left). The structures on the right depict the protein complexes required for completion of each step in BER initiated by OGG1.
Table 1.
Summary of Human Bifunctional DNA Glycosylases (with Associated β-Elimination) for Repair of Oxidatively Damaged DNA
| Gene symbol | Gene name | Gene ID | Uniprot accession number | Organelle expressed | Known substratea |
|---|---|---|---|---|---|
| OGG1 | 8-oxoguanine DNA glycosylase | 4968 | O15527 | Nucleus and mitochondria | 8-oxoG:C/T/G; me-FapyG:C; FapyG:C (4) 8-oxoA:C (43) urea (9) |
| NTHL1 (NTH1) | nth endonuclease III-like 1 (E. coli) | 4913 | P78549 | Nucleus and mitochondria | T or C-glycol; FapyA (4) 5,6-dihydro-U:G/A (9) 5-formyl-U (9) 5,6-dihydroxy-C (9) 5,6-dihydro-T (9) urea (9) 5-OH-U:G (9) 5-OH-C:G>A (9) 5-hydroxy-5,6,-dihydro-T (9) 8-oxoG:G (65) |
| NEIL3 | Nei endonuclease VIII-like 3 (E. coli) | 55247 | Q8TAT5 | Nucleus | Spiroiminodihydantoin (Sp):C (55) Guanidinohydantoin (Gh):C (55) FapyA (55) FapyG (55) 5-OH-U (55) 5-OH-C (55) Tg (55) |
Target base on left in mismatches.
Once the base is removed and the DNA backbone is hydrolyzed, the resulting gap is tailored by the 3′ phosphodiesterase activity of APE1 (3). It is suggested that OGG1 remains bound to the DNA upon base removal and strand scission, and the OGG1/DNA complex is recognized and bound by APE1, inducing OGG1 turnover and release and promoting APE1-induced gap tailoring (3). A similar APE1-mediated induction of enzyme turnover was observed for NTHL1 (60). OGG1 also appears to recruit XRCC1 to the lesion (Fig. 2) (3). Alternatively, it would be expected that the newly formed single-base gap is recognized by PARP1 (along with PARP2 and PARG), together with the XRCC1/LigIII heterodimer, promoting the recruitment of Polβ for the completion of repair (3). It has also been suggested that aprataxin may modulate this latter step in the pathway (39) by participating in the gap-tailoring step. However, it is likely that the protein complexes formed in the nucleus after strand cleavage will differ significantly as compared with protein complexes formed in the mitochondria, because PARP1 and PARP2 are predominantly nuclear enzymes.
With regard to OGG1-initiated BER, unique mechanisms of cell death in response to oxidative damage were observed when comparing nuclear and mitochondrial BER (70), supporting a role for PARP1 in nuclear BER initiated by OGG1, with no evidence for PARP1 activation in mitochondrial BER initiated by the mitochondrial form of OGG1. Similarly, it has been shown that XRCC1 will bind to the nicked DNA after 8-oxodG base removal by OGG1 and gap tailoring by APE1 (68). Once gap tailoring is complete, the DNA is then available for DNA synthesis mediated by Polβ and ligation by the XRCC1/LigIII heterodimer (3). These and additional BER proteins involved in repair of oxidatively damaged DNA are described in Table 4 (24).
Table 4.
Other Human BER Proteins Involved in the Repair of Oxidatively Damaged DNA
| Gene symbol | Gene name | Gene ID | Uniprot accession number | Organelle expressed |
|---|---|---|---|---|
| APEX1 (APE1) | APEX nuclease (Multifunctional DNA repair enzyme) 1 | 328 | P27695 | Endoplasmic reticulum nucleus, centrosome, and mitochondria |
| APEX2 (APE2) | APEX nuclease (apurinic/apyrimidinic endonuclease) 2 | 27301 | Q9UBZ4 | Nucleus and mitochondria |
| APTX | Aprataxin | 54840 | Q7Z2E3 | Chromatin and nucleus |
| DNA2 | DNA replication helicase 2 homologue (yeast) | 1763 | P51530 | Mitochondria |
| FEN1 | Flap structure-specific endonuclease 1 | 2237 | P39748 | Nucleus and mitochondria |
| LIG1 | Ligase I, DNA, ATP-dependent | 3978 | P18858 | Nucleus |
| LIG3 | Ligase III, DNA, ATP-dependent | 3980 | P49916 | Nucleus and mitochondria |
| NUDT1 | NUDT1 nudix (nucleoside diphosphate-linked moiety X)-type motif 1 | 4521 | P36639 | Cytoplasm |
| PARP1 | Poly (ADP-ribose) polymerase 1 | 142 | P09874 | Nucleus |
| PARP2 | Poly (ADP-ribose) polymerase 2 | 10038 | Q9UGN5 | Nucleus |
| PNKP | Polynucleotide kinase 3′-phosphatase | 11284 | Q96T60 | Nucleus |
| POLB | Polymerase (DNA directed), beta | 5423 | P06746 | Nucleus |
| POLG | Polymerase (DNA directed), gamma | 5428 | P51530 | Mitochondria |
| TDP1 | Tyrosyl-DNA phosphodiesterase 1 | 55775 | Q9NUW8 | Cytoplasm |
| XRCC1 | X-ray repair complementing defective repair in Chinese hamster cells 1 | 7515 | P18887 | Nucleus |
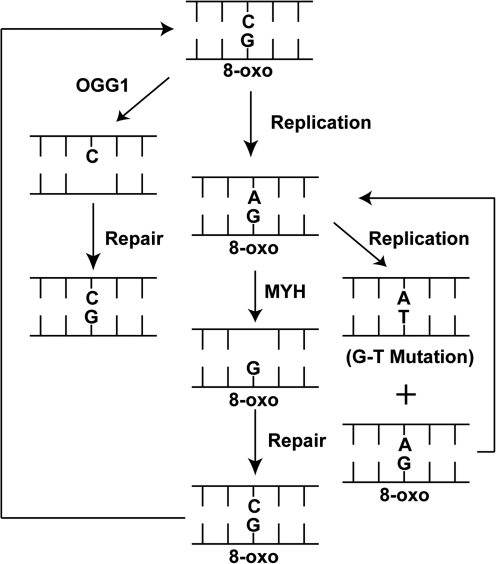
Many oxidative lesions such as those repaired by OGG1 are either replicated incorrectly by the replicative DNA polymerases (δ and ɛ) or are tolerated by low-fidelity DNA polymerases that conduct translesion DNA synthesis, as discussed earlier. Hence, failure to repair the DNA base lesion from oxidative damage before replication leads to a tremendous increase in DNA mutations. For example, for the replicative and translesion DNA polymerases, the 8-oxodG•A base pair is preferred, giving rise to an increase in G⇒T substitution mutations that arise from DNA synthesis past the lesion and insertion of dAMP opposite the 8-oxodG lesion. Fortunately, a second, compensatory BER pathway is initiated by the MYH glycosylase to prevent the onset of G⇒T substitution mutations that result from a failure to repair the 8-oxodG lesion before DNA replication. The classic example of the GO pathway (Fig. 3) readily diagrams and outlines the increase in mutations that could arise when OGG1- and/or MYH-mediated BER is absent. As shown, OGG1-mediated BER is designed to remove the offending lesion (8-oxodG), yielding the correct G•C base pair. MTH1 (NUTD1), a Nudix family enzyme, can also hydrolyze the oxidized 8-oxodGTP before incorporation into the DNA (not shown) (67). However, once 8-oxodG formation is induced or 8-oxodG is incorporated into the genome, replication continues past the lesion to form the mutagenic 8-oxodG•A base pair. This mismatch/lesion must be corrected to avoid the accumulation of G⇒T mutations resulting from a second round of replication (Fig. 3, right side). The activity of MYH (also referred to as MUTYH) is specific for adenine opposite the 8-oxodG lesion. This unique enzyme that mediates the removal of a “normal” base opposite a lesion functions as a monofunctional DNA glycosylase and is discussed in the next section.
FIG. 3.
The GO pathway. Diagrammatic representation of the repair and mutagenic consequences of 8-oxodG. Repair of 8-oxodG is mediated by OGG1-initiated BER (left). If not repaired, the replicative polymerases prefer to insert A opposite 8-oxodG. The incorrect A residue is repaired by MYH-initiated BER (center). However, if not repaired before a second round of replication (right), the 8-oxodG•A base pair is converted to another 8-oxodG•A base pair and an A•T base pair, inducing a G⇒T substitution mutation. Both the 8-oxodG•C base pair and 8-oxodG•A base pair can be repaired again, if needed (arrows).
Repair initiated by monofunctional DNA glycosylases
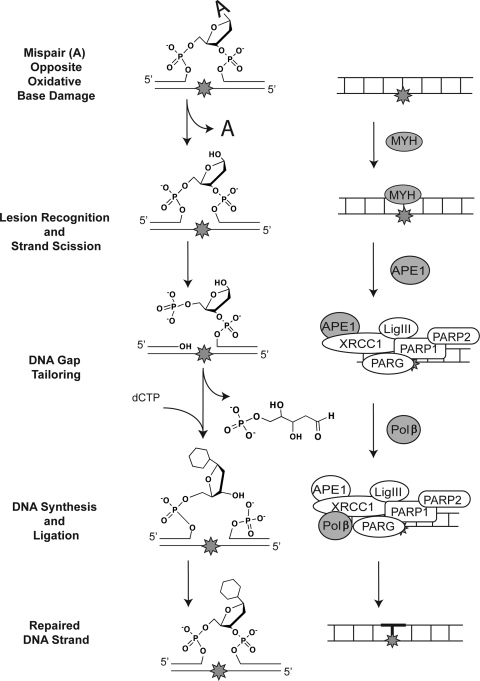
Monofunctional DNA glycosylases initiate a classic “short-patch” BER mechanism (Fig. 4) involving hydrolysis of the N-glycosidic bond and incision at the resulting abasic site (lesion recognition/strand scission) (3). The repair gap, containing a 3′-OH and a 5′-deoxyribose-phosphate (5′dRP) moiety at the margins, is tailored by the 5′-dRP lyase activity of Polβ (gap tailoring), and then Polβ fills the single nucleotide gap, preparing the strand for ligation by either DNA ligase I (LigI) or a complex of DNA ligase III (LigIII) and XRCC1 (DNA synthesis/ligation) (3). As described previously and in greater detail later, the transient complexes formed after lesion recognition/strand scission are mediated by XRCC1 together with PARP1 (plus PARP2 and PARG), acting as scaffolds for protein complex formation (3).
FIG. 4.
Monofunctional DNA glycosylase–initiated BER pathway. Repair of adenosine when opposite the oxidative base lesion 8-oxodG, as initiated by the monofunctional DNA glycosylase MYH. The chemistry of the lesion and the repair intermediates throughout the repair process are shown (center), highlighting the three essential steps for BER: lesion recognition/strand scission, gap tailoring, and DNA synthesis/ligation (left). The structures on the right depict the protein complexes required for completion of each step in BER initiated by MYH.
Of the 11 human DNA glycosylases, six are monofunctional DNA glycosylases, and each participates in the repair of oxidatively damaged DNA (Table 2). All are expressed in the nucleus, and of these, two (UNG and MYH) are also expressed in the mitochondria (Table 2). All of the monofunctional DNA glycosylases, with the exception of SMUG1, are posttranslationally modified by phosphorylation (UNG, TDG, MBD4, MPG, and MYH), acetylation (TDG and UNG), and/or SUMOylation (TDG) (3, 24, 74). In some cases, most notably TDG, PTMs drastically regulate enzymatic function (3). Table 2 indicates the oxidatively damaged DNA lesions repaired by the human monofunctional DNA glycosylases. A significant overlap in substrate specificity exists for all four “uracil-specific” DNA glycosylases (UNG, SMUG1, TDG, and MBD4), with the exception of SMUG1. Originally identified as a 5-hydroxymethyluracil DNA glycosylase (10), SMUG1 hydrolyzes lesions from both single- and double-stranded DNA. It is interesting to speculate whether SMUG1 can remove hmdC lesions identified in human Purkinje neurons (49) and whether SMUG1 expression affects the accumulation of hmdC lesions, because these readily deaminate to the SMUG1 substrate hmdU. With regard to the etheno-base adducts (see Fig. 1), MPG has been shown to recognize and remove 1,N6-ethenoadenine, N2,3-ethenoguanine, and 1,N2-ethenoguanine lesions, whereas the 3,N4-ethenocytosine and 1,N2-ethenoguanine lesions are repaired (excised) by TDG and MBD4 (32). MPG binds to but does not excise the 3,N4-ethenocytosine lesion (33). MYH (MUTYH) is unusual in that it recognizes the 8-oxodG•A base pair but removes the normal DNA base adenosine (Table 2), among other lesions. Interestingly, whereas all the DNA glycosylases discussed so far remove cytotoxic or genome-destabilizing DNA base lesions, MYH has the distinction of being the only glycosylase that removes a normal DNA base, yet is also the only one definitively linked to human disease (cancer) when defective (13), emphasizing the critical role of MYH in removing the premutagenic base, as shown in Fig. 3.
Table 2.
Summary of Human Monofunctional DNA Glycosylases for Repair of Oxidatively Damaged DNA
| Gene symbol | Gene name | Gene ID | Uniprot accession number | Organelle expressed | Known substratea |
|---|---|---|---|---|---|
| UNG | Uracil DNA glycosylase | 7374 | P13051 | Nucleus (UNG2) and mitochondria (UNG1) | ssU; U:G; U:A; 5-fluorouracil (4) 5,6-Dihydroxy-U:G (9) and 5-OH-U:G (9, 22) Isodialuric acid and Alloxan (22) |
| SMUG1 | Single-strand–selective monofunctional uracil-DNA glycosylase 1 | 23583 | Q53HV7 | Nucleus | ssU; U:G; U:A (4) 5-Fluorouracil:G and 5-chlorouracil:G (18) 5-Carboxyuracil:G (18) 5-(Hydroxymethyl)-U (9, 91) 5-Formyl-U and 5-hydroxyuracil (9) |
| TDG | Thymine DNA glycosylase | 6996 | Q13569 | Nucleus | U:G; T:G and ethenoC:G (4) 5-Fluorouracil and 5-fluorouracil (ss) (38) 5-Hydroxymethyluracil (38) hypoxanthine:G, 5-bromouracil and ɛC:A (38) Tg:G (96) and 5-formyl-U (9) |
| MBD4 | Methyl-CpG binding domain protein 4 | 8930 | O95243 | Nucleus | U or T in U/TpG; 5-meCpG (4) 5-Formyluracil (9) 5-(Hydroxymethyl)-U (9) Tg:G (96) |
| MPG (AAG) | N-methyl DNA glycosylase | 4350 | P29372 | Cytoplasm and nucleus | 3-meA; 7-meA; 3-meG; 7-meG; hypoxanthine and ethenoA; ethenoG (4) 1,N2-ɛG:C; U:G and ethanoadenine and 1-methylguanine (53) Etheno-A(ss); hypoxanthine(ss) and ssU (53) 8-OxoG:C (Mouse) (7) Cyanuric acid:CT>GA (9) |
| MUTYH (MYH) | mutY homologue (E. coli) | 4595 | Q9UIF7 | Nucleus and mitochondria | A:G; A:8-oxoG and C:A; 2-OH-A (4) 8-OxoA:G (72) |
Target base on left in mismatches.
Although the base lesions repaired by the monofunctional DNA glycosylases (Table 2) are all chemically different, the chemical makeup of the repair intermediates after repair initiation by these glycosylases is the same (Fig. 4). To demonstrate the functional, chemical, and protein interaction steps for BER mediated by a monofunctional DNA glycosylase, we show a detailed, step-by-step BER mechanism when MYH is the repair-initiating enzyme. The lesion recognition/strand scission step is initiated by MYH, recognizing the 8-oxodG•A base pair, and lesion removal is promoted by APE1 binding to the MYH/DNA complex. It has not been established whether MYH also recruits XRCC1 to the lesion site, as it does for MPG, NTH1, OGG1, and NEIL2 (1, 11). Although not yet formally tested, the requirement for Polβ in the gap tailoring and DNA synthesis step after MYH initiation of BER is expected (80). It has also been suggested that if repair occurs at or near broken DNA ends, then Ku can facilitate the required 5′-dRP lyase activity (75).
DNA Polβ appears to be unique in its ability (and structure) for correct insertion of C opposite the 8-oxodG lesion in vitro (48). Conversely, DNA polymerases λ and η will insert the correct C opposite the 8-oxodG lesion when stimulated by the auxiliary proteins RPA and PCNA (58), with preference over Polβ (57). However, a separate study suggested that only A was inserted opposite the 8-oxodG lesion, establishing a futile cycle of repair and synthesis until the 8-oxodG lesion is removed by OGG1-initiated BER or through a long-patch mechanism (40). Whereas Polβ-mediated repair involves insertion of a single nucleotide (Fig. 4), DNA polymerase λ–mediated repair initiated by MYH appears to prefer a long-patch mechanism in which MYH binds to the PCNA-bound 8-oxodG•A base pair, removes the A residue, and recruits APE1 to hydrolyze the DNA backbone, essentially as shown in Fig. 4 (with the exception of the added PCNA). However, in this proposed long-patch model, RPA and PCNA stay bound to the repair gap, recruiting DNA polymerase λ to fill the gap (past the lesion) and displace several bases (strand displacement), requiring FEN1 (and DNA ligase) for completion of repair (not shown). Although not yet determined, given the similarity in repair intermediate chemistry when comparing MYH-initiated BER and MPG-initiated BER, it is likely that the transient complex formed at the site of the repair intermediate for MYH initiated BER is similar to that of other BER processes involving monofunctional DNA glycosylases, such as MPG, and is likely to include PARP1, PARP2, and PARG (86).
Repair initiated by bifunctional DNA glycosylases with associated ß,δ-elimination
Finally, we come to the recently characterized BER subpathway initiated by NEIL1 and NEIL2, DNA glycosylases with an associated β,δ-elimination activity that initiates an APE-independent repair mechanism (19, 92) (Table 3). The currently known substrates for NEIL1 and NEIL2 also are detailed in Table 3. Whereas NEIL1 is reported to be phosphorylated (74), NEIL2 is acetylated (8, 74), the latter leading to inhibition of NEIL2 (8). Binding of NEIL2 to the lesion recruits XRCC1 to the DNA (11). It has not yet been established if the same process of XRCC1 recruitment is mediated by NEIL1. In Fig. 5, we draw XRCC1 with a dotted line to reflect that conjecture. The product of NEIL1- (and NEIL2-) mediated base removal is a 5′- and 3′-phosphate on the ends of the DNA in the single-base gap, resulting from hydrolysis of the glycosidic bond to release the base, cleavage 3′ to the abasic site through β-elimination and then cleavage 5′ to the abasic site through δ-elimination, releasing the trans-4-hydroxy-2,4-pentadienal (19, 92). The phosphatase PNKP is subsequently recruited to the site to remove the 3′ phosphate in the gap, completing the gap-tailoring step and readying the substrate for DNA synthesis by Polβ and ligation by XRCC1/LigIII (19, 92). We only speculate that PARP1, PARP2, and PARG facilitate complex recruitment through PAR synthesis once BER is initiated by either NEIL1 or NEIL2.
Table 3.
Summary of Human Bifunctional DNA Glycosylases (with Associated β,δ-Elimination) for Repair of Oxidatively Damaged DNA
| Gene symbol | Gene name | Gene ID | Uniprot accession number | Organelle expressed | Known substratea |
|---|---|---|---|---|---|
| NEIL1 | Nei endonuclease VIII-like 1 (E. coli) | 79661 | Q96FI4 | Nucleus, cytoplasm and mitochondria (mouse) | TgG; 5-OH-C; 5-OH-U:AT>G (4) Guanidinohydantoin (35) guanidinohydantoin (ss) (35) Iminoallantoin (35) Iminoallantoin (ss) (35) Spiroiminodihydantoin (35) Spiroiminodihydantoin (ss) (35) 5,6-Dihydro-T (9) 5,6-Dihydro-U:G/C/A>T (9) FapyG:C (9) 8-Oxo-G:C/G>T>A (9) FapyA:T (9) (5′R)-8,5′-Cyclo-2′-deoxyadenosine (42) (5′S)-8,5′-Cyclo-2′-deoxyadenosine (42) 8-Oxo-A:C (31) |
| NEIL2 | Nei-like 2 (E. coli) | 252969 | Q969S2 | Nucleus and cytoplasm | 5-OH-U:G>T>A; 5-OH-C (4) 5,6-Dihydro-U:G/A (9) 8-Oxo-G:C/A (9) 5,6-Dihydrothymine (9) Guanidinohydantoin (35) Guanidinohydantoin (ss) (35) Iminoallantoin (35) Iminoallantoin (ss) (35) Spiroiminodihydantoin (ss) (35) |
Target base on left in mismatches.
FIG. 5.
Bifunctional DNA glycosylase (through β,δ-elimination)–initiated BER pathway. Repair of oxidative base damage (e.g., (5′R)-8,5′-cyclo-2′-deoxyadenosine) initiated by the bifunctional DNA glycosylase NEIL1. The chemistry of the lesion and the repair intermediates throughout the repair process are shown (center), highlighting the three essential steps for BER: lesion recognition/strand scission, gap tailoring, and DNA synthesis/ligation (left). The structures on the right depict the protein complexes required for completion of each step in BER initiated by NEIL1.
Variable Modes of BER Dependent on the Chemistry of the Lesion or Intermediate
As we mention throughout this review, the chemical structures of the repair intermediates will dictate the proteins or protein complexes that are recruited for repair, triggering slight modifications of the classic BER pathway. In some cases, the base lesions, although a substrate for a monofunctional DNA glycosylase, may also be a direct substrate for APE1. For example, APE1 will hydrolyze oxidized cytosines directly, in the absence of a DNA glycosylase (3). The rest of the pathway would be similar to that as shown in Fig. 4.
Another modification or variable to the BER pathway is observed when the abasic site is oxidized. Oxidation of abasic sites from the chemotherapeutic agent bleomycin or γ-radiation yields several chemically different modified abasic lesions from damage-induced hydrogen abstraction at the C-1′, C-2′, C-4′, and C-5′ positions on the 2′-deoxyribose. The capacity for these lesions to be repaired by the BER machinery appears quite variable. Bleomycin-induced formation of the oxidized abasic lesion C-4-keto-C-1-aldehyde in DNA (oxidation at the C-4′ position) is readily repaired by APE1 and Polβ through a subpathway similar to that shown in Fig. 4. However, 2-deoxyribonolactone, formed by free radical modification of the C-1′ carbon of 2′-deoxyribose is not effectively repaired in the same manner. The 2-deoxyribonolactone lesions are hydrolyzed by APE1 in a manner similar to normal abasic sites. However, Polβ, which normally removes the 5′dRP moiety after APE1-mediated hydrolysis, cannot remove the hydrolyzed 2-deoxyribonolactone. Instead, the result is a covalent amide linkage between Polβ and the DNA, preventing further repair (21). Similar repair difficulties have been reported for other oxidized abasic sites. Neither the C-5′ oxidized abasic site [dioxybutane; 5′-(2-phosphoryl-1,4-dioxobutane] nor the C-2′ oxidized abasic site is a substrate for the 5′-dRP lyase activity of Polβ (34, 93).
Of course, any block to gap tailoring, for example, that would preclude or inhibit Polβ-mediated 5′-dRP lyase activity (or other intermediate steps in the pathway) would necessitate strand-displacement DNA synthesis and completion of BER via the long-patch pathway (3). For example, in cases in which the 5′dRP moiety is refractory to Polβ 5′-dRP lyase activity, DNA polymerase ɛ, δ, or β, coupled with proliferating cell nuclear antigen (PCNA) and replication factor C (RFC), synthesizes the DNA to fill the gap, resulting in a displaced DNA flap of two to 13 bases in length (3). This long-patch BER mechanism prevents the formation of the Polβ-DNA crosslink observed during repair of the 2-deoxyribonolactone (81). DNA synthesis and strand displacement by Polβ is stimulated by a combination of the structure-specific endonuclease FEN1 and PARP1. FEN1 then catalyzes the removal of this flap, leaving a nick that has been transferred two to 13 nucleotides downstream of the original damage site. DNA ligase I or a complex of DNA ligase III and XRCC1 conducts the final, nick-sealing, step in the pathway (3).
Mitochondrial Base Excision Repair
Mitochondrial DNA is damaged to a greater extent than nuclear DNA after oxidative stress (95), and if the mitochondrial DNA is damaged and not repaired, the defective mitochondria can generate an increased level of ROS, precipitating a cycle of oxidative-damage generation (59). Since the first report of mitochondrial BER by LeDoux and colleagues in 1992 (52), considerable effort has been put forth to characterize the mechanism of BER in mitochondria (51). Many of the BER proteins that function in nuclear BER are also expressed in mitochondria (Tables 1–4). Glycosylases from each of the three mechanistic groups are found in the mitochondria, including the bifunctional glycosylases (through β-elimination) OGG1 and NTHL1 (Table 1), the monofunctional glycosylases UNG1 and MYH (Table 2), and the bifunctional glycosylase (through β,δ-elimination) NEIL1 (Table 3). A shortened form of APE1 is expressed in the mitochondria, as is APE2 (Table 4). DNA ligase IIIα is expressed in both the nucleus and the mitochondria, but not XRCC1, whereas PARP1 is expressed in the mitochondria only in the presence of mitofilin (76). Although Polβ is not expressed in mitochondria (37), the mitochondrial DNA polymerase γ has the requisite DNA polymerase activity, as well as a 5′-dRP lyase function, implicating DNA polymerase γ in mitochondrial BER (56).
Finally, both FEN1 and DNA2 are expressed in the mitochondria and appear to promote long-patch BER for repair of oxidatively damaged DNA (82, 99). This suggests that all three subpathways (Figs. 2, 4, and 5) plus long-patch BER are used by the BER machinery in mitochondria. However, not all essential nuclear BER proteins are expressed in the mitochondria, and therefore, studies are still under way to work out the mechanistic details for the repair of the many base lesions found in mitochondrial DNA.
BER, Oxidative Damage, and Cancer
ROS function both as intracellular signaling molecules and as DNA-damaging agents (63), thereby complicating our understanding of the involvement of ROS in the onset of human disease. At elevated exposure levels, the DNA-damaging effects of ROS (Fig. 6) have a clear impact on cellular function, genome stability, and human health (16). For example, oxidative stress is recognized to play a significant role in the onset of cardiovascular and pulmonary diseases, such as atherosclerosis (88), and may have a role in the onset of triplet-repeat expansion in diseases such as Huntington's (20, 41). Over the past decade (or more), multiple examples suggested oxidative stress–mediated DNA damage as a possible environmental link to organ dysfunction, such as cataract and the onset of diabetes (20, 41). Further, oxidative-stress exposure either early in life or later in life is associated with age-dependent cognitive decline and an increase in the occurrence of neurodegenerative diseases such as Parkinson's, Alzheimer's, and familial amyotrophic lateral sclerosis (ALS) or Lou Gehrig's disease (41). Similarly, oxidative stress has been associated with aging and age-related disorders as well as diseases associated with inflammation (41).
FIG. 6.
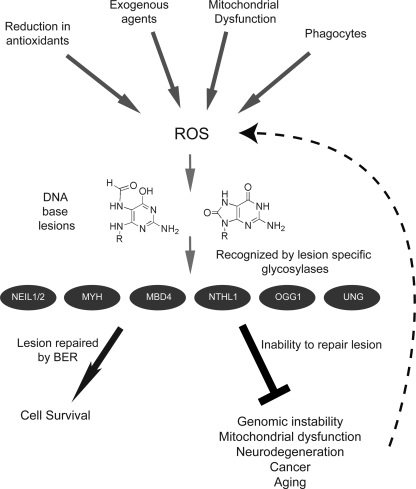
Model depicting the generation and consequences of ROS-mediated base damage. Reactive oxygen species are generated from multiple exogenous and endogenous sources, such as ultraviolet light, radiation, metal ions, cellular respiration, imbalance of cellular antioxidant systems, and phagocytes. ROS-generated DNA base lesions, such as 8-oxoG and FapyG, are subject to repair, initiated by lesion-specific DNA glycosylases (e.g., NEIL1, NEIL2, MYH, MBD4, NTHL1, OGG1, and UNG, as indicated). Recognition and repair of these lesions through the BER pathway results in enhanced cellular survival and genome maintenance. Inability to recognize or repair oxidative lesions leads to genomic instability, mitochondrial dysfunction, neurodegeneration, cancer, and aging. The consequences of unrepaired oxidative damage can result in a feedback loop generating additional ROS, leading to a cycle of increasing ROS and elevated genome instability.
In addition to disease states such as neurodegeneration that likely manifest from DNA damage–induced cell death and organ dysfunction, ROS and the associated oxidative DNA damage can induce gene mutations and genome instability as causative agents in the onset of cancer (41). For example, human exposure to elevated levels of the ROS-inducing agent arsenic is connected with an increased incidence of cancers of the lung, skin, bladder, and liver. It is proposed that oncogenesis from ROS exposure is through DNA damage–induced gene mutations or genome instability or both (41). Many of the highly prevalent oxidative DNA lesions such as the oxidized form of guanine (8-oxodG) are highly mutagenic (20, 30). As described earlier (see Fig. 3), BER is essential to prevent ROS or oxidative stress–induced DNA mutations. Failure to repair these base lesions can predispose to colon cancer, such as the colorectal adenoma phenotoype associated with a defect in the BER gene MYH (13). The increase in cancer incidence due to MYH mutations is certainly an extreme case, because the observed mutations lead to inactivation of MYH (13). However, this strongly suggests that any attenuation of DNA glycosylase activity has the potential to lead to an increase in DNA mutations and therefore an increase in cancer incidence. As described earlier, most DNA glycosylases are modified by PTMs, including phosphorylation, acetylation, and SUMOylation, among others (24, 74). Because the presence or absence of these PTMs has the potential to disrupt protein–protein interactions, to decrease protein stability, or to modify protein function, it will be essential, in future studies, to evaluate the in vivo functional significance of PTMs for DNA glycosylases, as well as other BER proteins as described earlier that are critical for repair of ROS-induced DNA damage.
Metabolic and oncogenic stress associated with cancer results in elevated ROS production (17, 90), driving additional genome damage and genome instability, reinforcing the requirement for BER in the repair of ROS-mediated DNA damage as a caretaker pathway in the defense against cancer (Fig. 6). Further complicating the cellular response to ROS and oxidative damage is the interconnection of DNA damage, DNA repair, and NAD+ metabolism. As we described recently, defective or incomplete BER results in hyperactivation of PARP1, triggering the loss of bioenergetic metabolites such as NAD+ and the onset of necrosis (86). Cell death, resulting from necrosis, has the unfortunate side effect of triggering the release of HMGB1 from the cell (86). Once released from the cell, HMGB1 functions as a RAGE ligand and inflammatory cytokine, enhancing ROS production. Whereas BER is an important process for the removal of mutagenic DNA lesions induced by ROS (Fig. 3), it is also clearly involved in preventing cell death and cellular degeneration (necrosis), in suppressing ROS-induced inflammation (by preventing release of HMGB1), and avoiding or eliminating lesions that can affect mitochondrial biogenesis (via loss of NAD+). As we describe in Fig. 6, the lesions induced by environmental and endogenous sources (ROS) can be repaired by the BER pathway. However, failure to repair the lesions effectively triggers a cycle of ROS generation that affects all functions of the cell, initiating a path to genome instability, cellular degeneration, mitochondrial dysfunction, cancer, and aging. This then promotes further genomic and mitochondrial DNA damage and cellular dysfunction, perpetuating the production of ROS and the requirement for BER-mediated lesion processing.
How the BER pathway facilitates the repair of these lesions to prevent the onset of cellular dysfunction is essential to understanding the connection between ROS, oxidative stress, DNA repair, and human disease. Recent evidence suggests that BER not only may be critical for the repair of DNA damage that results from acute, elevated exposures of ROS but may also be essential for the role of ROS in intracellular signaling (73). The epigenetic modulator enzyme lysine-specific demethylase 1 (LSD1; KDM1A) catalyzes the demethylation of proteins and histones, in particular, histone H3, removing the methyl group from lysine 9 (H3K9me2). LSD1 catalyzes this reaction, by using the cofactor FAD by an amine oxidase–mediated demethylation reaction in which FAD is reduced to FADH2 and then reoxidized to FAD by oxygen with the generation of H2O2. As recently reported, the local DNA damage resulting from LSD1-mediated demethylation triggers recruitment of the BER machinery and the onset of BER processing of local DNA damage induced by the H2O2, initiated by OGG1, at the site of H3K9me2 demethylation. Further, it is proposed that repair of the damage is coupled to transcription initiation (73). Therefore, it is conceivable that the BER machinery not only is critical for preventing ROS induced genome instability, but also, BER may be an essential component of ROS-mediated transcriptional regulation. Hence, seemingly disparate events mediate cellular function and dysfunction through a common thread, the generation of ROS, the onset of oxidative stress, and the induction of DNA damage repaired by BER.
Abbreviations Used
- 5dmC
5-methyl-deoxycytidine
- 5′-dRP
5′-deoxyribose phosphate
- 8-oxodA
8-oxo-7,8-dihydro-2′-deoxyadenosine
- 8-oxodG
8-oxo-7,8-dihydro-2′-deoxyguanosine
- 8-oxo-Gua
8-oxo-7,8-dihydroguanine
- AP
apurinic/apyrimidinic
- BER
base excision repair
- DDE
trans,trans-2,4-decadienal
- dHT
5,6-dihydro-thymine
- FapydA
4,6-diamino-5-formamidopyrimidine
- FapydG
2,6-diamino-4-hydroxy-5-formamidopyrimidine
- hmdC
5-hydroxymethyl-cytosine
- hmdU
5-hydroxymethyl-uracil
- HNE
trans-4-hydroxy-2-nonenal
- HR
homologous recombination
- IUPAC
International Union of Pure and Applied Chemistry
- LPO
lipid peroxidation
- LSD1
lysine-specific demethylase 1
- MMR
mismatch DNA repair
- MPP(+)
1-methyl-4-phenylpyridinium
- NER
nucleotide excision repair
- NHEJ
nonhomologous end joining
- OH5dC
5-hydroxy-2′-deoxycytidine
- OH5dU
5-hydroxy-2′-deoxyuridine
- PCNA
proliferating cell nuclear antigen
- PTMs
posttranslational modifications
- PUVA
psoralen and ultraviolet A
- RFC
replication factor C
- ROS
reactive oxygen species
- Th5
5-hydroxy-5,6-dihydro-thymine
Acknowledgments
This work was supported by grants from the National Institutes of Health (NIH) [CA132385; GM087798; CA148629] to RWS. Further, this publication was made possible by RI-INBRE grant P20RR016457 from the National Center for Research Resources (NCRR, NIH) awarded to KHA. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH. Support also was provided by the University of Pittsburgh Department of Pharmacology & Chemical Biology and a John S. Lazo Cancer Pharmacology Fellowship to EMG. Finally, we apologize to our colleagues for not citing each and every appropriate primary reference. The total number of references allowed in this review is limited.
Author Disclosure Statement
RWS is a scientific consultant for Trevigen, Inc. The remaining authors have no conflicts to disclose.
References
- 1.Akbari M. Solvang-Garten K. Hanssen-Bauer A. Lieske NV. Pettersen HS. Pettersen GK. Wilson DM., 3rd Krokan HE. Otterlei M. Direct interaction between XRCC1 and UNG2 facilitates rapid repair of uracil in DNA by XRCC1 complexes. DNA Repair (Amst) 2010;9:785–795. doi: 10.1016/j.dnarep.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albright CD. Salganik RI. Craciunescu CN. Mar MH. Zeisel SH. Mitochondrial and microsomal derived reactive oxygen species mediate apoptosis induced by transforming growth factor-beta1 in immortalized rat hepatocytes. J Cell Biochem. 2003;89:254–261. doi: 10.1002/jcb.10498. [DOI] [PubMed] [Google Scholar]
- 3.Almeida KH. Sobol RW. A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair. 2007;6:695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Almeida KH. Sobol RW. Increased specificity and efficiency of base excision repair through complex formation. In: Siede W, editor; Doetsch PW, editor; Kow YW, editor. DNA Damage Recognition. New York: Marcel Dekker; 2005. pp. 33–64. [Google Scholar]
- 5.Bartsch H. Nair J. Oxidative stress and lipid peroxidation-derived DNA-lesions in inflammation driven carcinogenesis. Cancer Detect Prevent. 2004;28:385–391. doi: 10.1016/j.cdp.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Belousova EA. Maga G. Fan Y. Kubareva EA. Romanova EA. Lebedeva NA. Oretskaya TS. Lavrik OI. DNA polymerases beta and lambda bypass thymine glycol in gapped DNA structures. Biochemistry. 2010;49:4695–4704. doi: 10.1021/bi901792c. [DOI] [PubMed] [Google Scholar]
- 7.Bessho T. Roy R. Yamamoto K. Kasai H. Nishimura S. Tano K. Mitra S. Repair of 8-hydroxyguanine in DNA by mammalian N-methylpurine-DNA glycosylase. Proc Natl Acad Sci U S A. 1993;90:8901–8904. doi: 10.1073/pnas.90.19.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhakat KK. Hazra TK. Mitra S. Acetylation of the human DNA glycosylase NEIL2 and inhibition of its activity. Nucleic Acids Res. 2004;32:3033–3039. doi: 10.1093/nar/gkh632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bjelland S. Seeberg E. Mutagenicity, toxicity and repair of DNA base damage induced by oxidation. Mutat Res. 2003;531:37–80. doi: 10.1016/j.mrfmmm.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Boorstein RJ. Cummings A., Jr Marenstein DR. Chan MK. Ma Y. Neubert TA. Brown SM. Teebor GW. Definitive identification of mammalian 5-hydroxymethyluracil DNA N-glycosylase activity as SMUG1. J Biol Chem. 2001;276:41991–41997. doi: 10.1074/jbc.M106953200. [DOI] [PubMed] [Google Scholar]
- 11.Campalans A. Marsin S. Nakabeppu Y. O'Connor TR. Boiteux S. Radicella JP. XRCC1 interactions with multiple DNA glycosylases: a model for its recruitment to base excision repair. DNA Repair (Amst) 2005;4:826–835. doi: 10.1016/j.dnarep.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 12.Cannon SV. Cummings A. Teebor GW. 5-Hydroxymethylcytosine DNA glycosylase activity in mammalian tissue. Biochem Biophys Res Commun. 1988;51:1173–1179. doi: 10.1016/s0006-291x(88)80489-3. [DOI] [PubMed] [Google Scholar]
- 13.Cheadle JP. Sampson JR. Exposing the MYtH about base excision repair and human inherited disease. Hum Mol Genet. 2003;12:R159–R165. doi: 10.1093/hmg/ddg259. [DOI] [PubMed] [Google Scholar]
- 14.Chetsanga CJ. Grigorian C. A dose-response study on opening of imidazole ring of adenine in DNA by ionizing radiation. Int J Radiat Biol Relat Stud Phys Chem Med. 1983;44:321–331. doi: 10.1080/09553008314551261. [DOI] [PubMed] [Google Scholar]
- 15.Cooke MS. Loft S. Olinski R. Evans MD. Bialkowski K. Wagner JR. Dedon PC. Moller P. Greenberg MM. Cadet J. Recommendations for standardized description of and nomenclature concerning oxidatively damaged nucleobases in DNA. Chem Res Toxicol. 2010;23:705–707. doi: 10.1021/tx1000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D'Errico M. Parlanti E. Dogliotti E. Mechanism of oxidative DNA damage repair and relevance to human pathology. Mutat Res. 2008;659:4–14. doi: 10.1016/j.mrrev.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Dang CV. Le A. Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res. 2009;15:6479–6483. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darwanto A. Theruvathu JA. Sowers JL. Rogstad DK. Pascal T. Goddard W., 3rd Sowers LC. Mechanisms of base selection by human single-stranded selective monofunctional uracil-DNA glycosylase. J Biol Chem. 2009;284:15835–15846. doi: 10.1074/jbc.M807846200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das A. Wiederhold L. Leppard JB. Kedar P. Prasad R. Wang H. Boldogh I. Karimi-Busheri F. Weinfeld M. Tomkinson AE. Wilson SH. Mitra S. Hazra TK. NEIL2-initiated, APE-independent repair of oxidized bases in DNA: evidence for a repair complex in human cells. DNA Repair (Amst) 2006;5:1439–1448. doi: 10.1016/j.dnarep.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.David SS. O'Shea VL. Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeMott MS. Beyret E. Wong D. Bales BC. Hwang JT. Greenberg MM. Demple B. Covalent trapping of human DNA polymerase beta by the oxidative DNA lesion 2-deoxyribonolactone. J Biol Chem. 2002;277:7637–7640. doi: 10.1074/jbc.C100577200. [DOI] [PubMed] [Google Scholar]
- 22.Dizdaroglu M. Karakaya A. Jaruga P. Slupphaug G. Krokan H. Novel activities of human uracil DNA N-glycosylase for cytosine-derived products of oxidative DNA damage. Nucleic Acids Res. 1996;24:418–422. doi: 10.1093/nar/24.3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dizdaroglu M. Kirkali G. Jaruga P. Formamidopyrimidines in DNA: mechanisms of formation, repair, and biological effects. Free Radic Biol Med. 2008;45:1610–1621. doi: 10.1016/j.freeradbiomed.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 24.DNA_Repair_Database. DNA repair database from 7 organisms. <http://dnapittcrew.upmc.com>. 2010. <http://dnapittcrew.upmc.com>
- 25.Dosanjh MK. Chenna A. Kim E. Fraenkel-Conrat H. Samson L. Singer B. All four known cyclic adducts formed in DNA by the vinyl chloride metabolite chloroacetaldehyde are released by a human DNA glycosylase. Proc Natl Acad Sci U S A. 1994;91:1024–1028. doi: 10.1073/pnas.91.3.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dosanjh MK. Roy R. Mitra S. Singer B. 1,N6-ethenoadenine is preferred over 3-methyladenine as substrate by a cloned human N-methylpurine-DNA glycosylase (3-methyladenine-DNA glycosylase) Biochemistry. 1994;33:1624–1628. doi: 10.1021/bi00173a002. [DOI] [PubMed] [Google Scholar]
- 27.Feig DI. Sowers LC. Loeb LA. Reverse chemical mutagenesis: identification of the mutagenic lesions resulting from reactive oxygen species-mediated damage to DNA. Proc Natl Acad Sci U S A. 1994;91:6609–6613. doi: 10.1073/pnas.91.14.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischhaber PL. Gerlach VL. Feaver WJ. Hatahet Z. Wallace SS. Friedberg EC. Human DNA polymerase kappa bypasses and extends beyond thymine glycols during translesion synthesis in vitro, preferentially incorporating correct nucleotides. J Biol Chem. 2002;277:37604–37611. doi: 10.1074/jbc.M206027200. [DOI] [PubMed] [Google Scholar]
- 29.Frenkel K. Goldstein MS. Duker NJ. Teebor GW. Identification of the cis-thymine glycol moiety in oxidized deoxyribonucleic acid. Biochemistry. 1981;20:750–754. doi: 10.1021/bi00507a014. [DOI] [PubMed] [Google Scholar]
- 30.Friedberg EC. Walker GC. Siede W. Wood RD. Schultz RA. Ellenberger T. 2nd. Washington, DC: ASM Press; 2006. DNA Repair and Mutagenesis; p. 164. [Google Scholar]
- 31.Grin IR. Dianov GL. Zharkov DO. The role of mammalian NEIL1 protein in the repair of 8-oxo-7,8-dihydroadenine in DNA. FEBS Lett. 2010;584:1553–1557. doi: 10.1016/j.febslet.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gros L. Ishchenko AA. Saparbaev M. Enzymology of repair of etheno-adducts. Mutat Res. 2003;531:219–229. doi: 10.1016/j.mrfmmm.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 33.Gros L. Maksimenko AV. Privezentzev CV. Laval J. Saparbaev MK. Hijacking of the human alkyl-N-purine-DNA glycosylase by 3,N4-ethenocytosine, a lipid peroxidation-induced DNA adduct. J Biol Chem. 2004;279:17723–17730. doi: 10.1074/jbc.M314010200. [DOI] [PubMed] [Google Scholar]
- 34.Guan L. Greenberg MM. Irreversible inhibition of DNA polymerase beta by an oxidized abasic lesion. J Am Chem Soc. 2010;132:5004–5005. doi: 10.1021/ja101372c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hailer MK. Slade PG. Martin BD. Rosenquist TA. Sugden KD. Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Repair (Amst) 2005;4:41–50. doi: 10.1016/j.dnarep.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 36.Hanes JW. Thal DM. Johnson KA. Incorporation and replication of 8-oxo-deoxyguanosine by the human mitochondrial DNA polymerase. J Biol Chem. 2006;281:36241–36248. doi: 10.1074/jbc.M607965200. [DOI] [PubMed] [Google Scholar]
- 37.Hansen AB. Griner NB. Anderson JP. Kujoth GC. Prolla TA. Loeb LA. Glick E. Mitochondrial DNA integrity is not dependent on DNA polymerase-beta activity. DNA Repair (Amst) 2006;5:71–79. doi: 10.1016/j.dnarep.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 38.Hardeland U. Bentele M. Jiricny J. Schar P. The versatile thymine DNA-glycosylase: a comparative characterization of the human, Drosophila and fission yeast orthologs. Nucleic Acids Res. 2003;31:2261–2271. doi: 10.1093/nar/gkg344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris JL. Jakob B. Taucher-Scholz G. Dianov GL. Becherel OJ. Lavin MF. Aprataxin, poly-ADP ribose polymerase 1 (PARP-1) and apurinic endonuclease 1 (APE1) function together to protect the genome against oxidative damage. Hum Mol Genet. 2009;8:4102–4117. doi: 10.1093/hmg/ddp359. [DOI] [PubMed] [Google Scholar]
- 40.Hashimoto K. Tominaga Y. Nakabeppu Y. Moriya M. Futile short-patch DNA base excision repair of adenine:8-oxoguanine mispair. Nucleic Acids Res. 2004;32:5928–5934. doi: 10.1093/nar/gkh909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 42.Jaruga P. Xiao Y. Vartanian V. Lloyd RS. Dizdaroglu M. Evidence for the involvement of DNA repair enzyme NEIL1 in nucleotide excision repair of (5′R)- and (5′S)-8,5′-cyclo-2′-deoxyadenosines. Biochemistry. 2010;49:1053–1055. doi: 10.1021/bi902161f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jensen A. Calvayrac G. Karahalil B. Bohr VA. Stevnsner T. Mammalian 8-oxoguanine DNA glycosylase 1 incises 8-oxoadenine opposite cytosine in nuclei and mitochondria, while a different glycosylase incises 8-oxoadenine opposite guanine in nuclei. J Biol Chem. 2003;278:19541–19548. doi: 10.1074/jbc.M301504200. [DOI] [PubMed] [Google Scholar]
- 44.Jovanovic SV. Simic MG. Mechanism of OH radical reaction with thymine and uracil derivatives. J Am Chem Soc. 1986;108:5968–5972. doi: 10.1021/ja00279a050. [DOI] [PubMed] [Google Scholar]
- 45.Kalam MA. Haraguchi K. Chandani S. Loechler EL. Moriya M. Greenberg MM. Basu AK. Genetic effects of oxidative DNA damages: comparative mutagenesis of the imidazole ring-opened formamidopyrimidines (Fapy lesions) and 8-oxo-purines in simian kidney cells. Nucleic Acids Res. 2006;34:2305–2315. doi: 10.1093/nar/gkl099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalivendi SV. Kotamraju S. Cunningham S. Shang T. Hillard CJ. Kalyanaraman B. 1-Methyl-4-phenylpyridinium (MPP+)-induced apoptosis and mitochondrial oxidant generation: role of transferrin-receptor-dependent iron and hydrogen peroxide. Biochem J. 2003;371:151–164. doi: 10.1042/BJ20021525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kasai H. Nishimura S. Hydroxylation of deoxyguanosine at the C-8 position by ascorbic acid and other reducing agents. Nucleic Acids Res. 1984;12:2137–2145. doi: 10.1093/nar/12.4.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krahn JM. Beard WA. Miller H. Grollman AP. Wilson SH. Structure of DNA polymerase beta with the mutagenic DNA lesion 8-oxodeoxyguanine reveals structural insights into its coding potential. Structure. 2003;1:121–127. doi: 10.1016/s0969-2126(02)00930-9. [DOI] [PubMed] [Google Scholar]
- 49.Kriaucionis S. Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kusumoto R. Masutani C. Iwai S. Hanaoka F. Translesion synthesis by human DNA polymerase eta across thymine glycol lesions. Biochemistry. 2002;41:6090–6099. doi: 10.1021/bi025549k. [DOI] [PubMed] [Google Scholar]
- 51.LeDoux SP. Druzhyna NM. Hollensworth SB. Harrison JF. Wilson GL. Mitochondrial DNA repair: a critical player in the response of cells of the CNS to genotoxic insults. Neuroscience. 2007;145:1249–1259. doi: 10.1016/j.neuroscience.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.LeDoux SP. Wilson GL. Beecham EJ. Stevnsner T. Wassermann K. Bohr VA. Repair of mitochondrial DNA after various types of DNA damage in Chinese hamster ovary cells. Carcinogenesis. 1992;13:1967–1973. doi: 10.1093/carcin/13.11.1967. [DOI] [PubMed] [Google Scholar]
- 53.Lee CY. Delaney JC. Kartalou M. Lingaraju GM. Maor-Shoshani A. Essigmann JM. Samson LD. Recognition and processing of a new repertoire of DNA substrates by human 3-methyladenine DNA glycosylase (AAG) Biochemistry. 2009;48:1850–1861. doi: 10.1021/bi8018898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee DH. Pfeifer GP. Translesion synthesis of 7,8-dihydro-8-oxo-2′-deoxyguanosine by DNA polymerase eta in vivo. Mutat Res. 2008;641:19–26. doi: 10.1016/j.mrfmmm.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu M. Bandaru V. Bond JP. Jaruga P. Zhao X. Christov PP. Burrows CJ. Rizzo CJ. Dizdaroglu M. Wallace SS. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc Natl Acad Sci U S A. 2010;107:4925–4930. doi: 10.1073/pnas.0908307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Longley MJ. Prasad R. Srivastava DK. Wilson SH. Copeland WC. Identification of 5′-deoxyribose phosphate lyase activity in human DNA polymerase and its role in mitochondrial base excision repair in vitro. Proc Natl Acad Sci U S A. 1988;95:12244–12248. doi: 10.1073/pnas.95.21.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maga G. Crespan E. Wimmer U. van Loon B. Amoroso A. Mondello C. Belgiovine C. Ferrari E. Locatelli G. Villani G. Hubscher U. Replication protein A and proliferating cell nuclear antigen coordinate DNA polymerase selection in 8-oxo-guanine repair. Proc Natl Acad Sci U S A. 2008;105:20689–20694. doi: 10.1073/pnas.0811241106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maga G. Villani G. Crespan E. Wimmer U. Ferrari E. Bertocci B. Hubscher U. 8-Oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature. 2007;447:606–608. doi: 10.1038/nature05843. [DOI] [PubMed] [Google Scholar]
- 59.Mandavilli BS. Santos JH. Van Houten B. Mitochondrial DNA repair and aging. Mutat Res. 2002;509:127–151. doi: 10.1016/s0027-5107(02)00220-8. [DOI] [PubMed] [Google Scholar]
- 60.Marenstein DR. Chan MK. Altamirano A. Basu AK. Boorstein RJ. Cunningham RP. Teebor GW. Substrate specificity of human endonuclease III (hNTH1): effect of human APE1 on hNTH1 activity. J Biol Chem. 2003;278:9005–9012. doi: 10.1074/jbc.M212168200. [DOI] [PubMed] [Google Scholar]
- 61.Marnett LJ. Oxy radicals, lipid peroxidation and DNA damage. Toxicology. 2002;181–182:219–222. doi: 10.1016/s0300-483x(02)00448-1. [DOI] [PubMed] [Google Scholar]
- 62.Marnett LJ. Oxyradicals and DNA damage. Carcinogenesis. 2000;21:361–370. doi: 10.1093/carcin/21.3.361. [DOI] [PubMed] [Google Scholar]
- 63.Martin KR. Barrett JC. Reactive oxygen species as double-edged swords in cellular processes: low-dose cell signaling versus high-dose toxicity. Hum Exp Toxicol. 2002;21:71–75. doi: 10.1191/0960327102ht213oa. [DOI] [PubMed] [Google Scholar]
- 64.Masaoka A. Matsubara M. Hasegawa R. Tanaka T. Kurisu S. Terato H. Ohyama Y. Karino N. Matsuda A. Ide H. Mammalian 5-formyluracil-DNA glycosylase, 2: role of SMUG1 uracil-DNA glycosylase in repair of 5-formyluracil and other oxidized and deaminated base lesions. Biochemistry. 2003;42:5003–5012. doi: 10.1021/bi0273213. [DOI] [PubMed] [Google Scholar]
- 65.Matsumoto Y. Zhang QM. Takao M. Yasui A. Yonei S. Escherichia coli Nth and human hNTH1 DNA glycosylases are involved in removal of 8-oxoguanine from 8-oxoguanine/guanine mispairs in DNA. Nucleic Acids Res. 2001;29:1975–1981. doi: 10.1093/nar/29.9.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McCarthy S. Somayajulu M. Sikorska M. Borowy-Borowski H. Pandey S. Paraquat induces oxidative stress and neuronal cell death; neuroprotection by water-soluble Coenzyme Q10. Toxicol Appl Pharmacol. 2004;201:21–31. doi: 10.1016/j.taap.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 67.Nakabeppu Y. Kajitani K. Sakamoto K. Yamaguchi H. Tsuchimoto D. MTH1, an oxidized purine nucleoside triphosphatase, prevents the cytotoxicity and neurotoxicity of oxidized purine nucleotides. DNA Repair (Amst) 2006;5:761–772. doi: 10.1016/j.dnarep.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 68.Nazarkina ZK. Khodyreva SN. Marsin S. Radicella JP. Lavrik OI. Study of interaction of XRCC1 with DNA and proteins of base excision repair by photoaffinity labeling technique. Biochemistry (Mosc) 2007;72:878–886. doi: 10.1134/s000629790708010x. [DOI] [PubMed] [Google Scholar]
- 69.Neeley WL. Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem Res Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 70.Oka S. Ohno M. Tsuchimoto D. Sakumi K. Furuichi M. Nakabeppu Y. Two distinct pathways of cell death triggered by oxidative damage to nuclear and mitochondrial DNAs. EMBO J. 2008;27:421–432. doi: 10.1038/sj.emboj.7601975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Olinski R. Zastawny T. Budzbon J. Skokowski J. Zegarski W. Dizdaroglu M. DNA base modifications in chromatin of human cancerous tissues. FEBS Lett. 1992;309:193–198. doi: 10.1016/0014-5793(92)81093-2. [DOI] [PubMed] [Google Scholar]
- 72.Parker AR. Sieber OM. Shi C. Hua L. Takao M. Tomlinson IP. Eshleman JR. Cells with pathogenic biallelic mutations in the human MUTYH gene are defective in DNA damage binding and repair. Carcinogenesis. 2005;26:2010–2018. doi: 10.1093/carcin/bgi166. [DOI] [PubMed] [Google Scholar]
- 73.Perillo B. Ombra MN. Bertoni A. Cuozzo C. Sacchetti S. Sasso A. Chiariotti L. Malorni A. Abbondanza C. Avvedimento EV. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 74.Phosphosite. PhosphositePlus. 2010. <http://www.phosphosite.org> <http://www.phosphosite.org>
- 75.Roberts SA. Strande N. Burkhalter MD. Strom C. Havener JM. Hasty P. Ramsden DA. Ku is a 5′-dRP/AP lyase that excises nucleotide damage near broken ends. Nature. 2010;464:1214–1217. doi: 10.1038/nature08926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rossi MN. Carbone M. Mostocotto C. Mancone C. Tripodi M. Maione R. Amati P. Mitochondrial localization of PARP-1 requires interaction with mitofilin and is involved in the maintenance of mitochondrial DNA integrity. J Biol Chem. 2009;284:31616–31624. doi: 10.1074/jbc.M109.025882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rydberg B. Qiu ZH. Dosanjh MK. Singer B. Partial purification of a human DNA glycosylase acting on the cyclic carcinogen adduct 1,N6-ethenodeoxyadenosine. Cancer Res. 1992;52:1377–1379. [PubMed] [Google Scholar]
- 78.Saparbaev M. Langouet S. Privezentzev CV. Guengerich FP. Cai H. Elder RH. Laval J. 1,N(2)-ethenoguanine, a mutagenic DNA adduct, is a primary substrate of Escherichia coli mismatch-specific uracil-DNA glycosylase and human alkylpurine-DNA-N-glycosylase. J Biol Chem. 2002;277:26987–26993. doi: 10.1074/jbc.M111100200. [DOI] [PubMed] [Google Scholar]
- 79.Singer B. Antoccia A. Basu AK. Dosanjh MK. Fraenkel-Conrat H. Gallagher PE. Kusmierek JT. Qiu ZH. Rydberg B. Both purified human 1,N6-ethenoadenine-binding protein and purified human 3-methyladenine-DNA glycosylase act on 1,N6-ethenoadenine and 3-methyladenine. Proc Natl Acad Sci U S A. 1992;89:9386–9390. doi: 10.1073/pnas.89.20.9386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sobol RW. Horton JK. Kuhn R. Gu H. Singhal RK. Prasad R. Rajewsky K. Wilson SH. Requirement of mammalian DNA polymerase-ß in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 81.Sung JS. DeMott MS. Demple B. Long-patch base excision DNA repair of 2-deoxyribonolactone prevents the formation of DNA-protein cross-links with DNA polymerase beta. J Biol Chem. 2005;280:39095–39103. doi: 10.1074/jbc.M506480200. [DOI] [PubMed] [Google Scholar]
- 82.Szczesny B. Tann AW. Longley MJ. Copeland WC. Mitra S. Long patch base excision repair in mammalian mitochondrial genomes. J Biol Chem. 2008;283:26349–26356. doi: 10.1074/jbc.M803491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tahiliani M. Koh KP. Shen Y. Pastor WA. Bandukwala H. Brudno Y. Agarwal S. Iyer LM. Liu DR. Aravind L. Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Takata K. Shimizu T. Iwai S. Wood RD. Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J Biol Chem. 2006;281:23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- 85.Tan X. Grollman AP. Shibutani S. Comparison of the mutagenic properties of 8-oxo-7,8-dihydro-2′-deoxyadenosine and 8-oxo-7,8-dihydro-2′-deoxyguanosine DNA lesions in mammalian cells. Carcinogenesis. 1999;20:2287–2292. doi: 10.1093/carcin/20.12.2287. [DOI] [PubMed] [Google Scholar]
- 86.Tang J. Goellner EM. Wang XW. Trivedi RN. St Croix CM. Jelezcova E. Svilar D. Brown AR. Sobol RW. Bioenergetic metabolites regulate base excision repair-dependent cell death in response to DNA damage. Mol Cancer Res. 2010;8:67–79. doi: 10.1158/1541-7786.MCR-09-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tardy-Planechaud S. Fujimoto J. Lin SS. Sowers LC. Solid phase synthesis and restriction endonuclease cleavage of oligodeoxynucleotides containing 5-(hydroxymethyl)-cytosine. Nucleic Acids Res. 1997;25:553–559. doi: 10.1093/nar/25.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vallyathan V. Shi X. The role of oxygen free radicals in occupational and environmental lung diseases. Environ Health Perspect. 1997;105(suppl 1):165–177. doi: 10.1289/ehp.97105s1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wallace SS. AP endonucleases and DNA glycosylases that recognize oxidative DNA damage. Environ Mol Mutagen. 1988;12:431–477. doi: 10.1002/em.2860120411. [DOI] [PubMed] [Google Scholar]
- 90.Weinberg F. Hamanaka R. Wheaton WW. Weinberg S. Joseph J. Lopez M. Kalyanaraman B. Mutlu GM. Budinger GR. Chandel NS. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wibley JE. Waters TR. Haushalter K. Verdine GL. Pearl LH. Structure and specificity of the vertebrate anti-mutator uracil-DNA glycosylase SMUG1. Mol Cell. 2003;1:1647–1659. doi: 10.1016/s1097-2765(03)00235-1. [DOI] [PubMed] [Google Scholar]
- 92.Wiederhold L. Leppard JB. Kedar P. Karimi-Busheri F. Rasouli-Nia A. Weinfeld M. Tomkinson AE. Izumi T. Prasad R. Wilson SH. Mitra S. Hazra TK. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 93.Wong RS. Sczepanski JT. Greenberg MM. Excision of a lyase-resistant oxidized abasic lesion from DNA. Chem Res Toxicol. 2010;23:766–770. doi: 10.1021/tx9003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wood RD. Mitchell M. Sgouros J. Lindahl T. Human DNA repair genes. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 95.Yakes FM. Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci U S A. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yoon JH. Iwai S. O'Connor TR. Pfeifer GP. Human thymine DNA glycosylase (TDG) and methyl-CpG-binding protein 4 (MBD4) excise thymine glycol (Tg) from a Tg:G mispair. Nucleic Acids Res. 2003;31:5399–5404. doi: 10.1093/nar/gkg730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang Y. Yuan F. Wu X. Taylor JS. Wang Z. Response of human DNA polymerase iota to DNA lesions. Nucleic Acids Res. 2001;29:928–935. doi: 10.1093/nar/29.4.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang Y. Yuan F. Wu X. Wang M. Rechkoblit O. Taylor JS. Geacintov NE. Wang Z. Error-free and error-prone lesion bypass by human DNA polymerase kappa in vitro. Nucleic Acids Res. 2000;28:4138–4146. doi: 10.1093/nar/28.21.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zheng L. Zhou M. Guo Z. Lu H. Qian L. Dai H. Qiu J. Yakubovskaya E. Bogenhagen DF. Demple B. Shen B. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol Cell. 2008;32:325–336. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zorov DB. Juhaszova M. Sollott SJ. Mitochondrial ROS-induced ROS release: an update and review. Biochim Biophys Acta. 2006;1757:509–517. doi: 10.1016/j.bbabio.2006.04.029. [DOI] [PubMed] [Google Scholar]