Abstract
Translational control has recently been recognized as an important facet of adaptive responses to various stress conditions. We describe the adaptation response of the yeast Saccharomyces cerevisiae to the loss of one of two mechanisms to target proteins to the secretory pathway. Using inducible mutants that block the signal recognition particle (SRP) pathway, we find that cells demonstrate a physiological response to the loss of the SRP pathway that includes specific changes in global gene expression. Upon inducing the loss of the SRP pathway, SRP-dependent protein translocation is initially blocked, and cell growth is considerably slowed. Concomitantly, gene expression changes include the induction of heat shock genes and the repression of protein synthesis genes. Remarkably, within hours, the efficiency of protein sorting improves while cell growth remains slow in agreement with the persistent repression of protein synthesis genes. Our results suggest that heat shock gene induction serves to protect cells from mislocalized precursor proteins in the cytosol, whereas reduced protein synthesis helps to regain efficiency in protein sorting by reducing the load on the protein translocation apparatus. Thus, we suggest that cells trade speed in cell growth for fidelity in protein sorting to adjust to life without SRP.
INTRODUCTION
All proteins destined for the secretory pathway must first be targeted to the endoplasmic reticulum (ER). In mammalian cells, this targeting reaction primarily occurs cotranslationally via the signal recognition particle (SRP) pathway. Both the components and the mechanism of SRP-dependent protein targeting are conserved in every organism studied to date from bacteria to eukaryotic cells. Without translocation, proteins would quickly accumulate in the cytosol, and the hydrophobic nature of many translocated membrane proteins would cause massive protein aggregation and severe stress for the cell.
SRP and the SRP-dependent protein-targeting pathway have been well characterized (reviewed by Brodsky, 1998; Walter and Johnson, 1994). In the yeast Saccharomyces cerevisiae, SRP consists of six protein subunits and a small RNA (Hann and Walter, 1991; Brown et al., 1994). Briefly, SRP-dependent targeting begins as nascent chains emerge from the ribosome, and those with ER-specific signal sequences are recognized and bound by SRP. The SRP–ribosome–nascent chain complex is then directed to the ER membrane through an interaction between SRP and the SRP receptor (SR), which consists of two proteins, SRα and SRβ, anchored to the ER membrane. The ribosome–nascent chain complex is released from SRP-SR and directed to the Sec61 membrane translocon, allowing cotranslational translocation of the protein across the ER membrane to proceed (Johnson and Van Waes, 1999).
In addition to the SRP pathway, many organisms have evolved alternative, SRP-independent protein-targeting pathways. In yeast, the core proteins of this pathway are Sec62, Sec63, Sec71, and Sec72. These proteins associate with the Sec61 translocon, forming a membrane complex required for this alternative translocation pathway (Deshaies and Schekman, 1989; Rothblatt et al., 1989; Deshaies et al., 1991). For SRP-independent targeting, chaperones are required to keep cytosolic precursor proteins in an unfolded, translocation-competent state. Proteins implicated for this role include the stress severity protein family A (SSA) chaperone family and Ydj1 (Chirico et al., 1988; Deshaies et al., 1988; Caplan et al., 1992). Directed by information contained in their hydrophobic signal sequences, targeting of some proteins, such as dipeptidyl aminopeptidase B (DPAP-B) or Kar2, is strongly SRP-dependent, whereas the targeting of others, such as carboxypeptidase Y, is SRP-independent (Brown et al., 1994; Ng et al., 1996).
The SRP pathway is essential in all organisms examined to date except the yeast S. cerevisiae (Hann and Walter, 1991). Deletion of any component of the SRP-targeting pathway displays indistinguishable phenotypes, indicating that each of these individual deletion mutations results in the disruption of the entire pathway. Yeast strains lacking the SRP pathway are exceedingly sick; they grow into heterogeneously sized colonies, growing three- to sixfold slower than isogenic wild-type strains (Hann and Walter, 1991; Ogg et al., 1992). Moreover, transcriptional shutoff of SRP pathway components results in an accumulation of untranslocated SRP-dependent proteins (Ogg et al., 1992; Brown et al., 1994). Thus, although SRP is not essential in S. cerevisiae, the loss of the SRP pathway has severe negative consequences for the cell.
Surprisingly, although depletion of SRP proteins causes an accumulation of many untranslocated precursor proteins, strains with genomic deletions of SRP genes do not display dramatic translocation defects of SRP-dependent proteins. Indeed, extended time courses with inducible depletion of SRP components demonstrated that cells “adapt” to the absence of the SRP-dependent pathway as monitored by the reduction of untranslocated precursor proteins (Ogg et al., 1992). Here we address the molecular basis of adaptation to begin to understand the adaptive response mounted by S. cerevisiae to survive the loss of SRP-mediated protein translocation.
MATERIALS AND METHODS
Strains Used in This Study
W303 (MATα, leu2-3-112, his3-11, trp1-1, ura3-1, can1-100, ade2-1); SMY246 (W303 [rho−]); SMY211 (W303, pDN66) (plasmid from Davis Ng, Penn State University, University Park, PA); SMY212 (W303, pGalSRP54); SMY226 (W303, hsf::LEU2, pDN66, pHF35 (HSF1C) (knockout construct from Sorger and Pelham, 1988); YTH119 (W303, MATα, srp102::URA3) from T. Hu, University of California, San Francisco, San Francisco, CA; SMY286 (YTH119, pTH123), SMY288 (YTH119, pSO462); SMY268 (W303 MATa, SEC63-prA HIS3, URA3) (sec63prA tag; Beckmann et al., 1997); SMY266 (W303 MATa, SEC63-prA, srp102::URA3, pSO462); SMY284a (W303 MATa, srp54::neo) from Gustavo Pesce, University of California, San Francisco; and SOY60 (W303, MATα, scr1::HIS3) (Ogg et al., 1992).
Plasmids Used in This Study
pGalSRP54 (Gal-SRP54, URA3, CEN4/ARS1) (Hann and Walter, 1991); pDN66 (Gal-SRP54dn, URA3, CEN4/ARS1) from Davis Ng, see below; pHF35 (HF/1-40Δ147 [HSF1C], TRP1, CEN6/ARSH4) (Sorger, 1990); pTH123 (SRP102-3xFlag, TRP1, CEN6/ARSH4) from Dr. T. Hu, University of California, San Francisco; pSO462 [srp102(K51I)-HA, TRP1, CEN6/ARSH4] (Ogg et al., 1998); pSM110 (pGalSRP54dn, TRP1, CEN6/ARSH4); and pSM131 (Gal-SRP54, TRP1, CEN6/ARSH4)
Construction of pDN66
The SRP54 G201A mutation was generated using the Kunkel method (Kunkel et al., 1987). The full-length SRP54 gene was inserted into the vector pRS313 (Sikorski and Heiter, 1989) at XbaI and BamHI to generate the phagemid pDN2. pDN2 single-stranded DNA was purified from phage produced from transformed CJ236 cells following infection with the helper phage VCSM13. Second strand synthesis was performed using a mutagenic primer changing glycine 201 to alanine (5′-GATACTTCAGCAAGGCATCA-3′). The resulting DNA was transformed into DH5α cells and the mutant plasmid (pDN50) was isolated from transformants and confirmed by DNA sequence analysis. pDN66 was constructed by subcloning a BstEII/SalI fragment from pDN50 to replace a similar fragment in pGALSRP54 (Hann and Walter, 1991).
Isotopic Labeling and Nonnative Immunoprecipitation
Metabolic labeling and immunoprecipitation assays were performed as described (Ng and Walter, 1996) except that cells were labeled for 7 min. All yeast cultures were grown and labeled at 30°C except for srp102(K51I) cells, which were grown and labeled at 23°C or 37°C as indicated. Monospecific polyclonal antisera were used for immunoprecipitation of endogenous protein. Anti-DPAP-B antiserum was generously provided by Tom Stevens (University of Oregon, Eugene, OR). Quantitation was performed with a Molecular Dynamics (Sunnyvale, CA) Storm 840 imager and ImageQuant software. Untranslocated precursor is represented as a ratio of precursor versus total protein recovered, which controls for expression changes of the substrate or loading differences.
Purification of the Sec63 Complex
The Sec63 complex was purified as described (Ogg et al., 1998) with the following modifications. Cells were grown to 0.3–0.8 OD600U/ml in medium lacking methionine followed by labeling at a density of 3 OD600U/ml with 30 μCi/OD600U of [35S] Promix cell-labeling mix (Amersham, Uppsala, Sweden) for 45 min to 1 h with aeration. Labeled cells were treated exactly as described except for the composition of the lysis buffer (50 mM HEPES-KOH pH 7.5, 200 mM sorbitol, 100 mM KOAc pH 7.5, 5 mM Mg(OAc2), 5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml pepstatin and leupeptin), and membranes were solubilized in digitonin (GHBD; 10% glycerol, 3% digitonin, 50 mM HEPES-KOH pH 7.5, 200 mM sorbitol, 400 mM KOAc pH 7.5, 5 mM Mg(OAc2), 5 mM diothiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml pepstatin and leupeptin). The detergent extracts were used immediately without freezing. Sec63prA protein complexes were purified in GHBD with 40 μl of Sepharose CL-4B and 0.5 μl-1 μl of IgG Sepharose 6 Fast Flow/OD600U cells (Amersham) for 3 h at 4°C with rotation. After extensive washing with GHBD, proteins were eluted from the IgG Sepharose with 100 mM glycine pH 2.0, trichloroacetic acid (TCA) precipitated, and analyzed by SDS-PAGE.
Genomic Arrays: Sample Preparation and Hybridization
Strains were grown to midlog phase in YPD or synthetic media as indicated. At the indicated time points, cells were centrifuged at room temperature and snap-frozen in liquid nitrogen. Total RNA was prepared by the SDS-hot phenol/bead lysis method (Kingston, 1997), and mRNA was isolated using the polyATtract system (Promega, Madison, WI) according to the manufacturer's instructions. Amino-allyl dUTP (aadUTP) was incorporated during reverse transcription of 2–3 μg of poly(A)+ RNA, primed with pd(T)12–18 (Amersham) and pdN6 (Life Technologies, Rockville, MD) as described (DeRisi et al., 1997), except the nucleotide final concentrations were 500 μM dATP, dCTP, and dGTP; 300 μM dTTP; and 200 μM aadUTP (#A0410; Sigma, St. Louis, MO). After reverse transcription, reactions were adjusted to 0.2 M NaOH, 0.1 M EDTA, and incubated for 15 min at 65°C for hydrolysis of RNA, followed by neutralization with Tris-HCl pH 7.4 to 0.33 M. Tris was removed from the reaction by washing with Centricon-30 microconcentrators (Amicon, Beverly, MA) as described (DeRisi et al., 1997). Monofunctional N-hydroxy succinimide-ester Cy3 or Cy5 (Amersham) was coupled to the cDNA via the incorporated aadUTP in 0.1 M sodium bicarbonate buffer pH 9.0 in the dark at room temperature for 1 h. The reactions were quenched by adjusting to 1.33 M hydroxylamine and incubating for 15 min at room temperature in the dark. Cy3 and Cy5 reactions were combined, and unincorporated dye was removed with the Qia-quick polymerase chain reaction purification kit (Qiagen, Chatsworth, CA) according to the manufacturer's instructions. cDNAs were hybridized to prepared microarrays as described (DeRisi et al., 1997) (see also http://www.microarrays.org/protocols.html).
Genomic Data Analysis and Categorization
Microarrays were visualized using a GenePix scanner (Axon Instruments, Foster City, CA), and fold changes in mRNA levels relative to control samples were determined using GenePix analysis software. Open reading frames (ORFs) of interest were placed into categories based on functional category descriptions in the Yeast Protein Database (http://www.proteome.com; Costanzo et al., 2000).
Quantitation of the Rate of [35S]Methionine Incorporation
Quantitation of the rate of [35S]methionine incorporation into protein was performed as described (Ogg and Walter, 1995) except that at each time point cells were plunged into ice-cold azide buffer (20 mM NaN3, 50 mM NaCl) and snap frozen in liquid nitrogen. The cells were then quick thawed, harvested, and washed once in azide buffer before lysis in TCA.
Online Supplemental Material
Datasets of the genomic expression experiments are available online at Molecular Biology of the Cell. The data are expression ratios formatted as text files that can be opened in various programs, including Microsoft Excel. The datasets include 1) “all srp ratio.txt”: the complete genomic data set with expression ratios of all SRP experiments, and 2) “704 ORFs.txt”: the subset of the complete data set that is described and categorized in Table 1.
Table 1.
Summary of 704 ORFs responsive to the loss of SRP
| Category | No. of ORFs | No. induced | No. repressed | % of total |
|---|---|---|---|---|
| Chaperone/heat shock | 31 | 30 | 1 | 4.4 |
| Protein synthesis | 77 | 1 | 76 | 11 |
| Mitochondrial/energy generationa | 36 | 1 | 35 | 5 |
| Metabolismb | 97 | 29 | 68 | 14 |
| Transcription | 18 | 7 | 11 | 2.5 |
| RNA processing | 6 | 5 | 1 | 0.8 |
| DNA replication, recombination, repair, structure | 18 | 3 | 15 | 2.5 |
| Protein modification | 11 | 1 | 10 | 1.6 |
| Protein degradation | 7 | 5 | 2 | 1 |
| Vesicular transport | 11 | 4 | 7 | 1.6 |
| Signaling | 7 | 1 | 6 | 1 |
| Cell wall/structural | 12 | 2 | 10 | 1.7 |
| Mating/budding | 4 | 0 | 4 | 0.6 |
| Cell cycle | 4 | 2 | 2 | 0.6 |
| Other | 5 | 1 | 4 | 0.7 |
| Uncharacterized ORFs | 360 | 148 | 212 | 51 |
| Total | 704 | 240 | 464 | 100% |
Twenty-three small molecule transporters, 74 involved in the metabolism of amino acids (20), carbohydrates (17), nucleotides (15), lipids/fatty acids (17), phosphate (2), and others (3).
Cells disrupted for SRP function rapidly lose the ability to grow on nonfermentable carbon sources, i.e., unless selective pressure is applied to the contrary, they become rho−. The reason for this tendency is unknown; it is not a prerequisite for survival as cells can be forced to retain mitochondrial function if they are continuously grown on nonfermentable carbon sources. We previously characterized protein translocation defects of rho− strains following SRP-depletion and found that these strains can also adapt (Ogg et al., 1992). We therefore conclude that a loss of respiratory function is not responsible for adaptation.
RESULTS
Cells Adapt to the Loss of the SRP Pathway by a Reversible, Physiological Process
To address the molecular basis of the adaptive response to the loss of SRP, we developed two independent means to disable the SRP pathway quickly and reversibly. The first approach depends on a plasmid-borne, galactose-inducible dominant negative allele of SRP54 (SRP54dn), one of the subunits of the signal recognition particle. The dominant negative allele in SRP54dn is a mutation in the second G-box domain (G201A) (Bernstein et al., 1989) that, by analogy to other GTPases, is predicted to interfere with GTP hydrolysis. Although the mechanism of action for Srp54dn remains to be characterized, the dominant negative effect is likely to arise from a block of GTP hydrolysis, resulting in Srp54dn locked onto the SRP receptor, sequestering the SRP receptor into an inactive pool (Rapiejko and Gilmore, 1992). After induction of SRP54dn, cells displayed phenotypes identical to SRP deletion strains, including their characteristic slow growth and variable colony size (data not shown).
We tested the effects of SRP54dn induction on protein translocation as a function of time with a pulse-labeling and immunoprecipitation experiment. Cells were grown in selective media containing raffinose, and then switched to galactose-containing media to induce SRP54dn or, as a control, SRP54. In galactose, the SRP54dn cells grow fourfold slower than the control strain (data not shown). At 0, 4, 8, 12, and 16 h after induction, cells were pulse-labeled, and an SRP-dependent protein substrate, Kar2, was immunoprecipitated and analyzed by SDS-PAGE. Translocation defects are monitored by following the lack of protein processing modifications normally made upon entry into the ER. For Kar2, translocation defects were inferred from the accumulation of a more slowly migrating precursor form, indicating the signal sequence has not been cleaved (Figure 1A, pre-Kar2). The precursor form of Kar2 reflects a defect in translocation rather than in processing as demonstrated previously (Ogg et al., 1992). To demonstrate that adaptation is not limited to a single substrate, we tested another SRP-dependent substrate, DPAP-B. For this protein, translocation defects are inferred from the appearance of a faster migrating unglycosylated precursor form (Figure 1, B and C, pre-DPAP-B). The translocation defect peaked at ∼4 h after SRP54dn induction with the accumulation of ∼60% untranslocated Kar2 (Figure 1A, lane 7), diminished at later time points, and persisted at ∼25% untranslocated Kar2 (Figure 1A, lanes 8–10). As expected, cells expressing wild-type SRP54 showed no growth or protein translocation defects indicating that the observed defect was not simply due to an overproduction of Srp54.
Figure 1.
Protein translocation returns to wild-type efficiency in the absence of functional SRP54 or SRβ over time. (A) Kar2 immunoprecipitation in the SRP54dn system. Translocation of Kar2 was compared in strains containing either a pGAL-SRP54wt plasmid (SMY212) or a pGAL-SRP54dn plasmid (SMY211). Cells were grown to midlog phase in raffinose-containing selective synthetic medium lacking uracil, and then shifted to the comparable galactose-containing media. Cells were labeled with [35S]methionine for 7 min, and harvested at the indicated times. Lysates at each time point were immunoprecipitated with anti-Kar2 and analyzed by SDS-PAGE followed by autoradiography. Lumenal (Kar2) and cytosolic precursor (pre-Kar2) forms are indicated. The amount of precursor protein relative to lumenal protein at each time point was quantified and graphed. The graph and error bars for the SRP54dn strain reflect the average and SD of nine experiments. (B) DPAP-B immunoprecipitation in the SRP54dn system. Experiments were carried out and analyzed as described for A. Lumenal (DPAP-B) and cytosolic precursor (pre-DPAP-B) forms are indicated. (C) DPAP-B immunoprecipitation in the SRβts system. Translocation of DPAP-B in a wild-type strain (SMY286, srp102::URA3, pTH123) or a strain containing the srp102(K51I) ts allele of SRP102 (SMY288, srp102::URA3, pSO462). Cells were grown to midlog phase in YPD at 23°C and then shifted to 37°C to induce the SRβts allele. Cells were labeled and immunoprecipitated as described in A.
DPAP-B showed a similar profile (Figure 1B). Four hours after induction of SRP54dn, as much as 90% of DPAP-B was detected as untranslocated pre-DPAP-B (Figure 1B, lane 7). Again, the amount of accumulated precursor protein diminished to nearly wild-type levels within 8 to 12 h. The cells' response to SRP loss was therefore biphasic: an immediate accumulation of untranslocated SRP-dependent precursor proteins (peaking around 4 h after SRP loss) followed by a reduction of untranslocated precursor proteins due to adaptation.
To assess the generality of adaptation, we used a second method to disrupt the SRP pathway. We took advantage of a strain in which the chromosomal copy of SRP102 (SRβ) has been disrupted but contains a plasmid with a temperature-sensitive allele, srp102(K51I) (Ogg et al., 1998). At 37°C, these cells grow approximately sixfold slower than wild-type cells (Ogg et al., 1998). As shown in Figure 1C, a shift to the nonpermissive temperature led to the accumulation of pre-DPAP-B after the 2 and 4 h time points, whereas at later time points precursor protein rapidly returned to levels close to those observed in wild-type cells (Figure 1C, lanes 9 and 10), reminiscent of the biphasic response observed after induction of the dominant negative allele of SRP54 (Figure 1, A and B).
Previous results indicated that adaptation is a physiological response and not due to a suppressor mutation. This argument was based on genetic evidence that, once backcrossed and sporulated, SRP or SRP receptor deletion strains that are constitutively adapted showed no evidence of inheritance of the adapted state (Ogg et al., 1992). To address this issue more directly, we took advantage of the inducible SRP54dn mutant to monitor adaptation over multiple rounds of switching the SRP pathway on and off. We induced expression of Srp54dn and monitored the effects on translocation of Kar2 as described above. As expected, we observed the transient accumulation of pre-Kar2 followed by adaptation (Figure 2, lanes 2 and 3). After blocking the SRP pathway for 16 h, the cells were switched back to growth under noninducing conditions for 24 h. After this time, we again induced Srp54dn expression and observed the initial accumulation of pre-Kar2, followed by adaptation over a 4- to 8-h period (Figure 2, lanes 7–10). Thus, the recovered cells behaved indistinguishably from wild-type cells that were never deprived of a functional SRP pathway. This result confirms that genetic suppression does not play a role in adaptation to the loss of the SRP pathway.
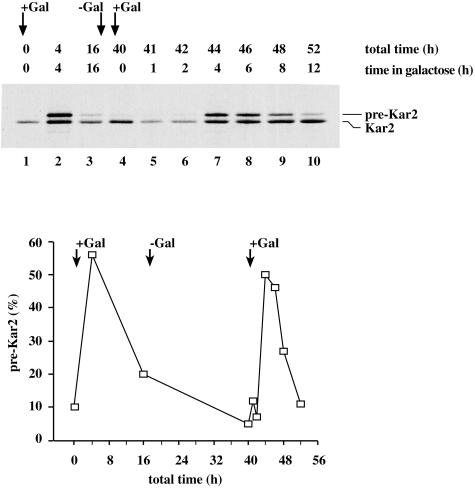
Figure 2.
Adaptation is a reversible, physiological process. Kar2 immunoprecipitation in the SRP54dn system. Cells (SMY211) were grown to midlog phase in raffinose-containing synthetic medium lacking uracil and shifted to galactose-containing medium. After 16 h, the cells were washed free of galactose and returned to raffinose-containing medium for a period of 24 h. After this recovery period, cells were again shifted to galactose-containing medium and allowed to grow for 12 h. Cells were labeled with [35S]methionine at the indicated time points, and Kar2 was immunoprecipitated as described in Figure 1. Cytosolic precursor forms (pre-Kar2) and lumenal forms (Kar2) are indicated. The relative amount of untranslocated protein at each time point was quantified and graphed as in Figure 1A. Differences in protein levels in lane 1, 4, 5, and 6 are due to experimental handling.
Composition of the Translocon Remains Unchanged after Adaptation
We next sought to identify physiological changes occurring in response to adaptation that are important for allowing cells to cope with the loss of the SRP pathway. In S. cerevisiae, the SRP-independent posttranslational translocation pathway has been well characterized (reviewed by Rapoport et al., 1996). Both translocation pathways are thought to use the same translocon composed of Sec61 and its associated subunits Sss1 and Sbh1, but different accessory proteins are required. For SRP-dependent translocation, these proteins include the heterodimeric SRP receptor, and for posttranslational translocation, these proteins include a complex of Sec63, Sec62, Sec71, and Sec72 (Rothblatt et al., 1989; Green et al., 1992; Panzner et al., 1995; Ng et al., 1996). Because most protein substrates studied show some degree of promiscuity in their choice of protein translocation pathways (Ng et al., 1996), we considered the possibility that in adapted cells, SRP-dependent proteins might be translocated posttranslationally with enhanced efficiency due to a structural change in the translocon itself. To explore this notion, we determined whether the composition of the translocon is changed in any quantitative or qualitative way in response to the loss of the SRP pathway.
To this end, we purified translocon complexes to examine their protein composition and abundance in wild-type and adapted cells. We used a strain containing a protein A-tagged version of Sec63 to allow for a one-step affinity isolation (Aitchison et al., 1995; Beckmann et al., 1997). We disrupted the SRP pathway either by expressing the SRP54dn allele or by a temperature shift of cells bearing the srp102(K51I) mutation, and allowed cells to adapt. Translocon complexes were purified by extracting microsomes with digitonin, a mild detergent that has been shown to preserve the integrity of the translocon (Panzner et al., 1995), and isolating the translocon complexes via protein A-tag binding to IgG Sepharose.
As expected, in the wild-type controls, Sec61, Sec62, Sec71, and Sec72 copurify with the Sec63 fusion protein and are the major proteins observed (Figure 3, lanes 2 and 4) With this gel system, we did not detect Sss1 or Sbh1 because of their smaller size. In the adapted cells, we see an indistinguishable pattern of proteins (Figure 3, compare lanes 2 and 3 and 4 and 5). Consistent with the similarities at the protein level, no up-regulation of mRNAs encoding these proteins was observed in adapted cells according to genomic expression array data (see supplemental genomics data). From these results, we conclude that neither the abundance nor the composition of the translocon is adjusted as cells adapt to the loss of the SRP pathway. These results suggest that if SRP-dependent proteins are translocated via the posttranslational pathway in adapted cells, they do so using translocon complexes present under normal growth conditions.
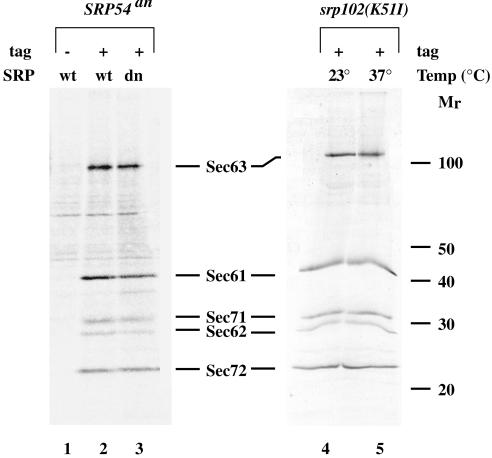
Figure 3.
The translocon in adapted cells is identical to that of wild-type cells. Sec63 complex proteins were purified from the following strains: SMY212 (lane 1, no tag); SMY268 bearing pSM131 (lane2, Sec64prA); SMY268 bearing pSM110 (lane 3, Sec63prA); srp102::URA3, SMY266 (lane 4 and 5, Sec63prA) as described in MATERIALS AND METHODS. Cells were grown in raffinose-containing medium to midlog phase and switched to growth in galactose-containing medium lacking methionine. Cultures were maintained in log phase in galactose-containing medium for 24 h (lanes 1–3). Alternatively, cultures were grown to midlog phase in YPD at 23°C (lane 4 and 5) and switched to 37°C for 12 h (lane 5). Cells were steady-state labeled with [35S]methionine for 45 min to 1 h at 30°C (lanes 1–3), 23°C (lane 4), or 37°C (lane 5), and membranes were isolated. Digitonin extracts of membranes were incubated with IgG Sepharose for 3 h at 4°C with rotation. The IgG Sepharose beads were washed and protein complexes were eluted with 100 mM glycine pH 2.0. The eluate was concentrated by TCA precipitation and analyzed by SDS-PAGE. The major protein components of the translocon are indicated.
Transcriptional Responses to the Loss of SRP
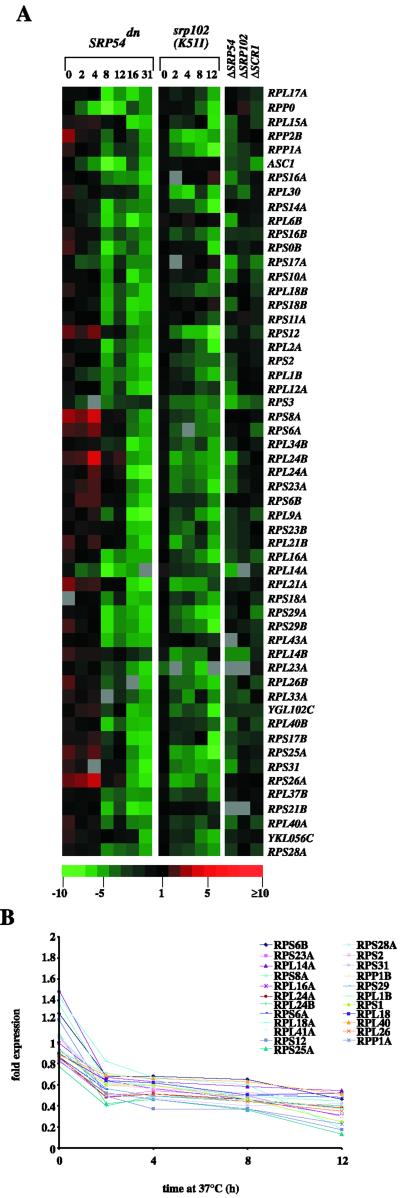
Expression levels of a limited number of chaperone proteins have been shown to be induced in a strain depleted of Srp54 (Arnold and Wittrup, 1994). To expand on this observation more comprehensively, we determined the global changes in the transcriptional program of the cell that accompany SRP loss and adaptation. For this purpose, we used DNA microarrays to screen the expression changes of all yeast ORFs under these conditions. The DNA microarrays were generated by polymerase chain reaction amplification of 6352 yeast ORFs and printing on a glass microscope slide (DeRisi et al., 1997). At various time points following disruption of the SRP pathway, mRNA was extracted from the cells, converted into cDNA, and fluorescently labeled. Reference samples were labeled with Cy3 (green), and experimental samples were labeled with Cy5 (red). For each time point, the experimental probes were mixed with the appropriate reference probes, and the mixture was hybridized to a microarray. The relative abundance of each mRNA was then measured by comparison of the relative intensity of the red and green signals, giving a measure of the relative expression of each ORF at various times during the process of SRP-depletion and adaptation. We represent relative expression levels visually with color blocks (Figures 4A and 6A). Shades of green represent levels of repression and shades of red represent induction relative to the reference strain.
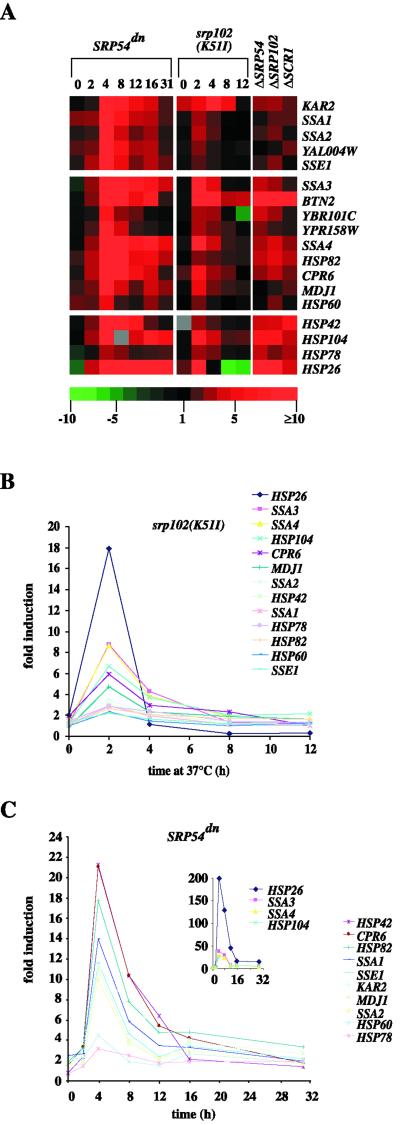
Figure 4.
Heat shock and chaperone gene transcription is induced during adaptation. Strains were grown to midlog phase in either YPD at 23°C (SRβts [SMY288]; versus SRβwt [SMY286]), or 30°C (ΔSRP54 [SMY284a]; ΔSRP102 (YTH119); ΔSCR1 (SOY60); all versus W303 rho−), or in synthetic raffinose-containing medium at 30°C SRP54dn (SMY211); versus SRP54wt [SMY212]). The SRβts strain and the SRβwt control strain were then both shifted to 37°C for the times indicated. The SRP54dn strain and the SRP54wt control strain were shifted to galactose-containing medium for the times indicated. It was critical to ensure that all strains were diluted as necessary to keep them continuously in log phase growth, and the medium used was derived from the same batch for each experiment. At the indicated time points, cells were centrifuged and snap-frozen in liquid nitrogen. Fluorescently labeled cDNA probes were made as described in MATERIALS AND METHODS. Cy3 (green) labeled probes are SRβwt at 23°C (0-h time point) or 37°C (2–12-h time points), W303 rho−, and SRP54wt in raffinose (0-h time point) or galactose (2–31-h time points). Cy5 (red) labeled probes are SRβts, SRP54dn, ΔSRP54, ΔSRP102, and ΔSCR1. For each time point or deletion, the differentially labeled probe pairs were mixed and hybridized to a microarray, and the relative abundance of each mRNA was measured by intensity of red or green fluorescence. The red/green fluorescence intensity ratio gives a measure of relative expression for each ORF as shown in the color scale. Brightest red color blocks indicate genes most highly induced relative to the control strain, brightest green blocks represent highest repression, black indicates no change in expression, and gray indicates no data. (A) Relative expression levels of selected genes encoding for heat shock proteins and chaperones are depicted with color blocks. (B) Relative expression levels of several heat shock and chaperone genes throughout the SRβts time course. (C) Relative expression levels of several heat shock and chaperone genes throughout the SRP54dn time course.
Using this approach, we analyzed the consequences of blocking the SRP pathway by either induction of SRP54dn or temperature shift of SRβts. To minimize variance due to the differences in growth conditions necessary to induce the SRP pathway mutations, we subjected reference strains to the same conditions to subtract out many of these differences. For example, the galactose-inducible SRP54dn cells were compared with galactose-inducible SRP54wt cells to control for both the carbon source shift and protein overexpression. Similarly, the SRβts cells were compared with SRβwt cells also grown at 37°C to control for the temperature shift. In both cases, we monitored transcriptional changes as a function of time, and comparison of the data from the two experimental systems allowed us to focus on major changes common to SRP loss. Thus, observed changes would be more likely to represent physiological responses to SRP loss rather than to reflect changes inherent in the changes of growth conditions. In addition to the time courses following SRP loss in the two inducible systems, we analyzed the long-term consequences of genomic deletions of three different components of the SRP pathway, Srp54, SRP RNA (encoded by SCR1), and SRβ.
For this study, we limited our analyses to ORFs that experienced at least twofold induction or repression in both time courses. We examined each time point and selected ORFs for which at least three time points from both experimental systems met the cutoff criteria. From the 6352 ORFs examined, 704 ORFs (11% of the total genome) met these criteria with two-thirds being repressed and one-third being induced (see supplemental genomics data). The ORFs were grouped according to cellular function, and these groups are summarized in Table 1.
Although a very broad spectrum of genes is either repressed or induced in response to the loss of the SRP pathway, changes in three major transcriptional programs stood out: 1) a large number of genes encoding chaperones and heat shock factors was induced (30 genes), 2) many genes encoding ribosomal proteins were repressed (76 genes), and 3) mitochondrial and/or energy generation genes are repressed (35 genes, for discussion of this category, see Table 1).
Chaperone/Heat Shock Induction
Induction of a limited number of heat shock proteins was previously observed upon SRP loss (Arnold and Wittrup, 1994). Our results showed that this transcriptional program was induced and included a large number of genes encoding chaperones and other heat shock proteins. Plotting expression levels of these genes versus time showed that the sharp, peak induction of these genes coincided with the peak of untranslocated proteins accumulated in the cytosol (2 h for the SRβts cells, 4 h for the SRP54dn cells, Figure 4, B and C). The steady-state levels of these mRNAs then decreased over time. In the case of SRP pathway depletion using a temperature shift of SRβts cells, the expression levels at late time points became comparable to those in a control strain also grown at 37°C, representing a sustained heat shock response (Figure 4B, 12 h). Sustained up-regulation, however, was also observed at the late time points after SRP54dn induction where experimental and control cells were maintained at 30°C (Figure 4C, 16 and 31 h). Representing the fully adapted state, the induction of chaperone/heat shock genes was also observed in several genomic deletions of SRP and SR components (Figure 4A, gene deletions in the three right-most columns).
To address how the heat shock/chaperone-inductive response corresponds in scope and magnitude to a genuine heat shock response, we compared our data to a published data set for a 39°C heat shock treatment (Roth et al., 1998), in which 263 genes were judged to have been induced relative to the control. We found that, upon the disruption of the SRP pathway, 10% of these genes were induced, at peak expression levels, at least threefold greater than in the heat shock experiment, 50% of these genes showed induction of similar magnitude, and 40% were not induced to heat shock levels. Thus, the chaperone and heat shock gene induction observed in response to the loss of the SRP pathway substantially overlaps with a heat shock response, yet it is not identical.
We next asked whether the observed induction of heat shock proteins would be sufficient for adaptation to the loss of SRP. If sufficient, adaptation should be facilitated in cells in which heat shock proteins are constitutively expressed at elevated levels. To test this hypothesis, we used a constitutively active form of Hsf1 (HSF1C), the transcription factor controlling genes with promoters containing a heat shock element. When expressed, Hsf1C is sufficient to cause a persistently high level of heat shock protein expression (greater than twofold higher than expression due to heat shock), without a need for elevated temperature or any other inducing stress (Sorger and Pelham, 1988; Sorger, 1990). We disrupted the SRP pathway in strains expressing Hsf1C by induction of SRP54dn and compared the amount of untranslocated protein with a wild-type strain after 4 h of SRP54dn expression (Figure 5). Even with its constitutively elevated level of heat shock proteins, however, the HSF1C strain showed translocation defects indistinguishable from those observed in wild-type strains. Given this result, we conclude that the elevated level of heat shock induction observed in this strain is not sufficient to result in or accelerate adaptation.
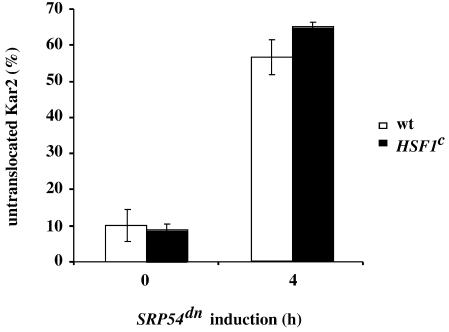
Figure 5.
Constitutive high expression of heat shock proteins and chaperones is not sufficient to adapt to the loss of SRP54. Accumulation of untranslocated Kar2 upon induction of SRP54dn was measured in a wild-type strain (SMY211; □; n = 4) and in a strain containing a constitutively active allele of HSF1 (SMY226 [HSF1C; hsf::LEU2, with pHF35]; ▪; n = 2). Cells were grown in galactose-containing medium for 4 h, labeled, and processed for immunoprecipitation with anti-Kar2 as described in Figure 1. The precursor form of Kar2 is plotted as percentage of total Kar2 immunoprecipitated.
We next wanted to determine whether elevated expression levels of heat shock proteins are necessary for adaptation. Because HSF1 is an essential gene, we used strains expressing a mutant version of HSF1 (HF/40Δ147-583) that is not inducible (Sorger, 1990). Analysis of HF/40Δ147-583 cells showed a marginal, if any, deficiency in the ability to adapt to the loss of the SRP pathway compared with an isogenic wild-type strain (data not shown). Similar experiments with knockouts or conditional alleles of individual chaperones (SSA1, SSA2, YDJ1ts, HSP104, HSP82, and HSP26/42 double knockout) showed either no effect or only very marginal effects on adaptation. Taken together, these results suggest that adaptation either relies on redundant signaling pathways or heat shock proteins that have not been tested or that the elevated levels of heat shock proteins are not required for adaptation.
Repression of Ribosome Biogenesis
The second and most comprehensive transcriptional program in response to SRP loss is the repression of genes responsible for protein synthesis. Seventy-one different ribosomal proteins, for example, are down-regulated at least twofold in response to the loss of SRβ. In addition, a variety of other genes encoding components of the protein synthesis machinery are repressed, including genes encoding elongation and initiation factors and rRNA and tRNA processing proteins. The same effect was observed upon induction of SRP54dn, when >100 genes encoding ribosomal proteins were down-regulated at least twofold during the time course (Figure 6, A and B). In contrast to the biphasic up-regulation of chaperone/heat shock genes described above, ribosomal protein genes show a monotonic repression profile following loss of SRP pathway function (Figure 6B).
Figure 6.
Ribosome biogenesis is repressed during adaptation. Strains, conditions, and color codes are exactly as described in Figure 4. (A) Relative expression levels of selected genes involved in ribosome biogenesis are depicted. (B) Relative expression levels of several ribosomal protein genes throughout the SRβ ts time course are plotted.
As expected, a significant reduction in the rate of total protein synthesis resulted as a consequence of the transcriptional repression of protein synthesis genes. After induction of SRP54dn, we pulse-labeled cells with [35S]methionine and measured incorporation of the radioactive amino acid over time into total protein. [35S]Methionine uptake was comparable between the wild-type and dominant negative strains (data not shown). We compared the incorporation rates after 4 h of disruption of the SRP pathway (unadapted cells) and after16 h (adapted cells). We observed a ninefold decrease in the rate of protein production after 16 h of SRP54dn expression compared with after 4 h, whereas the wild-type controls exhibited a threefold decrease presumably due to shift to galactose (data not shown). Thus, as predicted by the genomic expression data, protein synthesis is repressed in adapted cells.
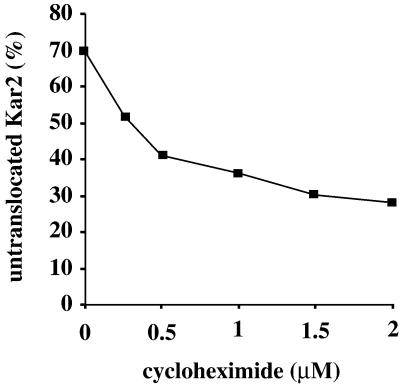
We next asked whether decreasing protein synthesis could suppress the translocation defects observed early after SRP pathway loss before cells become adapted. To this end, we treated SRP54dn cells with a range of sublethal cycloheximide concentrations (Ogg and Walter, 1995) to artificially cause a reduction in protein synthesis and monitored translocation defects at the 4-h time point after induction of SRP54dn. At the maximal cycloheximide concentration used (2 μM), [35S]methionine incorporation was reduced 19-fold compared with untreated cells (data not shown). As shown in Figure 7, we observed a significant dosage-dependent decrease in the relative amount of untranslocated proteins, suggesting that reduced protein synthesis can contribute to the cell's ability to adapt to the loss of SRP.
Figure 7.
Decreased protein synthesis caused by cycloheximide treatment can suppress translocation defects due to SRP54dn expression. SRP54dn cells were grown in galactose for 3 h, and cycloheximide was added to the concentrations indicated for an additional hour. Cells were labeled and processed for immunoprecipitation with anti-Kar2 as described in Figure 1.
DISCUSSION
In these studies, we have characterized the adaptive response to the loss of the SRP pathway in S. cerevisiae. Using inducible mutants, we have demonstrated that adaptation is a reversible, physiological response that occurs rapidly upon loss of SRP or SR function. Null mutants of SRP pathway components display the same adapted phenotype, demonstrating that adaptation is not due to the use of partial SRP function in our inducible systems. Although translocating proteins in adapted cells are likely to make extensive use of alternative targeting pathways, apparently no change in the wild-type state of the translocon is required to do so. However, we observed a highly complex set of transcriptional changes, including the induction of heat shock genes and the repression of genes involved in protein synthesis.
Concurrent with the accumulation of untranslocated proteins in the cytosol, we observed a large induction in heat shock gene expression, including chaperones implicated in protein translocation (such as the SSA genes encoding members of the HSP70 chaperone family). In addition to this inductive response, we also observed ribosomal repression upon disruption of the SRP pathway. Whereas the expression of ribosomal genes was consistently low throughout adaptation, heat shock gene induction peaked early during the response, concomitant with the amount of untranslocated proteins in the cell. Heat shock gene expression then persisted at lower levels throughout adaptation. Chaperones and heat shock proteins are likely to be required to maintain proteins in an unfolded state until they are either translocated in an SRP-independent manner or degraded in the cytosol. Thus, it is plausible that the spike in chaperone expression is necessary to accommodate an initially high load of untranslocated protein. Perhaps reduced protein synthesis diminishes the need for these proteins during the adaptation phase. In this view, heat shock proteins play a protective role rather than being instrumental for the process of adaptation per se: transient induction of chaperones may aid in clearing the cytosol of untranslocated proteins, whereas a persistent decrease in the cell's protein synthesis capacity may stop the problem at its source. Late in adaptation, cells may reach a translation rate at which they can accommodate all proteins that must enter the ER in the absence of SRP. This would explain why, over time, SRP− strains regain the ability to target SRP-dependent proteins to the ER, but still grow more slowly than wild-type strains.
Connections between regulation of ribosome biogenesis and the secretory pathway have been observed before. In Escherichia coli, for example, suppressor analysis of conditional sec mutations yielded primarily mutations that compromise protein synthesis, and it has been proposed that the decreased synthesis of precursor proteins relieves the lethal burden placed on the mutant Sec machinery (Oliver, 1985; Lee and Beckwith, 1986; Danese et al., 1995). In yeast, reduction in protein synthesis by cycloheximide treatment suppresses the temperature-sensitive effects of the SRP mutant, sec65-1 (Ogg and Walter, 1995). Furthermore, it was demonstrated that defects in the secretory pathway at any point from the ER membrane translocon to the trans-Golgi network cause a significant repression of ribosomal proteins and RNAs (Mizuta and Warner, 1994; Nierras and Warner, 1999). Translational regulation has also been suggested to play a role in cell survival during the unfolded protein response by reducing the protein load on the folding machinery during stress (Harding et al., 2000). It seems likely that repression of ribosome biogenesis is having a similar effect of supporting adaptation by reducing the protein load on alternative translocation pathways.
The exact mechanism of targeting and translocation of proteins in the absence of SRP remains unclear. Thus, we do not know whether it is the reduction in ribosomal capacity (as indicated by the genomic expression data), a reduced elongation rate, or both that allows for survival in the absence of the SRP pathway. Here, we have only shown that a reduction in translational elongation alone can partially alleviate translocation defects caused by the loss of SRP in nonadapted cells. Many proteins studied show some degree of flexibility in their choice of protein translocation pathway (Ng et al., 1996), and it seems plausible that protein translocation is accommodated posttranslationally in the absence of SRP. However, if the observed decrease in protein synthesis includes slowing elongation, it remains possible that some proteins may be translocated cotranslationally even in the absence of the SRP-targeting pathway. Parallels may exist in the mechanism of protein import into mitochondria where it has been argued that the relative kinetics of translation and import may allow a subset of protein import to occur cotranslationally (Lithgow, 2000).
Other models invoking SRP-independent cotranslational translocation are also conceivable. Recent studies suggest that a substantial fraction of large ribosomal subunits remains membrane bound after termination of protein synthesis (Potter and Nicchitta, 2000) and that translation of signal sequence-bearing proteins initiating on such membrane-bound ribosomal subunits can directly access the translocon in the absence of SRP receptor function (Seiser and Nicchitta, 2000). We have shown that the abundance of translocons does not change in response to the loss of the SRP pathway, yet based on the genomic expression data ribosomal capacity is reduced. Thus, the ratio of ribosomes to translocons in SRP-depleted cells is proportionally lower than in wild-type cells. It is thus conceivable that more translational initiation events occur on membrane-bound ribosomes, increasing the chance of proper targeting in the absence of a functional SRP pathway.
In this study, we focused on trends of only two transcriptional programs revealed by the genomic expression data. In light of the vast number of known genes and uncharacterized ORFs that are induced or repressed in the absence of the SRP pathway, it seems unlikely that the combined effects of chaperone up-regulation and decreased protein synthesis capacity describe the full extent of the adaptive process. Rather, a multiplicity of physiological changes may contribute to survival in the absence of the SRP pathway, including other responses revealed by the genomic expression data as well as processes regulated at the translational or posttranslational level. More sophisticated genetic tools will need to be used to provide focus on the key causal changes that allow cells to survive such a severe stress.
It is unclear what role, if any, protein degradation plays in the adaptive response. It is possible that what appears to be improved translocation efficiency in adapted cells is, in whole or part, due to increased specific degradation of accumulated precursor proteins. Pulse-chase experiments aiming to determine the half-lives of untranslocated proteins during adaptation have yielded divergent results, depending on the SRP disruption system chosen. After temperature shift of SRβts cells, for example, pre-Kar2 had similar half-lives throughout the adaptation time course (t1/2 = 89 min at 2 h; 94 min at 12 after temperature shift; Mutka and Walter, unpublished observations), suggesting that increased degradation of precursor proteins is not responsible for the apparent improvement of translocation efficiency observed in these cells. In contrast, upon induction of SRP54dn, we observed an increased rate of pre-Kar2 disappearance (t1/2 = 40 min at 4 h; 23 min at 16 after temperature shift; Mutka and Walter, unpublished observations) that was not accompanied by a corresponding increase in translocated protein, i.e., could not be accounted for by posttranslational protein translocation. This suggests that pre-Kar2 is degraded at an increased rate in the adapted cells. Thus, there may be different ways in which cells can cope with SRP loss that may depend on growth conditions or other factors.
With the sole exception of S. cerevisiae, the SRP pathway is essential in all organisms examined to date. Even in S. cerevisiae, however, it is clear from growth and protein translocation phenotypes, as well as from the vast number of gene expression changes characterized here, that the loss of the SRP pathway causes enormous stresses for the cell. Our data suggest that, in the absence of SRP, protein synthesis is repressed, which may be instrumental for allowing cell survival but at the same time giving rise to a much reduced growth rate. The cell may therefore trade speed for fidelity, as a compromise when the SRP pathway is no longer functional. Indeed, this recourse may be a very general principle that cells use for surviving a variety of stresses that, for cells growing in the wild, are likely to be transient. Translational regulation is now emerging as an important mechanism for surviving stresses, such as defects in the secretory pathway (Mizuta and Warner, 1994; Nierras and Warner, 1999) or accumulation of unfolded proteins (Harding et al., 2000) and, as argued here, may contribute to adaptation to the loss of the SRP pathway.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tianhua Hu, Gustavo Pesce, Davis Ng, Peter Sorger, Tom Stevens, and Roland Beckmann for plasmids, strains, and antibodies; Joe DeRisi and Holly Bennett for invaluable assistance with microarrays and data collection; and Steve Ogg for valuable discussions. We thank Carol Gross, Davis Ng, Jeff Cox, Maho Niwa, Ursula Rüegsegger, Jason Brickner, Max Heiman, and Chris Patil for critical reading of the manuscript. This work was supported by a Howard Hughes Predoctoral fellowship to S.M. and by a grant from the National Institute of Health to P.W. P.W. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Online version of this article contains data set material. Online version available at www.molbiolcell.org.
REFERENCES
- Aitchison JD, Rout MP, Marelli M, Blobel G, Wozniak RW. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J Cell Biol. 1995;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold CE, Wittrup KD. The stress response to loss of signal recognition particle function in Saccharomyces cerevisiae. J Biol Chem. 1994;269:30412–30418. [PubMed] [Google Scholar]
- Beckmann R, Bubeck D, Grassucci R, Penczek P, Verschoor A, Blobel G, Frank J. Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex [see comments] Science. 1997;278:2123–2126. doi: 10.1126/science.278.5346.2123. [DOI] [PubMed] [Google Scholar]
- Bernstein HD, Poritz MA, Strub K, Hoben PJ, Brenner S, Walter P. Model for signal sequence recognition from amino-acid sequence of 54k subunit of signal recognition particle. Nature. 1989;340:482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- Brodsky JL. Translocation of proteins across the endoplasmic reticulum membrane. Int Rev Cytol. 1998;178:277–328. doi: 10.1016/s0074-7696(08)62139-7. [DOI] [PubMed] [Google Scholar]
- Brown JD, Hann BC, Medzihradszky KF, Niwa M, Burlingame AL, Walter P. Subunits of the Saccharomyces cerevisiae signal recognition particle required for its functional expression. EMBO J. 1994;13:4390–4400. doi: 10.1002/j.1460-2075.1994.tb06759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan AJ, Cyr DM, Douglas MG. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell. 1992;71:1143–1155. doi: 10.1016/s0092-8674(05)80063-7. [DOI] [PubMed] [Google Scholar]
- Chirico WJ, Waters MG, Blobel G. 70K Heat shock related proteins stimulate protein translocation into microsomes. Nature. 1988;332:805–810. doi: 10.1038/332805a0. [DOI] [PubMed] [Google Scholar]
- Costanzo MC, et al. The yeast proteome database (YPD) and Caenorhabditis elegans proteome database (WormPD): comprehensive resources for the organization and comparison of model organism protein information. Nucleic Acids Res. 2000;28:73–76. doi: 10.1093/nar/28.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danese PN, Murphy CK, Silhavy TJ. Multicopy suppression of cold-sensitive sec mutations in Escherichia coli. J Bacteriol. 1995;177:4969–4973. doi: 10.1128/jb.177.17.4969-4973.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Koch BD, Werner WM, Craig EA, Schekman R. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature. 1988;332:800–805. doi: 10.1038/332800a0. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Sanders SL, Feldheim DA, Schekman R. Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature. 1991;349:806–808. doi: 10.1038/349806a0. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Schekman R. SEC62 encodes a putative membrane protein required for protein translocation into the yeast endoplasmic reticulum. J Cell Biol. 1989;109:2653–2664. doi: 10.1083/jcb.109.6.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green N, Fang H, Walter P. Mutant in three novel complementation groups inhibit membrane protein insertion into and soluble protein translocation across the endoplasmic reticulum membrane of Saccharomyces cerevisiae. J Cell Biol. 1992;116:597–604. doi: 10.1083/jcb.116.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann BC, Walter P. The signal recognition particle in S. cerevisiae. Cell. 1991;67:131–144. doi: 10.1016/0092-8674(91)90577-l. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- Johnson AE, Van Waes MA. The translocon: a dynamic gateway at the ER membrane. Annu Rev Cell Dev Biol. 1999;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]
- Kingston RE. In: Current Protocols in Molecular Biology. Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Vol. 1. New York: John Wiley & Sons, Inc; 1997. , pp 4.0.1–4.10.11. [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lee CA, Beckwith J. Suppression of growth and protein secretion defects in Escherichia coli secA mutants by decreasing protein synthesis. J Bacteriol. 1986;166:878–883. doi: 10.1128/jb.166.3.878-883.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lithgow T. Targeting of proteins to mitochondria. FEBS Lett. 2000;476:22–26. doi: 10.1016/s0014-5793(00)01663-x. [DOI] [PubMed] [Google Scholar]
- Mizuta K, Warner JR. Continued functioning of the secretory pathway is essential for ribosome synthesis. Mol Cell Biol. 1994;14:2493–2502. doi: 10.1128/mcb.14.4.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng D, Brown J, Walter P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane J. Cell Biol. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng D, Walter P. ER membrane protein complex required for nuclear fusion. J Cell Biol. 1996;132:499–509. doi: 10.1083/jcb.132.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierras CR, Warner JR. Protein kinase C enables the regulatory circuit that connects membrane synthesis to ribosome synthesis in Saccharomyces cerevisiae. J Biol Chem. 1999;274:13235–13241. doi: 10.1074/jbc.274.19.13235. [DOI] [PubMed] [Google Scholar]
- Ogg SC, Barz WP, Walter P. A functional GTPase domain, but not its transmembrane domain, is required for function of the SRP receptor beta-subunit. J Cell Biol. 1998;142:341–354. doi: 10.1083/jcb.142.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S, Poritz M, Walter P. The signal recognition particle receptor is important for growth and protein secretion in Saccharomyces cerevisiae. Mol Biol Cell. 1992;3:895–911. doi: 10.1091/mbc.3.8.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg SC, Walter P. SRP samples nascent chains for the presence of signal sequences by interacting with ribosomes at a discrete step during translation elongation. Cell. 1995;81:1075–1084. doi: 10.1016/s0092-8674(05)80012-1. [DOI] [PubMed] [Google Scholar]
- Oliver DB. Identification of five new essential genes involved in the synthesis of a secreted protein in Escherichia coli. J Bacteriol. 1985;161:285–291. doi: 10.1128/jb.161.1.285-291.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzner S, Dreier L, Hartmann E, Kostka S, Rapoport TA. Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell. 1995;81:561–70. doi: 10.1016/0092-8674(95)90077-2. [DOI] [PubMed] [Google Scholar]
- Potter MD, Nicchitta CV. Regulation of ribosome detachment from the mammalian endoplasmic reticulum membrane. J Biol Chem. 2000;275:33828–33835. doi: 10.1074/jbc.M005294200. [DOI] [PubMed] [Google Scholar]
- Rapiejko PJ, Gilmore R. Protein translocation across the endoplasmic reticulum requires a functional GTP binding site in the α-subunit of the signal recognition particle receptor. J Cell Biol. 1992;117:493–503. doi: 10.1083/jcb.117.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport TA, Jungnickel B, Kutay U. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- Roth FP, Hughes JD, Estep PW, Church GM. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation [see comments] Nat Biotechnol. 1998;16:939–945. doi: 10.1038/nbt1098-939. [DOI] [PubMed] [Google Scholar]
- Rothblatt JA, Deshaies RJ, Sanders SL, Daum G, Schekman R. Multiple genes are required for proper insertion of secretory proteins into the endoplasmic reticulum in yeast. J Cell Biol. 1989;72:61–68. doi: 10.1083/jcb.109.6.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiser RM, Nicchitta CV. The fate of membrane-bound ribosomes following the termination of protein synthesis. J Biol Chem. 2000;275:33820–33827. doi: 10.1074/jbc.M004462200. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Heiter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger PK. Yeast heat shock factor contains separable transient and sustained response transcriptional activators. Cell. 1990;62:793–805. doi: 10.1016/0092-8674(90)90123-v. [DOI] [PubMed] [Google Scholar]
- Sorger PK, Pelham HR. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell. 1988;54:855–864. doi: 10.1016/s0092-8674(88)91219-6. [DOI] [PubMed] [Google Scholar]
- Walter P, Johnson AE. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.