Abstract
DNA repair is essential for routine monitoring and repair of damage imparted to our genetic material by exposure to endogenous and exogenous carcinogens, including reactive oxygen species, UV light, and chemicals such as those found in cigarette smoke. Without DNA repair pathways, the continual assault on our DNA would be highly mutagenic and the risk of cancer increased. Paradoxically, the same pathways that help prevent cancer development are detrimental to the efficacy of DNA-damaging cancer therapeutics such as cisplatin. Recent studies demonstrate the inverse relationship between DNA repair capacity and efficacy of platinum-based chemotherapeutics: increased DNA repair capacity leads to resistance, while decreased capacity leads to increased sensitivities. Cisplatin's cytotoxic effects are mediated by formation of intrastrand DNA crosslinks, which are predominantly repaired via the nucleotide excision repair (NER) pathway. In an effort to personalize the treatment of cancers based on DNA repair capacity, we developed an ELISA-based assay to measure NER activity accurately and reproducibly as a prognostic for platinum-based treatments. Here we present an overview of DNA repair and its link to cancer and therapeutics. We also present data demonstrating the ability to detect the proteins of the pre-incision complex within the NER pathway from cell and tissue extracts. Antioxid. Redox Signal. 14, 2465–2477.
Introduction
The ability to respond to and repair damage to genetic material is a crucial activity for most living organisms. The impact of environmental exposures on living cells and organisms dates back to the 1930s, with DNA repair being first identified as a response to UV light in the mid 1960s in both bacteria and mammalian cells. Since that time, research on the fundamental mechanisms of DNA repair has continued unabated and has provided advances and insight into numerous biological and physiological systems and processes. The identification of mutant strains of Escherichia coli with increased sensitivity to UV light, the subsequent cloning of the responsible Uvr genes, and eventual reconstitution of the bacterial nucleotide excision repair (NER) pathway were at the forefront of molecular biology. The connection between transcription and DNA repair represented a significant finding, resulting in DNA repair as the selected “Molecule of the Year” in the mid 1990s (13). The identification of mismatch-mediated repair (MMR) as the causative factor in the etiology in hereditary nonpolyposis colorectal cancer (HNPCC) also highlighted the importance of maintaining genetic stability (35). Complete reconstitution of the protein factors involved in the NER pathway and the connection between DNA repair and cancer as observed in the rare genetic disease xeroderma pigmentosum (XP) and the identification of the breast and ovarian cancer genetic markers, BRCA1 and BRCA2, has once again pushed DNA repair into the forefront of medical research. More recently, the demonstration of synthetic lethality in cancer cells opens the possibility to target DNA repair pathways specifically towards more effective treatments for cancer (31). The complexity of most mammalian DNA repair pathways makes their assessment within biological samples difficult, and thus the integration of the knowledge we have gleaned from over 50 years of DNA repair research into the clinical setting has been slowed. However, we are critically poised with recent technological advances and the support of the medical community to apply our understanding of biological mechanisms towards combating diseases such as cancer.
DNA repair in human diseases and cancer
DNA repair is critical to genomic maintenance, and the impact of reduced DNA repair capacity influences a myriad of cellular processes. It is therefore not surprising that defects in genes that code for proteins involved in DNA repair lead to devastating physiological effects and diseases ranging from UV sensitivity and premature aging, to a predisposition to cancer and death. Ataxia telangiectasia (AT) is caused by mutation to ATM and manifests as developmental and neurological syndromes. ATM is a PI3K-like kinase activated in response to DNA damage, specifically DNA double-strand breaks (DSBs), and is involved in cell division signaling events (51). Fanconi anemia (FA) is clinically manifested by congenital defects, thumb abnormalities, bone marrow failure, and cancer. FA, unlike other diseases, seems to have a more complex association with DNA repair. It has been characterized by a general genomic instability and is caused by mutations in genes that regulate replication-dependent removal of interstrand DNA crosslinks. FA has been linked to defects in the BRCA2 gene and multiple DNA repair pathways, including DSB repair via homologous recombination, NER and translesion syntheisis (55). Defects in the nonhomologous end-joining pathway (NHEJ) via mutations in a number of genes, including DNA-PKcs, XLF/cernunnos and Artemis, result in immune deficiencies largely due to the inability to catalyze V(D)J recombination leading to reduced immune function (30). In addition, defects in NHEJ result in hypersensitivity to IR and levels of NHEJ proteins have been linked with both cancer progression and survival following treatment (7, 52). The role in carcinogenesis is likely manifested as decreased repair contributing to increased genetic instability while ultimate survival is often a function of the effectiveness of therapy, many of which are genotoxic and thus more effective in the absence of DNA repair.
Defects in DNA excision repair pathways also lead to devastating diseases and result in an increased predisposition to the development of cancer. XP was the first human disease linked to a defect in the NER pathway (11) and is characterized by extreme photosensitivity and a predisposition to cancers, especially those of the skin. In addition, XP heterozygosity was found to be associated with early onset familial lung cancer and was thus concluded to be a risk factor for lung cancer (32). Cockayne syndrome (CS), primarily a developmental disorder characterized by dwarfism, retinal degeneration, and photosensitivity, is caused by mutations in the CSA or CSB genes, which are essential for transcription-coupled repair via NER (TC-NER) (see below). Trichothiodystrophy (TTD), resulting from NER deficiency, is a rare genetic disease that manifests as dysmorphic facial characteristics, brittle hair, dry skin, and mental retardation. Defects in the base excision repair (BER) pathway were first associated with familial adenomatous polyposis (1) and has since been associated with Alzheimer's disease (57). Defects in MMR cause microsatellite instability with loss of function mutations genetically linked to HNPCC, an early age of onset form of colon cancer. Most HNPCC cases result from defects in one of two MMR genes, MSH2 and MLH1. In addition to these genetic and inheritable diseases, DNA repair defects have been identified in numerous cancers. Considering that genetic instability is a hallmark of most cancers, it has been suggested that, in fact, every cancer will display some deficiency in DNA repair (2).
Repair pathways
Several DNA repair mechanisms exist in cells, each performing a specialized repair for the type of DNA lesion or damage that is presented. The DNA damage repair pathways can be divided into two groups: those that repair modified or misincorporated bases mediated by an excision-type process and those that repair DNA strand breaks. DNA bases can be modified through endogenous or exogenous sources via alkylation, deamination, oxidation events, or exposure to UV light or environmental carcinogens such as those found in cigarette smoke. In addition, several chemotherapeutics target DNA for modification, which will be addressed later. DSBs can be caused by ionizing radiation, reactive oxygen species (ROS), and endogenous events such as V(D)J recombination.
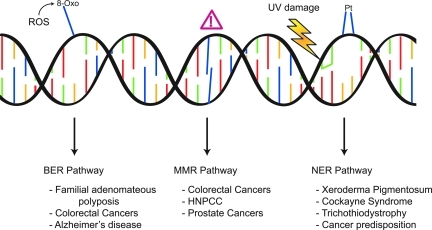
The excision repair pathways of BER, MMR, and NER are responsible for recognition and repair of lesions or adducts that require physical removal (Fig. 1). These pathways involve the initial recognition of the damage, followed by a mechanism of excision that is dictated by the particular type of lesion that occurs. Repair can involve removal of a single base as in the case of BER, to tens of bases in NER, and up to kilobases in MMR. Upon removal of the base(s), conventional DNA polymerases are recruited to fill in the resulting gap, and DNA ligases ultimately seal the resulting nick.
FIG. 1.
DNA damage excision repair pathways and the diseases related to dysfunctional repair of the lesions. The BER pathway recognizes and removes modified bases resulting from endogenous ROS, deamination, or alkylation. The MMR pathway is responsible for recognition and repair of misincorporated bases that occur during replication. The NER pathway repairs damage that occurs to DNA due to exogenous sources, primarily CPDs and 6-4 photoproducts, which form in response to UV exposure. In addition, the NER pathway is the primary repair pathway for repair of 1,2-d(GpG) and 1,3-d(GpNpG) cisplatin adducts. While all excision repair pathway deficiencies are associated with an increase risk and predisposition to cancer, BER deficiencies have been linked to Alzheimer's disease, and NER deficiencies are associated with XP, CS, and TTD. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
In BER, one of eleven DNA glycosylases recognizes and removes small, non-helix-distorting lesions, such as chemically modified bases (46). Left unrepaired, the resulting incorrect base pairing could lead to downstream mutation to the genome. Following DNA glycosylase base removal by cleavage of the N-glycosidic bond, resulting in an abasic site, the backbone is then nicked by an AP endonuclease, resulting in a gap that is filled in by DNA polymerase and sealed by DNA ligase (36). MMR is responsible for correcting replication errors that escape processing by the proofreading activity of DNA polymerases, and insertion/deletion mismatches, which also, typically occur during replication. While MMR in prokaryotes is fairly well understood, the molecular mechanism of MMR in eukaryotes remains unresolved (28). The specifics of the NER pathway will be described in depth in the next section.
DNA break repair pathways can be subdivided into high fidelity and low fidelity pathways. NHEJ repairs DSBs on chromosomes in an end-joining mechanism using limited to no sequence homology and is considered error prone. Homologous recombination (HR), on the other hand, requires significant regions of homology to repair DSBs accurately using sister chromatid sequence information and is thought to be highly accurate. Detailed descriptions of these pathways are covered in other articles within this Forum.
Nucleotide excision repair pathway
The NER pathway is one of the more versatile DNA damage repair pathways in the cell. It is responsible for the repair of most bulky DNA adducts, which are formed upon exposure of DNA to endogenous or exogenous DNA modifiers. The NER pathway repairs UV-induced lesions such as cyclobutane pyrimidine dimers (CPDs) and 6-4 photoproducts, as well as chemical modifications such as intrastrand adducts induced upon exposure to cis-diamminedichloroplatinum(II) (cisplatin). In NER, how the damage is initially encountered dictates the subpathway activated to repair the lesion (Fig. 2). The two subpathways of NER consist of transcription-coupled NER (TC-NER) (25), which repairs lesions encountered only during transcription, while the global genomic NER (GG-NER) pathway is responsible for repair independent of transcription. TC-NER and GG-NER pathways converge after the initial damage recognition step, resulting in the excision of a stretch of nucleotides on the damaged strand, followed by fill-in synthesis of the resulting gap. The details of these mechanisms are detailed below.
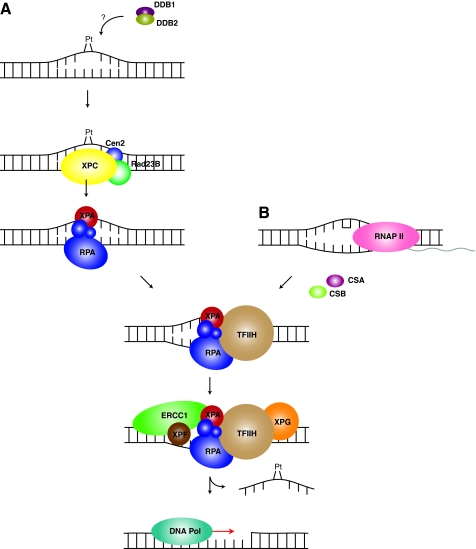
FIG. 2.
Overview of the NER pathway. (A) GG–NER pathway. Recognition of a platinum lesion triggers the XPC–Rad23B–Cen2 complex assembly at the site of damage. RPA and XPA are recruited, along with TFIIH to unwind the duplex and to keep the strands separated. The nucleases XPG and XPF-ERCC1 cleave the damaged strand 5′ and 3′ of the damage, releasing a ∼34 nt fragment of DNA. Subsequent fill-in of the resulting gap requires DNA polymerase, which re-synthesizes the excised sequence and DNA ligase I or III seals the nick between the newly synthesized DNA and the 5′ phosphate left behind after cleavage. (B) TC-NER pathway. Upon encountering a DNA lesion during transcription of a gene, RNAP II stalls, allowing for repair of the lesion to occur. This response is, in part, mediated by the transcription repair factors, CSA and CSB, which bypass the need for the GG-NER recognition factors. CSA and CSB recruit TFIIH to the site of damage, and the process proceeds in the same manner as for the GG-NER pathway. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Lesions that are less helix-distorting (i.e., CPDs and 1,2-d(GpG) cisplatin adducts) require assistance in their recognition via the DDB complex (DDB1-DDB2/XPE) (45). Upon recognition of such a lesion, DDB polyubiquitinates the XPC protein and DDB2/XPE, leading to stabilization of the XPC–DNA interaction and degradation of DDB2/XPE (54). Larger helix-distorting lesions, such as those caused by 1,3-d(GpNpG) cisplatin adducts or 6-4 photoproducts, can be recognized directly by the XPC complex (XPC–Rad23B–Cen2) (59) (Fig. 2). Following recognition of damage, the duplex must be opened around the site of the lesion, which is performed by the helicase activities of the TFIIH complex. While the exact mechanism of TFIIH opening of the duplex is uncertain, XPB and XPD are likely candidates for strand separation (12). Upon opening of the DNA, the single-strand DNA binding heterotrimer, RPA, and XPA are recruited, displacing the XPC complex. RPA binding to the undamaged single-stranded DNA prevents re-annealing of the bubble and protects the undamaged strand from nucleases (42, 43). Although the specific role of XPA is largely unconfirmed, it appears that, unlike RPA, XPA functions only in the NER pathway. XPA has been speculated to be required for verification of the damaged strand (10), in addition to providing a scaffold for formation of the pre-incision complex (22). The next proteins incorporated into the pre-incision complex are the nucleases XPF-ERCC1 and XPG, which nick the DNA 5′ and 3′, respectively, of the site of damage, displacing a ∼34 nucleotide piece of DNA. XPF-ERCC1 incision 5′ of the damage precedes the 3′ incision by XPG, and coordination of these incisions and repair synthesis minimize the presence of single stranded DNA intermediates (53). Conventional DNA polymerase machinery fills in the resulting gap and the nick is ligated by DNA ligase I or III (5, 18).
Activation of the TC-NER subpathway leads to an accelerated and rapid response compared to that from GG-NER (18). As RNA polymerase II (RNAP II) encounters a lesion in the coding strand of an active gene, recruitment of TC-NER specific factors bypasses the need of the XPC–Rad23B–Cen2 or DDB damage recognition complexes (47). While less mechanistic detail is understood for the TC-NER pathway, two transcription repair factors, CSA and CSB, are required for efficient repair, recruiting TFIIH to the site of damage. CSB serves as a repair coupling factor that recruits other NER proteins and chromatin remodeling factors to the site of damage. CSA, on the other hand, recruits XAB2 and other chromatin remodeling factors. Following the recruitment of TFIIH to the damage, the TC-NER pathway follows the GG-NER pathway with formation of the pre-incision complex, followed by incision and removal of a stretch of nucleotides containing the damaged DNA.
NER and the response to chemotherapeutics
Mounting evidence suggests an intimate relationship between DNA repair capacity and efficacy of DNA damaging chemotherapeutics, such as cisplatin. Cisplatin continues to be the first line therapy for several cancers, including head and neck, testicular, ovarian, cervical, lung, and colorectal cancers (15). While cisplatin treatment leads to activation of DNA damage signaling pathways, increased ROS and altered gene expression profiles, the cytotoxic effects are largely attributed to the intrastrand crosslink formation on DNA. The most predominant lesion formed upon treatment with cisplatin is the 1,2-d(GpG) intrastrand crosslink, which is recognized and repaired by the NER cellular machinery. The demonstration of the connection between decreased DNA repair and sensitivity to cisplatin came from analysis of XP patients, who have underlying defects in genes encoding proteins involved in NER. It was observed that cells derived from XP patients, who generally develop cancer due to their genetic defects, tend to be hypersensitive to DNA damaging agents such as cisplatin (9). It was later demonstrated that extracts prepared from XP cells were ineffective in repairing UV-induced DNA adducts, and eventually this work was extended to cisplatin lesions (19).
Hypersensitivity to cisplatin as a function of DNA repair capacity has been observed in vivo and in vitro in model systems (6, 17). A lung cancer adenocarcinoma model showed that upon reduction of XPA protein by antisense RNA technology there was a corresponding increase in sensitivity to cisplatin (61). The relationship of NER capacity and cisplatin sensitivity has been further demonstrated in testicular cancers, which display an acute chemosensitivity, in part due to the reduced NER capacity of these cells, which has been attributed to reduced levels of XPA (26). Similarly, it has been shown that overexpression of NER genes is associated with cisplatin resistance in ovarian, glioma, bladder, and lung cancer cells (61). Inhibition of NER via antisense RNA technology targeting the XPA gene revealed increased survival in a mouse xenograft model of human ovarian cancer demonstrating the utility of inhibiting the NER pathway for treatment of cisplatin resistance in ovarian cancer (48). The overexpression of another NER protein, ERCC1, in non-small cell lung cancer (NSCLC) correlates with cisplatin resistance, and single nucleotide polymorphisms (SNPs) have been identified in ERCC1 that represent the first steps in individualizing therapy based on NER status (50). Recent studies have shown that ERCC1 expression levels, assessed by immunohistochemistry, can serve as a prognostic marker for determining which NSCLC patients are likely to respond to cisplatin adjuvant therapy following tumor resection (38, 39). This result remains controversial as a result of technical issues with the antibodies used (8, 37, 40) and highlights the importance of validating the assays and reagents selected for assessment of physiological function and activity in vivo.
Assessment of DNA repair capacity—From biology to treatment
The current technologies for monitoring DNA repair capacity from tissue samples are cumbersome and challenging. There are, however, several assays used for the purpose of identifying the levels of proteins in DNA repair pathways or the ability of a cell to repair DNA damage. Unfortunately, each of these methods has serious downfalls related to the amount of material required, type of equipment needed, and the lack of direct assessment of activity. The host cell reactivation (HCR) assay and comet assays (27, 29) require culturing of a patient's tumor cells prior to the analysis of repair activity. Global expression or proteomics-based assays and DNA sequencing for SNPs or common genetic variants are good for identifying biomarkers, however are less effective at assessing the activity associated with a biological pathway. Immunohistochemical analysis of tumor biopsies or tissues only identifies the presence or absence of a protein and not necessarily the functionality of the protein itself or the pathways in which it participates. Direct measurement of NER activity from cell extracts, while a significant advance in the DNA repair field, requires large quantities of proteins (4). Here we present the initial development of a solid-phase ELISA assay from which to measure NER repair activity/capacity. The ultimate goal is to be able to detect the levels of repair from patient biopsy samples in an effort to more effectively treat cancer patients.
Materials and Methods
Materials
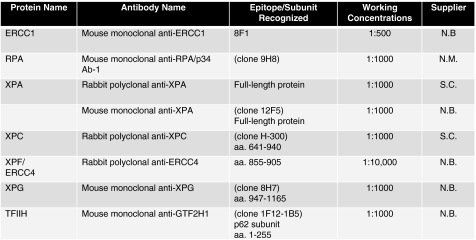
Oligonucleotides were purchased from Integrated DNA Technologies (Coralville, Iowa) and were PAGE-purified on 12% denaturing (8 M urea) gels. Standard nucleotides were purchased from Promega (Madison, WI), dUTP, biotin-dUTP, and streptavidin-coated 96-well plates were purchased from Roche Diagnostics (Mannheim, Germany). Klenow fragment (exo-) and restriction endonucleases were obtained from New England Biolabs (Ipswich, MA). cis-Diamminedichloroplatinum (II) (cisplatin) was purchased from Sigma (St. Louis, MO). Primary antibodies were obtained from NeoMarkers (Fremont, CA), Novus Biologicals (Littleton, CO), and Santa Cruz Biotechnology (Santa Cruz, CA) (Fig. 3). Secondary antibodies: goat anti-mouse IgG was purchased from Santa Cruz (Santa Cruz, CA) and goat anti-rabbit IgG was purchased from BioRad (Hercules, CA). All other reagents were obtained from standard suppliers.
FIG. 3.
Primary antibodies used in this study. Antibodies to each protein component of the pre-incision complex are listed along with the epitope of recognition (where available), and the working concentration used. Suppliers are: N.B., Novus Biologicals; N.M., NeoMarkers; S.C., Santa Cruz Biotechnology. In studies where whole cell extracts (WCEs) were used, the monoclonal XPA antibody (clone 12F5) provided an increased signal:noise ratio, and was thus used for all WCE analyses.
Oligonucleotides and DNA substrates
Two oligonucleotides were used in these studies: Biotin-60mer (5′-biotin-CCCTTCTTTC TCTTCCCCCTCTCCTTCTTGGCCTCTTCCTTCTTCCCCTT-3′) and 60mer-complement (5′-GGGGAGGAAAGGGAAGGGGAAGGAAGAGGCCAAGAAGGAGAGGGGGAAGAGAA-3′). Biotin-60mer was platinated (where indicated in the text) at 1 μM to produce a 1,2-d(GpG) intrastrand crosslink in the presence of 10 μM cisplatin for 16 h in the dark at 37°C. Double-stranded, platinated, 60mer DNA was produced by annealing platinated biotin-60mer to 60mer-complement in the presence of 50 mM Tris pH 7.5, 10 mM MgOAc, 5 mM DTT, by heating the reaction to 95°C for 10 min and allowing the mixture to cool to room temperature.
Longer double-stranded DNA fragments were produced by restriction endonuclease digestion of a pBS+ plasmid (Agilent Technologies (formerly Stratagene), La Jolla, CA) that was pre-treated with cisplatin at a 25:1 molar ratio of cisplatin:plasmid DNA. The plasmid was digested with ApaLI to generate fragments of 497, 1461, and 1246 base pairs with 5′ overhangs. The overhangs were filled in with biotin-dUTP (to allow for attachment to the ELISA plates), dATP, dCTP, and dGTP using Klenow fragment (exo-) according to manufacturer's protocol.
Protein expression and purification
The human RPA expression vector was kindly provided by Marc Wold, University of Iowa, and was purified as previously described (20). XPA protein was overexpressed and purified as previously described (21).
Whole cell extract preparation
Extracts from HeLa cells were prepared according to Wood et al. (60). Briefly, a 1 L cell pellet, from a suspension culture, was resuspended in a hypotonic buffer and lysed by homogenization. Extracts were fractionated by ammonium sulfate precipitation before being dialyzed into 25 mM K-HEPES pH 7.9, 0.1 M KCl, 12 mM MgCl2, 1 mM EDTA, 17% (v/v) glycerol, and 2 mM DTT. Protein concentrations were determined by Bradford assay (BioRad) and extracts were frozen at −80°C for long-term storage.
Tumor extract preparation
Tumor biopsies, obtained from the Indiana University Lilly tissue bank, were used to prepare extracts. ∼100 mg of tissue was incubated with 400 μl hypotonic buffer (10 mM Tris pH 8.0, 15 mM NaCl, 1.5 mM MgCl2, 5 mM DTT) supplemented with 10 μg each, leupeptin and pepstatin, and 2.5 mM PMSF. The tissues were homogenized, followed by a 30-min incubation on ice in the presence of 0.4 M NaCl. Lysates were centrifuged at 13,000 rpm for 30 min at 4°C. The supernatant was dialyzed against 50 mM Tris, pH 8.0, 50 mM NaCl, 1.5 mM MgCl2, 10 % (v/v) glycerol, 2 mM DTT, and the protein concentrations were determined by Bradford assay (BioRad). Extracts were stored at −80°C.
ELISA assays
Biotinylated 60mer DNA was pre-bound to streptavidin coated 96-well plates at 200 fmol (for ssDNA or dsDNAs substrates) or 50 ng (for digested plasmid fragments) in 2% BSA-TBST (20 mM Tris pH 7.5, 170 mM NaCl, 0.5% (v/v) Tween-20, 2% (w/v) bovine serum albumin (BSA)). Purified proteins or whole cell extracts (WCEs) were incubated with the DNA in 2% BSA-TBST for 1 h to allow for proteins to bind to the DNA and the BSA to block all nonspecific sites. Wells were washed three times with 2% BSA-TBST, followed by the addition of the primary antibodies, which were incubated for 1 h at dilutions of 1:500 – 1:10,000 (Fig. 3). The primary antibody was removed and the wells were washed an additional three times with 2% BSA-TBST before adding the secondary antibody, which was incubated for 1 h at 1:1000 dilutions in 2% BSA-TBST. The horseradish peroxidase (HRP)-conjugated secondary antibody was removed, and the wells washed three times with 2% BSA-TBST. 50 μl of 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (Pierce), a colorimetric substrate for HRP, was added to each well and conversion of the TMB substrate to a blue-colored product was monitored over time in a kinetic reads mode at 370 nm using a SpectraMax M5 plate reader and analyzed using SoftMax Pro software (Molecular Devices, Sunnyvale, GA). The absorbance was monitored over time and the initial velocity calculated. A depiction of the assay is presented in Figure 4. Assays were performed in duplicate or triplicate, and single representative data sets are shown in the figures. Data from purified protein titrations were fit to a standard two-state binding model.
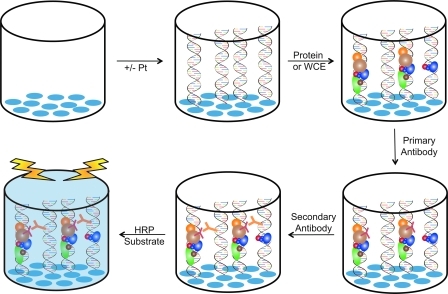
FIG. 4.
Conceptual view of the ELISA assay. Streptavidin-coated (blue ovals) 96-well plates are incubated with platinated or non-platinated biotinylated DNA. Protein (purified, recombinant protein or WCEs) is incubated with DNA to allow time for binding to occur. A primary antibody, specific to the protein of interest, is added (red Y-shape), followed by subsequent wash steps to alleviate nonspecific interactions. A secondary antibody is added (orange Y-shape), which is conjugated to the HRP enzyme. Wells are washed again to remove nonspecific interactions, and the HRP-substrate is added for detection. HRP converts the chromagenic substrate, yielding a blue-colored solution. The development of the color is measured over time, and the initial velocity of the conversion is proportional to the level of protein in the assay. (To see this illustration in color the reader is referred to the web version of this article at www.liebertonline.com/ars).
Results
Detection of purified RPA and XPA binding to ssDNA in an ELISA assay
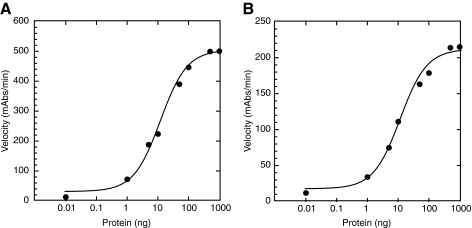
Studies aimed at monitoring NER proteins in a solid-phase ELISA were initiated with the optimization and detection of two DNA binding proteins, RPA and XPA, which are central to the formation of the incision complex at a site of DNA damage. Purified proteins were incubated with and bound to biotinylated 60-nucleotide ssDNA. 200 fmol of biotin-60mer ssDNA oligonucleotide were attached to the streptavidin-coated 96-well plates via the biotin–streptavidin interaction. Protein was titrated from 1 ng to 1000 ng and incubated with the DNA to determine a detection limit of protein and to validate the quantitative nature of the assay. Signal from these assays is highly dependent on the antibodies used and the ability of the protein to interact with the DNA tethered to the plate. Optimization of these assays involved examination of several antibody combinations, as well as alternative suppliers for the best signal:noise ratios. Experiments involving detection of RPA showed robust signal even at the lowest amount of protein tested, 1 ng (Fig. 5A). The XPA protein was readily detected in our assay as well (Fig. 5B), however the maximum signal obtained from XPA was significantly less than that obtained from RPA. There are several reasons for the decrease in signal obtained for the XPA–DNA interaction, including the specificities of the antibodies used and the affinity of the protein-DNA complexes. The affinity of XPA for ssDNA is on the order of ∼100-fold weaker than that of RPA (23, 44), which would lead to an overall decrease in the ability of XPA to interact with the ssDNA as well as increase the potential for dissociation of XPA molecules that are bound to the ssDNA during successive wash cycles. Nonetheless, we are able to detect as low as ∼10 ng of XPA binding to the 60-mer ssDNA substrate.
FIG. 5.
Detection of purified RPA and XPA on ssDNA substrate in ELISA assay. (A) Titration of purified RPA protein onto the biotin-60mer ssDNA substrate. (B) Titration of purified XPA protein onto the biotin-60mer ssDNA substrate. In both experiments, conversion of the chromagenic substrate was monitored over time (mAbs/min) and plotted vs. amount of protein added to the reaction.
Detection of purified RPA and XPA binding to platinated-dsDNA in an ELISA assay
Ultimately, detection of NER proteins in a biologically significant context will need to be performed in the presence of a double-stranded damaged DNA substrate. To this end, we designed two forms of cisplatin-damaged dsDNA substrates. The first is a synthetic oligonucleotide, which can be platinated at a single site (biotin-60mer), then annealed to an oligonucleotide of complementary sequence (60mer-complement). The second design involved global platination of the pBS+ plasmid DNA of ∼3200 base pairs, which was subsequently treated with a restriction endonuclease, ApaLI, to generate relatively large (∼400–1400 base pair) fragments upon which to assemble NER complexes. These plasmid fragments were biotinylated via fill-in synthesis of the resulting 5′ overhang for attachment to the streptavidin-coated 96-well plates. Biotin incorporation was achieved through the use of a biotinylated-dUTP nucleotide. We initially examined the ability to detect purified RPA and XPA proteins in the context of these damaged dsDNA substrates.
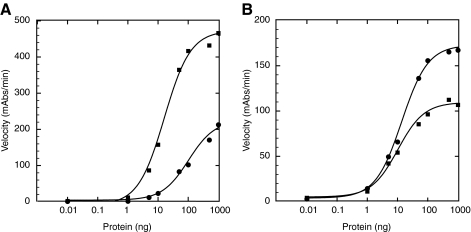
To assess damaged DNA bound, we first used the synthetic 60 base pair double-stranded oligonucleotide containing a single cisplatin lesion. In these experiments, 200 fmol of dsDNA was pre-bound to the streptavidin-coated 96-well plate, and increasing amounts of purified proteins were titrated and incubated with the immobilized damaged dsDNA substrates. RPA detection on this substrate was robust (Fig. 6A), however the decreased ability of RPA to interact with this substrate is apparent in the larger concentrations of RPA required to saturate binding and the increase in the amount of RPA required to reach 50% of maximum detectable binding. Approximately 10x more RPA is required to reach 50% maximum binding in the presence of the 60 base pair damaged dsDNA substrate relative to the 60-nucleotide ssDNA substrate (compare Figs. 6A and 5A). Similarly, we examined the detection limit of RPA binding to larger stretches of dsDNA that had been globally platinated at multiple sites. 50 ng of globally platinated dsDNA was immobilized on streptavidin-coated 96-well plates, and increasing amounts of purified RPA protein was incubated with the DNA. In the presence of multiple platinated sites and larger stretches of DNA, saturated binding was observed with 1000 ng of purified RPA and the detection limit was reduced to 1 ng of RPA (Fig. 6A).
FIG. 6.
Detection of purified RPA and XPA on dsDNA in ELISA assay. (A) Titration of purified RPA protein onto platinated, dsDNA substrates of 60 base pairs (circles) or plasmid fragments ∼400–1400 base pairs in length (squares). (B) Titration of purified XPA protein onto platinated, dsDNA substrates of 60 base pairs (circles) or plasmid fragments ∼400–1400 base pairs in length (squares).
Detection of purified XPA was also examined using the two different damaged-dsDNA (single- and multiple-site platinum lesions) substrates. XPA binding to these substrates demonstrated that as little as ∼10–20 ng of purified protein is sufficient for detection in the ELISA (Fig. 6B). Interestingly, detection of XPA binding to either substrate was very similar in saturation of binding as well as in the minimum amount of protein required. Next, we examined the ability to detect RPA and XPA binding to a platinum-damaged dsDNA substrate when the purified proteins are mixed in the same reaction. Purified RPA and XPA were mixed in equal amounts, then incubated with the 60 base pair, platinated dsDNA, to determine if the RPA–XPA interaction interferes with detection of either protein. Under these conditions, both proteins were detected in the ELISA assay, requiring ∼10 ng of XPA and ∼30–40 ng of RPA (Fig. 7). The increased amount of RPA required is not surprising given that RPA is ∼3x the size of XPA and the RPA–XPA interaction forms a 1:1 complex (33, 56). The results of these experiments suggest that the antibodies chosen for these studies do not recognize epitopes in the protein–protein interface, and will be suitable for further investigation of the NER incision complex formation.
FIG. 7.
Detection of purified RPA–XPA complex on dsDNA in ELISA assay. RPA and XPA were pre-mixed with equal amounts of protein. The protein complex was then titrated onto 60 base pair platinated dsDNA and detected with both the RPA (circles) and XPA (squares) antibodies separately.
Detection of NER proteins from cellular extracts binding to platinated-dsDNA in an ELISA assay
While detection of purified proteins in our ELISA-based assay is critical in determining our detection limits under ideal conditions, an important extension is the ability to detect RPA and XPA, as early indicators of NER complex formation, from WCEs. Extracts were prepared from HeLa cells and total protein levels were determined by the Bradford method. In these experiments, 50 ng of globally platinated, biotinylated dsDNA was pre-bound to a streptavidin-coated 96-well plate, and WCEs were added at 100, 10, 1, and 0 μg total protein. Due to the lower detectable signal that was observed for XPA from WCEs (data not shown), a different primary and secondary antibody combination was used to detect XPA in these experiments (see Fig. 3). While maximum signal from RPA was considerably higher than that for XPA, both proteins were readily detected with greater than 2-fold signal:noise ratio using 100 μg of total WCE protein (Fig. 8). RPA and XPA detection from WCEs further demonstrates the feasibility of detecting each protein of the NER incision complex assembly on platinum-damaged dsDNA in an ELISA format.
FIG. 8.
Detection of RPA and XPA from cell extracts on dsDNA in ELISA assay. Whole cell extracts prepared from HeLa cells were incubated with large fragments of platinated dsDNA. RPA (top graph) and XPA (bottom graph) were detected separately from the total amount of protein (μg) that was incubated in the ELISA assay. Note that the monoclonal antibody for XPA was used in these experiments (see Fig. 3).
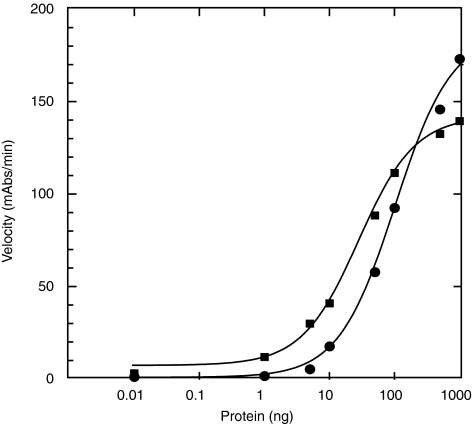
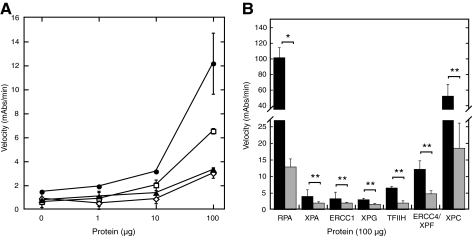
Using the same WCEs, we also examined detection of the additional NER incision complex proteins, as detection of each of these proteins is essential and would suggest formation of incision complexes on our surrogate damaged DNA substrates. In addition to RPA and XPA, we examined the ability to detect TFIIH, ERCC1, XPF/ERCC4 and XPG. Detection of each of these proteins was successful in our ELISA assay with at least 3-fold signal:noise from 100 μg of WCE total protein (Fig. 9A). Detection of ERCC1, XPG, TFIIH, and XPF/ERCC4 is consistent with complete formation of incision complexes under our assay conditions.
FIG. 9.
Detection of other NER pre-incision complex proteins and platinum preference for binding to dsDNA. (A) Detection of ERCC4/XPF, TFIIH, XPG, and ERCC1 from WCEs. Extracts were prepared from HeLa cells and the indicated amount of total protein was incubated with large fragments of platinated or nonplatinated dsDNA. (∙) ERCC4/XPF, (□) TFIIH, (▴) XPG, and (◊) ERCC1. (B) Detection of platinum preference for NER proteins: RPA, XPA, ERCC1, XPG, TFIIH, ERCC4/XPF, and XPC. Binding to platinated dsDNA (black) and non-platinated dsDNA (gray). *p = 0.01, **p > 0.05.
Damaged DNA specificity of NER proteins
In addition to being able to detect the presence or absence of a particular NER protein, it is also necessary to establish a preference for these proteins to associate with damaged dsDNA compared to undamaged dsDNA. To do this, we performed experiments using WCEs and globally platinated or unplatinated biotinylated dsDNA. The undamaged dsDNA was generated following the same protocols as for the globally platinated DNA, simply leaving out the initial platination procedure. We also examined detection of XPC, the damage recognition protein in these assays. As Figure 9B shows, in all cases, the NER proteins showed a higher association with platinated DNA compared to unplatinated DNA, however, the preference observed for ERCC1 and XPG was less than 2-fold. Clearly, further optimization of the assay is thus required, though it is interesting that both are nucleases with structure-specific affinity for DNA (34).
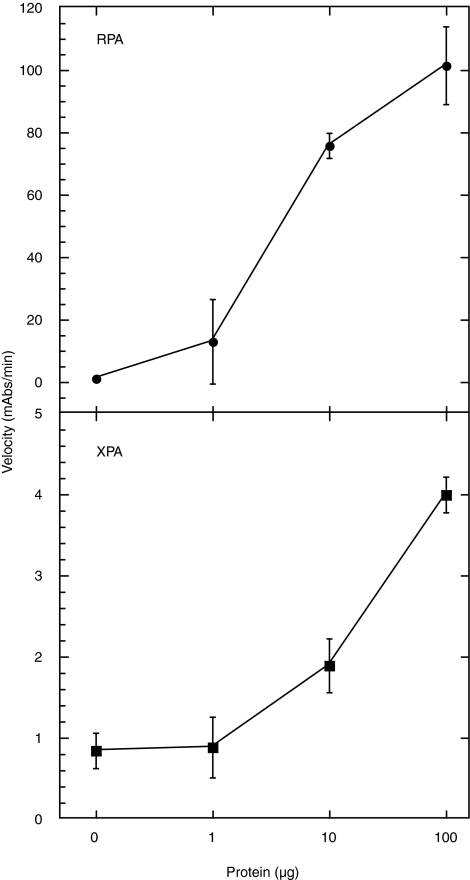
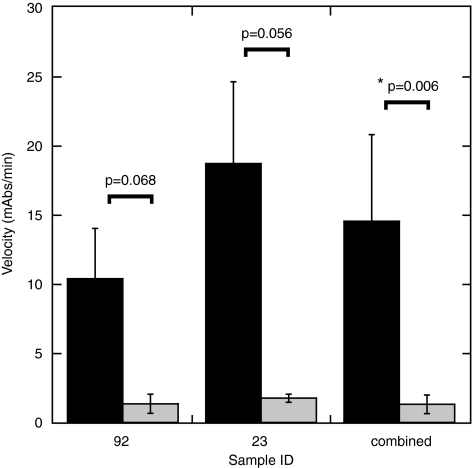
We obtained two patient biopsies from the Indiana University Lilly tissue bank to test our detection limits of the RPA protein from a biological source. The biopsies were obtained from normal lung tissue (sample 92) and from a neuroendocrine tumor (sample 23). The experiments were performed from two separate extractions from the original biopsy. The biopsies tested were dissected into two parts of approximately equal mass and treated independently as described in the methods. Each extraction yielded enough total protein for ∼5–10 independent analyses using our ELISA assay. Tissue extracts were incubated in the presence of globally platinated or unplatinated biotinylated dsDNA in the same manner as was done for the WCE detection of the NER proteins. We were able to detect RPA from 100 μg of total protein extract with greater than 5-fold specificity for the platinated dsDNA substrate (Fig. 10). This represents a significant advance towards implementing our assay in the detection of NER activity from patient tumor samples.
FIG. 10.
Detection of RPA from whole tissue extracts. Extracts from sample numbers 92 (normal lung tissue), 23 (Neuorendocrine carcinoma) were prepared and incubated with 50 ng of plasmid-digested fragments of platinated (black) or nonplatinated (gray) dsDNA. The combined data from the two samples is also presented. The * indicates P < 0.01 and a statistically significant difference. For the individual analyses the statistical assessment did not reach the threshold for a significant difference; however, the power of the t-test was below the desired power of 0.800 (0.476 for sample 92 and 0.550 for sample 23). Less than desired power indicates a lower chance of detecting a difference when one actually exists, and negative results should be interpreted cautiously.
Discussion
Defects in DNA repair capacity, specifically those associated with the NER pathway, have been linked to an increase in cancer susceptibility and chemotherapeutic response in lung, esophageal, cervical, and ovarian cancers. Despite this link, practical assays for measuring NER activity have been technically challenging, prohibiting their application in a clinical setting. As a result, there is continuing need to be able to measure, quantitatively, an individual's DNA repair capacity. More precisely, assessment of the DNA repair capacity or functionality of the NER machinery of an individual tumor would assist physicians in predicting the outcome of certain chemotherapeutic treatments administered to their patients. This type of personalized medicine is gaining popularity for cancer treatment and therapies. We recognize the inherent differences between individual's responses to chemotherapeutics, and that not all tumors or cancers (even of the same type) can be treated in the same way. In addition, a more personalized approach to cancer therapy could lead to overall more effective treatment of cancers.
Currently available clinical assays to detect defects in DNA repair capacity include: DNA sequencing of SNPs, HCR assays, and comet assays (24, 27). These methods have limitations, and none directly measures the ability of an individual patient's cancer cells to repair specific adducts or lesions caused by chemotherapeutic regimens. DNA sequencing provides information based on known mutations or defects, however, it does not provide information about gene expression, or protein levels and functionality. The HCR assay examines peripheral blood lymphocytes collected from patients, which are then cultured and treated with the DNA damaging agent of choice (27). The effect of the damaging agent is subsequently determined by viability screening. This assay assumes that the DNA repair capacity of these cells is identical or similar to that of the patient's cancerous cells, which have been transformed at some level through mutation or up/downregulation of protein(s) or pathway(s). Thus, normal healthy cells are not necessarily the best surrogates or models for cancer cells. This is especially true in the case of acquired resistance that develops following treatment. Comet assays provide a quantitative assessment of the amount of strand breakage induced by a treatment or how much remains after a given time period, allowing for repair of the damage; however, they are limited to strand breakage and are not effective in assessment of BER, NER, or MMR (41). Maintenance of cells and the specialized equipment required for analysis of these assays prevents routine use of these assays in the clinical setting for screening of patients for therapeutic options.
In an effort to develop a reproducible, quantitative, and practical clinical assay to monitor DNA damage repair by the NER pathway, we have developed an ELISA-based assay to detect the formation of NER incision complexes on damaged DNA substrates. This work demonstrates an ability to use an ELISA-based assay to detect all of the required proteins for in vitro incision of a platinum adduct, specifically, RPA, XPA, TFIIH, XPF/ERCC4, ERCC1, and XPG, as well as the initial damage recognition protein, XPC. We have demonstrated the ability to detect these proteins from both purified protein sources and 100 μg of total protein from WCEs of HeLa cells, which are proficient in NER, using specific antibodies. Additionally, we have shown detection of the RPA component of NER from patient samples with similar amounts of total protein. While statistical significance was demonstrated in a portion of the data presented, further optimization and larger sample sizes will result in more statistically significant differences observed between detection of pre-incision complex formation as well as detection of the downstream incision event on damaged versus undamaged substrates.
RPA participates in many cellular processes, including DNA replication, repair, and recombination (58). Because of this, RPA is the most abundant single-stranded DNA binding protein in eukaryotic cells, with ∼500,000 molecules of RPA present in HeLa cells (49). Detection of RPA in our ELISA assay was very robust, while detection of XPA results in significantly lower signals (Figs. 8 and 9). This observation is consistent with the lower affinity of XPA for damaged DNA substrates relative to that of RPA, and the observation that XPA protein levels are about half that of RPA in HeLa cells, and even lower in testicular cells (3). While further optimization is required for platinum-damaged specific binding of XPF and XPG, the fact that these are structure-specific nucleases associated with NER suggests that the signal we are detecting is a reflection of fully formed pre-incision complexes. However, further assessment of antibody pairs may alleviate the lower sensitivity, and more stringent assay conditions may ensure that cleavage by these nucleases is not occurring in the detection phase of the assay. If cleavage occurs, we would anticipate dissociation of the pre-incision complex, thus lowering our detection of NER proteins bound to DNA.
The use of ATP or slowly hydrolysable analogs holds the potential to extend the utility of the assay. Research has demonstrated that ATP is required for complete coordinated assembly of the pre-incision complex, particularly the unwinding of the duplex driven by the helicase activities of the TFIIH complex to allow dual incision (16). Future experiments therefore will include optimization of ATP addition and ATP-analogue addition to the reagents used in the ELISA assay. Another important consideration with this type of assay is the use of appropriate controls. In the results presented, assays were controlled with the comparison of binding to cisplatin-damaged vs. undamaged DNA substrates. At this time, this is the most appropriate control because, as we have previously discussed, the level of NER activity in other tissues or from blood samples may have inherently different innate levels of NER activity, yet the mechanism of NER is specific to repair of damaged DNA. For analysis of relative activity, the inclusion of a positive control on which to base maximal activity will be employed. This will allow the assessment of activity in different laboratories under different conditions to be compared and ultimately determine the usefulness in a variety of settings.
Ultimately, the success of this assay in the clinic will be demonstrated by the ability to directly measure incision of the DNA damage by the NER incision complex. While progress has been made in the development of this assay, optimization of our methods to improve the signal:noise and specificity for damaged DNA is paramount. The amount of protein required may be reduced with further optimization of assay conditions or platforms, protein extraction procedures and reagents used, especially the antibodies used for protein detection and generation of damaged DNA substrates. Currently, our efforts in developing a clinical assay have centered on the NER pathway, and repair of cisplatin lesions as cisplatin is the first line treatment in NSCLC; however, the ideas and concepts of this assay could potentially be expanded to examine the repair of lesions caused by other DNA damaging agents used in the treatment of cancers. Furthermore, while this method focuses on defects in NER, a future adaptation of this assay could examine deficiencies in other DNA repair pathways such as BER, MMR, and NHEJ. In the absence of being able to monitor direct incision, future studies will also examine any correlations to the levels of specific proteins detected and the sensitivity of patient samples to cisplatin treatment. At this time, we are unable to correlate our results of RPA levels and the sensitivity of the patient samples, as future experiments are underway with the two tissue samples discussed in this article, and revealing the sensitivity of the patient at this time would be premature.
While the initial assay development has been centered on a solid-phase ELISA-based assay, efforts are concurrently underway to expand into new technological frontiers. Microfluidics technologies would improve upon our current assay allowing for nano-liter sample volumes, significantly overcoming obstacles to the initial sample size from which we generate extracts and the total amount of reagents used. In addition, xMAP technology, a solution/bead-based technology, which uses flow cytometry-based fluorescence detection coupled with ‘labeled’ beads (with a ligand of choice), could be employed to improve our signals and detection limits (14). Beads can be modified to display nucleic acids, antibodies, or proteins, and each bead type can be fractionated or sorted via flow cytometry. These newer technologies and improvements to our current methodologies should launch this work into a viable clinical assay for the benefit of cancer patients.
Abbreviations Used
- AT
ataxia telangiectasia
- BER
base excision repair
- BSA
bovine serum albumin
- cisplatin
cisdiamminedichloroplatinum(II)
- CS
Cockayne syndrome
- DSB
double-strand break
- dsDNA
double-stranded DNA
- EDTA
disodium ethylenediamine tetraacetate
- ELISA
enzyme-linked imunosorbent assay
- GG-NER
global-genomic nucleotide excision repair
- HCR
host cell reactivation
- HNPCC
hereditary nonpolyposis colorectal cancer
- HRP
horseradish peroxidase
- MgOAc
magnesium acetate
- MMR
mismatch-mediated repair
- NER
nucleotide excision repair
- NHEJ
nonhomologous end-joining
- NSCLC
non-small cell lung cancer
- PMSF
phenylmethylsulfonyl fluoride
- RNAP II
RNA polymerase II
- ROS
reactive oxygen species
- SNPs
short nucleotide polymorphisms
- ssDNA
single-stranded DNA
- TC-NER
transcription-coupled nucleotide excision repair
- TTD
trichothiodystrophy
- WCE
whole cell extract
- XP
xeroderma pigmentosum
Acknowledgments
We thank Dr. Shadia Jalal for help with generating tumor extracts of proteins and the members of the Turchi Lab for their helpful discussions. This research was funded by NIH awards CA128628 and CA082741.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Al–Tassan N. Chmiel NH. Maynard J. Fleming N. Livingston AL. Williams GT. Hodges AK. Davies DR. David SS. Sampson JR. Cheadle JP. Inherited variants of MYH associated with somatic G:C–>T:A mutations in colorectal tumors. Nat Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 2.Alberts B. Redefining cancer research. Science. 2009;325:1319. doi: 10.1126/science.1181224. [DOI] [PubMed] [Google Scholar]
- 3.Araujo SJ. Nigg EA. Wood RD. Strong functional interactions of TFIIH with XPC and XPG in human DNA nucleotide excision repair, without a preassembled repairosome. Mol Cell Biol. 2001;21:2281–2291. doi: 10.1128/MCB.21.7.2281-2291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Araujo SJ. Tirode F. Coin F. Pospiech H. Syvaoja JE. Stucki M. Hubscher U. Egly JM. Wood RD. Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes Devel. 2000;14:349–359. [PMC free article] [PubMed] [Google Scholar]
- 5.Batty DP. Wood RD. Damage recognition in nucleotide excision repair of DNA. Gene. 2000;241:193–204. doi: 10.1016/s0378-1119(99)00489-8. [DOI] [PubMed] [Google Scholar]
- 6.Berek JS. Bertelsen K. du Bois A. Brady MF. Carmichael J. Eisenhauer EA. Gore M. Grenman S. Hamilton TC. Hansen SW. Harper PG. Horvath G. Kaye SB. Luck HJ. Lund B. McGuire WP. Neijt JP. Ozols RF. Parmar MKB. Piccart–Gebhart MJ. van Rijswijk R. Rosenberg P. Rustin GJS. Sessa C. Thigpen JT. Trope C. Tuxen MK. Vergote I. Vermorken JB. Willemse PHB. Advanced epithelial ovarian cancer: 1998 consensus statements. Ann Oncol. 1999;10:87–92. doi: 10.1023/a:1008323922057. [DOI] [PubMed] [Google Scholar]
- 7.Beskow C. Skikuniene J. Holgersson A. Nilsson B. Lewensohn R. Kanter L. Viktorsson K. Radioresistant cervical cancer shows upregulation of the NHEJ proteins DNA-PKcs, Ku70 and Ku86. Br J Cancer. 2009;101:816–821. doi: 10.1038/sj.bjc.6605201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biswas EE. Zhu FX. Biswas SB. Stimulation of RTH1 nuclease of the yeast Saccharomyces cerevisiae by replication protein A. Biochemistry. 1997;36:5955–5962. doi: 10.1021/bi962890u. [DOI] [PubMed] [Google Scholar]
- 9.Bradbury PA. Kulke MH. Heist RS. Zhou W. Ma C. Xu W. Marshall AL. Zhai R. Hooshmand SM. Asomaning K. Su L. Shepherd FA. Lynch TJ. Wain JC. Christiani DC. Liu G. Cisplatin pharmacogenetics, DNA repair polymorphisms, and esophageal cancer outcomes. Pharmacogenet Genomics. 2009;19:613–625. doi: 10.1097/FPC.0b013e32832f3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camenisch U. Dip R. Schumacher SB. Schuler B. Naegeli H. Recognition of helical kinks by xeroderma pigmentosum group A protein triggers DNA excision repair. Nat Struct Mol Biol. 2006;13:278–284. doi: 10.1038/nsmb1061. [DOI] [PubMed] [Google Scholar]
- 11.Cleaver J. Defective repair replication of DNA in xeroderma pigmentosum. Nature. 1968;218:652–656. doi: 10.1038/218652a0. [DOI] [PubMed] [Google Scholar]
- 12.Coin F. Oksenych V. Egly JM. Distinct roles for the XPB/p52 and XPD/p44 subcomplexes of TFIIH in damaged DNA opening during nucleotide excision repair. Mol Cell. 2007;26:245–256. doi: 10.1016/j.molcel.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 13.Culotta E. Koshland DE. Molecule of the Year—DNA repair works its way to the top. Science. 1994;266:1926–1929. doi: 10.1126/science.7801115. [DOI] [PubMed] [Google Scholar]
- 14.Dunbar SA. Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection. Clin Chim Acta. 2006;363:71–82. doi: 10.1016/j.cccn.2005.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Einhorn LH. Curing metastatic testicular cancer. Proc Natl Acad Sci USA. 2002;99:4592–4595. doi: 10.1073/pnas.072067999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans E. Moggs JG. Hwang JR. Egly J. Wood RD. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 1997;35:2157–2167. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferry KV. Hamilton TC. Johnson SW. Increased nucleotide excision repair in cisplatin-resistant ovarian cancer cells. Role of ERCC1-XPF. Biochem Pharmacol. 2000;60:1305–1313. doi: 10.1016/s0006-2952(00)00441-x. [DOI] [PubMed] [Google Scholar]
- 18.Fousteri M. Mullenders LH. Transcription-coupled nucleotide excision repair in mammalian cells: Molecular mechanisms and biological effects. Cell Res. 2008;18:73–84. doi: 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- 19.Hansson J. Wood RD. Repair synthesis by human cell extracts in DNA damaged by cis- and trans-diamminedichloroplatinum(II) Nucleic Acids Res. 1989;17:8073–8091. doi: 10.1093/nar/17.20.8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henricksen LA. Umbricht CB. Wold MS. Recombinant replication protein A: Expression, complex formation, and functional characterization [published erratum in J Biol Chem 1994 Jun 10;269:16519] J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 21.Hermanson IL. Turchi JJ. Overexpression and purification of human XPA using a Baculovirus expression system. Protein Expression Purif. 2000;19:1–11. doi: 10.1006/prep.2000.1224. [DOI] [PubMed] [Google Scholar]
- 22.Hess MT. Schwitter U. Petretta M. Giese B. Naegeli H. Bipartite substrate discrimination by human nucleotide excision repair. Proc Natl Acad Sci USA. 1997;94:6664–6669. doi: 10.1073/pnas.94.13.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hey T. Lipps G. Krauss G. Binding of XPA and RPA to damaged DNA investigated by fluorescence anisotropy. Biochemistry. 2001;40:2901–2910. doi: 10.1021/bi002166i. [DOI] [PubMed] [Google Scholar]
- 24.Himmelstein MW. Boogaard PJ. Cadet J. Farmer PB. Kim JH. Martin EA. Persaud R. Shuker DE. Creating context for the use of DNA adduct data in cancer risk assessment: II. Overview of methods of identification and quantitation of DNA damage. Crit Rev Toxicol. 2009;39:679–694. doi: 10.1080/10408440903164163. [DOI] [PubMed] [Google Scholar]
- 25.Kantor G. Barsalou L. Hanawalt P. Selective repair of specific chromatin domains in UV-irradiated cells from xeroderma pigmentosum complementation group C. Mutat Res. 1990;235:171–180. doi: 10.1016/0921-8777(90)90071-c. [DOI] [PubMed] [Google Scholar]
- 26.Koberle B. Masters JRW. Hartley JA. Wood RD. Defective repair of cisplatin-induced DNA damage caused by reduced XPA protein in testicular germ cell tumours. Curr Biol. 1999;9:273–276. doi: 10.1016/s0960-9822(99)80118-3. [DOI] [PubMed] [Google Scholar]
- 27.Li C. Wang LE. Wei Q. DNA repair phenotype and cancer susceptibility—A mini review. Int J Cancer. 2009;124:999–1007. doi: 10.1002/ijc.24126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 29.Liao W. McNutt MA. Zhu WG. The comet assay: A sensitive method for detecting DNA damage in individual cells. Methods. 2009;48:46–53. doi: 10.1016/j.ymeth.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 30.Lieber MR. The mechanism of double-strand DNA breakrepair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin SA. Lord CJ. Ashworth A. DNA repair deficiency as a therapeutic target in cancer. Curr Opin Genet Dev. 2008;18:80–86. doi: 10.1016/j.gde.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 32.Matakidou A. Eisen T. Fleischmann C. Bridle H. Houlston RS. Evaluation of xeroderma pigmentosum XPA, XPC, XPD, XPF, XPB, XPG and DDB2 genes in familial early-onset lung cancer predisposition. Int J Cancer. 2006;119:964–967. doi: 10.1002/ijc.21931. [DOI] [PubMed] [Google Scholar]
- 33.Matsuda T. Saijo M. Kuraoka I. Kobayashi T. Nakatsu Y. Nagai A. Enjoji T. Masutani C. Sugasawa K. Hanaoka F. DNA repair protein XPA binds replication protein A (RPA) J Biol Chem. 1995;270:4152–4157. doi: 10.1074/jbc.270.8.4152. [DOI] [PubMed] [Google Scholar]
- 34.Matsunaga T. Park CH. Bessho T. Mu D. Sancar A. Replication protein A confers structure-specific endonuclease activities to the XPF-ERCC1 and XPG subunits of human DNA repair excision nuclease. J Biol Chem. 1996;271:11047–11050. doi: 10.1074/jbc.271.19.11047. [DOI] [PubMed] [Google Scholar]
- 35.Modrich P. Mismatch repair, genetic stability, and cancer. Science. 1994;266:1959–1960. doi: 10.1126/science.7801122. [DOI] [PubMed] [Google Scholar]
- 36.Mol CD. Parikh SS. Putnam CD. Lo TP. Tainer JA. DNA repair mechanisms for the recognition and removal of damaged DNA bases. Annu Rev Biophys Biomol Struc. 1999;28:101–128. doi: 10.1146/annurev.biophys.28.1.101. [DOI] [PubMed] [Google Scholar]
- 37.Mu D. Sancar A. Model for XPC-independent transcription-coupled repair of pyrimidine dimers in humans. J Biol Chem. 1997;272:7570–7573. doi: 10.1074/jbc.272.12.7570. [DOI] [PubMed] [Google Scholar]
- 38.Olaussen KA. Dunant A. Fouret P. Brambilla E. Andre F. Haddad V. Taranchon E. Filipits M. Pirker R. Popper HH. Stahel R. Sabatier L. Pignon J. Tursz T. Le Chevalier T. Soria J. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355:983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 39.Olaussen KA. Mountzios G. Soria JC. ERCC1 as a risk stratifier in platinum-based chemotherapy for nonsmall-cell lung cancer. Curr Opin Pulm Med. 2007;13:284–289. doi: 10.1097/MCP.0b013e32816b5c63. [DOI] [PubMed] [Google Scholar]
- 40.Olaussen KA. Soria JC. Validation of ERCC1-XPF immunodetection—Letter. Cancer Res. 2010;70:3851–3852. doi: 10.1158/0008-5472.CAN-09-4352. [DOI] [PubMed] [Google Scholar]
- 41.Olive PL. The comet assay. An overview of techniques. Methods Mol Biol. 2002;203:179–194. doi: 10.1385/1-59259-179-5:179. [DOI] [PubMed] [Google Scholar]
- 42.Patrick SM. Turchi JJ. Replication protein A (RPA) binding to duplex cisplatin-damaged DNA is mediated through the generation of single-stranded DNA. J Biol Chem. 1999;274:14972–14978. doi: 10.1074/jbc.274.21.14972. [DOI] [PubMed] [Google Scholar]
- 43.Patrick SM. Turchi JJ. Stopped-flow kinetic analysis of replication protein A-binding DNA. Damage recognition and affinity for single-stranded DNA reveal differential contributions of k(on) and k(off) rate constants. J Biol Chem. 2001;276:22630–22637. doi: 10.1074/jbc.M010314200. [DOI] [PubMed] [Google Scholar]
- 44.Patrick SM. Turchi JJ. Xeroderma pigmentosum complementation group A protein (XPA) modulates RPA-DNA interactions via enhanced complex stability and inhibition of strand separation activity. J Biol Chem. 2002;277:16096–16101. doi: 10.1074/jbc.M200816200. [DOI] [PubMed] [Google Scholar]
- 45.Payne A. Chu G. Xeroderma pigmentosum group E binding factor recognizes a broad spectrum of DNA damage. Mutat Res. 1994;310:89–102. doi: 10.1016/0027-5107(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 46.Robertson AB. Klungland A. Rognes T. Leiros I. DNA repair in mammalian cells: Base excision repair: The long and short of it. Cell Mol Life Sci. 2009;66:981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarasin A. Stary A. New insights for understanding the transcription-coupled repair pathway. DNA Repair (Amst) 2007;6:265–269. doi: 10.1016/j.dnarep.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 48.Selvakumaran M. Pisarcik DA. Bao R. Yeung AT. Hamilton TC. Enhanced cisplatin cytotoxicity by disturbing the nucleotide excision repair pathway in ovarian cancer cell lines. Cancer Res. 2003;63:1311–1316. [PubMed] [Google Scholar]
- 49.Seroussi E. Lavi S. Replication protein A is the major single-stranded DNA binding protein detected in mammalian cell extracts by gel retardation assays and UV cross-linking of long and short single-stranded DNA molecules. J Biol Chem. 1993;268:7147–7154. [PubMed] [Google Scholar]
- 50.Seve P. Dumontet C. Chemoresistance in non-small cell lung cancer. Curr Med Chem Anti-Canc Agents. 2005;5:73–88. doi: 10.2174/1568011053352604. [DOI] [PubMed] [Google Scholar]
- 51.Shiloh Y. ATM and related protein kinases: Safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 52.Someya M. Sakata K. Matsumoto Y. Yamamoto H. Monobe M. Ikeda H. Ando K. Hosoi Y. Suzuki N. Hareyama M. The association of DNA-dependent protein kinase activity with chromosomal instability and risk of cancer. Carcinogenesis. 2006;27:117–122. doi: 10.1093/carcin/bgi175. [DOI] [PubMed] [Google Scholar]
- 53.Staresincic L. Fagbemi AF. Enzlin JH. Gourdin AM. Wijgers N. Dunand-Sauthier I. Giglia–Mari G. Clarkson SG. Vermeulen W. Scharer OD. Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 2009;28:1111–1120. doi: 10.1038/emboj.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takedachi A. Saijo M. Tanaka K. The DDB2 complex-mediated ubiquitylation around DNA damage is oppositely regulated by XPC and Ku, and contributes to the recruitment of XPA. Mol Cell Biol. 2010;30:2708–2723. doi: 10.1128/MCB.01460-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thompson LH. Hinz JM. Cellular and molecular consequences of defective Fanconi anemia proteins in replication-coupled DNA repair: Mechanistic insights. Mutat Res. 2009;668:54–72. doi: 10.1016/j.mrfmmm.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang M. Mahrenholz A. Lee SH. RPA stabilizes the XPA-damaged DNA complex through protein–protein interaction. Biochemistry. 2000;39:6433–6439. doi: 10.1021/bi000472q. [DOI] [PubMed] [Google Scholar]
- 57.Weissman L. de Souza–Pinto NC. Stevnsner T. Bohr VA. DNA repair, mitochondria, and neurodegeneration. Neuroscience. 2007;145:1318–1329. doi: 10.1016/j.neuroscience.2006.08.061. [DOI] [PubMed] [Google Scholar]
- 58.Wold MS. Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. [Review] [190 refs] Ann Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 59.Wood RD. DNA damage recognition during nucleotide excision repair in mammalian cells. Biochimie. 1999;81:39–44. doi: 10.1016/s0300-9084(99)80036-4. [DOI] [PubMed] [Google Scholar]
- 60.Wood RD. Robins P. Lindahl T. Complementation of the xeroderma pigmentosum DNA repair defect in cell-free extracts. Cell. 1988;53:97–106. doi: 10.1016/0092-8674(88)90491-6. [DOI] [PubMed] [Google Scholar]
- 61.Wu XM. Fan W. Xu SW. Zhou YK. Sensitization to the cytotoxicity of cisplatin by transfection with nucleotide excision repair gene xeroderma pigmentosun group a antisense RNA in human lung adenocarcinoma cells. Clin Cancer Res. 2003;9:5874–5879. [PubMed] [Google Scholar]