Abstract
Objective
To examine five available software packages for the assessment of abdominal adipose tissue with magnetic resonance imaging, compare their features and assess the reliability of measurement results.
Design
Feature evaluation and test–retest reliability of softwares (NIHImage, SliceOmatic, Analyze, HippoFat and EasyVision) used in manual, semi-automated or automated segmentation of abdominal adipose tissue.
Subjects
A random sample of 15 obese adults with type 2 diabetes.
Measurements
Axial T1-weighted spin echo images centered at vertebral bodies of L2–L3 were acquired at 1.5 T. Five software packages were evaluated (NIHImage, SliceOmatic, Analyze, HippoFat and EasyVision), comparing manual, semi-automated and automated segmentation approaches. Images were segmented into cross-sectional area (CSA), and the areas of visceral (VAT) and subcutaneous adipose tissue (SAT). Ease of learning and use and the design of the graphical user interface (GUI) were rated. Intra-observer accuracy and agreement between the software packages were calculated using intra-class correlation. Intra-class correlation coefficient was used to obtain test–retest reliability.
Results
Three of the five evaluated programs offered a semi-automated technique to segment the images based on histogram values or a user-defined threshold. One software package allowed manual delineation only. One fully automated program demonstrated the drawbacks of uncritical automated processing. The semi-automated approaches reduced variability and measurement error, and improved reproducibility. There was no significant difference in the intra-observer agreement in SAT and CSA. The VAT measurements showed significantly lower test–retest reliability. There were some differences between the software packages in qualitative aspects, such as user friendliness.
Conclusion
Four out of five packages provided essentially the same results with respect to the inter- and intra-rater reproducibility. Our results using SliceOmatic, Analyze or NIHImage were comparable and could be used interchangeably. Newly developed fully automated approaches should be compared to one of the examined software packages.
Keywords: magnetic resonance imaging, abdominal adipose tissue, software evaluation, image segmentation, visceral adipose tissue, subcutaneous adipose tissue
Introduction
Increased central (abdominal) adiposity, and in particular, increased visceral adipose tissue (VAT), is related to numerous metabolic aberrations (including insulin resistance, type 2 diabetes mellitus, hepatic steatosis, hypertension and hyperlipidemia).1–7 Many direct and indirect methods have been used to quantify body fat, such as anthropometric measurements8 (for example, waist circumference), underwater weighing,8,9 bioelectric impedance,10 dual-energy X-ray absorptiometry,11,12 computed tomography (CT)13–16 and magnetic resonance imaging (MRI). However, at present, most published research endorses either CT or MRI as the best in vivo methods for accurate and reliable data about the amount and distribution of abdominal adipose tissue. Of these, MRI has the advantage of not exposing the subject to ionizing radiation. This is especially useful in research projects that require serial whole-body measurements to examine the response to diet or exercise.7,17
MRI assessment of adipose tissue has been validated in phantoms,18,19 animals14,20,21 and human cadavers.22,23 Abdominal adipose tissue measurement by in vivo MRI has been shown to be in good agreement with values produced by dissection and chemical analysis.17,20,22,24 Whole-body MRI data make it possible to assess the influence of weight loss and other interventions on regional body composition.25–28
Identifying and segmenting MR images accurately, and measuring abdominal VAT, subcutaneous adipose tissue (SAT) and total adipose tissue (TAT) precisely are of key importance. In the literature, MRI data appear as either area values (cm2) obtained from a single image or multiple images, or as volume values (cm3) derived using the tissue area measurements from multiple images. Recent studies suggest using single-slice images in the upper abdomen (L1–L3) to minimize scanning and data analysis time and cost. The results show that single images are a good surrogate (P<0.01–<0.05 5,6,29–33) for volume measurements of VAT or TAT.
A wide variety of software packages and methodologies has been described in the published literature on MR body fat measurements.14,17,34–39 Most approaches are composed of two steps: in the first step, based on the histogram a threshold between adipose and lean tissues is determined manually or automatically and, in the second step, the areas of SAT and VAT are delineated. The segmentation routine can also include data pre-processing steps like de-noising and smoothing of the MR images to facilitate segmentation.40
Semi-automated approaches support the user by determining a histogram-based threshold,30,41–46 boundary enhancement,47 histogram-based region growing41,47 or clustering.40 Hybrid algorithms, which incorporate anatomical knowledge with segmentation algorithms,32,33,38,39 have been successfully used to segment high contrast, artifact-free images. Unfortunately, the intensity variation at tissue boundaries in abdominal MR images is gradual and not sharp causing poor differentiation of tissue interfaces. The complex visceral anatomy of the abdomen still requires substantial operator involvement for the delineation of VAT.
Recently, two fully automated approaches for the segmentation of abdominal adipose and lean tissues have been published.34–37 Positano et al.34–36 proposed a fuzzy c-means approach. An active contour algorithm was applied to identify and estimate the volumes of VAT and SAT. Liou et al.37 determined a threshold slice by slice based on the average signal intensity of the imaged slice. The segmentation consisted of a four-step procedure in which masks for the body contour, non-fat tissue, TAT, VAT and SAT were applied to determine the tissue boundaries. The results were visually examined by a physician and compared to a manual segmentation approach.
To our knowledge, no study has systematically determined the difference between software packages that are commonly used to measure abdominal adipose tissue. Being able to apply a fast, reliable and accurate routine to segment abdominal adipose tissue is a key element in obtaining reliable data. This study examines the software packages that were cited most frequently for the estimation of abdominal adipose tissue area. The goal is to compare and contrast their features and to compare the reproducibility and reliability of the quantitative measurement results. The focus is on the software packages; the study will not compare various MR methodologies (for example, extrapolation of single-slice versus multi-slice MRI acquisitions), MRI sequences or examine the influence of the measurement site (L1–L2, L2–L3, L4–L5, umbilicus), subsampling or the position of the volunteer with regard to the isocenter of the scanner during the data acquisition.
Methods
Subjects
A random sample of 15 obese adults with type 2 diabetes from the Fatty Liver Ancillary Study to the Action For Health in Diabetes (Look AHEAD) trial was selected. The sample consisted of 10 men and 5 women; 5 African-American (2 men) and 10 Caucasian (3 men) subjects, aged 53–76 years (mean age 65 years), body mass index (BMI) 25.8–45.5 kg/m2 (mean BMI 34.5 kg/m2), waist circumference 93.9–131 cm (mean 111cm). Look AHEAD is a multicenter randomized clinical trial to examine the effects of a lifestyle intervention designed to achieve and maintain weight loss over the long term on the incidence of cardiovascular disease (CVD) events and mortality. The design of the trial has been previously published.48 The single-center Fatty Liver Ancillary study looked at non-alcoholic fatty liver disease within this group of subjects. All volunteers provided written, informed consent to participate. Both the ancillary study and the Look AHEAD trials were approved by the Western Institutional Review Board.
MRI acquisition
Axial T1-weighted spin echo images (8 slices, 10 mm thickness, 1 mm interslice distance, repetition time/echo time = 150/4.6 ms, acquisition matrix 256 × 256, field of view 53.0cm × 53.0cm) centered at vertebral bodies of L2–L3 were acquired with a Philips (Philips Medical System Best, The Netherlands) Gyroscan NT Intera 1.5T scanner using the body coil with the patient in supine position. A breath–hold technique was applied to avoid breathing-induced artifacts. An example of the anatomic position of the axial slices on a mid-sagittal image is shown in Figure 1. The observers rated the quality of all MR images on a scale of 1–6 (6 being optimal signal-to-noise ratio, 1 being unreadable). The average MR image quality was 4.6 (range 3 to 6).
Figure 1.
Location of magnetic resonance scan slices to estimate abdominal fat. Eight slices were obtained.
Selection of software packages
Software packages were identified by using Internet search engines (for example, Google, Yahoo, AltaVista), as well as a review of the published MRI studies and reference sections. Keywords were used to limit the search to imaging software used in the evaluation of abdominal adipose tissue in MRI. In addition, MRI researchers were consulted to identify any additional MRI analysis software packages. The final selection was based on frequency of citation in the reviewed literature, availability and cost.
Features examined
First, we made note of the URL of each software package in our survey. The year each package was first released and the version used were included in our study as an index of the program’s stability, popularity and maturity. We noted whether a package is freeware, including in-house software that was made available upon request, shareware (software which is made available with a request to donate money to the authors) or commercially available. The software and operating system requirements for each software package were noted.
Next we evaluated each package on nine key features. The completeness of instructions for installation, download and set up was rated, and the installation medium was noted. The source for instructions and documentation for program package was observed. In our ratings, we differentiated between the completeness and the helpfulness of the user’s manual. We distinguished between the learning to use (including understanding how routines are performed) and operating the packages. Completeness of the software manual, the helpfulness of the manual, the ease of installation, the ease of learning how to use the software, using the software and the graphical user interface (GUI) design were rated on a five-point scale, where 1 was the most difficult or bad performance and 5 was very easy or very good performance. Two observers independently evaluated each package and the average rating for each feature was calculated. Ratings were also given for features tested and the MR image quality.
All packages are GUI based, and we rated the design and user friendliness of the GUI on a five-point scale. Image display features and image manipulation tools as well as the availability to store the segmented images and region of interest (ROI) measurements were documented. We examined segmentation routines and the average time an observer needed to segment 10 MR images was measured.
Finally, the intra- and inter-rater reproducibility of the cross-sectional area (CSA), SAT, VAT and TAT measurements were examined. The results obtained using the software packages were compared with each other.
Image analysis
The image data were transferred to personal computers in Digital Imaging and Communication in Medicine (DICOM) format. The platform for analysis depended upon the software package used. SliceOmatic, HippoFat, Analyze were installed on a personal computer equipped with an Intel Pentium 4 CPU, 2.4 GHz, 1 GB of RAM, the Philips EasyVision was installed on a Sparc Sun Ultra2 workstation and NIHImage was used on a Mac Powerbook G4, 1.5 GHz, 512 MB of RAM.
The three central slices out of eight acquired MR images were selected from each subject’s data set to define SAT, VAT, TAT and CSA. Given the high grayscale variation within the image regions representing fat tissue and the grayscale values of some other tissues like liver, the determination of an adequate threshold of the signal intensity was performed in each patient individually to distinguish between adipose and lean tissues on MR images. The images were then segmented into adipose and lean tissues. ROIs describing the whole CSA, the SAT and the VAT were created, and the respective areas were measured. CSA was defined as the sum of all pixels inside a line tracing the skin. Abdominal SAT area was defined as the area of adipose tissue between the skin and the outermost aspect of the abdominal muscle wall. All adipose tissue pixels within the intra-abdominal cavity at the innermost aspect of the abdominal and oblique wall musculature and the anterior aspect of the vertebral body were considered VAT. Retroperitoneal adipose tissue compartments were excluded from the VAT measurement, using anatomical points such as ascending and descending colon, aorta and inferior vena cava. For images in which the kidneys appeared, an oblique line was drawn from the aorta and inferior vena cava to the anterior border of the kidney and continued to the abdominal wall. High-intensity non-fat pixels arising from bone marrow, liver and fatty intestinal contents within the VAT were avoided or manually removed when possible. TAT was calculated by adding SAT and VAT (TAT = SAT+VAT); therefore, some small intramuscular deposits of adipose tissue were not accounted for. Similar to other published reports,5,7,18,22,25,29,33–37,40–45,47,49–63 VAT and SAT were not further subdivided.
Intra- and inter-observer reliability and reproducibility of tissue characterization
To assess the reproducibility of this procedure, images of all volunteers were analyzed by two observers independently. In addition, one observer performed the segmentations on two separate occasions 3 months apart. Both observers were blinded to the results obtained earlier. Next, all software packages were compared against the NIHImage software package, which was chosen as the standard to compare against because it is free and widely available. Each observer used and rated the software packages in the same, pre-set order.
Statistical analysis
A series of tests were conducted to assess the reliability of the five MRI software packages among the 15 selected subjects. Intra- and inter-rater reliability were conducted for each software package individually using the intra-class correlation coefficient (ICC).64 In the intra-observer analyses, two measurements of the primary rater were compared. In the inter-observer analyses, the average of the primary raters’ results were compared to a second rater. For the inter-test comparison, the average of the primary raters’ measurements were compared among each of the programs. For this analysis, one-way analysis of variance was also used to calculate ICC. The coefficient of variability (CV), another index of reliability, was also calculated to assess the variation between replicate measurements. A low CV (for example, <10%) corresponds to a high ICC, indicating good precision in the measurement of each respective software package.64
Finally, graphical assessment against a line of perfect agreement (for example 45° line) was used across all three series of analyses. All statistical analysis was performed using Stata version 8.2 (Stata Corp LP, College Station, TX, USA).
Results
Software selection
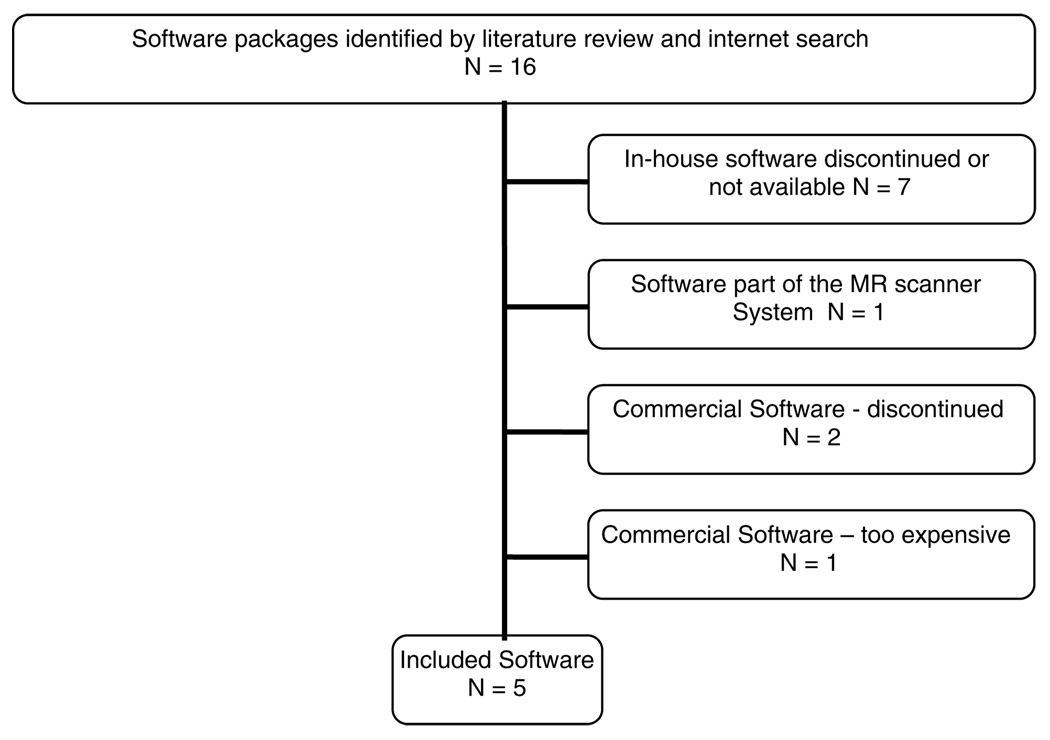
The selection process is reflected in Figure 2. The original search identified 16 programs. Seven in-house programs were excluded because they were not available2,7,18,33,53,65–69 or had been discontinued. One commercial program was not available to us because it was part of an MR scanner system’s software. Two cited commercial packages were outdated.70,71 Finally, one commercial package was too costly ($4800) and was outside the budget of this project. Thus, a total of five software packages were examined as part of this study.
Figure 2.
Identification and selection of software packages.
Features examined
Table 1 presents an overview of the evaluation of the five software packages included in this study. NIHImage was the first software package each observer used, followed by SliceOmatic, Analyze, HippoFat and the Philips EasyVision workstation.
Table 1.
Summary of features and ratings of each software package
| Features | NIHImage | SliceOmatic | Analyze | HippoFat | EasyVision |
|---|---|---|---|---|---|
| Address | http://rsb.info.nih.gov/nih-image/Default.html | http://www.tomovision.com | http://www.Analyzedirect.com/ | positano@ifc.cnr.it | http://www.medical.philips.com/main/products/itproductsolutions/ |
| First release of software | 1987 | 1990 | ∼25 years | October 2004 | Unknown (Version 4.4.1. 2000) |
| Availability | Freeware | Commercial (academic license $3600) | Commercial (academic license $1500) | In house | Philips’ scanner software |
| Platform/operating system | Mac Os 9/X | Windows | Windows, Mac Os, Linux, Unix | Windows, Linux, Unix | Sparc Sun Ultra2 |
| Installation | |||||
| Installation medium | FTP or HTTP download | CD+dongle | CD | File via email | Pre-installed |
| Ease of installation* | 4 | 3 | 5 | 4 | NA |
| Installation pre-requirements | None | None | None | IDL 6.1 virtual machine | None |
| Documentation/support | User’s manual/online FAQ/ mailing list | User’s manual/online help/ workshops | User’s manual/online help/ workshops | User’s manual/email support | User’s manual |
| Manual—completeness* | 2 | 5 | 5 | 2 | 2.5 |
| Manual—helpfulness* | 2 | 2 | 4 | 2 | 5 |
| Usage | |||||
| Ease of learning to use software* | 2 | 2.5 | 2.5 | 2 | 4 |
| Ease of using the software* | 3 | 4 | 3 | 1 | 4 |
| Graphical user interface* | 3 | 4.5 | 2.5 | 2 | 2.5 |
| Data input | Most medical image formats | Most medical image formats | Most medical image formats | DICOM | NA |
| Image display | |||||
| Image manipulation tools | Many | Many | Many options | Windowing, zooming | Windowing, zooming |
| Output of regions, sizes, result saving | Many options | Many options | Segmented image and results | Results | Segmented image |
| Statistical output | Yes | Yes | Yes | No | No |
| Segmentation/ROI routines | Many | Many | Many | Fuzzy c-means clustering | Manual drawing |
| Average time for measuring 10 slices (min) | 322.5 | 32 | 100 | 120 | 97.5 |
Abbreviations: DICOM, Digital Imaging and Communication in Medicine; NA, not applicable: feature was not tested; ROI, region of interest.
Numeric ratings were on a scale of 1–5; 5 indicated ease with the feature or very good performance, 1 indicated insufficient performance or difficulty with the feature.
Platform/operating system
Analyze and HippoFat support multiple platforms, NIHImage only runs on Mac OS 9.2. 2 or OsX and SliceOmatic solely runs on Windows. The 4.4.1.2 version of Philips EasyVision used in this study was installed on a Sparc Sun Ultra 2 workstation, whereas newer versions of the Philips software are Windows based. HippoFat was the only software that required the ITT Visual Information Solution, Boulder, Co, USA (IDL) Virtual Machine (http://www.rsinc.com/idlvm/index.asp). IDL is a higher level commercial programming package for data analysis, visualization and cross-platform application development.
Installation and documentation
Analyze, HippoFat, NIH-Image and SliceOmatic provide instructions for installing the software. There was some variability in the ease of installment for these programs, with SliceOmatic getting the lowest rating on this feature (3 of 5). Since the EasyVision workstation was pre-installed, no installation instructions are provided for this system.
All packages provide a user’s manual; the HippoFat manual was very short and concise. NIHImage also offers online discussion groups. NIHImage and SliceOmatic provide web sites that answer frequently asked questions. Analyze, HippoFat and SliceOmatic offer email support. The commercial packages Analyze and SliceOmatic also offer user training workshops for a fee.
Ease of learning and use
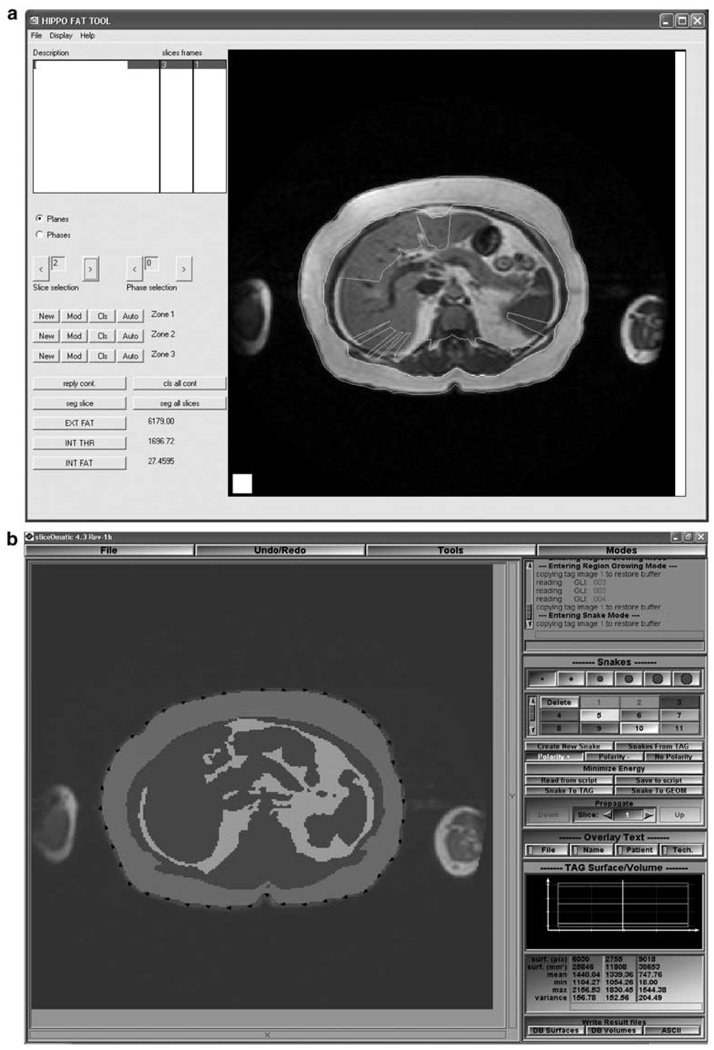
The ease of learning to use each software package ranged from 2 to 4, with EasyVision getting the highest rating. All other packages were considered harder to learn since they offered more options and had longer, more complex manuals. For ease of use, the scores ranged from 1 to 4. Both observers rated HippoFat the lowest because the fully automated segmentation did not provide acceptable results. Especially VAT and surrounding non-fat tissue were often misclassified. The subsequent manual correction of the segmentation was very tedious and time consuming. Finally, the GUI ratings ranged from 2 to 4. Examples of the highest and lowest rated GUI are shown in Figures 3a and b.
Figure 3.
Screenshots of (a) lowest rated graphical user interface (GUI) (HippoFat) and (b) highest rated GUI (SliceOmatic).
Input of data and image display
None of the software packages required pre-processing of the data to reduce noise levels. All packages could open and visualize the DICOM image data, except NIHImage, which did not recognize our specific data set and the data had to be imported in a ‘raw’ format. All packages displayed the DICOM header information as well as the image.
The software packages automatically picked a windowing and brightness that were appropriate for the data but these settings could also be manually modified. All program packages offered zooming, rotating and scaling of the image, except HippoFat, which had only limited options. NIH-Image, Analyze and SliceOmatic could save the measurements as either square centimeter or square millimeter as well as the segmented image in various formats. HippoFat only stored the measurement, while EasyVision could not be saved in an appropriate format (text file or segmented image) at all and the results had to be copied manually.
Segmentation routines
The segmentation procedure for each software package varies slightly. We tried to use a simple, fast and accurate approach for each package. NIHImage provides several segmentation approaches. We choose to separate SAT and VAT areas manually and then set a threshold value for separation of lean and fat tissues to the lowest point of bright pixels corresponding to adipose tissue. In the second step, ROIs were drawn using the tracing tool. The whole abdominal circumference and the outer margin of the abdominal cavity were determined to measure CSA and SAT. VAT was manually selected with the abdominal cavity. SliceOmatic is a package with numerous ROI routines. We selected a region-growing routine to measure SAT and VAT. The CSA was selected using a ‘snake’ (a rough manual outline will ‘fit’ the body contour based on the difference between pixel values of the body versus the background). Analyze provides a large variety of ROI-drawing routines. We manually separated SAT from VAT and used the ‘Auto Trace’ routine to select adipose tissue based on the pixel values. HippoFat is fully automated. It uses a fuzzy c-means clustering approach. The automated segmentation was rarely accurate and a manual correction was necessary for every one of our images. The software only measures adipose tissue, the area of lean tissue and, therefore, CSA could not be evaluated. The Philips EasyVision workstation does not provide many options, so we choose to perform a manual circumscribe SAT, VAT and CSA to measure these areas.
Time
The average processing time was about 135 min to segment 10 images, with time decreasing as the observer became more familiar with the utilization of the software interface. As a rule, it took the novice user longer to learn and use the software packages, and there was substantially more variability in the time to analyze 10 images. For the more experienced user, there was little variability in time to analyze 10 images. This analysis time did seem to be related to the order in which the software packages were evaluated.
Reliability and reproducibility of abdominal adipose area estimations
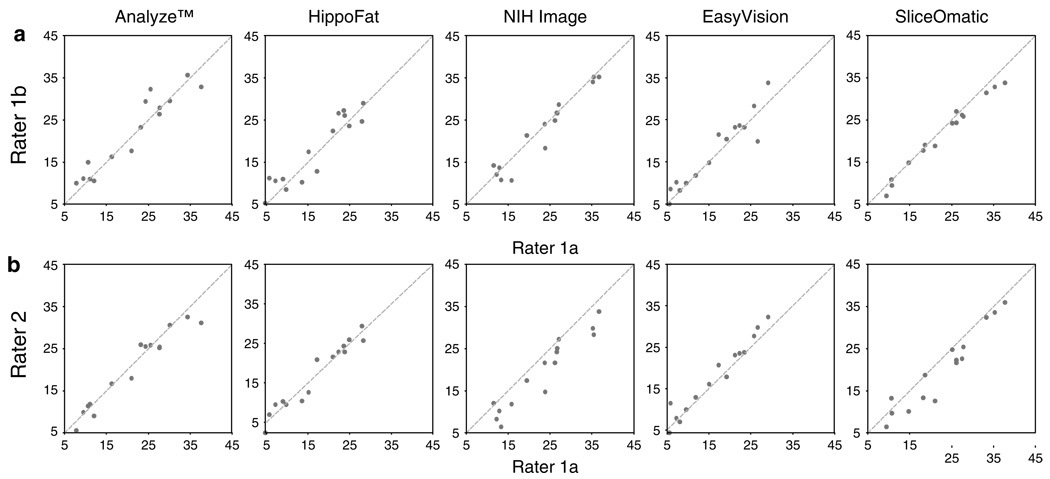
The intra-rater reliability for each software package is shown in Table 2 and Figure 4a. Overall, the reliability was excellent, ranging from a low of 0.934 for VAT intra-rater reproducibility of HippoFat to a high of 0.999 for CSA intrarater reproducibility of NIHImage and SliceOmatic. VAT and TAT measurements showed highest agreement using SliceOmatic, while HippoFat had the highest correlation for SAT.
Table 2.
Intra-rater reproducibility for readings by observer 1
| NIHImage | SliceOmatic | Analyze | HippoFat | EasyVision | |
|---|---|---|---|---|---|
| CSA agreement ICC | 0.999 | 0.999 | 0.953 | NA | 0.995 |
| CSA agreement CV | 0.356 | 0.203 | 1.519 | NA | 0.959 |
| TAT agreement ICC | 0.977 | 0.987 | 0.952 | 0.980 | 0.961 |
| TAT agreement CV | 4.174 | 2.791 | 5.039 | 4.251 | 4.893 |
| SAT agreement ICC | 0.996 | 0.997 | 0.998 | 0.999 | 0.984 |
| SAT agreement CV | 2.415 | 1.777 | 1.545 | 0.894 | 3.563 |
| VAT agreement ICC | 0.963 | 0.979 | 0.949 | 0.934 | 0.944 |
| VAT agreement CV | 6.851 | 5.217 | 8.147 | 13.447 | 8.843 |
Abbreviations: CSA, cross-sectional area; CV, coefficient variation; ICC, intraclass correlation coefficient; NA, not applicable; SAT, subcutaneous adipose tissue; TAT, total adipose tissue; VAT, visceral adipose tissue.
Figure 4.
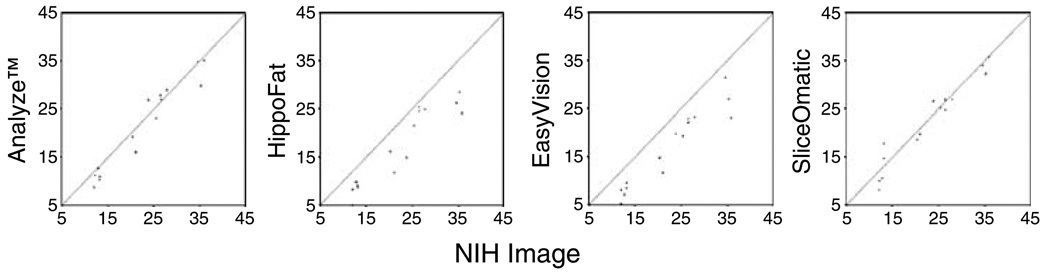
Scatter plots show (a) intra-rater and (b) inter-rater reproducibility of visceral adipose tissue (VAT) measurements in 10 cm2 achieved using Analyze, HippoFat, EasyVision, SliceOmatic and NIHImage.
The inter-observer reproducibility (Table 3 and Figure 4b) was highest for SliceOmatic for TAT and VAT. NIHImage was best with regard to CSA measurements. The agreement between the two observers was highest when using Analyze with regard to SAT measurements. In general, intra- and inter-rater reliability were lowest for VAT across all programs.
Table 3.
Inter-rater reproducibility: intra-class correlation coefficient for the two readings of observer 1 (1a and 1b) compared to observer 2
| Analyze |
HippoFat |
NIHImage |
EasyVision |
SliceOmatic |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1a | 1b | 1a | 1b | 1a | 1b | 1a | 1b | 1a | 1b | |
| CSA | 0.975 | 0.989 | NA | NA | 0.999 | 1.000 | 0.996 | 0.996 | 0.999 | 0.999 |
| TAT | 0.948 | 0.984 | 0.960 | 0.939 | 0.896 | 0.939 | 0.963 | 0.972 | 0.980 | 0.985 |
| SAT | 0.998 | 0.997 | 0.929 | 0.925 | 0.990 | 0.993 | 0.977 | 0.992 | 0.994 | 0.997 |
| VAT | 0.966 | 0.952 | 0.973 | 0.909 | 0.873 | 0.927 | 0.964 | 0.942 | 0.958 | 0.965 |
Abbreviations: CSA, cross-sectional area; NA, not applicable, since HippoFat cannot measure non-fat areas; SAT, subcutaneous adipose tissue; TAT, total adipose tissue; VAT, visceral adipose tissue.
The results of the intra-class correlation comparing NIHImage to the other software packages are displayed in Table 4 and Figure 5. The values displayed are the results of a comparison of the average over two measurements of observer 1. These comparisons varied more widely. Analyze and NIHImage are closest with respect to SAT, while SliceOmatic correlates best to NIHImage for TAT and VAT measurements.
Table 4.
Inter-test comparison of results from other software packages to those from NIHImage
| SliceOmatic | Analyze | HippoFat | EasyVision | |
|---|---|---|---|---|
| CSA agreement with NIHImage | 0.991 | 0.994 | NA | 0.997 |
| TAT agreement with NIHImage | 0.968 | 0.965 | 0.935 | 0.829 |
| SAT agreement with NIHImage | 0.997 | 0.997 | 0.964 | 0.983 |
| VAT agreement with NIHImage | 0.967 | 0.959 | 0.752 | 0.750 |
Abbreviations: CSA, cross-sectional area; NA, not applicable; SAT, subcutaneous adipose tissue; TAT, total adipose tissue; VAT, visceral adipose tissue. Results shown are comparisons of the average over two measurements of observer 1.
Figure 5.
Scatter plot showing the inter-test comparison of visceral adipose tissue (VAT) in 10 cm2 using the average over two measurements obtained by observer 1 with Analyze, HippoFat, EasyVision and SliceOmatic compared to the averaged results obtained using NIHImage.
Discussion
All the GUI-based packages we examined require minimal prerequisite computer skills and were perceived as very user friendly. We consider SliceOmatic a very good choice for segmentation of abdominal adipose tissue on MR images. It performed very well for all observed areas and the processing time was low for both observers. Since the agreement of the experts on the segmentation results proved to be equivalent for all examined software packages, we find it hard to recommend one software package solely based on the reproducibility. Analyze was rated lower because the multitude of options this package provides made it hard to determine an optimal segmentation routine. SliceOmatic offers a number of additional features, but the segmentation routine was easily determined and performed. HippoFat was rated slightly lower although the basic use is easy and straightforward, but the manual correction of mislabeled areas was very tedious.
The time it took to segment the MR images varied between the two observers and, again, NIHImage performed poorer than other software packages, most likely due to the fact that it was the first package we examined. Both observers had the lowest processing times using SliceOmatic. Although EasyVision required a manual segmentation, the processing time was equal to the time required to perform the semi-automated segmentation with Analyze. Using Analyze or NIHImage was perceived as more complicated and time consuming than segmenting images with SliceOmatic.
The amount of effort and time laboratory members are willing to commit to understanding and choosing between particular segmentation algorithms and software packages is an important consideration. The trade-off between manual interaction and performance is a significant factor. Manual interaction can improve accuracy by incorporating prior anatomical knowledge of the operator. The type of interaction can vary from complete manual delineating of adipose tissue to thresholding or selection of a seed point for region growing. In this study, the Philips EasyVision workstation was the only software package that did not offer any of these approaches, but the manual segmentation was straightforward and did not require more time than some of the computer-aided approaches, although the reproducibility of the VAT was not as good as the one of Analyze or SliceOmatic. Analyze, NIHImage and SliceOmatic required some user interaction for specifying initial parameters, like a threshold or the manual separation of SAT and VAT. The values of these parameters can significantly affect performance and results. An automated and unsupervised approach would allow data sets to be processed in a short time without any user interaction and would be inherently free from any variability introduced by the human observer. Positano et al.36 reported a high correlation (SAT: r=0.9917, P<0.0001; VAT: r=0.09601, P=0.0001) between the unsupervised method they proposed and manual segmentation of 20 patients (32 images each) using their software package HippoFat. They reported that the unsupervised method slightly overestimated the SAT and underestimated the VAT. Using the same software package for the automated segmentation of the MR images examined in this study resulted in unsatisfactory segmentation. This was most likely due to the complex structure of the viscera, patient movement, partial volume averaging, tissue inhomogeneity, poor contrast and poor signal-to-noise ratio or the image acquisition protocol, although the image quality did not affect any other software package to this extent. The results show that it requires substantial operator involvement for the re-delineation of VAT. Sometimes the SAT selection had to be adjusted as well. While the unsupervised algorithm itself was fairly quick, both observers perceived the manual corrections as time consuming and tedious.
Financial constraints might also affect the decision. NIH-Image and HippoFat were two free software packages. Many NIHImage users have added plug-ins, which are also free and can be downloaded. The upside of acquiring commercial software is the better support and a very comprehensive manual, which both Analyze and SliceOmatic offer.
Nonetheless the intra- and inter-rater reproducibility were very good for all packages. We decided to use NIHImage as the standard for our comparisons. We report intra-class correlation comparing NIHImage to all other packages, since it is a freely available program package, which has been frequently used to measure adipose tissue compartments. Using the pairwise correlations (data not shown) among all programs, we obtained similar results. The reproducibility of our measurements was affected by the quality of the image. Because of a smaller partial volume effect, the SAT could be best distinguished from the lean tissue and the inter-observer reproducibility is highest. The intra- and inter-rater reproducibility of VAT measurements were slightly lower. This might be due to the fact that signal of intestinal content with short T1 and bone marrow could be interpreted as VAT. Another issue is that the small amount of intramuscular and paravertebral adipose tissues may influence the accuracy of the measurement of VAT. However, it is still controversial whether such compartments of adipose tissue should be included in VAT. We tried to exclude these adipose tissues in VAT. Since our aim was the analysis of large amounts of image data such as in epidemiological studies, a simplified adipose tissue classification and ignoring small numbers of high-intensity non-fat pixels could be justified. When comparing the results of the five packages, we noted that the slightly inferior results for NIHImage, especially the low VAT reproducibility, might be due to the fact that it was the first software package the observers used. This can account for a larger number of misclassified pixels and as a result a higher variability of measurement results. It was, therefore, found more difficult to understand and it took longer to establish a routine, whereas the other packages benefited from the knowledge we had acquired using NIHImage.
Limitations to this study must be acknowledged. The reliability and reproducibility of MR assessment of adipose tissue might vary across degrees of adiposity, but only obese individuals were included in this study. However, we are reassured that in cadaver and animal studies of lean and obese subjects, the MRI areas of adipose tissue correlated well with dissection results.20,22 We evaluated only one fully automated program, one free software package and two commercially available programs, whereas a number of other programs are available. Most in-house packages had been developed to process MR images and estimate adipose tissue, none of the free software packages or the commercial packages were similarly specialized. Since we did not expect to find large difference between the commercial image processing packages, we selected the commercially available programs based on the cost and frequency of citations they received in other publications assessing abdominal adipose tissue. At the time this project started, only a few fully automated approaches had been published and had been made available. Recently published algorithms, like the one developed by Liou et al.37 may perform well or better and should be validated. Furthermore, we did not validate the software packages through use of a phantom, animal or cadaver model, but relied on previously published studies.17,20,22,24 Instead we compared the segmentation of semi-automated, automated and manual delineation approaches, none of which guarantees a perfect true model.
The fact that the results obtained with NIHImage and the results obtained with the four other software packages respectively correlate highly, supports our opinion that each of the examined software packages will allow for rapid, reliable and accurate measurements of abdominal SAT and VAT. We conclude that rather than trying to fully automate the segmentation process, the aim should be to support the medical expert with an image-based, objective segmentation tool, but without giving up the visual control of the expert and the expert’s possibility to interact.
In summary, four packages provided essentially the same results with respect to the inter- and intra-rater reproducibility, only the fully automated approach (HippoFat) proved slightly inferior. The choice of which package to adopt would depend on financial resources of the users as well as the user’s computer skills. NIHImage is free and can essentially perform most routines that Analyze and SliceOmatic offer. SliceOmatic has been widely used for MR image segmentation and is quick and reliable. Analyze requires a little more computer knowledge, but it is a very powerful software package that can be used for a large variety of image analysis purposes. The use of the EasyVision software is limited to a certain group of users since it is associated with a Philips scanner. To be able to compare measurements in longitudinal studies, researchers should consider reporting the reliability of their data, especially if more than one observer is evaluating the MR images. Although it may be beneficial to use the same software package for repeated measurements, our results using SliceOmatic, Analyze or NIHImage were comparable and could be used interchangeably. New fully automated approaches, like the one published by Liou et al.37 should be compared to the semi-automated measurements, since they could shorten the time and effort dramatically and perhaps provide even better reproducibility of the results.
Acknowledgements
The study was supported in part by NIH/NIDDK Grant RO1-DK060427 and UO1-DK57149. We thank Charlette Diggs, Project Coordinator for the Fatty Liver Ancillary Study, Kathleen Kahl and Terry Brawner, MR technicians, and the participants for their time and commitment.
References
- 1.Banerji MA, Faridi N, Atluri R, Chaiken RL, Lebovitz HE. Body composition, visceral fat, leptin, and insulin resistance in Asian Indian men. J Clin Endocrinol Metab. 1999;84:137–144. doi: 10.1210/jcem.84.1.5371. [DOI] [PubMed] [Google Scholar]
- 2.Thomas EL, Hamilton G, Patel N, O’Dwyer R, Dore CJ, Goldin RD, et al. Hepatic triglyceride content and its relation to body adiposity: a magnetic resonance imaging and proton magnetic resonance spectroscopy study. Gut. 2005;54:122–127. doi: 10.1136/gut.2003.036566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diehl AM, Li ZP, Lin HZ, Yang SQ. Cytokines and the pathogenesis of non-alcoholic steatohepatitis. Gut. 2005;54:303–306. doi: 10.1136/gut.2003.024935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sironi AM, Gastaldelli A, Mari A, Ciociaro D, Postano V, Buzzigoli E, et al. Visceral fat in hypertension: influence on insulin resistance and beta-cell function. Hypertension. 2004;44:127–133. doi: 10.1161/01.HYP.0000137982.10191.0a. [DOI] [PubMed] [Google Scholar]
- 5.Kuk JL, Church TS, Blair SN, Ross R. Does measurement site for visceral and abdominal subcutaneous adipose tissue alter associations with the metabolic syndrome? Diabetes Care. 2006;29:679–684. doi: 10.2337/diacare.29.03.06.dc05-1500. [DOI] [PubMed] [Google Scholar]
- 6.Kuk JL, Nichaman MZ, Church TS, Blair SN, Ross R. Liver fat is not a marker of metabolic risk in lean premenopausal women. Metabolism. 2004;53:1066. doi: 10.1016/j.metabol.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 7.Machann J, Thamer C, Schnoedt B, Haap M, Haring H-U, Claus D, et al. Standardized assessment of whole body adipose tissue topography by MRI. J Magn Reson Imaging. 2005;21:455–462. doi: 10.1002/jmri.20292. [DOI] [PubMed] [Google Scholar]
- 8.Durnin JVG, Womersley J. Body fat assessment from total body density and its estimate from skinfold thicknesses: measurements on 481 men and women aged from 16 to 72 years. Br J Nutr. 1974;32:77–87. doi: 10.1079/bjn19740060. [DOI] [PubMed] [Google Scholar]
- 9.Garrow JS, Stalley S, Diethelm R, Pittet P, Hesp R, Halliday D. A new method for measuring the body density of obese adults. Br J Nutr. 1979;42:173–183. doi: 10.1079/bjn19790105. [DOI] [PubMed] [Google Scholar]
- 10.Segal KR, Gutin B, Presta E, Wang J, Van Itallie TB. Estimation of human body composition by electrical impedance methods: a comparative study. J Appl Physiol. 1985;58:1565–1571. doi: 10.1152/jappl.1985.58.5.1565. [DOI] [PubMed] [Google Scholar]
- 11.Svendsen OL, Hassager C, Christiansen C. Age- and menopause-associated variations in body composition and fat distribution in healthy women as measured by dual-energy X-ray absorptiometry. Metabolism. 1995;44:369–373. doi: 10.1016/0026-0495(95)90168-x. [DOI] [PubMed] [Google Scholar]
- 12.Mazess RB, Barden HS, Bisek JP, Hanson J. Dual-energy X-ray absorptiometry for total-body and regional bone-mineral and soft-tissue composition. Am J Clin Nutr. 1990;51:1106–1112. doi: 10.1093/ajcn/51.6.1106. [DOI] [PubMed] [Google Scholar]
- 13.Sohlström A, Wahlund LO, Forsum E. Adipose tissue distribution as assessed by magnetic resonance imaging and total body fat by magnetic resonance imaging, underwater weighing and body-water dilution in healthy women. Am J Clin Nutr. 1993;58:830–838. doi: 10.1093/ajcn/58.6.830. [DOI] [PubMed] [Google Scholar]
- 14.Ross R, Leger L, Guardo R, De Guise J, Pike BG. Adipose tissue volume measured by magnetic resonance imaging and computerized tomography in rats. J Appl Physiol. 1991;70:2164–2172. doi: 10.1152/jappl.1991.70.5.2164. [DOI] [PubMed] [Google Scholar]
- 15.Lovejoy JC, Smith SR, Rood JC. Comparison of regional fat distribution and health risk factors in middle-aged white and African American women: the healthy transitions study. Obes Res. 2001;9:10–16. doi: 10.1038/oby.2001.2. [DOI] [PubMed] [Google Scholar]
- 16.Smith SR, Lovejoy JC, Greenway F, Ryan D, deJonge L, de la Bretonne J, et al. Contributions of total body fat, abdominal subcutaneous adipose tissue compartments, and visceral adipose tissue to the metabolic complications of obesity. Metabolism. 2001;50:425–435. doi: 10.1053/meta.2001.21693. [DOI] [PubMed] [Google Scholar]
- 17.Ross R. Magnetic resonance imaging provides new insights into the characterization of adipose and lean tissue distribution. Can J Physiol Pharmacol. 1996;74:778–785. [PubMed] [Google Scholar]
- 18.Donnelly LF, O’Brien KJ, Dardzinski BJ, Poe SA, Bean JA, Holland SK, et al. Using a phantom to compare MR techniques for determining the ratio of intraabdominal to subcutaneous adipose tissue. AJR Am J Roentgenol. 2003;180:993–998. doi: 10.2214/ajr.180.4.1800993. [DOI] [PubMed] [Google Scholar]
- 19.Peng Q, McColl RW, Wang J, Chia JM, Weatherall PT. Water-saturated three-dimensional balanced steady-state free precession for fast abdominal fat quantification. J Magn Reson Imaging. 2005;21:263–271. doi: 10.1002/jmri.20252. [DOI] [PubMed] [Google Scholar]
- 20.Fowler PA, Fuller MF, Glasbey CA, Cameron GG, Foster MA. Validation of the in vivo measurement of adipose tissue by magnetic resonance imaging of lean and obese pigs. Am J Clin Nutr. 1992;56:7–13. doi: 10.1093/ajcn/56.1.7. [DOI] [PubMed] [Google Scholar]
- 21.Ishikawa M, Koga K. Measurement of abdominal fat by magnetic resonance imaging of OLETF rats, an animal model of NIDDM. Magn Reson Imaging. 1998;16:45–53. doi: 10.1016/s0730-725x(97)00221-x. [DOI] [PubMed] [Google Scholar]
- 22.Abate N, Burns D, Peshock RM, Garg A, Grundy SM. Estimation of adipose tissue mass by magnetic resonance imaging: validation against dissection in human cadavers. J Lipid Res. 1994;35:1490–1496. [PubMed] [Google Scholar]
- 23.Foster MA, Hutchison JM, Mallard JR, Fuller M. Nuclear magnetic resonance pulse sequence and discrimination of high- and low-fat tissues. Magn Reson Imaging. 1984;2:187–192. doi: 10.1016/0730-725x(84)90004-3. [DOI] [PubMed] [Google Scholar]
- 24.Purnell JQ, Kahn SE, Schwartz RS, Brunzell JD. Relationship of insulin sensitivity and ApoB levels to intra-abdominal fat in subjects with familial combined hyperlipidemia. Arterioscler Thromb Vasc Biol. 2001;21:567–572. doi: 10.1161/01.atv.21.4.567. [DOI] [PubMed] [Google Scholar]
- 25.Gray DS, Fujioka K, Colletti PM, Kim H, Devine W, Cuyegkeng T, et al. Magnetic-resonance imaging used for determining fat distribution in obesity and diabetes. Am J Clin Nutr. 1991;54:623–627. doi: 10.1093/ajcn/54.4.623. [DOI] [PubMed] [Google Scholar]
- 26.Ross R, Goodpaster B, Kelley D, Boada F. Magnetic resonance imaging in human body composition research. From quantitative to qualitative tissue measurement. Ann N Y Acad Sci. 2000;904:12–17. doi: 10.1111/j.1749-6632.2000.tb06415.x. [DOI] [PubMed] [Google Scholar]
- 27.Ross R, Rissanen J. Mobilization of visceral and subcutaneous adipose tissue in response to energy restriction and exercise. Am J Clin Nutr. 1994;60:695–703. doi: 10.1093/ajcn/60.5.695. [DOI] [PubMed] [Google Scholar]
- 28.Ross R, Rissanen J, Pedwell H, Clifford J, Shragge P. Influence of diet and exercise on skeletal muscle and visceral adipose tissue in men. J Appl Physiol. 1996;81:2445–2455. doi: 10.1152/jappl.1996.81.6.2445. [DOI] [PubMed] [Google Scholar]
- 29.Abate N, Garg A, Coleman R, Grundy SM, Peshock RM. Prediction of total subcutaneous abdominal, intraperitoneal, and retroperitoneal adipose tissue masses in men by a single axial magnetic resonance imaging slice. Am J Clin Nutr. 1997;65:403–408. doi: 10.1093/ajcn/65.2.403. [DOI] [PubMed] [Google Scholar]
- 30.Han TS, Kelly IE, Walsh K, Greene RM, Lean ME. Relationship between volumes and areas from single transverse scans of intraabdominal fat measured by magnetic resonance imaging. Int J Obes Relat Metab Disord. 1997;21:1161–1166. doi: 10.1038/sj.ijo.0800530. [DOI] [PubMed] [Google Scholar]
- 31.Kuk JL, Lee S, Heymsfield SB, Ross R. Waist circumference and abdominal adipose tissue distribution: influence of age and sex. Am J Clin Nutr. 2005;81:1330–1334. doi: 10.1093/ajcn/81.6.1330. [DOI] [PubMed] [Google Scholar]
- 32.Shen W, Punyanitya M, Wang Z, Gallagher D, St-Onge MP, Albu J, et al. Visceral adipose tissue: relations between single-slice areas and total volume. Am J Clin Nutr. 2004;80:271–278. doi: 10.1093/ajcn/80.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas EL, Bell JD. Influence of undersampling on magnetic resonance imaging measurements of intra-abdominal adipose tissue. Int J Obes Relat Metab Disord. 2003;27:211–218. doi: 10.1038/sj.ijo.802229. [DOI] [PubMed] [Google Scholar]
- 34.Gastaldelli A, Sironi AM, Ciociaro D, Positano V, Buzzigoli E, Giannessi D, et al. Visceral fat and beta cell function in nondiabetic humans. Diabetologia. 2005;48:2090–2096. doi: 10.1007/s00125-005-1891-3. [DOI] [PubMed] [Google Scholar]
- 35.Gastaldelli SA, Mari A, Ciociaro D, Positano V, Buzzigoli E, Pettiti LM, et al. Visceral fat accumulation and beta-cell function in nondiabetic subjects. Diabetes. 2004;53 (Suppl):A571. [Google Scholar]
- 36.Positano V, Gastaldelli A, Sironi AM, Santarelli MF, Lombardi M, Landini L. An accurate and robust method for unsupervised assessment of abdominal fat by MRI. J Magn Reson Imaging. 2004;20:684–689. doi: 10.1002/jmri.20167. [DOI] [PubMed] [Google Scholar]
- 37.Liou TH, Chan WP, Pan LC, Lin PW, Chou P, Chen CH. Fully automated large-scale assessment of visceral and subcutaneous abdominal adipose tissue by magnetic resonance imaging. Int J Obes (Lond) 2006;5:844–852. doi: 10.1038/sj.ijo.0803216. [DOI] [PubMed] [Google Scholar]
- 38.Yang G, Myerson S, Chabat F, Pennell DJ, Firmin DN. Automatic MRI adipose tissue mapping using overlapping mosaics. MAGMA. 2002;14:39–44. doi: 10.1007/BF02668185. [DOI] [PubMed] [Google Scholar]
- 39.Jin Y, Imielinska CZ, Laine AF, Udupa J, Shen W, Heymsfield SB. Segmentation and evaluation of adipose tissue from whole body MRI scans. MICCAI’03; Montreal, Canada: 2003. Nov, pp. 635–642. Part I, [Google Scholar]
- 40.Ilea D, Ghita O, Robinson K, Sadleirb R, Lynch M, Brennan D, et al. Proceeedings of Optimization of Electrical and Electronic Equipment; Brasov, Romania: 2004. May 20–21, pp. 227–232. [Google Scholar]
- 41.Elbers JM, Haumann G, Asscheman H, Seidell JC, Gooren LJ. Reproducibility of fat area measurements in young, non-obese subjects by computerized analysis of magnetic resonance images. Int J Obes Relat Metab Disord. 1997;21:1121–1129. doi: 10.1038/sj.ijo.0800525. [DOI] [PubMed] [Google Scholar]
- 42.Gronemeyer SA, Steen RG, Kauffman WM, Reddick WE, Glass JO. Fast adipose tissue (FAT) assessment by MRI. Magn Reson Imaging. 2000;18:815–818. doi: 10.1016/s0730-725x(00)00168-5. [DOI] [PubMed] [Google Scholar]
- 43.Poll LW, Wittsack HJ, Koch JA, Willers R, Scherer A, Kapitza C, et al. Quantification of total abdominal fat volumes using magnetic resonance imaging. Eur J Med Res. 2002;7:347–352. [PubMed] [Google Scholar]
- 44.Poll L, Wittsack HJ, Willers R, Modder U, Heinemann L, Kapitza C, et al. Correlation between anthropometric parameters and abdominal fat volumes assessed by a magnetic resonance imaging method in patients with diabetes. Diabetes Technol Ther. 2004;6:844–849. doi: 10.1089/dia.2004.6.844. [DOI] [PubMed] [Google Scholar]
- 45.Poll WL, Wittsack HJ, Koch JA, Willers R, Cohnen M, Kapitza C, et al. A rapid and reliable semiautomated method for measurement of total abdominal fat volumes using magnetic resonance imaging. Magn Reson Imaging. 2003;21:631–636. doi: 10.1016/s0730-725x(03)00093-6. [DOI] [PubMed] [Google Scholar]
- 46.Song T, An J, Chen Q, Lee V, Laine V. Assessment of adipose tissue from whole body 3T MRI scans. 27th Annual International Conference IEEE Engineering in Medicine and Biology Society (EMBS); Shanghai, China: 2005. 2005. Sep 1–4, [DOI] [PubMed] [Google Scholar]
- 47.Brennan DPW, Robinson K, Ghita O, O’Brien J, Sadleir R, Eustace S. Rapid automated measurement of body fat distribution from whole-body MRI. AJR. Am J Roentgenol. 2005;185:418–423. doi: 10.2214/ajr.185.2.01850418. [DOI] [PubMed] [Google Scholar]
- 48.Ryan DH, Espeland MA, Foster GD, Haffner SM, Hubbard VS, Johnson KC, et al. Look AHEAD (Action for Health in Diabetes): design and methods for a clinical trial of weight loss for the prevention of cardiovascular disease in type 2 diabetes. Control Clin Trials. 2003;24:610–628. doi: 10.1016/s0197-2456(03)00064-3. [DOI] [PubMed] [Google Scholar]
- 49.Barnard ML, Schwieso JE, Thomas EL, Bell JD, Saeed N, Frost G, et al. Development of a rapid and efficient magnetic resonance imaging technique for analysis of body fat distribution. NMR Biomed. 1996;9:156–164. doi: 10.1002/(SICI)1099-1492(199606)9:4<156::AID-NBM412>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 50.Bonora E, Micciolo R, Ghiatas AA, Lancaster JL, Alyassin A, Muggeo M, et al. Is it possible to derive a reliable estimate of human visceral and subcutaneous abdominal adipose tissue from simple anthropometric measurements? Metabolism. 1995;44:1617–1625. doi: 10.1016/0026-0495(95)90084-5. [DOI] [PubMed] [Google Scholar]
- 51.Busetto L, Tregnaghi A, Bussolotto M, Sergi G, Beninca P, Ceccon A, et al. Visceral fat loss evaluated by total body magnetic resonance imaging in obese women operated with laparoscopic adjustable silicone gastric banding. Int J Obes Relat Metab Disord. 2000;24:60–69. doi: 10.1038/sj.ijo.0801086. [DOI] [PubMed] [Google Scholar]
- 52.Changani KK, Nicholson A, White A, Latcham JK, Reid DG, Clapham JC. A longitudinal magnetic resonance imaging (MRI) study of differences in abdominal fat distribution between normal mice, and lean overexpressers of mitochondrial uncoupling protein-3 (UCP-3) Diabetes Obes Metab. 2003;5:99–105. doi: 10.1046/j.1463-1326.2003.00249.x. [DOI] [PubMed] [Google Scholar]
- 53.Concepcion L, Marti-Bonmati L, Aliaga R, Delgado F, Morillas C, Hernandez A. Abdominal fat assessment by magnetic resonance: comparison with biometric profiles and cardiovascular risk markers. Med Clin (Barc) 2001;117:366–369. doi: 10.1016/s0025-7753(01)72117-3. [DOI] [PubMed] [Google Scholar]
- 54.Gautier JF, Mourier A, de Kerviler E, Tarentola A, Bigard AX, Villette JM, et al. Evaluation of abdominal fat distribution in noninsulin-dependent diabetes mellitus: relationship to insulin resistance. J Clin Endocrinol Metab. 1998;83:1306–1311. doi: 10.1210/jcem.83.4.4713. [DOI] [PubMed] [Google Scholar]
- 55.Janssen I, Ross R. Effects of sex on the change in visceral, subcutaneous adipose tissue and skeletal muscle in response to weight loss. Int J Obes Relat Metab Disord. 1999;23:1035–1046. doi: 10.1038/sj.ijo.0801038. [DOI] [PubMed] [Google Scholar]
- 56.Kamel EG, McNeill G, Van Wijk MC. Change in intra-abdominal adipose tissue volume during weight loss in obese men and women: correlation between magnetic resonance imaging and anthropometric measurements. Int J Obes Relat Metab Disord. 2000;24:607–613. doi: 10.1038/sj.ijo.0801204. [DOI] [PubMed] [Google Scholar]
- 57.Kanaley JA, Giannopoulou I, Tillapaugh-Fay G, Nappi JS, Ploutz-Snyder LL. Racial differences in subcutaneous and visceral fat distribution in postmenopausal black and white women. Metabolism. 2003;52:186–191. doi: 10.1053/meta.2003.50024. [DOI] [PubMed] [Google Scholar]
- 58.Kanaley JA, Sames C, Swisher L, Swick AG, Ploutz-Snyder LL, Steppan CM, et al. Abdominal fat distribution in pre- and postmenopausal women: the impact of physical activity, age, and menopausal status. Metabolism. 2001;50:976–982. doi: 10.1053/meta.2001.24931. [DOI] [PubMed] [Google Scholar]
- 59.Lee S, Janssen I, Ross R. Interindividual variation in abdominal subcutaneous and visceral adipose tissue: influence of measurement site. J Appl Physiol. 2004;97:948–954. doi: 10.1152/japplphysiol.01200.2003. [DOI] [PubMed] [Google Scholar]
- 60.Ohsuzu F, Kosuda S, Takayama E, Yanagida S, Nomi M, Kasamatsu H, et al. Imaging techniques for measuring adipose-tissue distribution in the abdomen: a comparison between computed tomography and 1.5-tesla magnetic resonance spin-echo imaging. Radiat Med. 1998;16:99–107. [PubMed] [Google Scholar]
- 61.Ross R, Janssen I, Dawson J, Kungl AM, Kuk JL, Wong SL, et al. Exercise-induced reduction in obesity and insulin resistance in women: a randomized controlled trial. Obes Res. 2004;12:789–798. doi: 10.1038/oby.2004.95. [DOI] [PubMed] [Google Scholar]
- 62.Ryu JE, Craven TE, MacArthur RD, Hinson WH, Bond MG, Hagaman AP, et al. Relationship of intraabdominal fat as measured by magnetic resonance imaging to postprandial lipemia in middle-aged subjects. Am J Clin Nutr. 1994;60:586–591. doi: 10.1093/ajcn/60.4.586. [DOI] [PubMed] [Google Scholar]
- 63.Sobol W, Rossner S, Hinson B, Hiltbrandt E, Karstaedt N, Santago P, et al. Evaluation of a new magnetic resonance imaging method for quantitating adipose tissue areas. Int J Obes. 1991;15:589–599. [PubMed] [Google Scholar]
- 64.Szklo MNF. Epidemiology: Beyond the Basics. Gaithersberg, MD: Aspen Publishers Inc.; 2000. Chapter 8: quality assurance and control; pp. 343–404. [Google Scholar]
- 65.Chan DC, Watts GF, Barrett PH. Comparison of intraperitoneal and posterior subcutaneous abdominal adipose tissue compartments as predictors of VLDL apolipoprotein B-100 kinetics in overweight/obese men. Diabetes Obes Metab. 2003;5:202–206. doi: 10.1046/j.1463-1326.2003.00261.x. [DOI] [PubMed] [Google Scholar]
- 66.Chan DC, Watts GF, Sussekov AV, Barrett PH, Yang Z, Hua J, et al. Adipose tissue compartments and insulin resistance in overweight-obese Caucasian men. Diabetes Res Clin Pract. 2004;63:77–85. doi: 10.1016/j.diabres.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 67.Machann J, Thamer C, Schnoedt B, Stefan N, Stumvoll M, Haring H-U, et al. Age and gender related effects on adipose tissue compartments of subjects with increased risk for type 2 diabetes: a whole body MRI/MRS study. MAGMA. 2005;18:128–137. doi: 10.1007/s10334-005-0104-x. [DOI] [PubMed] [Google Scholar]
- 68.Thamer C, Machann J, Haap M, Stefan N, Heller E, Schnodt B, et al. Intrahepatic lipids are predicted by visceral adipose tissue mass in healthy subjects. Diabetes Care. 2004;27:2726–2729. doi: 10.2337/diacare.27.11.2726. [DOI] [PubMed] [Google Scholar]
- 69.Tintera J, Harantova P, Suchanek P, Dvorakova A, Adamova M, Hajek M, et al. Quantification of intra-abdominal fat during controlled weight reduction: assessment using the water-suppressed breath – hold MRI technique. Physiol Res. 2004;53:229–234. [PubMed] [Google Scholar]
- 70.Miyazaki Y, Glass L, Triplitt C, Wajcberg E, Mandarino LJ, DeFronzo RA. Abdominal fat distribution and peripheral and hepatic insulin resistance in type 2 diabetes mellitus. Am J Physiol Endocrinol Metab. 2002;283:E1135–E1143. doi: 10.1152/ajpendo.0327.2001. [DOI] [PubMed] [Google Scholar]
- 71.Seppala-Lindroos A, Vehkavaara S, Hakkinen AM, Goto T, Westerbacka J, Sovijarvi A, et al. Fat accumulation in the liver is associated with defects in insulin suppression of glucose production and serum free fatty acids independent of obesity in normal men. J Clin Endocrinol Metab. 2002;87:3023–3028. doi: 10.1210/jcem.87.7.8638. [DOI] [PubMed] [Google Scholar]