Abstract
Differential gene expression plays a critical role in the development and physiology of multicellular organisms. At a ‘systems level’ (e.g. at the level of a tissue, organ or whole organism), this process can be studied using gene regulatory network (GRN) models that capture physical and regulatory interactions between genes and their regulators. In the past years, significant progress has been made toward the mapping of GRNs using a variety of experimental and computational approaches. Here, we will discuss gene-centered approaches that we employed to characterize GRNs and describe insights that we have obtained into the global design principles of gene regulation in complex metazoan systems.
Keywords: transcription factor, differential gene expression, gene-centered, gene regulatory network, yeast one-hybrid assay, Caenorhabditis elegans
INTRODUCTION
In multicellular of organisms, most genes need to be expressed in a specific spatiotemporal manner, in order to guide development and to maintain post-developmental physiology. For instance, the stem cell factors Oct4, Sox2, Klf4 and c-Myc are expressed early in development and function to preserve the pluripotency of uncommitted stem cells. Their expression is sufficient to preserve or induce this unique cellular property [1]. On the other hand; a different group of genes is activated upon cellular differentiation. One of the classical examples is the Pax6 gene that is required for eye development in a variety of organisms [2]. Likewise, in Drosophila embryos, the spatiotemporal expression of gap and pair-rule genes is crucial for defining segmentation patterns and development (reviewed in: [3]). Finally, yet another group of genes is expressed in specialized, fully differentiated tissues to enable post-developmental functions in physiology throughout the lifetime of an organism. A textbook example is proinsulin, the insulin precursor, which is specifically expressed in the pancreas and regulates the level of glucose in the blood after food intake.
As exemplified above, differential gene expression is a highly regulated and controlled process. It occurs at a first level by the action of transcription factors (TFs): proteins that physically interact with cis-acting genomic regions to control expression of their target genes. TFs can either repress or activate transcription and many can do both, depending on the cellular context. In addition to TFs, chromatin modifications (e.g. histone acetylation, methylation, etc. [4]), microRNAs (reviewed in: [5]), RNA binding proteins, mRNA stability, export and splicing, and post-translational modifications also contribute to differential gene expression. However, it is transcriptional regulation which first and foremost determines where and when a gene is expressed, whereas other types of regulation often modulate and dampen gene expression, rather than to determine it.
The human genome encodes ∼1500 TFs [6] and 600 microRNAs [7]. For most of these regulators, their function is completely unknown. Indeed, even in large community efforts such as the ENCODE project, only a handful have been comprehensively studied [8]. Furthermore, increasingly more non-protein-coding nucleotides are being associated with a regulatory function in the 3.2 Gb human genome [8]. Thus, the comprehensive delineation of the mechanisms that control differential gene expression at a genome scale, or systems level in humans is as of yet a daunting task.
Systems level studies of differential gene expression are greatly advanced by the use of genetically tractable model organisms such as the fruitfly Drosophila melanogaster and the nematode Caenorhabditis elegans. We have focused on C. elegans because it is a relatively simple animal with a fixed lineage of only 959 cells. In addition, the C. elegans genome is fully sequenced and annotated, and is compact compared to the human genome: even though both contain ∼20 000 genes, the 100 Mb worm genome is 30 times smaller [9, 10]. Consequently, ∼26% of the worm genome is exonic, compared with 1–2% in humans. In addition, the majority of intergenic regions are shorter than 2 kb [11], and introns are much shorter with a median length of 65 bp, whereas the median length of human introns is 3 kb [12]. Thus, the potential regulatory genomic ‘space’ that needs to be considered in studies of differential gene expression is much smaller. The C. elegans genome also encodes fewer TFs (∼940) and microRNAs (∼150) than the human genome [13–15]. Finally, studies about the mechanisms of differential gene expression at a systems level are greatly facilitated by the fact that C. elegans is a transparent animal. By using reporters such as the green fluorescent protein (GFP) one can elucidate where and when genes are expressed in living animals, and determine how different perturbations affect gene expression [16–20].
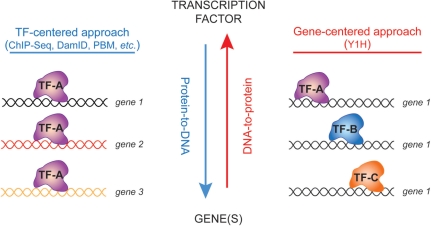
Differential gene expression can be studied at a systems level using gene regulatory networks (GRNs) that model physical and regulatory interactions between genes and their trans regulators (Figure 1) [21]. Physical TF-DNA interactions can be delineated using two conceptually different but highly complementary approaches (Figure 2). TF-centered, ‘protein-to-DNA’, methods start with a TF or set of TFs of interest and identify genomic DNA fragments that these TF(s) interact with. Chromatin-immunoprecipitation (ChIP) and DamID are the most widely used TF-centered methods [22, 23]. ChIP has been particularly powerful for the identification of TF–DNA interactions in homogeneous systems such as yeast, and in mammalian tissue culture cells, including primary cells or stem cells. Although powerful, it is technically difficult to systematically apply ChIP to most TFs in heterogeneous and complex metazoan systems such as intact worms. This is because many TFs are expressed at low levels, and some may be expressed in a limited number of cells, or during a narrow developmental interval. Furthermore, antibodies that are suitable for ChIP assays are only available for a handful of worm TFs. On the other hand, gene-centered, ‘DNA-to-protein’, methods start with one or more regulatory DNA fragments and identify the TFs that can interact with these fragments [21]. Here, we will discuss our efforts on the delineation of gene-centered GRNs in C. elegans and describe some of the insights that we have obtained into the global mechanisms of differential gene expression.
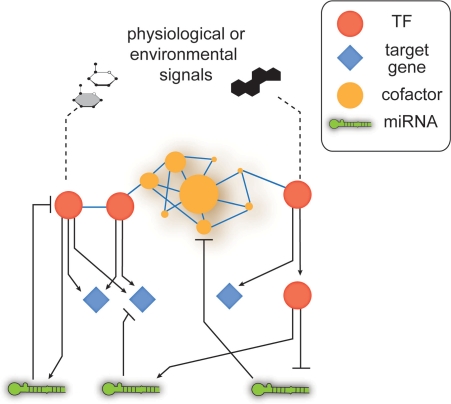
Figure 1:
Cartoon illustrating hypothetical interactions in a GRN, which combines physical and regulatory interactions between TFs and their target genes. In addition to TFs, other regulators need to be incorporated such as microRNAs and transcriptional cofactors.
Figure 2:
GRNs can be delineated using two conceptually different, but highly complementary approaches. TF-centered methods start with a TF of interest and identify DNA fragments this TF binds to. Gene-centered methods start with a set of DNA fragments and identify the TFs these fragments interact with.
GATEWAY-COMPATIBLE YEAST ONE-HYBRID ASSAYS
Seminal work from Eric Davidson and colleagues has characterized the wiring of endo-mesodermal gene regulation in the sea urchin embryo. This work epitomizes the concept of gene-centered regulatory network mapping because it focused on cis-regulatory DNA elements to understand where and when genes are expressed during development, followed by the identification of TFs that may regulate this process [24]. The endo-mesodermal network has been delineated over the course of many years because the work required laborious assays that were not amenable to use in high-throughput settings in complex animals. In addition, the interactions identified are not necessarily direct, as physical associations are not immediately revealed. To provide a gene-centered method that can detect physical interactions between sets of genes and multiple TFs in a relatively short amount of time, we have developed a high-throughput version of the yeast one-hybrid (Y1H) system [25]. With this method, interactions identified are strictly physical, as Y1H assays do not immediately reveal in vivo regulatory consequences (see also below).
Y1H assays are conceptually similar to yeast two-hybrid (Y2H) assays that have been used to identify thousands of protein–protein interactions in many systems, including C. elegans [26, 27]. Instead of using two hybrid proteins (a protein bait and a protein prey), the Y1H system uses a DNA bait and a single hybrid protein prey (Figure 3). The Y1H system was first developed to facilitate the identification of proteins that can bind to multiple copies of a short DNA sequences of interest [28, 29]. However, the comprehensive mapping of GRNs needs to be unbiased, as most small cis-regulatory DNA elements that control gene expression are not yet discovered. Instead, larger genomic fragments that likely harbor many of these elements such as gene promoters and enhancers need to be interrogated. Furthermore, the original Y1H system used conventional, restriction-enzyme-based cloning methods for DNA bait generation and was therefore not amenable for system-level analyses. To alleviate these limitations, we have developed a Y1H system that uses Gateway recombinational cloning to rapidly transfer multiple DNA baits into Y1H Destination vectors in parallel [25]. We have demonstrated that this Y1H system can be used with both small elements and with large, complex DNA fragments such as gene promoters.
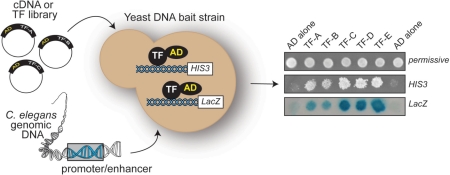
Figure 3:
Gateway-compatible Y1H assays provide a convenient gene-centered method for GRN mapping. A yeast DNA bait strain contains two reporter constructs integrated into its genome. Each construct contains the same DNA fragment, but different reporter genes, such as HIS3 and LacZ. A cDNA or TF library can be used as a prey resource, or collections of TFs can be tested individually. The panel on the right shows the readout of a Y1H experiment. AD alone indicates the negative control used to assess DNA bait background growth on media lacking histidine and containing 3-AT, and background bait coloring on a βGal assay. Different interacting TFs confer different interaction phenotypes; i.e. light blue versus dark blue.
Gateway-compatible Y1H assays start with a set of DNA fragments (DNA baits) of interest (Figure 3). Briefly, the DNA bait is Gateway-cloned upstream of two Y1H reporter genes (HIS3 and LacZ) and the two DNA bait::reporter constructs are integrated into the yeast genome (double integration). This ensures that DNA baits are chromatinized which minimizes background and, therefore, reduces false positive interactions. To enable the identification of a wide variety of DNA binding proteins, including transcriptional repressors, a strong heterologous activation domain (AD – Figure 3) is added to the prey proteins. If the prey protein contains a DNA binding domain that can interact with the DNA bait, reporter gene expression is activated. Activation of HIS3 expression is assessed on media lacking histidine and containing 3-aminotriazole (3-AT), a competitive inhibitor of the His3 enzyme. Activation of LacZ is assessed by a colorimetric (‘blue-white’) assay (Figure 3). So far, we have used both a cDNA library and TF library as prey resources in our Y1H screens. It was important to include a low-complexity TF library because TFs that are expressed at low levels or in only a few cells in an organism are difficult to retrieve from screens that employ high-complexity, non-normalized cDNA libraries. Recently, we have made additional adaptations of the Y1H system. For instance, we have developed a smart-pooling method that is based on the Steiner triple system, which allows direct testing of multiple TFs that are allocated to specific pools, and the immediate identification of interacting TFs by deconvolution [14]. This assay eliminates the need for extensive prey sequencing (as with library screens) and therefore reduces the cost per screen and increases the throughput. One can also perform mating or transformation experiments with arrays of TFs to directly test TF–DNA interactions (an example of five TFs is shown in Figure 3) [14]. In addition to predicted TFs, we have also retrieved several proteins that do not possess a recognizable DNA binding domain. These proteins robustly interact with their Y1H targets in yeast, and for 11 of them we have confirmed direct promoter interactions by ChIP from yeast using an anti-AD antibody [30, 31]. The retrieval of these proteins suggests that not all regulatory TFs have been uncovered, and that there may be as of yet unrecognized DNA-binding folds. The observation that the yeast Arg5,6 enzyme can bind DNA supports this notion [32]. It is possible that these interactions may not necessarily be directly with DNA, instead they could be with other (chromatin) proteins that specifically bind C. elegans gene promoters in yeast. However, we believe that this is not very likely as we rarely retrieve known or predicted chromatin proteins or transcriptional cofactors. In sum, identifying novel TFs is a unique feature of gene-centered methods.
GENE-CENTERED C. ELEGANS REGULATORY NETWORKS
Thus far, we have used gene-centered Y1H assays to delineate several medium-scale GRNs [30, 31, 33] (Arda et al., submitted). As discussed below, these networks have already provided insights into the design principles of regulatory circuits. In addition, they have enabled us to estimate the sensitivity and specificity of the assay.
Y1H assays can efficiently delineate tissue-specific GRNs. By using a set of genes expressed in the C. elegans digestive tract or involved in its development, we identified many digestive tract TFs. Similarly, our neuronal GRN was enriched for neuronally expressed TFs [30, 31]. Recently, we also mapped a metabolic GRN, which shows that process-specific GRNs can also be characterized by Y1H assays (Arda et al., submitted).
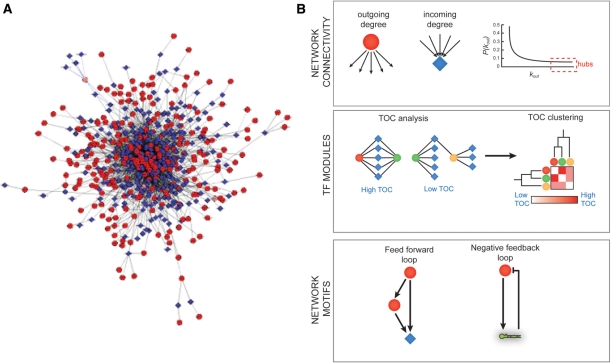
GRNs are bipartite as they contain directed interactions between two types of nodes: genes and regulators. GRNs can be visualized using publicly available tools such as Cytoscape and N-browse [34, 35]. The resulting graph models are highly complex, particularly when large numbers of protein–DNA interactions are incorporated, which makes them difficult to interrogate by eye (Figure 4A). Instead, a variety of computational and mathematical tools for network analysis need to be used. These tools can inform us about the properties of the network as a whole, or can identify important network neighborhoods ('modules') or overrepresented network building blocks ('motifs') (Figure 4). At the level of whole networks, measures that are often used are the degree and degree distribution [36]. The degree is defined as the connectivity of individual nodes, i.e. the number of TFs bound by a DNA fragment or the number of DNA fragments bound by a TF. These are referred to as incoming and outgoing degree, respectively. The degree distribution provides information about the overall network connectivity. In the majority of biological networks, the degree distribution follows a power law, rather than a normal distribution: most nodes have a relatively low degree, but a small number are disproportionally highly connected [37]. These TFs and promoters are referred to as ‘hubs’. TF hubs interact with many genes, from many different tissues or organs. They are often essential, indicating their overall importance in gene regulation and development [30, 38].
Figure 4:
GRNs can be analyzed at the whole network level, for instance by determining individual and global connectivities. (A) A compiled GRN of ∼200 C. elegans gene promoters (based on our published data). This model reflects the complexity of systems level gene regulation. Circles—TFs, diamonds, diamonds—target gene promoters, edges—protein–DNA interactions as determined by Y1H assays. This visualization was done using Cytoscape v.2.6 software with ‘random layout’ settings, which randomly distributes the nodes in a given network. (B) Top—analysis of incoming or outgoing degrees of individual nodes reveals network hubs. Middle—TOC analysis and clustering can reveal TF modules. Bottom—examples of network motifs that are overrepresented in GRNs. Circles—TFs, diamonds—target gene promoters, hairpin structure—miRNA gene promoters.
Network modules are highly interconnected network neighborhoods. Such modules can contain functionally related genes and TFs. Several measures can be used to identify network modules, including topological overlap coefficient (TOC) analysis. TOC analysis followed by TOC clustering can be used to identify TF modules that are based on similarities between TFs in terms of the target genes they interact with (Figure 4). We have previously identified TF modules in a neuronal GRN and found that one of these was enriched for paired-type homeodomain TFs [31]. Interestingly, these TFs associate predominantly with genes that are exclusively neuronal and these TFs are themselves neuronally expressed. Moreover, the neuronal expression of paired-type homedomain TFs is conserved between C. elegans and mice. This example illustrates how TF modules can be used to functionally annotate sets of TFs at a systems level.
Network motifs are small building blocks composed of two or more interactions (or ‘edges’) that are overrepresented in GRNs compared to randomized networks [39]. Thus, such motifs represent successful mechanisms of gene regulation. One of the best-studied motifs is the feed-forward loop in which a TF regulates another TF and both share a downstream target [40] (Figure 4). This motif is found in GRNs from bacteria, yeast and C. elegans [30, 39]. We recently delineated the first genome-scale GRN that not only includes TFs and their target genes, but also incorporates microRNAs [33]. This network contains interactions between microRNA promoters and TFs, and predicted interactions between microRNAs and the 3′UTRs of TF-encoding mRNAs. Interestingly, we found that this network contains a novel type of network motif: a feedback loop in which microRNAs and TFs reciprocally regulate each other (Figure 4). The microRNAs and TFs that participate in these loops have a high flux-capacity: a combined high in-and outgoing degree. This indicates that such motifs may provide adaptable and robust gene expression programs (reviewed in: [15]). Feedback loops are not overrepresented in pure transcriptional networks, and therefore microRNAs may provide an important GRN component for feedback regulation. It is likely that other types of interactions may also contribute to different types of network motifs, including protein–protein interactions [41], and perhaps interactions involving RNA binding proteins.
Table 1:
Advantages and limitations of TF versus gene-centered methods
| TF-centered methods | Gene-centered methods |
|---|---|
| • Start with a TF and find all possible genomic regions the TF can bind. | • Start with a genomic fragment (e.g. a gene promoter) and find TFs that can interact with the DNA fragment. |
| • Interactions captured in vivo. | • Interactions captured are ‘in levuro’, i.e. not in vivo, but not in vitro either. |
| • Powerful for the identification of protein–DNA interactions in unicellular or homogenous systems. | • Powerful for the identification of protein–DNA interactions that pertain to complex tissues or processes. |
| • Powerful for capturing interactions when TF activity depends on post-transcriptional modifications. | • May miss interactions that involve TFs that are post- transcriptionally modified. |
| • Limited by the endogenous expression level and/or spatio-temporal expression pattern of TFs, i.e. TFs that are expressed at high levels, or in a large number of cells can be assayed. | • Not limited by the endogenous expression level and/or spatio- temporal state of TFs, interactions found are ‘condition- independent’. |
| • Limited by the availability of TF-specific antibodies. | • No antibody is required. |
| • Cannot yet detect heterodimers. |
TF BINDING SITES
Y1H data provide a unique resource for the identification of TF binding sites; short sequences within the larger DNA bait with which the TF directly associates. By using our data and a combination of available computational algorithms, we have delineated the consensus binding site for several C. elegans TFs. First, we identified the binding site for ZTF-2, a novel pharyngeal TF [30]. We found that this site greatly resembles a predicted pharyngeal regulatory motif that is found in the promoters of genes whose expression is enriched in the C. elegans pharynx. We subsequently showed that ZTF-2 represses gene expression by binding to this element [30]. Second, we identified the binding site for FLH-1 and FLH-2, two TFs that interact with several microRNA promoters in Y1H assays. We found that FLH-1 and FLH-2 redundantly repress early embryonic expression of these microRNAs by binding to this element [33, 42]. Finally, we identified an extended binding site for the Snail-type TF CES-1 and demonstrated that this site is both necessary and sufficient for CES-1 binding [43].
LIMITATIONS OF Y1H ASSAYS: FALSE NEGATIVES AND FALSE POSITIVES
Y1H assays provide information regarding interactions between TFs and genomic DNA fragments that can occur. When carried out appropriately, the interactions retrieved are highly robust, which means that the technical false positive rate is low. Y1H assays ideally need to be carried out using DNA baits that do not have a high background reporter gene activity (e.g. because they interact with a yeast transcriptional activator), readouts from both reporter assays should be considered and interactions identified need to be retested in fresh yeast cells. However, even when carried out appropriately there are limitations to Y1H assays as not all interactions retrieved are by definition biologically meaningful (biological false positives), and interactions may be missed (false negatives).
In our combined GRNs, we identified TFs from all major families, including nuclear hormone receptors (NHRs), C2H2 zinc fingers, homeodomains and basic helix-loop-helix (bHLH) proteins [30, 31, 33]. This demonstrates that the Y1H system does not have an inherent bias for or against particular types of TFs. However, a current limitation of the method is that it can only detect monomeric or homodimeric TFs. Many TFs bind DNA as obligatory dimers [44], and, therefore, implementations of the Y1H system will need to be developed to alleviate this limitation. Other TFs that may be missed include those that need to be post-translationally modified before they can interact with DNA, for instance by phosphorylation. Finally, TFs that will be missed include those that are underrepresented in cDNA libraries or for which a full-length open reading frame has not yet been cloned (i.e. they are not yet present in TF libraries). So far, we have detected interactions for ∼25% of all predicted C. elegans TFs in Y1H assays, which is unprecedented for any multicellular organism.
Y1H-based TF-DNA interactions can be validated in vivo in C. elegans using a variety of methods. By performing ChIP and functional assays such as comparing promoter activity or target gene expression in the presence or absence of an interacting TF, we have demonstrated that numerous TF-target gene interactions that we identified do occur in vivo. However, we do not expect all physical interactions to have an observable biological consequence. It has been shown in other systems that TFs can be physically associated with chromatin/DNA without regulatory consequences [45, 46]. In addition, it is important to note that validation assays are not foolproof: interactions that occur in only a few cells or during a short (developmental) time period will be difficult to detect in whole animal assays. Indeed, we used prior knowledge to validate microRNA promoter interactions for the TFs DAF-3 and LIN-26: for DAF-3 we used dauer animals, where we know DAF-3 is active, and for LIN-26 we analyzed embryos as loss of lin-26 confers an embryonic phenotype [33].
CONCLUSION
Gene-centered methods such as Gateway-compatible Y1H assays provide powerful tools for GRN mapping. For complete, high-quality GRNs the data obtained with these methods need to be integrated with other data types such as those obtained by TF-centered methods. In the future, it will be important to incorporate multiple types of regulatory molecules and to connect GRNs to signaling networks. Finally, it is a major future challenge to go beyond static, Boolean GRN models and to incorporate cellular states, dynamics and interaction affinities as well.
Key Points.
Gene-centered methods (DNA-to-protein) such as the Y1H system provide a powerful tool for the identification of TF-target gene interactions. Instead of starting with individual TFs and identifying interacting DNA fragments (protein-to-DNA), Y1H assays start with DNA fragments of interest, and can identify interactions between many DNA fragments and many TFs simultaneously, and in a high-throughput manner.
GRNs can be analyzed using computational and mathematical tools to gain insights into the global principles of differential gene expression that cannot be obtained by studying single genes.
In the future, it will be necessary to improve gene-centered mapping efforts and obtain more comprehensive GRNs by integrating these networks with other types of networks, such as protein–protein interaction networks and signaling networks. In addition, the different aspects of these interactions, such as interaction affinities, should be incorporated to reflect the dynamic state of systems.
FUNDING
National Institutes of Health (DK068429 and DK071713) and by a grant from the Ellison Medical Foundation. The National Institutes of Health grant No. EY017589 and GM076102.
Acknowledgements
The authors thank John Reece-Hoyes Lesley MacNeil, Pedro Batista and Job Dekker for discussions and critical reading of the manuscript.
Biographies
H. Efsun Arda is a Graduate Student in the Interdisciplinary Graduate Program and Program in Gene Function and Expression at the University of Massachusetts Medical School. She studies gene regulatory networks controlling C. elegans metabolism
Albertha J. M. (Marian) Walhout is an Associate Professor in the Program in Gene Function and Expression and Program in Molecular Medicine at the University of Massachusetts Medical School. She studies gene regulatory networks that are involved in a variety of metazoan systems
References
- Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Ikeo K. Pax6: mastering eye morphogenesis and eye evolution. Trends Genet. 1999;15:371–7. doi: 10.1016/s0168-9525(99)01776-x. [DOI] [PubMed] [Google Scholar]
- St Johnston D, Nusslein-Volhard C. The origin of pattern and polarity in the Drosophila embryo. Cell. 2009;68:201–19. doi: 10.1016/0092-8674(92)90466-p. [DOI] [PubMed] [Google Scholar]
- Rando OJ, Chang HY. Genome-wide views of chromatin structure. Annu Rev Biochem. 2009;78:245–71. doi: 10.1146/annurev.biochem.78.071107.134639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10:252–63. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, et al. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The C. elegans Genome Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–18. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–45. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Dupuy D, Li Q, Deplancke B, et al. A first version of the Caenorhabditis elegans promoterome. Genome Res. 2004;14:2169–75. doi: 10.1101/gr.2497604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieth J, Lawson D. Overview of gene structure (January 18, 2006) WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.65.1, http://www.wormbook.org.
- Reece-Hoyes JS, Deplancke B, Shingles J, et al. A compendium of C. elegans regulatory transcription factors: a resource for mapping transcription regulatory networks. Genome Biol. 2005;6:R110. doi: 10.1186/gb-2005-6-13-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeirssen V, Deplancke B, Barrasa MI, et al. Matrix and Steiner-triple-system smart pooling assays for high-performance transcription regulatory network mapping. Nat Methods. 2007;4:659–64. doi: 10.1038/nmeth1063. [DOI] [PubMed] [Google Scholar]
- Martinez NJ, Walhout AJ. The interplay between transcription factors and microRNAs in genome-scale regulatory networks. Bioessays. 2009;31:435–45. doi: 10.1002/bies.200800212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, et al. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–5. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Hunt-Newbury R, Viveiros R, Johnsen R, et al. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 2007;5:e237. doi: 10.1371/journal.pbio.0050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Shingles J, Dupuy D, et al. Insight into transcription factor gene duplication from Caenorhabditis elegans Promoterome-driven expression patterns. BMC Genomics. 2007;8:27. doi: 10.1186/1471-2164-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Reece-Hoyes JS, et al. Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Res. 2008;18:2005–15. doi: 10.1101/gr.083055.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove CA, De Masi F, Barrasa MI, et al. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell. 2009;138:314–27. doi: 10.1016/j.cell.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout AJM. Unraveling transcription regulatory networks by protein-DNA and protein-protein interaction mapping. Genome Res. 2006;16:1445–54. doi: 10.1101/gr.5321506. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Delrow J, Henikoff S. Chromatin profiling using targeted DNA adenine methyltransferase. Nat Genet. 2001;27:304–8. doi: 10.1038/85871. [DOI] [PubMed] [Google Scholar]
- Kim TH, Ren B. Genome-wide analysis of protein-DNA interactions. Annu Rev Genomics Hum Genet. 2006;7:81–102. doi: 10.1146/annurev.genom.7.080505.115634. [DOI] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, et al. A genomic regulatory network for development. Science. 2002;295:1669–78. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- Deplancke B, Dupuy D, Vidal M, et al. A Gateway-compatible yeast one-hybrid system. Genome Res. 2004;14:2093–101. doi: 10.1101/gr.2445504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout AJM, Sordella R, Lu X, et al. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science. 2000;287:116–22. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–3. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MM, Reed RR. Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature. 1993;364:121–6. doi: 10.1038/364121a0. [DOI] [PubMed] [Google Scholar]
- Li JJ, Herskowitz I. Isolation of the ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science. 1993;262:1870–4. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- Deplancke B, Mukhopadhyay A, Ao W, et al. A gene-centered C. elegans protein-DNA interaction network. Cell. 2006;125:1193–205. doi: 10.1016/j.cell.2006.04.038. [DOI] [PubMed] [Google Scholar]
- Vermeirssen V, Barrasa MI, Hidalgo C, et al. Transcription factor modularity in a gene-centered C. elegans core neuronal protein-DNA interaction network. Genome Res. 2007;17:1061–71. doi: 10.1101/gr.6148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DA, Zhu H, Zhu X, et al. Regulation of gene expression by a metabolic enzyme. Science. 2004;306:482–4. doi: 10.1126/science.1096773. [DOI] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Barrasa MI, et al. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev. 2008;22:2535–49. doi: 10.1101/gad.1678608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HL, Gunsalus KC. Browsing multidimensional molecular networks with the generic network browser (N-browse) Curr Protoc Bioinformatics. 2008;9:9.11. doi: 10.1002/0471250953.bi0911s23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi AL, Oltvai ZN. Network biology: understanding the cell's; functional organization. Nat Rev Genet. 2004;5:101–13. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- Yu H, Greenbaum D, Xin Lu H, et al. Genomic analysis of essentiality within protein networks. Trends Genet. 2004;20:227–31. doi: 10.1016/j.tig.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Milo R, Shen-Orr S, Itzkovitz S, et al. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–7. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci USA. 2003;100:11980–5. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeger-Lotem E, Sattath S, Kashtan N, et al. Network motifs in integrated cellular networks of transcription-regulation and protein-protein interaction. Proc Natl Acad Sci U S A. 2004;101:5934–9. doi: 10.1073/pnas.0306752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow MC, Martinez NJ, Olsen P, et al. The FLYWCH transcription factors FLH-1, FLH-2 and FLH-3 repress embryonic expression of microRNA genes in C. elegans. Genes Dev. 2008;22:2520–34. doi: 10.1101/gad.1678808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Deplancke B, Barrasa MI, et al. The C. elegans Snail homolog CES-1 can activate gene expression in vivo and share targets with bHLH transcription factors. Nucleic Acids Res. 2009;37:3689–98. doi: 10.1093/nar/gkp232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove CA, Walhout AJ. Transcription factor functionality and transcription regulatory networks. Molecular BioSystems. 2008;4:309–14. doi: 10.1039/b715909a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, MacArthur S, Bourgon R, et al. Transcription factors bind thousands of active an inactive regions in the Drosophila blastoderm. PLoS Biol. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–6. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]