Abstract
Nature is replete with examples of diverse cell types, tissues and body plans, forming very different creatures from genomes with similar gene complements. However, while the genes and the structures of proteins they encode can be highly conserved, the production of those proteins in specific cell types and at specific developmental time points might differ considerably between species. A full understanding of the factors that orchestrate gene expression will be essential to fully understand evolutionary variety. Transcription factor (TF) proteins, which form gene regulatory networks (GRNs) to act in cooperative or competitive partnerships to regulate gene expression, are key components of these unique regulatory programs. Although many TFs are conserved in structure and function, certain classes of TFs display extensive levels of species diversity. In this review, we highlight families of TFs that have expanded through gene duplication events to create species-unique repertoires in different evolutionary lineages. We discuss how the hierarchical structures of GRNs allow for flexible small to large-scale phenotypic changes. We survey evidence that explains how newly evolved TFs may be integrated into an existing GRN and how molecular changes in TFs might impact the GRNs. Finally, we review examples of traits that evolved due to lineage-specific TFs and species differences in GRNs.
Keywords: transcription factors, gene regulatory network, evolution, lineage-specific genes
LINEAGE-SPECIFIC EXPANSION OF TRANSCRIPTION FACTOR FAMILIES
The existence of complex multicellular organisms presents us with a profound challenge: How, mechanistically, did they evolve from their single celled precursors? What drives their continued diversification? What underlies the ability to create a myriad of cell types that interact and cooperate in countless different ways? Answering these great puzzles will require a focus on the issue of genome orchestration—that is, the mechanisms through which initially identical cells can generate the unique protein and functional profiles that define different cell types. Transcription factors (TFs) are key players in this process, serving as early gatekeepers in the steps that determine each cell’s unique protein complement. TFs therefore play critical roles in essentially every function throughout development, from the proliferation and differentiation of stem cells to the maintenance of differentiated cells and tissues in the adult organisms.
TFs typically cooperate to activate or repress the expression of target genes, creating gene regulatory networks (GRNs) that vary in identity in different cells. Because they regulate fundamental processes, TFs are often assumed to be highly conserved across species. This is indeed the case for many of the best-studied TF proteins [1]. However, certain TFs—particularly those encoded by large and evolutionarily active gene families—provide dramatic exceptions to this general rule. Because of their interconnectivity, small differences in the identity and interplay of TFs can have substantial impact on GRNs. Therefore, although cis-regulatory mutations clearly contribute in significant ways to species-specific differences [2] alterations in the structure or expression patterns of TF proteins are also likely to be major determinants of evolution and biodiversity [3]. In the discussion to follow, we focus on the properties and evolutionary histories of TFs and on studies that provide a basis for understanding the potential role their variation plays in determining GRNs and organismal phenotypes. We will use the term ‘transcription factor’ to refer to proteins that bind directly to DNA thereby affecting the expression of target genes. We should point out that transcriptional regulators, such as co-activators or—repressors exist, that do not directly bind to DNA but rather assemble to other TFs bound to DNA.
An overview of TF family evolution
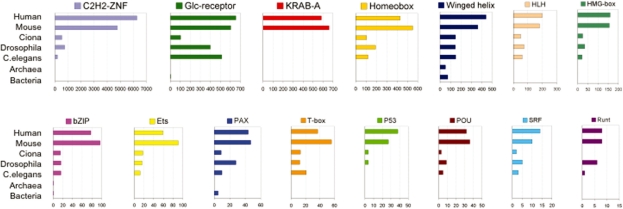
TFs act by binding to specific sites in genomic DNA, typically in or near the genes whose expression they control. Most TFs are composed of multiple sub-structures or ‘domains’, each specialized to make a different contribution to the TF's; overall function. Most importantly, each TF contains one or more specific type(s) of DNA-binding domain (DBD) that determine(s) its recognition of specific DNA target sequences. TF proteins can therefore be usefully classified on the basis of DBD type. Although TF numbers vary substantially from genome to genome, the number of distinct DBD types is relatively small, and most DBDs are ancient [4]. For instance, ‘winged-helix’ and ‘zinc ribbon’ domains are found in all three of life's; superkingdoms (bacteria, archaea, eukarya). Other DBDs are specific to certain evolutionary lineages; examples include the ribbon–helix–helix domain (which is specific to bacteria and archaea) and C2H2-ZNFs, Homeobox box, and T-Box domains which are found only in eukaryotic species (Figure 1). Within the eukaryotes, TF families that are specific to, or prevalent in, certain lineages can also be found. For example, the glucocorticoid receptor-like DBD (a domain of nuclear hormone receptors) is massively expanded in Caenorhabditis elegans, the SRF like DBD is almost exclusively found in plants, and ZN2/Cys6 zinc fingers are fungi-specific [4]. Unfortunately, the incomplete annotation of TFs and DBDs in most sequenced genomes prevents a full assessment of their prevalence and evolutionary histories. In fact, the complete repertoire of TF genes in the human genome was only recently determined [5], and the TF content of other deeply sequenced model organisms is still not entirely known. Nevertheless, an overall picture of TF gene structure, representation and evolution can be gleaned from available EST and genome sequence data.
Figure 1:
DBD repertoire in different taxa. The examples shown correspond to the largest mammalian domain families. For bacteria and archaea, mean numbers are given. All metazoan genomes from which these data were taken are completely sequenced with the exception of Ciona intestinalis, which is currently a draft assembly at 11× coverage. Data are taken from Pfam, Superfamily and ref. [20]. Note that the scale of the x-axis corresponding to the number of proteins of each type in different genomes varies between plots.
Over the course of evolution, different types of DBDs have partnered with other protein domains to create new TFs with distinct functionalities. C2H2-ZNF proteins, for instance, have partnered with one or more KRAB domains in vertebrates, which allows them to interact with the co-factor KAP1 to recruit histone deactelylase complexes to their target genes [6]. The basic helix–loop–helix (bHLH) TFs provide a second example. Their helix–loop–helix motif enables them to form homo- or heterodimers, but some TFs of this class have incorporated additional dimerization domains that can inhibit partnering with other proteins [7]. By incorporating these types of protein interaction domains, the TFs form new dimerization or ‘interactome’ networks in which combined TFs, or TFs and co-factors, determines target choice and regulation [8].
Different types of these novel TFs have been maintained and expanded into large gene families in specific taxonomic orders. The pairing of the C2H2-ZNF DBD with different types of chromatin-interacting motifs provides an excellent example. For instance, the combination of the C2H2 domains with a ZAD motif is found only in insects [9], proteins consisting of C2H2 partnered with FAX motifs predominate in amphibians [10], and the pairing of C2H2-ZNFs with a KRAB domain dominates in vertebrate genomes [11]. In each of these three cases, once the successful domain partnership was established, gene numbers were expanded to create large families of similarly structured genes.
Expansion of TF families
A substantial fraction of genes in every genome that has been studied encode TF proteins, ranging from ∼300 TF genes in E. coli to ∼2000 in the human genome [5, 12]. The genomes of multi-cellular organisms tend to have more TFs per gene [12], suggesting more complex regulatory systems. This dramatic evolutionary increase in TF number cannot yet be entirely reconstructed. However, it is clear that most TF genes are members of gene families that have expanded through DNA sequence duplication events of several different types.
In the history of vertebrate evolution, evidence suggests that two whole-genome duplications (WGD) took place [13, 14], each leading to an initial complete doubling of all genomic information. In some vertebrate lineages (for instance in certain amphibians), further WGDs occurred, creating multiploid species [15]. Subsequent deletions or smaller-scale duplications, however, eliminated many of the new gene copies or created additional copies leading to very complicated family histories. Segmental duplications (SDs), in which smaller segments of DNA are duplicated, and retrotransposition, in which DNA copies of processed gene transcripts are inserted into the genome, have also played roles in gene family expansions. SDs are often generated in tandem, creating arrays of similar genes that are concentrated in specific chromosome regions [16]. Once generated, these tandem duplications provide templates for further duplication and deletion events, via mechanisms involving illegitimate recombination [16]. A recent study has demonstrated accelerated rates of gene duplications and deletions compared to nucleotide substitution in primates, suggesting that this mode of gene family expansion and evolution may have been particular important during human evolution [17].
Indeed, genes that reside in tandem SDs are frequently lineage-specific, and many encode protein functions that are known to be subject to positive selection. Genes involved in immune defense, reproduction, intercellular communication, neural development and transcription are particularly enriched in families that have expanded through tandem SD [18]. These families include genes encoding immunoglobulin G receptors, protocadherins, Cytochrome P450 enzymes, olfactory receptors, RAB, PRAME, the primate-specific FAM90A protein family, as well as the NANOG, homeobox (HOX) and KRAB-ZNF families of TFs [19]. Some TFs have been added to the human TF repertoire by duplication during the last 35-40 million years (Myr) of primate history including predicted FOXD4-, STAT5-, TGIF2L-, RHOXF2- and GTF2-like genes (Nowick et al., in preparation; Table 1). One of the most prolific classes of SD-borne gene families, which has also given rise to a disproportionate large number of recently evolved primate TF genes, encode proteins of the KRAB-ZNF type [20].
Table 1:
Recently duplicated human TFs
| Class | Number | Name |
|---|---|---|
| KRAB-ZNF | 54 | PRDM7, PRDM9, ZFP112, ZKSCAN3, ZKSCAN4, ZNF100, ZNF135, ZNF181, ZNF208, ZNF254, ZNF28, ZNF285A, ZNF286A, ZNF300, ZNF302, ZNF320, ZNF321, ZNF33A, ZNF33B, ZNF354A, ZNF354B, ZNF37A, ZNF417, ZNF419, ZNF440, ZNF443, ZNF468, ZNF479, ZNF493, ZNF552, ZNF585A, ZNF585B, ZNF587, ZNF600, ZNF611, ZNF658, ZNF676, ZNF679, ZNF688, ZNF69, ZNF705A, ZNF705D, ZNF705F, ZNF720, ZNF733, ZNF734, ZNF738, ZNF761, ZNF765, ZNF773, ZNF799, ZNF845, ZNF98, ZNF99 |
| other ZnF-C2H2 | 7 | ZBTB12, ZC3H11A, ZNF322A, ZNF322B, ZNF834, ZXDA, ZXDB |
| Other | 7 | MLL3, PRKRIR, RHOXF2, RHOXF2B, TERF1, TGIF2LX, TGIF2LY |
| Forkhead | 6 | FOXD4, FOXD4L1, FOXD4L3, FOXD4L5, FOXD4L6, FOXO3 |
| bHLH | 3 | GTF2H2, GTF2IRD2, GTF2IRD2B |
| Beta-scaffold-STAT | 2 | STAT5A, STAT5B |
| Beta-scaffold-HMG | 2 | HMG1L1, SP100 |
Locations of the complete set of human TFs [5] were intersected with coordinates of recent segmental duplications in the human genome ([81]; http://humanparalogy.gs.washington.edu, 30 March 2007). Only TFs with an official gene symbol (HGNC symbol) are listed.
An interesting question is if these recent gene copies are functional. If complete open reading frames including at least part of the regulatory elements are duplicated it is likely that the copy can be translated into protein. Newly generated gene copies are typically redundant, and in some cases the extra gene dosage could be harmful to the organism. As a result, most gene duplicates degenerate to become pseudogenes within a few million years [21–23]. To escape elimination from the genome new genes must therefore improve organismal fitness within a very short period of evolutionary time. Therefore, gene copies can follow two general paths: they may subfunctionalize (specialize in a subset of functions performed by the ancestral gene), or they may eventually neofunctionalize (take on novel biological roles).
A common path to functional divergence of gene duplicates involves structural change in protein-coding sequence. For example, coding sequences of about 10% of the lineage-specific duplicates in the human genome show signs of positive selection [24], indicating a drive for the evolution of new protein functions in the duplicated genes. In the case of TFs, new functions can be acquired by changes in the DBD, which can alter the affinity of the TF for DNA binding sequences. Another possibility is sequence divergence in the protein interaction domains, which can affect partnerships with other TFs, cofactors, or chromatin modifying proteins. Some types of TF genes give rise to alternative splice forms that encode distinct domain compositions, and gain or loss of splice sites in these genes can generate isoforms that are unique to particular paralogs [20] or to orthologous genes in different species [25]. Like other types of gene duplicates, new copies of TF genes can also gain or lose functional roles through regulatory changes that alter either the pattern or timing of their expression, translation or the stability of RNA or protein in different tissues. Such alterations in TF expression can substantially reshape the GRNs in which that TF participates. These facts raise the question: how are new or diverged TFs integrated into existing GRNs and what are the functional consequences of that integration for the organism?
Different TF families appear to vary quite dramatically in their inherent susceptibility to evolutionary changes of all kinds. In mammalian genomes, KRAB-ZNFs are on an exceptional evolutionary ‘fast lane’ that sets them apart from most other TFs—and indeed most other types of genes. This distinction can be seen clearly even in comparisons between closely related species. For instance, KRAB-ZNF genes exhibit particularly striking copy number differences in different mammals [26, 27] and display substantial protein sequence divergence even in comparisons between humans and chimpanzees [26]. Our recent studies have confirmed and extended these findings, providing specific examples of species-specific changes in KRAB-ZNF copy number, DBD-sequence divergence and gene expression patterns between the two primate species ([28, 29]; Nowick et al., in preparation). What makes some TF families so much more flexible than others? As we will discuss in the next sections, the position of a TF in the GRN and its evolutionary history dramatically influence it’s susceptibility to change.
STRUCTURE, CHARACTERISTICS AND EVOLVABILITY OF GRNs
The ‘network’ structure of GRNs
Many different types of networks exist in natural and manmade systems, ranging from the biological (e.g. protein–protein interaction networks, GRNs and metabolomic networks) to the technical (e.g. the World Wide Web, citations of scientific publications, or Facebook). The players (nodes) are different in each type of network, but many of these networks have small-world characteristics, in that every node is connected to every other node by only a small number of links (or edges). This property is commonly achieved by a small number of central nodes (hubs) with many connections [30]. Networks typically grow over time by adding nodes. Interestingly new nodes seem to prefer to connect to nodes that already have many connections [30] (e.g. publications that are already famous tend to be cited more frequently by new ones).
The nodes in a GRN represent the genes coding for TFs or TF target genes while the links indicate the transcriptional regulation of the target by a TF. Like many biological networks GRNs have a hierarchical and modular organization (Figure 2). Different scientific fields have developed their own concepts and classification systems for the description of network elements and focus on different aspects of the networks. This section gives a brief overview of these elements (without attempting to be complete) to provide a basis for the discussion of the evolutionary outcome of molecular GRN changes in the following section.
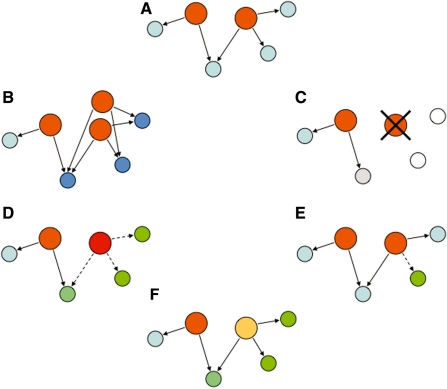
Figure 2:
Schematics of a GRN. Illustration of a typical GRN structure. The nodes represent TFs (orange, large circles) and their targets (blue, small circles). The links represent the regulation of target genes by TFs, indicated by the direction of the arrow. Links between TFs are shown in bold. TFs usually regulate multiple target genes and targets can be regulated by several TFs. Examples for a feed-forward loop (left) and for a bi-fan motif (right) are shown by black-green double arrows. Nodes with many links are called hubs. Subsets of highly interconnected nodes form distinct but interconnected network modules (shaded). GRNs are hierarchically organized. Tiers are labeled according to the different concepts used in the text. Top layer, kernels or initial TFs affect most other modules in the network and are often involved in initiating certain functions or pathways. Bottom layer, differentiation batteries, or terminal TFs act more downstream and usually function in differentiation programs.
The field of network theory focuses, among other issues, on the definition of patterns in a network and mechanisms for network growth and topology change. The investigation of different natural and man-made networks has revealed that certain patterns of connections between nodes occur more frequent than expected by chance [31]. Such so-called ‘network motifs’ form the simplest building blocks of networks and different types of networks are characterized by certain sets of motifs. In GRNs for example the feed forward loop and the bi-fan motif are overrepresented [31]. Feed forward loops consist of two TFs, one regulating the other and both regulating the same target gene, and can function to accelerate or delay the gene regulation of the target [32]. Bi-fan motifs are pairs of TFs that regulate the expression of two common target genes by binding cooperatively to their promoters [33]. Larger subsets of nodes and links are organized into modules. All modules then collectively constitute the network. Modules can be thought of fulfilling one function [34], but many modules are interconnected and nested within each other [4].
In the field of transcriptional regulation, a higher order structural classification for the hierarchy of TFs in a GRN has been developed [34]. The model is based on experimental evidence in yeast and organizes TFs in a stratified nature of three distinct layers: the top, core, and bottom layers. TFs within a layer are highly interconnected and share similar properties. While the TFs of the different layers regulate distinct sets of targets genes, the three layers are also connected by a central skeleton, a feed-forward structure that utilizes the TFs of the top layer to regulate TFs of the core layer, and TFs of the core layer to regulate TFs of the bottom layer. The core layer is characterized by the highest number of TFs and hubs and is important for signal propagation for the regulation of almost all targets. Another study on TF networks in yeast demonstrated that bi-fan motifs typically form extended structures, bi-fan arrays, in which the TF pair regulates a large number of common targets [33]. One major source for the evolution of these bi-fan arrays are TF duplications [33].
Developmental biologists have proposed a concept that concentrates on the aspect of the timely order of events in developmental pathways. In this system modules are classified as kernels, plug-ins, input-output switches and differentiation batteries [35]. Modules can be thought of fulfilling one specific function [36]. Kernels are the initial modules of the network that impact most other parts of the network. They are, for instance, involved in the initiation of the development of certain body parts. In contrast, differentiation batteries may play a role in terminal steps of the differentiation of body parts and do generally not affect other parts of the network. Further, the concept of ‘terminal selector genes’ (terminal TFs) and ‘terminal selector motifs’ (referring to cis-regulatory motifs) has been suggested [37]. These terminal features represent the distal ends of a developmental network that determines, for instance, the exact identity of a neuron during the post-mitotic differentiation step.
Interactions in networks can be directional: links going into a node are called incoming edges, while links going out of a node are referred to as outgoing edges. Some TFs (‘hub TFs’) are characterized by a relatively high number of outgoing but a smaller number of incoming edges. This property reflects the fact that these TFs regulate a large number of targets but are themselves regulated by only a small number of other TFs. For instance in the yeast GRN, <5 TFs regulate 93% of all target genes [38]. Other TFs, which regulate fewer targets, serve as ‘fine tuners’. Target genes can possess a few to many incoming links. The sum of the incoming edges on a target gene defines the combinatorial effect of TFs on the target’s overall regulation.
Another important aspect of biological networks is their dynamics. In a given network, the activity of and the connections between nodes can be quickly adjusted if conditions change. This has been demonstrated nicely for the yeast GRN [39]. While some TFs were expressed in all five of the conditions investigated in this study, about half of the TFs in this network were uniquely expressed in only one condition. Moreover, depending on the conditions, extensive rewiring took place so that only 10% of the links (‘hot links’, mostly regulating house keeping functions) were found active in all conditions. The authors showed further that endogenous processes, such as cell cycle or sporulation, are regulated in a more complex way, involving more cross-talk and feed forward loops between the TFs. Processes in response to external stimuli, such as stress response, on the other hand are optimized for speed. In these pathways, the GRN changes such that each target is regulated by a smaller number of TFs, which transmit their regulatory signals more directly with fewer intermediate regulators [39].
In addition to these short-term network changes, biological networks can further change over longer evolutionary time periods. A comparison of GRNs across different prokaryotes revealed that TFs are less conserved than their targets and that lineage-specific TFs are constantly added to form species-specific GRNs [40]. Both, phylogenetic distance and similarity in the lifestyle of the organisms determine how similar the networks are between the species [40]. A detailed discussion of evolutionary changes in network structure will follow in the next chapters. However, we wish to emphasize that in the context of GRNs TFs can fulfill different functions depending on the conditions, and that TFs that serve as hubs in one condition might not be hubs in another.
Hierarchical structure of a network increases adaptability
The hierarchical organization of GRNs predicts distinct classes of mutational outcomes. Mutations of the most central players (e.g. hub or core TFs) and of initial modules (e.g. kernels or top TFs), are expected to have the most dramatic impact because they influence many other genes in the network [30, 35]. Computational modeling supports this prediction, and bolsters the inference that hub TFs evolve more slowly because their mutation tends to disrupt critical features of the network [41]. In agreement with these findings, core and ‘top-layer’ TFs are generally more highly conserved [34] and the removal of hub genes from the yeast network is often lethal [42]. In metazoans, mutations in kernel TFs, are typically highly consequential and often lethal [35]. These observations explain why kernels are highly conserved between species and metazoans have broadly similar body plans [35].
However, if mutations in hub TFs are not detrimental, they can serve as drivers of species differences. This was nicely demonstrated by Crombach and Hogeweg [43] by computational modeling of digitally simulated organisms. Starting with reasonable assumptions about network behavior they showed that hub genes, or genes with direct input to a hub, act as ‘evolutionary sensors’, and that duplications or deletions of those genes can impact the GRN substantially, allowing the digital organisms to adapt to a changing environment. Further, comparisons between orthologous TFs in two yeast species demonstrated that TFs with high connectivity or ‘betweenness’ (which is an indicator of a central location in the network) have higher dN/dS values [44], pointing to positively selected change in central network nodes. In another study, higher variability in gene expression levels were observed for top-layer TFs compared to their counterparts in the core or bottom layers between individual yeasts and interpreted as favorable for triggering phenotypic adaptations (27).
While the importance of hub TFs seems intuitive and has been supported by several studies (as discussed above) this view has been challenged by a study by Evangelisti and Wagner [45]. These authors compared the frequency of different types of changes between highly connected TFs and sparsely connected TFs in yeast. They observed no differences in dN/dS values or duplication rates of the TFs, nor differences in the duplication rate of the TF’s target genes, arguing that the network is not more sensitive to changes in highly connected TFs. Another study compared GRNs across different prokaryotes and found no correlation between the degree of connectiveness of a TF and its conservation [40]. Therefore, these concepts remain a matter of intense debate, and worthy subjects of further experimentation.
In contrast to hubs, mutations in TFs with fewer connections or involved in terminal differentiation processes, can be predicted to have more subtle effects because they do not affect other modules [35]. Loss or gain of a terminal TF could lead, for instance, to loss of a specific neuron-type or the evolution of a novel one, but would unlikely compromise other viable functions of the organism [37]. Such changes appear to occur more frequently than changes in central or initial network components and can also be a source for individual or species differences. For example, when genes of the olfactory pathways were compared between three worm species, developmental TFs showed the highest diversity and divergence [46].
Do orthologous TFs regulate orthologous targets? Studies in yeast, worm and fly suggest that they do, if sequence similarity is high [42]. Despite this fact, GRNs of distant taxa can differ dramatically. For example, a recent analysis of gene co-expression networks in humans and mice revealed that only the global aspects of network architecture are conserved between these species [47]. Less than 10% of co-expressed gene pairs are shared between the human and mouse network, and genes that are hubs in one species are not necessarily hubs in the other [47]. This dramatic flux in the co-expressed gene sets that make up local network features could not occur without extensive changes in network integration. Altered network integration has even been observed between orthologous human and chimpanzee genes [48] and TFs [29] in brain co-expression networks.
The hierarchical organization of the GRN therefore allows for both drastic and subtle phenotypic changes via modification of central and terminal TFs, respectively. But, has this organization evolved by natural selection or is it simply a by-product of the mechanisms by which networks grow and change? Models based on the simple reasonable assumptions that duplications are the major force for network growth and that divergence of duplicated genes leads to re-wiring, have demonstrated that modules and hubs can emerge spontaneously from these simple dynamical rules alone [49]. However, no matter which forces underlie network architecture, GRN evolvability seems to benefit from it [49]. Furthermore, it is not the identity, but the position of a TF in the network that is the main determinant of its effects on the organism. While many changes might be neutral and have no affect on the GRN, a few small changes in a top-layer or hub TF might completely alter GRN architecture and possibly lead to speciation as at least one modeling study has shown [50]. Or, as Oliveri and Davidson [36] put it: TFs are in principle interchangeable, the architecture of their connections in the network is the critical issue for function.
IMPACT OF MOLECULAR CHANGES ON GRNs
Types of molecular changes
Different molecular changes affect network structure in different ways (Figure 3). It is typically assumed that a duplicated TF initially inherits all links of its parent TF which will lead to a stronger regulation of all the target genes. Deletion of a TF interrupts the transcriptional regulation of downstream genes by this TF. Protein sequence changes in a TF (trans) can be consequential for all genes regulated by this TF. In contrast, a sequence change in the promoter of a target gene (cis) affects only the link between the TF and this target. Expression change of a TF (caused by cis change in its promoter or trans change upstream of this TF) can in turn alter the expression of all its target genes.
Figure 3:
Predicted impact of molecular changes on GRN. Variations of a simple model of a network consisting of two TFs (orange/red/yellow, large circles), four target genes (blue/green/gray, small circles) and their interactions (black arrows). (A) Putative ancestral GRN. (B) Duplication of a TF. New TF inherits all links of parent TF which leads to a stronger regulation of the target genes. (C) Deletion of a TF. Two targets of this TF are not regulated anymore, while the third target is now only regulated by the other TF. (D) Protein sequence change in a TF (trans). Regulation of all targets is changed and can be stronger or weaker. Targets also regulated by other TFs are less affected. (E) Sequence change in the promoter of one target gene (cis). Only the link between the TF and this target is affected and leads to different regulation. (F) Expression change of one TF (caused by cis change in its promoter or trans change upstream of this TF). Expression of all target genes is affected. The effect on targets regulated also by other TFs is smaller.
However, since genes are typically regulated by the combinatoric effect of several TFs [51], in most cases the outcome of a TF change of any type will be an altered instead of a complete breakdown of the regulation of downstream genes. Genes regulated by many other TFs are expected to be less affected than genes regulated by a fewer number of TFs. GRNs may be particularly sensitive to hub TF mutations, since any change in the balance between cooperating and competing TFs could affect the expression of hundreds of downstream target genes.
Although there are many examples of TFs with deeply conserved expression patterns, changed timing, levels, or location of TF gene expression are clearly linked to morphology differences between species. For example, variation in the onset and amplitude of BMP4 expression is one of the major determinants of beak morphology in Darwin finches and other birds [52, 53]. Another dramatic example is provided by the varied Pitx1 gene expression on pelvic development in three-spined sticklebacks and other vertebrate species [54]. Many other examples support the importance of subtle changes of TF gene expression on the evolution of morphological traits [2].
Another level of complexity in gene regulation is added by the fact that TFs can dimerize via interaction domains. TF families that frequently form homo- or heterodimers to bind DNA include the bHLH, bZIP, and nuclear receptors (NRs) [8]. Gene duplication events have added family members to each of these groups in different taxa. Subsequent divergence of the paralogs by loss or gain of interaction domains, as well as the evolution of new alternative splice forms that include (or miss) specific interaction domains have altered the ability of the proteins to dimerize with different partners [8]. For example, the protein SHP is one of the hubs in the NR interactome network. SHP is a member of the NR family but has lost its DBD, and SHP-containing dimers are therefore not able to bind to DNA. In this way, SHP has evolved as a negative switch that can shut down gene regulation by the NR interactome network [55]. Similar mechanisms to quickly interfere with gene regulation if conditions change have been postulated for the bHLH interactome network [7].
Network growth and integration of new TFs into existing networks
As outlined above, some TF families have expanded by gene duplication events throughout evolution, or specifically in certain lineages. These duplications play a major role in network growth [56] and contribute significantly to species-specific topologies of the network [40]. How are these novel TFs integrated into an existing network? Since the new ‘daughter’ TF inherits all of the parent’s interactions, daughters of TFs with many connections are more likely to have many connections as well. In yeast and Escherichia coli more than two-thirds of the TFs have at least one target in common with their paralog [56]. However, for most duplicated genes, the duplication event is followed by a period of rapid DNA sequence [24] and gene expression divergence [57]. In fact, it has been shown computationally that, starting from two connected nodes, these two basic assumptions—gene duplication and divergence—are sufficient to produce networks with very similar characteristics to real biological networks (Duplication–Divergence model [58]).
Duplications can range from small-scale (encompassing single genes), over SDs (encompassing genomic regions up to several megabases) to whole genome duplications. The fate of the duplicated gene seems to depend, at least partially, on the type of duplication through which it was created [59] and on its functional class [60, 61]. While genes created by small scale or SD need to be selectively advantageous for the organism to become fixed in the population, and functionally diverge for both parent and daughter copies to be preserved, genes created by WGD duplications must survive rearrangements and gene loss rather than be fixed as an individual gene [59]. Due to these distinct characteristics of the two different duplication modes, it can be predicted that functional groups of genes might differ in their preference for one or the other duplication mode. For example, dosage-sensitive genes and genes that encode members of protein complexes are unlikely to get fixed after SD [59]. Accordingly, TFs were significantly underrepresented among SD genes in yeast [59, 60]. In contrast to this finding, our recent studies demonstrated that one family of TFs, KRAB-ZNFs are overrepresented in SDs in the human genome (Table 1; Nowick et al., manuscript in preparation). These findings suggest that KRAB-ZNFs have properties that distinguish them from the bulk of TF genes; for example, it seems likely that proteins of this type may be able to bind DNA targets without the need to partner with other, stabilizing TFs. While still speculative, this possibility is supported by the fact that most proteins of this class have long DBDs and, in cases where target genes are known, exceptionally long DNA recognition sequences [20].
The different modes of duplications further differ in their evolutionary impact on the GRN. In case of WGD, the complete GRNs with all of their genes and links are duplicated. In general, it was observed in yeast that WGD paralogs more frequently than SD paralogs share the same interaction partners [60]. On the other hand, WGD paralogs share fewer TF binding sites and diverge more quickly in expression pattern [60, 62]. The WGD that occurred in yeast about 100 million years ago, was followed by extensive rewiring of the daughter GRNs that gave rise to two relatively independent sub-networks with different functionalities [63]. The duplicated modules typically show evidence of subfunctionalization, or a ‘division of labor’, rather than behaving as redundant copies. For example, one daughter pathway may react under high glucose while the other one operates under low glucose levels [63]. Additional examples of such functional innovations are described in the following chapter.
Small scale duplications may include a gene with only an incomplete set of regulatory sequences, or can be dispersed in that the paralog locates to a different genomic location. Consequently, such paralogs frequently demonstrate different expression patterns than parental genes [61]. Rapid expression divergence of genes duplicated by such small-scale events has been found in several taxonomic groups. For instance, it has been demonstrated that recently generated paralogs diverge quickly in their expression patterns in human tissues, presumably gaining and losing co-expressed partners during this process [64]. This study also found that one gene copy was typically distinguished by many connections, while the other paralog had only a few. Because loss of function is more likely than gain, it can be presumed that this pattern usually reflects the loss of connections by one paralog. In yeast and flies, duplicated genes have been observed to change more often in expression between species compared to single copy genes [57] and lineage-specific duplications are thought to contribute disproportionately to expression divergence between human and mouse [57].
Recent studies on KRAB-ZNF genes provide examples for rapid sequence divergence of TFs ([28, 65]; Nowick et al., manuscript in preparation). These genes, that primarily duplicate within SDs also diverge rapidly in expression patterns and therefore represent an exception to the general rule mentioned above. We have shown that KRAB-ZNFs located in neighboring positions within genomic clusters (typically, tandemly duplicated paralogs) display different expression patterns in human tissues ([20]; but see [66, 67]) and even recently duplicated KRAB-ZNF gene copies can display distinct expression patterns (Nowick et al., manuscript in preparation). We have also demonstrated that the most recently duplicated human KRAB-ZNFs (generated during the last 35–40 million years) show signs of positive selection, specifically in the DNA binding domains ([28, 68], Nowick et al., manuscript in preparation). These sequence variations predict that some parent and recent daughter genes encode proteins with distinctly different DBDs, and therefore very likely have different network connections.
Evolutionary rates of duplicated genes also depend on their interaction partners. If both paralogs interact with the same proteins via the same binding domain in the other protein (the same interface), their interactions are mutually exclusive. In this case, paralogs often evolve different expression patterns, or they acquire sequence changes to gain new interaction partners [69]. On the other hand, proteins that can interact with multiple proteins at the same time (multi-interface) typically evolve more slowly [69].
EXAMPLES OF PHENOTYPIC CHANGES DRIVEN BY TF AND GRN CHANGES
Analysis of species differences at a system biology level (e.g. GRN-based analysis), is still a new endeavor. However, system-level approaches will be essential to understanding how changes in DNA sequence or gene expression patterns contribute to the process through which new species evolve and how changes in the transcription factor repertoire might translate into species-specific biology. In the following sections, we discuss examples for the evolution of species differences due to TF duplication and GRN changes.
New traits originating from TF duplication
The role that gene duplication can play in the evolution of new traits is nicely illustrated by recent studies of the fate of duplicate copies of the TF, Runt. The network connection between Runt and hedgehog existed in the common ancestor of cephalochordates and vertebrates and is important for cartilage formation. The duplication of Runt and acquisition of new network partners of this new gene copy early during vertebrate evolution opened the door for development of teeth and skeleton in fish and tetrapods [70].
Both sub- and neofunctionalization are seen in recently duplicated members of the nuclear hormone receptor family of TFs [71]. One example is the mineralocorticoid receptor (MR, encoded by the gene NR3C2) and the glucocorticoid receptor (GC, NR3C1), which originated by duplication from an ancestral corticoid receptor ∼450 million years ago. The ancestral receptor can be activated by the hormones aldosterone and cortisol. It can be shown that a few protein sequence mutations in GC resulted in loss of its sensitivity to aldosterone, giving rise to a cortisol-specific receptor. Interestingly, MR has a strong preference for the tetrapod-specific hormone aldosterone that was not around at the time of the ancestral corticoid receptor. This indicates that once aldosterone evolved it co-opted the old receptor into a new functional partnership that controls electrolyte homeostasis.
Although it involved a study of protein–protein interaction networks and not a GRN, another interesting example involves the evolution of the apoptosis pathway. Mining existing eukaryotic genomes for orthologous genes, Castro and colleagues [72] tracked the origin of the human apoptosis pathway. They make a compelling case for the recruitment of BCL2 and the caspase gene family from evolutionarily older DNA repair and chromosome stability pathways around the time of origin of metazoa. In this way, a pathway was born that allowed multi-cellular organisms to eliminate damaged cells that threaten the health of the organism. Lineage-specific duplications then added individual genes creating increasing complexity and species diversity in the apoptosis pathway. This study provides an exemplary model for future studies to investigate the evolution of other pathways.
Species differences in GRN wiring
The so-called ‘micromer’ cells, which form the larval skeleton of sea urchins, provide an example of the origin of a new cell type. These cells form a mesodermal cell lineage that is unique to sea urchin larvae, but they are also involved in endomesoderm specification. How then, do other echinoderms without micromere cells form endo- and mesoderm? A comparison of the GRN responsible for endomesoderm specification in sea urchin and sea star reveals that even though the same TFs are involved, extensive re-wiring has taken place [73]. For example in sea urchin, the TF, Tbrain, is exclusively expressed in micromeres, while in sea star it is expressed in the endomesoderm. The Otxβ1/2 promoter has undergone sequence changes between the two species and is regulated by Tbrain in sea urchin but by Blimp1 in sea star. In the sea urchin, FoxA represses mesoderm formation, whereas in sea star, this function is fulfilled by GataE. These TFs were already in place in the ancestor of both species, but have been co-opted to fulfill the same function in different developmental contexts.
In the last case discussed, closely related species changed the way TFs are wired into GRNs in order to achieve similar developmental outcomes, but such changes can also generate the kinds of phenotypic differences that distinguish species. For example, while many components of the GRN necessary for embryonic stem cell differentiation are conserved between humans and mouse, the WNT and BMP pathways show species-specific differences [74]. Differences in the identity of co-expressed genes, (which point to altered TF interactions), support the inference that the WNT pathway is involved in maintenance of pluripotency and stem cell differentiation in mouse, while the human WNT pathway contributes more strongly to the differentiation of these cells. Similarly, mouse BMP is necessary for maintenance of pluripotency, while human BMP induces trophoectoderm differentiation. Pluripotency in human stem cells is in turn maintained through signaling of ACTIVIN/NODAL.
Important clues to network evolution can be gleaned from the comparison of homologous GRNs in closely related species. For example, differences in modules of co-expression networks between humans and chimpanzees have been identified in several brain regions [29, 48]. Interestingly, consistent with the different evolutionary histories of the brain regions examined, the largest number of differences in connections between the human and chimpanzee modules were found in the cerebral cortex, while other brain areas were more similar between both species [48]. Prominent among the genes that have gained connections in the human compared to the chimpanzee prefrontal cortex network are genes that influence energy metabolism [48]. Our recent study has linked a specific TF module to the upregulation of brain energy metabolism genes in humans [29]. This module is enriched for recently duplicated KRAB-ZNF genes, raising the intriguing possibility that the newly evolved TFs reshaped the primate GRN in a way that supported increased energy metabolism in the human prefrontal cortex. These findings are especially interesting in the light of the exceptional energy demands of the larger human brain [75, 76] and the critical role that the prefrontal cortex plays in behaviors and skills that are especially developed in humans.
Differences in GRN modules can also define differences between related cell types in the same species. The differentiation into different neuronal cell types, e.g. of the 118 anatomically defined classes of neurons in C. elegans, is probably determined by similar but cell type-specific modules of very simple structure and logic [37]. Similarly, Winden et al. [77] suggest that cell diversity, for instance differences between major cell types in the human brain, astrocytes, microglia, oligodendrocytes and GABAergic and glutamatergic neurons, is achieved by several quantitative, continuous characteristics like the expression ratio of co-expressed genes. Differences in hub genes have been characterized as the most important factors for this cell type determination. It is likely that similar mechanisms support the differentiation other cell types. It is easy to imagine that the modular structure of GRNs makes it relatively easy to incorporate new lineage-specific TFs with the potential of altering or creating new cell types, and enabling diversity, for example of neuronal subtypes, immune system cell types and novel cell–cell interactions. The rapidly evolving KRAB-ZNF genes, many of which are expressed in immune and neuronal cells [20], are excellent candidate TFs that could play key roles in shaping cell-type diversity in these systems.
CONCLUSIONS
Ultimately, the phenotypic consequences of changes in TF genes can only be understood in the context of regulatory networks. Due to the complex architecture and dynamics of GRNs, there is no simple mapping from genome to phenotype. A network change can be either drastic or imperceptible, depending on the nature of the change and the position of the TF within the network. While the hubs and kernels of GRNs are usually relatively conserved, terminal genes and differentiation batteries are evolution’s playground. It seems likely that the most rapidly evolving classes of TFs, like the KRAB-ZNF family in mammals [20, 65], the ZAD-ZNF family in insects [9, 78] and the nuclear receptor family in nematodes [4], all of which have experienced extensive expansion, pruning and divergence during the course of evolution, perform critical but flexible roles at the periphery of regulatory networks.
Many studies have demonstrated a surprising lack of conservation of network links across species [29]. Are these links indeed species-specific links or artifacts? The functional importance of any given link in a GRN is difficult to determine, and it is often hard to infer how many nodes or links could be removed before a GRN would break down. Just as most mutations leading to protein sequence [79] and gene expression [80] changes are neutral or nearly neutral, it may turn out that most mutations affecting network structure are also neutral. In the future it will be necessary to develop statistical instruments that can detect positive or negative selection on network links. But progress will also depend on improved annotation of TF genes in a larger number of species, together with high-throughput experimental methods to determine their target genes, partnerships, competitions, as well as their expression and variation between species.
Progress on each of these fronts will bolster our understanding of the functions of each player in a given GRN, and allow us to develop and test improved models of network structure and function that will illuminate the connections between evolutionary changes in biological networks and species-specific biology.
Key points.
Different families of transcription factors (TFs) have expanded in different evolutionary lineages.
The hierarchical organization of the gene regulator network (GRN) allows for different degrees of phenotypic change.
The impact of molecular changes in TFs depends on the nature of the change and the position of the TF in the GRN.
GRN differences may explain many species-specific traits, but predicting the outcome or functional importance of any given change is still difficult given the current state of knowledge regarding gene function.
FUNDING
This work was supported by a grant from the U.S. National Institutes of Health, National Institute of General Medical Sciences, grant number GM78368 (awarded to L.S.).
ACKNOWLEDGEMENTS
We thank Tim Gernat, Elbert Branscomb, Aron Branscomb, Sarah London and Christopher Balakrishnan for helpful critiques of the article.
Biographies
Katja Nowick is a post-doctoral researcher in Lisa Stubbs’ group. Her research interests are human and primate evolution with a focus on differences in gene regulation.
Lisa Stubbs is Professor of Cell and Developmental Biology at the University of Illinois. Her research focuses on comparative genomics and the evolution of regulatory networks, with emphasis on the rapidly evolving KRAB-ZNF transcription factor family.
References
- Ranz JM, Machado CA. Uncovering evolutionary patterns of gene expression using microarrays. Trends Ecol Evol. 2006;21:29–37. doi: 10.1016/j.tree.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA. The locus of evolution: evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- Babu MM, Luscombe NM, Aravind L, et al. Structure and evolution of transcriptional regulatory networks. Curr Opin Struct Biol. 2004;14:283–91. doi: 10.1016/j.sbi.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, et al. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10:252–63. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- Vissing H, Meyer WK, Aagaard L, et al. Repression of transcriptional activity by heterologous KRAB domains present in zinc finger proteins. FEBS Lett. 1995;369:153–7. doi: 10.1016/0014-5793(95)00728-r. [DOI] [PubMed] [Google Scholar]
- Amoutzias GD, Robertson DL, Oliver SG, et al. Convergent evolution of gene networks by single-gene duplications in higher eukaryotes. EMBO Rep. 2004;5:274–9. doi: 10.1038/sj.embor.7400096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoutzias GD, Robertson DL, Van de Peer Y, et al. Choose your partners: dimerization in eukaryotic transcription factors. Trends Biochem Sci. 2008;33:220–9. doi: 10.1016/j.tibs.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Chung HR, Lohr U, Jackle H. Lineage-specific expansion of the zinc finger associated domain ZAD. Mol Biol Evol. 2007;24:1934–43. doi: 10.1093/molbev/msm121. [DOI] [PubMed] [Google Scholar]
- Nietfeld W, Conrad S, van Wijk I, et al. Evidence for a clustered genomic organization of FAX-zinc finger protein encoding transcription units in Xenopus laevis. J Mol Biol. 1993;230:400–12. doi: 10.1006/jmbi.1993.1158. [DOI] [PubMed] [Google Scholar]
- Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4:231. doi: 10.1186/gb-2003-4-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nimwegen E. Scaling laws in the functional content of genomes. Trends Genet. 2003;19:479–84. doi: 10.1016/S0168-9525(03)00203-8. [DOI] [PubMed] [Google Scholar]
- Kuraku S, Meyer A, Kuratani S. Timing of genome duplications relative to the origin of the vertebrates: did cyclostomes diverge before or after? Mol Biol Evol. 2009;26:47–59. doi: 10.1093/molbev/msn222. [DOI] [PubMed] [Google Scholar]
- Escriva H, Manzon L, Youson J, et al. Analysis of lamprey and hagfish genes reveals a complex history of gene duplications during early vertebrate evolution. Mol Biol Evol. 2002;19:1440–50. doi: 10.1093/oxfordjournals.molbev.a004207. [DOI] [PubMed] [Google Scholar]
- Fisher MT, Nagarkatti M, Nagarkatti PS. Aryl hydrocarbon receptor-dependent induction of loss of mitochondrial membrane potential in epididydimal spermatozoa by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) Toxicol Lett. 2005;157:99–107. doi: 10.1016/j.toxlet.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Eichler EE. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet. 2006;7:552–64. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- Hahn MW, Demuth JP, Han SG. Accelerated rate of gene gain and loss in primates. Genetics. 2007;177:1941–9. doi: 10.1534/genetics.107.080077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuth JP, Hahn MW. The life and death of gene families. Bioessays. 2009;31:29–39. doi: 10.1002/bies.080085. [DOI] [PubMed] [Google Scholar]
- Nowick K, Huntley S, Stubbs L. Rapid expansion and divergence suggest a central and distinct role for KRAB-ZNF genes in vertebrate evolution. In: Yoshida K, editor. Focus on Zinc Finger Protein Research. Research Signpost. 2009. pp. 13–29. [Google Scholar]
- Huntley S, Baggott DM, Hamilton AT, et al. A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 2006;16:669–77. doi: 10.1101/gr.4842106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. The evolution of functionally novel proteins after gene duplication. Proc Biol Sci. 1994;256:119–24. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–5. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Ohno S. Gene duplication and the uniqueness of vertebrate genomes circa 1970-1999. Semin Cell Dev Biol. 1999;10:517–22. doi: 10.1006/scdb.1999.0332. [DOI] [PubMed] [Google Scholar]
- Han MV, Demuth JP, McGrath CL, et al. Adaptive evolution of young gene duplicates in mammals. Genome Res. 2009;19:859–67. doi: 10.1101/gr.085951.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FC, Chen CJ, Ho JY, et al. Identification and evolutionary analysis of novel exons and alternative splicing events using cross-species EST-to-genome comparisons in human, mouse and rat. BMC Bioinformatics. 2006;7:136. doi: 10.1186/1471-2105-7-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CD, Fledel-Alon A, Williamson S, et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–7. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Bustamante C, Clark AG, et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AT, Huntley S, Tran-Gyamfi M, et al. Evolutionary expansion and divergence in the ZNF91 subfamily of primate-specific zinc finger genes. Genome Res. 2006;16:584–94. doi: 10.1101/gr.4843906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowick K, Gernat T, Almaas E, et al. Differences in human and chimpanzee gene expression patterns define an evolving network of transcription factors in brain. Proc Natl Acad Sci USA. 2009;106:22358–63. doi: 10.1073/pnas.0911376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi AL, Oltvai ZN. Network biology: understanding the cell's; functional organization. Nat Rev Genet. 2004;5:101–13. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Milo R, Shen-Orr S, Itzkovitz S, et al. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–7. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci USA. 2003;100:11980–5. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JJ, Thornton JM. Evolutionary models for formation of network motifs and modularity in the Saccharomyces transcription factor network. PLoS Comput Biol. 2007;3:1993–2002. doi: 10.1371/journal.pcbi.0030198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jothi R, Balaji S, Wuster A, et al. Genomic analysis reveals a tight link between transcription factor dynamics and regulatory network architecture. Mol Syst Biol. 2009;5:294. doi: 10.1038/msb.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Davidson EH. Development. Built to run, not fail. Science. 2007;315:1510–1. doi: 10.1126/science.1140979. [DOI] [PubMed] [Google Scholar]
- Hobert O. Regulatory logic of neuronal diversity: terminal selector genes and selector motifs. Proc Natl Acad Sci USA. 2008;105:20067–71. doi: 10.1073/pnas.0806070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelzim N, Bottani S, Bourgine P, et al. Topological and causal structure of the yeast transcriptional regulatory network. Nat Genet. 2002;31:60–3. doi: 10.1038/ng873. [DOI] [PubMed] [Google Scholar]
- Luscombe NM, Babu MM, Yu H, et al. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431:308–12. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- Madan Babu M, Teichmann SA, Aravind L. Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J Mol Biol. 2006;358:614–33. doi: 10.1016/j.jmb.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Stewart AJ, Seymour RM, Pomiankowski A. Degree dependence in rates of transcription factor evolution explains the unusual structure of transcription networks. Proc Biol Sci. 2009;276:2493–501. doi: 10.1098/rspb.2009.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Greenbaum D, Xin Lu H, et al. Genomic analysis of essentiality within protein networks. Trends Genet. 2004;20:227–31. doi: 10.1016/j.tig.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Crombach A, Hogeweg P. Evolution of evolvability in gene regulatory networks. PLoS Comput Biol. 2008;4:e1000112. doi: 10.1371/journal.pcbi.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovelin R, Phillips PC. Evolutionary rates and centrality in the yeast gene regulatory network. Genome Biol. 2009;10:R35. doi: 10.1186/gb-2009-10-4-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelisti AM, Wagner A. Molecular evolution in the yeast transcriptional regulation network. J Exp Zool B Mol Dev Evol. 2004;302:392–411. doi: 10.1002/jez.b.20027. [DOI] [PubMed] [Google Scholar]
- Jovelin R, Dunham JP, Sung FS, et al. High nucleotide divergence in developmental regulatory genes contrasts with the structural elements of olfactory pathways in Caenorhabditis. Genetics. 2009;181:1387–97. doi: 10.1534/genetics.107.082651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaparas P, Marino-Ramirez L, Bodenreider O, et al. Global similarity and local divergence in human and mouse gene co-expression networks. BMC Evol Biol. 2006;6:70. doi: 10.1186/1471-2148-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Horvath S, Geschwind DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc Natl Acad Sci USA. 2006;103:17973–8. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sole RV, Valverde S. Spontaneous emergence of modularity in cellular networks. J R Soc Interface. 2008;5:129–33. doi: 10.1098/rsif.2007.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Tusscher KH, Hogeweg P. The role of genome and gene regulatory network canalization in the evolution of multi-trait polymorphisms and sympatric speciation. BMC Evol Biol. 2009;9:159. doi: 10.1186/1471-2148-9-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon B, Tjian R. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 2000;14:2551–69. doi: 10.1101/gad.831000. [DOI] [PubMed] [Google Scholar]
- Abzhanov A, Protas M, Grant BR, et al. Bmp4 and morphological variation of beaks in Darwin's; finches. Science. 2004;305:1462–5. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- Wu P, Jiang TX, Suksaweang S, et al. Molecular shaping of the beak. Science. 2004;305:1465–6. doi: 10.1126/science.1098109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MD, Bell MA, Kingsley DM. Parallel genetic origins of pelvic reduction in vertebrates. Proc Natl Acad Sci USA. 2006;103:13753–8. doi: 10.1073/pnas.0604706103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoutzias GD, Pichler EE, Mian N, et al. A protein interaction atlas for the nuclear receptors: properties and quality of a hub-based dimerisation network. BMC Syst Biol. 2007;1:34. doi: 10.1186/1752-0509-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichmann SA, Babu MM. Gene regulatory network growth by duplication. Nat Genet. 2004;36:492–6. doi: 10.1038/ng1340. [DOI] [PubMed] [Google Scholar]
- Li WH, Yang J, Gu X. Expression divergence between duplicate genes. Trends Genet. 2005;21:602–7. doi: 10.1016/j.tig.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Vazquez A, Flammini A, Maritan A, et al. Modeling of protein interaction networks. ComPlexUs. 2003;1:38–44. [Google Scholar]
- Davis JC, Petrov DA. Do disparate mechanisms of duplication add similar genes to the genome? Trends Genet. 2005;21:548–51. doi: 10.1016/j.tig.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Guan Y, Dunham MJ, Troyanskaya OG. Functional analysis of gene duplications in Saccharomyces cerevisiae. Genetics. 2007;175:933–43. doi: 10.1534/genetics.106.064329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casneuf T, De Bodt S, Raes J, et al. Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana. Genome Biol. 2006;7:R13. doi: 10.1186/gb-2006-7-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N. Comparative analysis indicates regulatory neofunctionalization of yeast duplicates. Genome Biol. 2007;8:R50. doi: 10.1186/gb-2007-8-4-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. Functional partitioning of yeast co-expression networks after genome duplication. PLoS Biol. 2006;4:e109. doi: 10.1371/journal.pbio.0040109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WY, Albert R, Albert I, et al. Rapid and asymmetric divergence of duplicate genes in the human gene coexpression network. BMC Bioinformatics. 2006;7:46. doi: 10.1186/1471-2105-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadepally HD, Burger G, Aubry M. Evolution of C2H2-zinc finger genes and subfamilies in mammals: species-specific duplication and loss of clusters, genes and effector domains. BMC Evol Biol. 2008;8:176. doi: 10.1186/1471-2148-8-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'G;een H, Squazzo SL, Iyengar S, et al. Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs. PLoS Genet. 2007;3:e89. doi: 10.1371/journal.pgen.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel MJ, Guelen L, de Wit E, et al. Human heterochromatin proteins form large domains containing KRAB-ZNF genes. Genome Res. 2006;16:1493–1504. doi: 10.1101/gr.5391806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson RO, Thomas JH. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 2009;5:e1000325. doi: 10.1371/journal.pgen.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PM, Lu LJ, Xia Y, et al. Relating three-dimensional structures to protein networks provides evolutionary insights. Science. 2006;314:1938–41. doi: 10.1126/science.1136174. [DOI] [PubMed] [Google Scholar]
- Hecht J, Stricker S, Wiecha U, et al. Evolution of a core gene network for skeletogenesis in chordates. PLoS Genet. 2008;4:e1000025. doi: 10.1371/journal.pgen.1000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean AM, Thornton JW. Mechanistic approaches to the study of evolution: the functional synthesis. Nat Rev Genet. 2007;8:675–88. doi: 10.1038/nrg2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro MA, Dalmolin RJ, Moreira JC, et al. Evolutionary origins of human apoptosis and genome-stability gene networks. Nucleic Acids Res. 2008;36:6269–83. doi: 10.1093/nar/gkn636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman VF, Yankura KA, McCauley BS. Evolution of gene regulatory network architectures: examples of subcircuit conservation and plasticity between classes of echinoderms. Biochim Biophys Acta. 2009;1789:326–32. doi: 10.1016/j.bbagrm.2009.01.004. [DOI] [PubMed] [Google Scholar]
- Sun Y, Li H, Liu Y, et al. Evolutionarily conserved transcriptional co-expression guiding embryonic stem cell differentiation. PLoS One. 2008;3:e3406. doi: 10.1371/journal.pone.0003406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokoloff L. Quantitative measurements of cerebral blood flow in man. Methods Med Res. 1960;8:253–61. [PubMed] [Google Scholar]
- Varki A, Altheide TK. Comparing the human and chimpanzee genomes: searching for needles in a haystack. Genome Res. 2005;15:1746–58. doi: 10.1101/gr.3737405. [DOI] [PubMed] [Google Scholar]
- Winden KD, Oldham MC, Mirnics K, et al. The organization of the transcriptional network in specific neuronal classes. Mol Syst Biol. 2009;5:291. doi: 10.1038/msb.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HR, Schafer U, Jackle H, et al. Genomic expansion and clustering of ZAD-containing C2H2 zinc-finger genes in Drosophila. EMBO Rep. 2002;3:1158–62. doi: 10.1093/embo-reports/kvf243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. Preponderance of synonymous changes as evidence for the neutral theory of molecular evolution. Nature. 1977;267:275–6. doi: 10.1038/267275a0. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Weiss G, Lachmann M, et al. A neutral model of transcriptome evolution. PLoS Biol. 2004;2:E132. doi: 10.1371/journal.pbio.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Ventura M, She X, et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]