Abstract
The reconstitution of translation initiation complexes from purified components is a reliable approach to determine the complete set of essential canonical initiation factors and auxiliary proteins required for the 40S ribosomal subunit to locate the initiation codon on individual mRNAs. Until now, it has been successful mostly for formation of 48S translation initiation complexes with viral IRES elements. Among cap-dependent mRNAs, only globin mRNAs and transcripts with artificial 5′ leaders were amenable to this assembly. Here, with modified conditions for the reconstitution, 48S complexes have been successfully assembled with the 5′ UTR of beta-actin mRNA (84 nucleotides) and the tripartite leader of adenovirus RNAs (232 nucleotides), though the latter has been able to use only the scanning rather then the shunting model of translation initiation with canonical initiation factors. We show that initiation factor 4B is essential for mRNAs that have even a rather moderate base pairing within their 5′ UTRs (with the cumulative stability of the secondary structure within the entire 5′ UTR < −13 kcal/mol) and not essential for beta-globin mRNA. A recombinant eIF4B poorly substitutes for the native factor. The 5′ UTRs with base-paired G residues reveal a very sharp dependence on the eIF4B concentration to form the 48S complex. The data suggest that even small variations in concentration or activity of eIF4B in mammalian cells may differentially affect the translation of different classes of cap-dependent cellular mRNAs.
Translation initiation is now recognized to play an important role in gene expression in eukaryotes in the course of viral infection, nutritional adaptation, stress, apoptosis, development and cell division (11). However, in most cases, the molecular mechanisms for this translational control remain mysterious. This is largely accounted for by our poor understanding of the molecular events that underlie the basic mechanisms of translation initiation in eukaryotes, especially in mammals. As mammalian systems are poorly amenable to genetic approaches, alternative ways need to be elaborated to address this problem. To this end, it was suggested several years ago to use an approach that is based on the assembly of translation initiation complexes from individual translational components combined with toeprinting (22). (Toeprinting is based on the primer extension inhibition of reverse transcription from an oligodeoxynucleotide, which is hybridized 3′ to the initiation codon of an mRNA. The arrest of reverse transcriptase always occurs at the same positions, +16 to +18 nucleotides 3′ of A in the AUG initiation codon.) This approach has proven to be fruitful in the elucidation of the function of several viral internal ribosome entry sites (IRES) (11). Surprisingly, it has proven to be unsuccessful with cap-dependent transcripts if their 5′ UTRs comprised even short stem-loop structures enriched in G-C pairs (26). Among mammalian mRNAs with natural 5′ UTRs, only globin mRNAs have been reported to result in high yields of the 48S complexes (23, 27).
For standard cap-dependent mRNAs, the scanning machinery is thought to require the following set of components: the 40S ribosomal subunit, initiation factors eIF1, eIF1A, eIF2, eIF3, eIF4A, eIF4B, eIF4F, the initiator tRNA, ATP, and GTP. The experiments performed with rabbit beta-globin mRNA have demonstrated that this set of components (eIF1, eIF1A, eIF4A and eIF4B were used as recombinant proteins) is sufficient to place the 40S subunit at the AUG codon and the 48S complex thus formed is competent for further incorporation into the final 80S initiation complex (23, 24). The data obtained in our laboratory have shown that the same reconstitution protocol does not result in assembly of 48S complexes with either 5′ UTRs of natural mRNAs (such as mammalian beta-actin mRNA or late adenoviral mRNAs) or even simpler model 5′ UTRs. In all these cases, the toeprint signal was hardly visible or not detected at all. These unexpected negative results prompted us to undertake an extensive search for an missing essential initiation factor(s) and modified experimental conditions which would allow the assembly of 48S translation initiation complexes with natural 5′ UTRs other than the 5′ UTR of beta-globin mRNA. Otherwise, our inability to assemble initiation complexes with such mRNAs would limit application of the reconstitution technique to cap-dependent mRNAs that possess even more complex 5′ UTRs.
Here, we report a successful reconstitution of 48S complexes with mRNAs carrying the 5′ UTR of tripartite leader of late adenoviral mRNAs (232 nucleotides long) and beta-actin mRNA (84 nucleotides). We show that unlike viral IRES elements studied to date and the beta-globin mRNA, initiation factor eIF4B is absolutely essential to reconstitute 48S complexes with mRNAs that have a rather moderate base pairing involving G residues within their 5′ UTRs. Its quality (eIF4B is poorly stored), method of preparation (recombinant or native factor) and concentration have a paramount importance to succeed in the reconstitution of 48S complexes with such 5′ UTRs.
MATERIALS AND METHODS
Plasmid constructs.
Plasmid pbG was described earlier (6). The plasmid with cDNA containing the tripartite leader (tripartite leader) of late mRNAs of adenovirus Ad2 was a kind gift of B. Naroditsky. Plasmid pTbG containing the tripartite leader fused to the beta-globin mRNA sequence at the 3′ end and to the T7 promoter at the 5′ end was obtained from this plasmid by PCR with 5′GTAATACGACTCACTATAGGACTCTCTTCCGCATCGCTGTC3′ and 5′AAGCTTCTTGCGACTGTGACTGGTTAG3′ as forward and reverse primers, respectively, followed by blunt end-ligation of this PCR product to that obtained by PCR from pbG with primers 5′ACTTGCAATCCCCCAAAACAG3′ and TTACGAGCTCAAGGGGCTTCATG. The resulting fusion product was finally blunt end-ligated into pUC18 at the KpnI and Ecl136II sites. Construct pTd1 was obtained from pTbG by excision of the Sca-XhoI fragment from tripartite leader, blunt-ending of the large fragment and religation. Constructs pTd2 and and pTd3 were prepared in the same way after excision of the PflMI-XhoI and PflMI-HindIII fragments, respectively.
Constructs pTd4, pTd5, pTd6, pT3Td3, and pbTd5 were prepared by PCR of the entire plasmid pTd3 with 5′ AATGGTGCATCTGTCCAGTG 3′, and 5′ CTGGCCCTCGCAGACAG 3′, 5′ CCTGCGAGGGCCAG 3′, and 5′ CCTATAGTGAGTCGTATTACG 3′, 5′ AAGCTTACTTGCAATCC 3′, and 5′ AGCGATGCGGAAGAGAG 3′, 5′ GATTTCCAGAGCTTACTTGC 3′, and 5′ GCAGACAGCGATGC 3′, 5′ GCCAGCAATCCCCCAAAAC 3′, and 5′ CCTCGCAGTTGTGTCAAAAGC 3′ as forward and reverse primers, respectively, followed by ligation of the ends of the resulting linear PCR products. Plasmid pbTd3 and pTd3b were obtained from pTd3 by oligonucleotide-directed mutagenesis with 5′ GTAATACGACTCACTATAGGACACTTGCTTTTGACACAACTGTGTACTCTCTTCCGCATCGCTGTC 3′ and 5′ GCTGTCTGCGAGGGCCAGACACTTGCTTTTGACACAACTGTGTTTACTTGCAATCCCCCAAAAC 3′, respectively, with the GeneEditor system (Promega) as recomended by manufacturer.
Plasmid pAbG encoding the 5′ UTR of human beta-actin mRNA fused to the beta-globin mRNA coding sequence was obtained as follows. A PCR product was prepared from human brain cDNA (a kind gift of E. Nadezhdin) by PCR with primers 5′ GTAATACGACTCACTATAGGACCGCCGAGACCGCGTCC 3′, and 5′ CCCACGATGGAGGGGAAGAC 3′. The product was ligated into pUC18 at SmaI. Another PCR product was obtained from plasmid pbG with primers 5′ CCCATGGTGCATCTGTCCAG 3′, and 5′ TTACGAGCTCAAGGGGCTTCATG 3′. This product comprising the coding sequence of beta-globin mRNA with the NcoI site around the initiation codon was ligated into pUC18 at the SmaI site. The NcoI-BamHI fragment was then excised from the resulting construct and ligated at the NcoI-BamHI sites into the first plasmid containing the beta-actin sequence.
Plasmids pTSbG and pTd1S, derivatives of pTbG and pTd1 containing a low-energy stem at the HindIII site (position 204 in pTbG), were obtained by self-annealing of the oligonucleotide 5′ AGCTT(CGCGGATCCGCG)5A 3′ followed by insertion of the resulting duplex containing HindIII sticky ends into the HindIII site of pTbG and pTd1, respectively.
In vitro transcription.
Plasmids pbG, pAbG, pTbG and all their derivatives were linearized prior to transcription by digestion at the EcoRI site located in the 3′-terminal part of globin coding sequence which is present in all constructs. All the transcripts used in our study were cotranscriptionaly capped. The transcription for the production of capped RNA was carried out as described previously (4, 9).
Preparation of 40S ribosomal subunits, initiation factors, and Met-tRNAiMet.
The 40S ribosomal subunits were prepared from cytoplasmic extracts of ascites cells Krebs-2 rather than from rabbit reticulocyte lysate to avoid any possible contamination of the subunits with endogenous beta-globin mRNA according to the protocol described in (19). Factors eIF2, eIF3, and eIF4A were prepared from rabbit reticulocyte lysate as described previously (19) with inclusion of FPLC as a final step of purification. This was especially important for eIF3 which is normally contaminated with many other initiation factors including eIF4F, and eIF4B. For this, the chromatographic MonoQ HR 5/5 column (Amersham Biosciences) and a KCl gradient from 100 to 500 mM was used. Recombinant eIF4B was expressed and isolated according to the procedure described in (22). The native eIF4B was isolated from HeLa cell cytoplasmic extract kindly provided by R. Luhrmann with the protocol described before (19) and additionally purified by fast protein liquid chromatography (FPLC) with a KCl gradient from 100 to 500 mM. Apparently homogeneous eIF4B eluted from the MonoQ column at 310 mM. eIF4F was isolated as recommended (10).
Recombinant eIF1 and eIF1A were prepared as described before (23, 27). eIF4H was isolated from ribosomal salt wash of rabbit reticulocyte lysate with the protocol described previously (30). Aminoacylation of tRNAiMet in the total calf liver tRNA (Novagen) was carried out as described previously (22) with aminoacyl-tRNA synthetase fraction isolated from Escherichia coli MRE600. Plasmid pTRM-1 containing the sequence of tRNAiMet was obtained from T. Pestova and C. Hellen. In vitro transcribed unmodified tRNAiMet was prepared as recommended (25) followed by its aminoacylation with the aminoacyl-tRNA synthetase from E. coli MRE600.
Assembly and analysis of 48S translation initiation complexes.
Ribosomal 48S initiation complexes were assembled in a reaction volume of 20 μl as described earlier (22, 23). The reaction mixture (20 μl) contained 40S subunits (2.5 pmol), an mRNA (0.5 pmol), eIF1 (0.5 μg), eIF1A (0.5 μg), eIF2 (2 μg), eIF3 (3 μg), eIF4F (0.5 μg), eIF4A (0.5 μg), eIF4B (up to 0.5 μg, see text), eIF4H (see text), Met-tRNAi (5 pmol), ATP (1 mM), and GTP (0.4 mM). 48S complexes were analyzed either by sucrose gradient sedimentation (22) or by primer extension as described previously (22, 23) with a primer 5′-TCACCACCAACTTCTTCCAC-3′ complementary to nucleotides 114 to 133 of the rabbit beta-globin mRNA sequence. Electrophoresis of the resulting cDNA was performed by denaturing 6% polyacrylamide gel electrophoresis (PAGE). Radioactive bands were visualized, and relative amounts of radioactivity in the bands were determined (when appropriate) with a Phosphorimager (Molecular Dynamics). Assembly and toeprinting of 48S complexes in nuclease-treated rabbit reticulocyte lysate (RRL) was performed as described (5).
Determination of the eIF4B content in the rabbit reticulocyte lysate by quantitative Western blotting.
Five microliters of rabbit reticulocyte lysate (Promega) along with different amounts of purified native eIF4B were subjected to 10% PAGE. Proteins were transferred to nitrocellulose membranes under semidry conditions (Semi-Phor, Hoefer Scientific Instruments), and then membranes were processed as described earlier (22) with polyclonal mice eIF4B antiserum diluted 1,000-fold. The antibodies were raised in mice with recombinant eIF4B prepared as indicated in (22). Bands were revealed with an ECL system (Amersham Biosciences). After scanning the bands, the amount of eIF4B in the aliquot of RRL was determined with the calibration plot obtained from control bands of eIF4B (run on the same gel).
RESULTS
Inefficient assembly of 48S complexes with cap-dependent mRNAs that have some base pairing within their 5′ UTRs with the original protocol of reconstitution.
Our initial interest was focused on the tripartite leader of late adenoviral RNAs. This mRNA is believed to use both the classical scanning mechanism of translation initiation and the shunting mode (33). The latter mechanism has been reported to operate preferentially in human adenovirus infected or heat shock-stressed cells (33). RNA transcripts carrying tripartite leader are efficiently translated in rabbit reticulocyte lysate (data not shown). In our design, the tripartite leader and all other 5′ UTRs used in this study were fused to the beta-globin mRNA sequence at position 26 of beta-globin mRNA (Fig. 1A). Bearing in mind that beta-globin mRNA is actively reconstituted into stable 48S complexes (23, 24, 27), such a design was intended to minimize any possible negative effect of other nontested coding sequences on the reconstitution of these complexes and any possible variation in stability of 48S complexes dependent on the nucleotide context around the initiation codon.
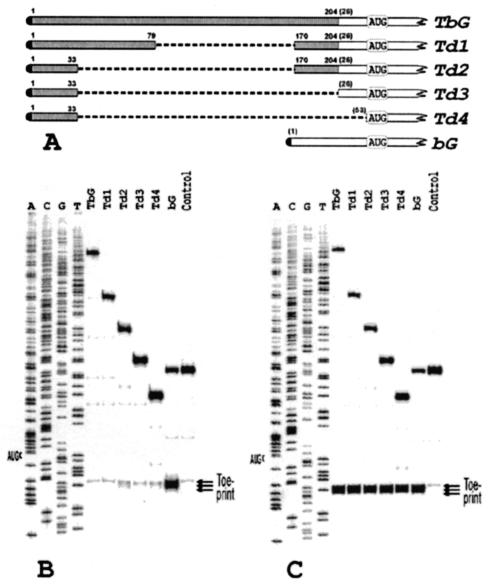
FIG. 1.
(A) A schematic representation of the tripartite leader (TPL) of adenovirus 2 and its deletion derivatives fused to beta-globin mRNA. Shaded and open bars represent the tripartite leader and beta-globin sequences, respectively. Figures without parentheses above the bars show the terminal nucleotides of the tripartite leader that are still present in the constructs. Those in parentheses denote nucleotides from the beta-globin sequence. (B) Primer extension analysis (toeprinting) of 48S initiation complexes assembled with the transcripts shown in A. Lane control corresponds to the complex reconstituted in the absence of Met-tRNAi and eIF2. The reconstitution was performed with total tRNA (where only tRNAiMet was aminoacylated) and recombinant eIF4B. A dideoxynucleotide sequence of pTbG generated with the same primer was run in parallel. For other details see the text. (C) Toeprint analysis of 48S initiation complexes formation in rabbit reticulocyte lysate in the presence of GMP-PNP with the same set of transcripts. In lane control, the assembly was inhibited by addition of 30 mM Mg2+.
Figure 1B shows that the transcript with the complete tripartite leader (construct TbG in Fig. 1A) is inactive in forming 48S complexes with the original protocol of reconstitution (22, 23). As the unique structure of tripartite leader may have some specific initiation factor requirements, a series of tripartite leader deletions of increasing length was prepared (see Fig. 1A). In this way, the specific structure of tripartite leader was destroyed and converted into regular 5′ UTRs. Before reconstitution, the mRNAs were assayed for formation of 48S complexes in the rabbit reticulocyte lysate system and were found to be as active as beta-globin mRNA (Fig. 1C). However, only construct Td2 demonstrated slight formation of the 48S complex in the purified system. Other constructs were almost inactive (Fig. 1B). From these data, one may also infer that the yield of the complex does not correlate with the size of a 5′ UTR, as construct Td3 was less active than the longer construct Td2 (compare lane Td2 with Td3 in Fig. 1B).
Among these constructs, Td3 was arbitrarily selected for further studies. In this beta-globin mRNA derivative, 25 5′-terminal nucleotides of the 5′ UTR of beta-globin mRNA are replaced with 33 5′-terminal nucleotides of the tripartite leader (Fig. 1A). Addition of the missing 5′-terminal section of the beta-globin mRNA 5′ UTR to Td3 (constructs bTd3 and Td3b, Fig. 2A) did not result in a significant restoration of the 48S complex formation (Fig. 2B). It was clear that the inability of Td3 to form the 48S complex was accounted for by these 35 nucleotide residues derived from the tripartite leader. This segment of Td3 was then arbitrarily divided into the left (5′-terminal) and the right (3′-terminal) parts. Each part was separately deleted from Td3, and the resulting transcripts (Td5 and Td6, Fig. 2A) were assayed for assembly of 48S complexes. As shown in Fig. 2B, the RNA lacking the left part of the segment (Td5) was absolutely inactive in 48S complex formation, whereas the yield of the complex for RNA Td6 with the deleted right part was similar to that for the control beta-globin mRNA (compare lanes Td5, Td6, and bG in Fig. 2B). It was concluded, therefore, that the G-rich sequence GCGAGGGCCAGAGC causes a strong negative effect on the reconstitution of the 48S complex from purified components.
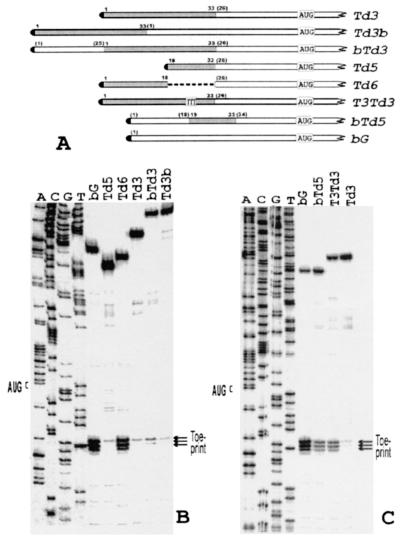
FIG. 2.
(A) A schematic representation of construct Td3 and its different modifications. Boxed TTT in construct T3Td3 denote a substitution of three consecutive G residues in construct Td3 for three T residues. Other designations are as in Fig. 1A. (B) Toeprint analysis of 48S initiation complexes assembled on mRNAs bG, Td3, Td5, Td6, bTd3, and Td3b. A dideoxynucleotide sequence generated with the same primer corresponds to construct Td3. (C) Toeprint analysis of 48S initiation complexes reconstituted with model mRNAs bTd5 and T3Td3. For comparison, the complexes assembled on Td3 and bG were analyzed in parallel. A dideoxynucleotide sequence generated with the same primer corresponds to construct Td3.
When the 5′ UTR of beta-globin mRNA was interrupted with the sequence GCGAGGGCC at position 23 (construct bTd5), the yield of 48S complexes, compared to the intact 5′ UTR of bG, substantially decreased (compare lanes bG and bTd5 in Fig. 2C). On the other hand, substitution of just three G residues for three T residues in the inhibitory sequence of Td3 (construct T3Td3 in Fig. 2A) resulted in a substantial increase of the toeprint signal (Fig. 2C, compare lanes Td3 with T3Td3).
The reconstitution in a complete RRL gave a strong toeprint signal for Td3 (Fig. 1C), suggesting the presence in RRL of an essential component specific for G-rich sequences which was missing in the purified system. Searching for this component in the ribosomal high-salt wash fraction was unsuccessful. The crude ribosomal salt wash fractions either did not stimulate the reconstitution or caused an inhibitory effect. The UV-induced protein cross-linking in RRL of transcripts Td3 and Td6 that were labeled on G residues resulted in exactly the same pattern of cross-linked proteins (data not shown). These data taken together argued against the presence in RRL of a special protein that helps the ribosome to overcome this negative effect by specific binding to the G-rich sequence. This suggested that the structure of the 5′ UTR of Td3 by itself could not be overcome by the canonical initiation factors used in these experiments.
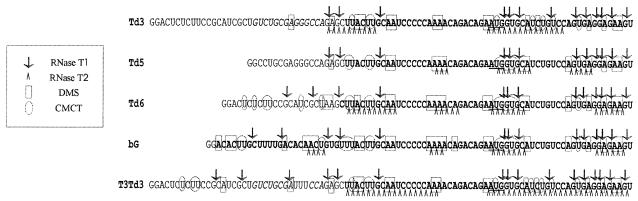
To analyze this structure, chemical and enzymatic probing of 5′-terminal parts of Td3, Td5, Td6, and beta-globin mRNA (bG, taken as a control), was performed. These probing data are described in detail in a separate report (6). Some of them are presented in Fig. 3. The probing revealed not only a complete protection of nucleotide residues within the inhibitory sequence of the 5′ UTR of Td3 but also protection of some nucleotides upstream of it. No protection of the 5′-terminal nucleotides was revealed either in Td6 or in the 5′ UTR of beta-globin mRNA. The 5′ UTR of Td6 appeared to be accessible to nuclease or chemical attack over its entire length with the exception of a short sequence immediately preceding the initiation codon. The same was true for the 5′ UTR of beta-globin mRNA. When three G residues in the inhibitory sequence of construct Td3 were substituted for three U's, resulting in construct T3Td3, the 5′-terminal nucleotides become accessible for probing reagents (compare probing for Td3 and T3Td3, Fig. 3) and this correlates with a higher efficiency of incorporation of T3Td3 mRNA into 48S complexes (Fig. 2C). Thus, after the mutation GGG→UUU, the 5′-terminal section of the 5′ leader is no longer involved in base pairing.
FIG. 3.
Accessibility of nucleotide residues in the 5′ termini of constructs bG, Td3, Td5, Td6, and T3Td3 to chemical and enzymatic attack (for details see 6). The sequence corresponding to the beta-globin mRNA is shown in bold. In our hands, the C- and U-tracts are poorly attacked by RNases and chemicals, and hence their secondary structure status is not clear.
Surprisingly, however, computer modeling of the secondary structure of Td3 did not result in any conventional stem-loop structure within its 5′ UTR. The overall stability of the secondary structure comprising the entire 5′ UTR did not exceed −13 kcal/mol and, according to published data, should have been easily overcome by the RNA unwinding apparatus (31, 32). This demonstrates that even moderate changes in base pairing within 5′ UTRs involving G residues produce rather strong effects on the ability of the 40S ribosomal subunit to locate the initiation codon.
These results strengthened our impression that the original protocol for reconstitution of 48S initiation complexes was insufficient to overcome even a moderate and imperfect base pairing (presumably involving G residues) within the 5′ UTR of an mRNA and focused our efforts on studies of the RNA unwinding process and factors that mediate it.
Initiation factor eIF4B is essential for the reconstitution of 48S initiation complexes with mRNAs that have some base pairing within their 5′ UTRs.
The inability of mRNAs to form 48S complexes that have base pairing within their 5′ UTRs may be accounted for by either the existence of an additional RNA-helicase (or additional partners of eIF4A) in mammalian cells (14 and references cited therein) or some imperfection of the helicase reaction in the purified system. As mentioned above, a search for additional factors to facilitate such mRNAs incorporation into 48S complexes was unsuccessful. Therefore, the attention was focused on the known components of the helicase system, factors eIF4F, eIF4A, and eIF4B. Increasing concentrations of eIF4F and eIF4A (both factors were native) did not result in any improvement of the toeprint signal (data not shown).
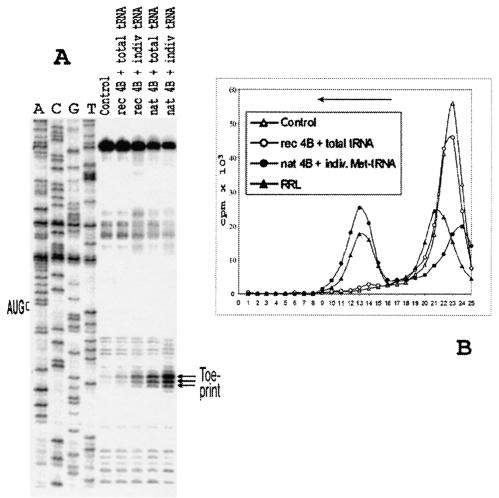
Two important changes were found to be instrumental to overcome the base pairing within the 5′ UTR of Td3. The total tRNA (where only its minor fraction is represented by Met-tRNAiMet) was replaced by in vitro transcribed tRNAiMet that had been shown to adequately substitute for the natural tRNAiMet in the 48S complex formation (25). This permitted us to avoid a sequestration of RNA-binding initiation factors by an excess of tRNA (26). The recombinant eIF4B was substituted for by native eIF4B purified to homogeneity from HeLa cells. These two changes resulted in an efficient reconstitution of the 48S complex with Td3 as revealed by both primer extension inhibition (Fig. 4A) and sucrose gradient centrifugation (Fig. 4B).
FIG. 4.
(A) Toeprint analysis of 48S initiation complexes assembled on mRNA Td3 with various combinations of eIF4B and Met-tRNAi preparations. rec 4B and nat 4B, recombinant and native eIF4B, respectively. Total tRNA corresponds to the total calf liver tRNA (Novagen), where only tRNAiMet was amynoacylated; indiv. tRNA corresponds to individual Met-tRNAi. A dideoxynucleotide sequence of pTd3 generated with the same primer is shown to the left. (B) Sucrose gradient sedimentation of 48S complexes assembled under different conditions on mRNA Td3. The mRNA was capped and labeled by incorporation of [32P]UTP by T7 transcription reaction. Curve control corresponds to the complex reconstituted in the absence of Met-tRNAiMet and eIF2. The direction of sedimentation is shown by the arrow.
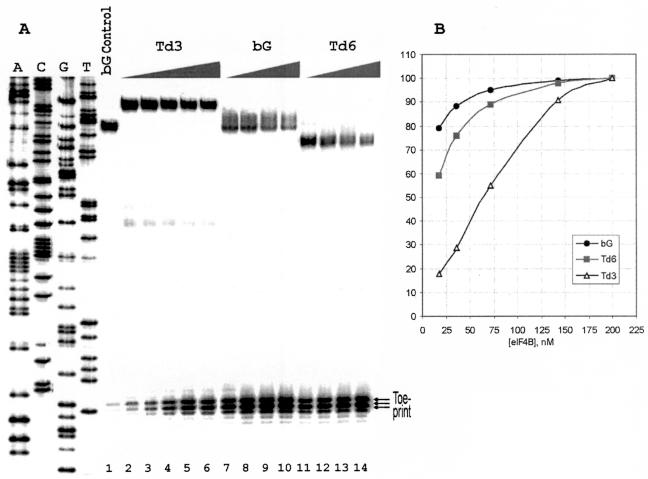
These data prompted us to compare the intact 5′ UTR of beta-globin mRNA, the 5′ UTR of transcript Td6 which is lacking the inhibitory sequence and the 5′ UTR of Td3 for their eIF4B requirement. To this end, the yield of the 48S complex was assessed for these transcripts in dependence of concentration of eIF4B in the reaction mixture. The corresponding toeprints are presented in Fig. 5A. The data of this eIF4B titration experiment plotted in Fig. 5B show a sharp dependence of the yield of the 48S complex on variations in eIF4B concentration for Td3 and a much lower requirement for eIF4B with the Td6 and bG constructs. This is in contrast with the requirement for eIF4F (Table 1). Omission of eIF4F dramatically reduces the formation of 48S complexes for both Td3 and bG mRNAs (Td6 was not tested), though the assembly with transcript Td3 is again affected somewhat more severely.
FIG. 5.
(A) Toeprinting of 48S complexes assembled on mRNAs Td3, bG, and Td6 with addition of different amounts of native eIF4B. The amounts of eIF4B per standard volume of the reconstitution mixture (20 μl) were as follows: lanes 2 and 7, 11 to 0.025 μg, lanes 3 and 8, 12 to 0.05 μg, lanes 4 and 9, 13 to 0.1 μg, lanes 5 and 10, 14 to 0.2 μg, and lane 6, 0.3 μg. The yields of 48S complexes for Td6 and bG with 0.3 μg of eIF4B were the same as with 0.2 μg of eIF4B, and the respective lanes are not presented in the figure. No exogenous eIF4B was added to the samples corresponding to lanes 2, 7, and 11. The indicated amount of eIF4B in these samples is accounted for by contamination of affinity purified eIF4F with eIF4B. It was estimated by quantitative Western blotting as 0.05 μg of eIF4B/1 μg of eIF4F. Lane bG control corresponds to the 48S complex reconstituted with mRNA bG in the absence of Met-tRNAiMet and eIF2. A dideoxynucleotide sequence was generated from pTd3. (B) Dependence of formation of 48S complexes assembled on mRNAs bG, Td3, and Td6 on the molar concentration of eIF4B in the reconstitution mixture. The yield of 48S complexes (radioactivity in toeprint bands) was determined with phosphorimager from data presented in A and presented as the ratio to the maximal yield of the complexes for each mRNA.
TABLE 1.
Effect of single omission of initiation factors of the helicase complex on the yield of 48S complexes assembled on mRNAs bG and Td3
| Factor omitted | Relative toeprinting efficiency (%)
|
|
|---|---|---|
| bG | Td3 | |
| None | 100 | 100 |
| eIF4Aa | 73 | 25 |
| eIF4Bb | 80 | 20 |
| eIF4F | 13 | <10c |
In this case, the system contained ∼110 nM eIF4A as a eIF4F subunit compared to its usual concentration of 550 nM of free eIF4A and 110 nM of eIF4A from eIF4F.
It was not possible to exclude eIF4B completely due to its slight contamination of the eIF4F preparation (final concentration ∼20 nM compared to regular 300 nM).
It's difficult to define an exact value because the relative error is quite large when the signal is only slightly above background.
It should be stressed that the concentration of eIF4B used throughout this work did not exceed that in rabbit reticulocyte lysate. This conclusion was inferred from determination of eIF4B concentration in the commercial RRL (Promega) by quantitative Western blotting (see Materials and Methods). The Promega RRL routinely used in many laboratories for translation assays contained eIF4B at a concentration estimated to be 250 to 300 nM (data not shown). This value suggests that eIF4B is a rather abundant initiation factor in mammalian cells. The maximal amount of eIF4B in our reconstitution system did not exceed 0.5 μg/20 μl of the reconstitution mixture which corresponds to an eIF4B concentration of about 300 nM.
Initiation factor eIF4H cannot substitute for eIF4B in the reconstitution of the 48S complex with transcript Td3.
eIF4H was isolated as a factor that stimulated translation initiation of beta-globin mRNA under reduced concentration of eIF4B in the reconstituted reticulocyte lysate system (30). eIF4H was also found to assist eIF4F and eIF4A in unwinding of model RNA-RNA and RNA-DNA duplexes in the absence of eIF4B or under its reduced concentrations (31). Its amino acid sequence is homologous to the N-terminal part of eIF4B containing the RRM domain. To some extent, this small factor (25 kDa) may be regarded as eIF4B lacking its C-terminal section comprising the second unspecific RNA-binding site (20). It was therefore of interest to analyze the effect of eIF4H in our modified reconstitution system.
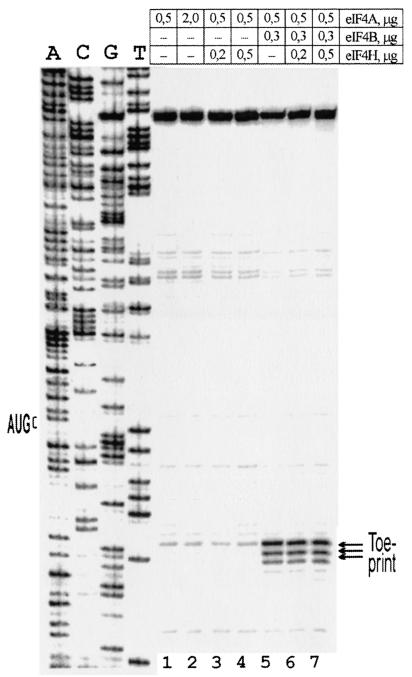
Figure 6 shows the effect of native eIF4H addition to the assembly system with mRNA Td3. No effect of eIF4H was found either with a complete omission of eIF4B or partial eIF4B depletion. These data indicate that eIF4H cannot substitute for eIF4B in unwinding of the 5′ UTR of our model mRNAs under these assay conditions. However, this does not rule out the possibility that eIF4H may still play some role in the unwinding of 5′ UTRs with specific secondary structures. Figure 6 (lanes 1 and 2) also shows that in the absence of eIF4B, a fourfold increase in the content of eIF4A does not cause any positive effect on the intensity of the toeprint. This is in contrast with data obtained with artificial RNA-RNA duplexes where an increase of eIF4A concentration stimulated RNA-unwinding even in the absence of eIF4B (31).
FIG. 6.
Effect of substitution of eIF4B for eIF4H on the reconstitution of 48S complexes with mRNA Td3. Recombinant and native eIF4H gave similar results. The data in the figure were obtained with native eIF4H. Amounts of factors eIF4A, eIF4B, and eIF4H indicated above the lanes are given per standard volume of the reconstitution mixture (20 μl). A dideoxynucleotide sequence of pTd3 is presented to the left.
Assembly of 48S complexes with 5′ UTRs from beta-actin and late adenoviral mRNAs.
Once the conditions of 48S complexes reconstitution for the model structured 5′ UTRs were established, it was pertinent to verify whether the modified protocol worked in the case of natural structured 5′ UTRs. For this, two 5′ UTRs were tested—the 5′ UTR from beta-actin mRNA (84 nucleotides) and the complete tripartite leader from late adenoviral mRNAs (>200 nucleotides) which, as described above, initiated these studies (Fig. 7A). The secondary structure of the 5′ UTR of beta-actin mRNA is not known. However, as its G+C content is ≈76%, there is no doubt about significant base pairing within this 5′ UTR involving G and C residues. In addition, when selecting the beta-actin mRNA, we aimed to adopt a new natural reference (instead of beta-globin mRNA) for future studies of IRES-less mRNAs with the same assembly technique.
FIG. 7.
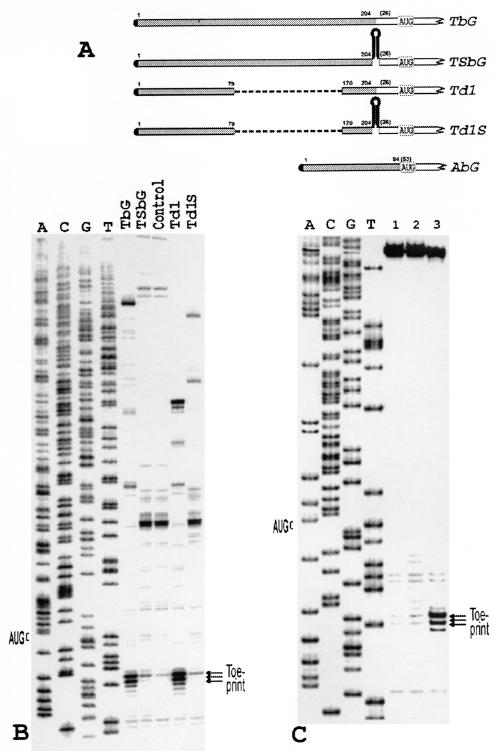
(A) A schematic representation of constructs TbG, Td1, its derivatives with a low-energy stem-loop structures inserted into position 204 of the tripartite leader, and construct AbG. Shaded bars represent the tripartite leader or beta-actin sequences. Other designations are as in Fig. 1A. (B) Reconstitution of the 48S complex with the mRNAs TbG, Td1, TSbG, and Td1S. The individual in vitro-transcribed tRNAiMet and native eIF4B were used in this experiment. Control shows the assembly in the absence of Met-tRNAiMet and eIF2. A dideoxynucleotide sequence was obtained from pTbG. (C) Assembly of the 48S complex with the mRNA carrying the 5′ UTR of human beta-actin mRNA. 1, control (no Met-tRNAiMet, no eIF2); 2, reconstitution with the total tRNA and recombinant eIF4B; 3, reconstitution with in vitro-transcribed individual Met-tRNAiMet and native eIF4B (0.5 μg/20 μl). A dideoxynucleotide sequence generated from pAbG with the same primer was run in parallel.
The beta-actin mRNA may be regarded as a standard cap-dependent mammalian mRNA. As before, the 5′ UTR of beta-actin mRNA was linked to the beta-globin coding sequence (Fig. 7A), a standard coding sequence used in all experiments described in this report. Unlike the 5′ UTR of beta-actin mRNA, the secondary structure of the tripartite leader has been studied in detail (34). It represents a single-stranded 5′-terminal sequence followed by a series of short stem-loops collectively forming a flower-like structure. Neither of these 5′ UTRs was able to direct formation of 48S complexes with the original protocol of reconstitution.
As shown in Fig. 7B and C, substitution of total tRNA for individual tRNAiMet, and recombinant eIF4B for the native one resulted in the 48S complex formation for both RNAs. It was of a particular interest to verify whether the tripartite leader allowed the use of only the normal scanning process or else whether some shunting might occur in our reconstitution system. To this end, a very stable stem-loop structure (>50 kcal/mol) was inserted at position 204 of the tripartite leader and at the analogous position of its deletion derivative Td1 (Fig. 7A), which is incapable of using the shunting model (33). Such a modification in both cases dramatically suppressed the formation of the 48S complex (Fig. 7B). This strongly suggests that the shunting mechanism on the tripartite leader involves specific auxiliary proteins not represented in the canonical initiation factors. Consequently, this purified assembly system with the tripartite leader described in this study offers a unique opportunity to analyze in detail the molecular mechanism of shunting, at least in the case of adenovirus mRNAs.
DISCUSSION
We hold the point of view that no cellular process may be regarded as elucidated before it is reproduced in vitro with totally purified components. The method of assembly of translation initiation complexes combined with toeprinting is a powerful tool to determine which components are essential for a particular mRNA to form such complexes and which are less important or dispensable. If the reconstitution does not occur, the method allows one to identify a missing activity. It is especially valuable for mammalian cells which are poorly amenable to genetic approaches.
In this report, we demonstrate with purified components a successful reconstitution of the 48S translation initiation complex with mRNAs that have some base pairing within their 5′ UTRs. The complexes have been reconstituted on mRNAs bearing either model 5′ UTRs or those from natural mRNAs (beta-actin mRNA, late adenoviral mRNAs). We have shown that initiation factor eIF4B is absolutely essential for assembly of this complex with such mRNAs and not essential for beta-globin mRNA. Its quality (eIF4B is poorly stored and does not tolerate repeated freezing and thawing) and source of isolation are of paramount importance to allow the 40S ribosomal subunit to locate the initiation codon within even moderately structured 5′ leaders of mRNAs. Recombinant eIF4B substitutes poorly for the native factor, presumably, due to some misfolding of the protein, lack of posttranslational modification (phosphorylation) or both. This may account for an observation (G. Rogers, personal communication) that recombinant eIF4B poorly stimulates the RNA-helicase activity of eIF4A. It should be emphasized that the reconstitution of 48S complexes described in this work have been performed under concentrations of eIF4B not exceeding the physiological one. Thus, at least for mRNAs used in this study, no helicase activity in addition to eIF4A is required for the 40S subunit to locate the initiation codon within their 5′ UTRs!
In previous publications, a successful assembly of 48S complexes has been described mostly for viral IRESs. Some of them (cricket paralysis virus, hepatitis C virus, and some pestivirus RNAs) do not require eIF4B at all (11). The best studied to date, the EMCV IRES (22, Pisarev et al., submitted for publication) reveals only marginal stimulation of the 48S complex assembly by eIF4B. The same may be true for other homologous IRESs from cardio- and aphthoviruses. As shown in this report, beta-globin mRNA may be incorporated into the 48S complex even in the presence of a tiny amount of this factor. In some sense, the structural features of the 5′ UTR of this mRNA ensure a high translation initiation potential and may be regarded as close to ideal. This fact questions the practice of with beta-globin mRNA as a general reference, in particular for further development of the reconstitution technique with IRES-less mRNAs. These data as well as the widespread opinion that eIF4B plays an auxiliary (albeit important) role in the translation helicase reaction give some excuse why the importance of quality and concentrations of eIF4B have been neglected when trying to assemble 48S complexes with cap-dependent natural mRNAs other than beta-globin mRNA.
It should be noted that like other mRNAs tested in this work, the beta-globin mRNA remains strongly dependent on eIF4F in the reconstitution of 48S complexes. This fact confirms the essential role of eIF4F in the delivery of the helicase eIF4A onto 5′ UTRs of many if not all cap-dependent mRNAs and some viral IRESs (21).
There is ample literature on the organization and functioning of mRNA-unwinding apparatus in general, and on the structure and function of eIF4B, in particular (13). Surprisingly, we still have no idea how eIF4B works. In previous investigations, the function of eIF4B in unwinding of the secondary structure of 5′ UTRs was studied either in a reconstituted cell-free system (3) where it is difficult to assess contribution of unknown factors or in model systems (2, 15, 16, 28, 29, 31, 32). Some of these model systems included artificial RNA or RNA-DNA duplexes and factors eIF4A, eIF4F and eIF4B taken in 100 to 1,000 molar excess over the substrate (31, 32). These studies proved to be very fruitful to assess the contribution of individual components of helicase machinery to the efficiency of the unwinding process. However, the studies with model duplexes do not allow us to judge what will happen if we proceed to a more physiologic system with more realistic ratios of the translational components and mRNA substrates.
In our system, the contribution of unwinding factors to the mRNA accommodation on the 40S subunit is analyzed as a function of the formation of the 48S complex in the presence of all necessary translational components. It was shown that in the absence of eIF4B, eIF4F and eIF4A were not able to cope with even a moderate base pairing within the 5′ UTR of an mRNA. As noticed above, this moderate base pairing is not necessarily reflected in the folding of 5′ UTR sequences into conventional stem-loop structures. In contrast, in some of the model experiments mentioned above, not only eIF4F and eIF4A but even eIF4A alone is able to unwind to some extent stem-loop structures whose calculated energy seems to be lower than that for our model 5′ UTRs (31, 32). Thus, our data clearly demonstrate that the results obtained with artificial RNA/RNA or RNA/DNA duplexes and those obtained with the reconstitution system are not the same.
One of the most important finding of this study is a sharp eIF4B concentration dependence for the formation of the 48S complex with structured 5′-leaders. The maximal stimulation is achieved only at the physiological concentration of eIF4B. No such dependence is observed for the 5′ UTR of beta-globin mRNA. All these observations taken together allow us to speculate that eIF4B not only stimulates binding of RNA and ATP to eIF4A, but may change the very mechanism of the unwinding reaction, in agreement with suggestions made in (2, 31).
The data on eIF4B dependence for the 48S complex assembly for structured 5′ leaders are provocative enough to speculate that even small variations in concentration or activity of eIF4B in mammalian cells may differentially affect the translation of various cellular mRNAs (by influencing the processivity with which their 5′ UTRs are unwound). At present, divergence in efficiency of translation regulation is attributed mainly to eIF4F (eIF4E) (8). That eIF4B along with eIF4F may preferentially stimulate in vivo translation of mRNAs with structured 5′-leaders has been noted in some, albeit not many publications (1, 18). eIF4F in conjunction with eIF4B may ensure a finer tuning of translational efficiency of different mRNAs in development, during cell cycle or action of external stimuli in agreement with previous reports (7, 12, 17).
Unfortunately, not much is known about eIF4B regulation and whether its synthesis is controlled by the same signal transduction pathway as for eIF4E. It is known, however, that eIF4B is active in its phosphorylated form (this may partially explain the inability of recombinant eIF4B to efficiently stimulate the 48S complex formation) and that this phosphorylation is directly correlated with the up- and downregulation of translation in development, serum stimulation, stress conditions, etc. (7, 12, 17, 18). The effect of eIF4B phosphorylation on the formation of 48S complexes has been beyond the scope of this paper. This is a subject of our further studies. However, it is important to emphasize that these variations in eIF4B activity will probably have little or no effect on the translation initiation driven by viral IRESs (at least those studied to date), and short unstructured 5′ leaders of cellular mRNAs.
The principal goal of these investigations was to find a missing activity that would allow us to assemble 48S complexes on some cap-dependent mammalian mRNAs (known for their elevated GC content) other than beta-globin mRNA. The mRNAs described in this study need only canonical translation initiation factors. We do not exclude that some other 5′ end-dependent mRNAs may require additional protein factors. For instance, as shown in this paper, the tripartite leader of late adenoviral mRNAs may use with canonical initiation factors only the classical scanning mechanism of translation initiation rather than the shunting model. Certainly, tremendous work is needed to determine factor requirements for other natural cellular or viral mRNAs. The work in this direction is in progress. However, we believe that this report will strengthen the optimism about unique possibilities of the reconstitution method to elucidate complex mechanisms of translation initiation in mammalian cells.
Acknowledgments
We are grateful to M. Hentze for the suggestion to use HeLa cell extracts for the isolation of mammalian initiation factors and to R. Luhrmann for offering this extract. We thank T. Pestova and C. Hellen for providing us with plasmid pTRM-1 and M. Belozersky for permission to use the FPLC equipment in his laboratory.
This work was supported in part by grants 99-04-49230 and 02-04-48798 from the Russian Foundation for Basic Researches to I.N.S. and by NIH grant GM26796 to W.C.M.
REFERENCES
- 1.Altmann, M., P. P. Muller, B. Wittmer, F. Ruchti, S. Lanker, and H. Trachsel. 1993. A Saccharomyces cerevisiae homologue of mammalian translation initiation factor 4B contributes to RNA helicase activity. EMBO J. 12:3997-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bi, X., Ren, J., and D. J. Goss, 2000. Wheat germ translation initiation factor eIF4B affects eIF4A and eIFiso4F helicase activity by increasing the ATP binding affinity of eIF4A. Biochemistry 39:5758-5765. [DOI] [PubMed] [Google Scholar]
- 3.Browning, K. S., S. R. Lax, J. Humphreys, J. M. Ravel, S. A. Jobling, and L. Gehrke. 1988. Evidence that the 5′-untranslated leader of mRNA affects the requirement for wheat germ initiation factors 4A, 4F, and 4G (4B). J. Biol. Chem. 263:9630-9634. [PubMed] [Google Scholar]
- 4.Dasso, M. C., and R. J. Jackson. 1989. Efficient initiation of mammalian mRNA translation at a CUG codon. Nucleic Acids Res. 17:6485-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dmitriev, S. E., A. V. Pisarev, M. P. Rubtsova, Y. E. Dunaevsky, and I. N. Shatsky. 2003. Conversion of 48S translation preinitiation complexes into 80S initiation complexes as revealed by toeprinting. FEBS Lett. 533:99-104. [DOI] [PubMed] [Google Scholar]
- 6.Dmitriev, S. E., I. M. Terenin, M. P. Rubtsova, and I. N. Shatsky. 2003. Minor secondary structure variations in the 5′-untranslated region of the beta-globin mRNA changes the concentration requirements for eIF2. Mol. Biol. (Moscow) 37:494-503. [PubMed] [Google Scholar]
- 7.Gallie, D. R., H. Le, C. Caldwell, R. L. Tanguay, N. X. Hoang, and K. S. Browning. 1997. The phosphorylation state of the translation initiation factors is regulated developmentally and following heat shock in wheat. J. Biol. Chem. 272:1046-1053. [DOI] [PubMed] [Google Scholar]
- 8.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4F initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 9.Gray, N. K., S. Quick, B. Goossen, A. Constable, H. Hirling, L. Kuhn, and M. W. Hentze. 1993. Recombinant iron-regulatory factor functions as an iron-responsive-element-binding protein, a translational repressor and an aconitase. A functional assay for translational repression and direct demonstration of the iron switch. Eur. J. Biochem. 218:657-667. [DOI] [PubMed] [Google Scholar]
- 10.Grifo, J. A., S. M. Tahara, M. A. Morgan, A. J. Shatkin, and W. C. Merrick 1983. New initiation factor activity required for globin mRNA translation. J. Biol. Chem. 258:5804-5810. [PubMed] [Google Scholar]
- 11.Hellen, C. U. T., and P. Sarnow. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593-1612. [DOI] [PubMed] [Google Scholar]
- 12.Hershey, J. W. B. 1991. Translational control in mammalian cells. Annu. Rev. Biochem. 60:717-755. [DOI] [PubMed] [Google Scholar]
- 13.Hershey, J. W. B., and W. C. Merrick. 2000. The pathway and mechanism of initiation of protein synthesis, p. 33-88. In N. Sonenberg et al. (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Iost, I., Dreyfus, M., and P. Linder. 1999. Ded1p, a DEAD-box protein, required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J. Biol. Chem. 274:17677-17683. [DOI] [PubMed] [Google Scholar]
- 15.Lawson, T. G., B. K. Ray, J. T. Dodds, J. A. Grifo, R. D. Abramson, W. C. Merrick, D. F. Betsch, H. L. Weith, and R. E. Thach. 1986. Influence of the 5′-proximal secondary structure on the translational efficiency of eukaryotic mRNAs and on their interaction with initiation factors. J. Biol. Chem. 261:13979-13989. [PubMed] [Google Scholar]
- 16.Lawson, T. G., K. A. Lee, M. M. Maimone, R. D. Abramson, T. E. Dever, W. C. Merrick, R. E. Thach. 1989. Dissociation of double-stranded polynucleotide helical structures by eukaryotic initiation factors, as revealed by a novel assay. Biochemistry 28:4729-4734. [DOI] [PubMed] [Google Scholar]
- 17.Le, H., K. S. Browning, and D. R. Gallie. 1998. The phosphorylation state of the wheat translation initiation factors eIF4B, eIF4A, and eIF2 is differentially regulated during seed development and germination. J. Biol. Chem. 273:20084-20089. [DOI] [PubMed] [Google Scholar]
- 18.Manzella, J. M., W. Rychlik, R. E. Rhoads, J. W. Hershey, and P. J. Blackshear. 1991. Insulin induction of ornithine decarboxylase. Importance of mRNA secondary structure and phosphorylation of eukaryotic initiation factors eIF4B and eIF4E. J. Biol. Chem. 266:2383-2389. [PubMed] [Google Scholar]
- 19.Merrick, W. C. 1979. Purification of protein synthesis initiation factors from rabbit reticulocytes. Methods Enzymol. 60:101-108. [DOI] [PubMed] [Google Scholar]
- 20.Methot, N., A. Pause, Hershey, J. W. B., and N. Sonenberg. 1994. The translation initiation factor eIF4B contains an RNA-binding region that is distinct and independent from its ribonucleoprotein consensus sequence. Mol. Cell. Biol. 14:2307-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pause, A., N. Methot, Y. Svitkin, W. C. Merrick, and N. Sonenberg. 1994. Dominant negative mutants of mammalian translation initiation factor eIF4A define a critical role for eIF4F in cap-dependent and cap-independent initiation of translation. EMBO J. 13:1205-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pestova, T. V., C. U. Hellen, and I. N. Shatsky. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16:6859-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pestova, T. V., S. I. Borukhov, and C. U. Hellen. 1998. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394:854-859. [DOI] [PubMed] [Google Scholar]
- 24.Pestova, T. V., I. B. Lomakin, J. H. Lee, S. K. Choi, T. E. Dever, and C. U. Hellen. 2000. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403:332-335. [DOI] [PubMed] [Google Scholar]
- 25.Pestova, T. V., and C. Hellen. 2001. Preparation and activity of synthetic unmodified mammalian tRNAiMet in initiation of translation in vitro. RNA. 7:1496-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pestova, T. V., and V. G. Kolupaeva. 2002. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 16:2906-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pisarev, A. V., M. A. Skabkin, A. A. Thomas, W. C. Merrick, L. P. Ovchinnikov, and I. N. Shatsky. 2002. Positive and negative effects of the major mammalian messenger ribonucleoprotein p50 on binding of 40S ribosomal subunits to the initiation codon of beta-globin mRNA. J. Biol. Chem. 277:15445-15451. [DOI] [PubMed] [Google Scholar]
- 28.Ray, B. K., T. G. Lawson, J. C. Kramer, M. H. Cladaras, J. A. Grifo, R. D. Abramson, W. C. Merrick, and R. E. Thach. 1985. ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J. Biol. Chem. 260:7651-7658. [PubMed] [Google Scholar]
- 29.Ray, B. K., T. G. Lawson, R. D. Abramson, W. C. Merrick, and R. E. Thach. 1986. Recycling of messenger RNA cap-binding proteins mediated by eukaryotic initiation factor 4B. J. Biol. Chem. 261:11466-11470. [PubMed] [Google Scholar]
- 30.Richter-Cook, N. J., T. E. Dever, J. O. Hensold, and W. C. Merrick. 1998. Purification and characterization of a new eukaryotic protein translation factor. Eukaryotic initiation factor 4H. J. Biol. Chem. 273:7579-7587. [DOI] [PubMed] [Google Scholar]
- 31.Rogers, G. W., N. J. Richter, W. F. Lima, and W. C. Merrick. 2001. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 276:30914-30922. [DOI] [PubMed] [Google Scholar]
- 32.Rozen, F., I. Edery, K. Meerovitch, T. E. Dever, W. C. Merrick, and N. Sonenberg. 1990. Bidirectional RNA helicase activity of eukaryotic translation initiation factors 4A and 4F. Mol. Cell. Biol. 10:1134-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yueh, A., and R. J. Schneider. 1996. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 10:1557-1567. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, Y., P. J. Dolph, and R. J. Schneider. 1989. Secondary structure analysis of adenovirus tripartite leader. J. Biol. Chem. 264:10679-10684. [PubMed] [Google Scholar]