Abstract
The eukaryotic translation initiation factor 4E (eIF4E) acts as both a key translation factor and as a promoter of nucleocytoplasmic transport of specific transcripts. Traditionally, its transformation capacity in vivo is attributed to its role in translation initiation in the cytoplasm. Here, we demonstrate that elevated eIF4E impedes granulocytic and monocytic differentiation. Our subsequent mutagenesis studies indicate that this block is a result of dysregulated eIF4E-dependent mRNA transport. These studies indicate that the RNA transport function of eIF4E could contribute to leukemogenesis. We extended our studies to provide the first evidence that the nuclear transport function of eIF4E contributes to human malignancy, specifically in a subset of acute and chronic myelogenous leukemia patients. We observe an increase in eIF4E-dependent cyclin D1 mRNA transport and a concomitant increase in cyclin D1 protein levels. The aberrant nuclear function of eIF4E is due to abnormally large eIF4E bodies and the loss of regulation by the proline-rich homeodomain PRH. We developed a novel tool to modulate this transport activity. The introduction of IκB, the repressor of NF-κB, leads to suppression of eIF4E, elevation of PRH, reorganization of eIF4E nuclear bodies, and subsequent downregulation of eIF4E-dependent mRNA transport. Thus, our findings indicate that this nuclear function of eIF4E can contribute to leukemogenesis by promoting growth and by impeding differentiation.
The eukaryotic translation initiation factor 4E (eIF4E) promotes cellular growth and transformation. In fact, eIF4E levels are used as prognostic indicators of clinical outcome in human cancers, including breast cancer and head and neck squamous cell carcinomas (6, 28, 29, 45). Even moderate overexpression of eIF4E leads to dysregulated cellular proliferation and malignant transformation in immortalized cell lines (21-23). Biochemically, eIF4E acts at the rate-limiting step of cap-dependent translation initiation where it binds the methyl 7 guanosine (m7G) cap present on the 5′ end of mRNAs and recruits the given transcript to the ribosome (36). The traditional view is that eIF4E transforms cells by promoting inappropriate translation of mRNAs important to growth (36).
Recent findings suggest that eIF4E has functions in addition to its well-defined role in translation. Up to 68% of eIF4E is found in multiprotein nuclear structures referred to as eIF4E nuclear bodies (15, 20, 24, 37). In the nucleus, eIF4E promotes the selective transport of specific mRNAs, such as cyclin D1, from the nucleus to the cytoplasm without affecting housekeeping mRNAs such as GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and actin or altering levels of cyclin D1 transcripts (20, 35, 43, 44). This activity of eIF4E responds to physiological stresses such as interferon (43). Similar to its cytoplasmic function, eIF4E requires its m7G cap-binding activity for its mRNA transport function (4). The molecular mechanism of how eIF4E-sensitive transcripts are transported, what the features are that impart sensitivity, how many transcripts are regulated in this manner, and whether eIF4E directly transports mRNAs or participates in a process required for transport is not known. However, for ease of terminology we will refer to this general phenomenon as eIF4E-dependent mRNA transport. To monitor this process, we used cyclin D1 mRNA, since it is the best-characterized target transcript, but it is important to consider that other transcripts are also modulated by eIF4E. Interestingly, whereas the W73A eIF4E mutant cannot form an active translation complex (36), it still functions in cyclin D1 mRNA transport and still transforms cell lines (4). Thus, the ability of eIF4E to transform cell lines lies, at least in part, in its mRNA transport function. We hypothesize that eIF4E-dependent mRNA transport could be dysregulated in human malignancies, and in this way its nuclear functions could contribute to its transformation potential in vivo.
In mammalian cells, eIF4E nuclear bodies coincide with those associated with the promyelocytic leukemia protein PML and the proline-rich homeodomain protein PRH, also known as the hematopoietically expressed homeodomain Hex (1, 4, 20, 42-44). PRH is expressed in limited tissues in adults, including myeloid cell, lung, thyroid, and liver tissues (14, 27) and thus is positioned as a tissue-specific regulator of eIF4E function (44). PRH is required for hematopoiesis in a variety of organisms, including zebrafish, Xenopus, chicken, mice, and humans (5, 25, 30, 41, 47). In vitro, PRH uses a conserved eIF4E binding site to directly interact with eIF4E (44), causing a conformational change in the protein (unpublished observations). In cell culture, PRH overexpression inhibits eIF4E-dependent transformation by directly interacting with eIF4E and subsequently inhibiting its mRNA transport function (44). Although PRH is found in both the nucleus and the cytoplasm, the nuclear fraction of PRH is required to inhibit eIF4E-dependent mRNA transport and subsequent transformation (44).
Because PRH plays a major role in myelopoiesis, we investigated whether disruption of PRH-mediated suppression of eIF4E transport activity contributes to hematologic malignancy. To test this theory, we examined a series of primary human leukemia specimens. We demonstrate here that eIF4E levels are upregulated and PRH is downregulated in a subset of acute myelogenous leukemia (AML) and chronic myelogenous leukemia (CML) specimens. Furthermore, there is a striking alteration in the subcellular distributions of these proteins in the leukemic versus normal specimens. eIF4E-dependent cyclin D1 transport is upregulated in the leukemia specimens, leading to increased levels of cyclin D1 proteins. In seeking to determine a mechanism for upregulation of eIF4E, we examined the role of NF-κB, since this important transcription factor is commonly activated in primary leukemia specimens (11) and is implicated in pathways related to eIF4E regulation (36). We demonstrate that inhibition of NF-κB by introduction of IκB leads to reduced eIF4E levels, increased PRH levels, restoration of normal nuclear architecture, and downregulation of eIF4E-mediated mRNA transport. Further, we demonstrate that eIF4E overexpression impedes differentiation, and this activity is at least in part due to its function in mRNA transport. These findings implicate dysregulation of eIF4E-dependent mRNA transport in leukemogenesis.
MATERIALS AND METHODS
Cell isolation and culture.
AML blood cells and normal bone marrow cells were isolated and processed as described previously (12, 17). Briefly, primary AML and CML cells were obtained from the peripheral blood of patients at the Markey Cancer Center, University of Kentucky Medical Center. Normal bone marrow was obtained as waste material after pathological analysis, surgical marrow harvest, or from the National Disease Research Interchange. All tissues were obtained with the approval of the Institutional Review Board and appropriate informed consent. Marrow cells were depleted of erythrocytes by suspending them in 150 mM NH4Cl plus 10 mM NaHCO3 for 5 min, followed by two washes with phosphate-buffered saline (PBS). Blood cells were subjected to Ficoll-Paque (Pharmacia Biotech) density gradient separation to isolate the mononuclear white blood cell compartment. Resulting leukocytes from marrow or blood were then used for immunoaffinity selection as needed. For CD34+ cell selection, the Miltenyi immunoaffinity device (varioMACS) was used according to the manufacturer's instructions. In some cases, leukocytes were cryopreserved at a concentration of ∼5 × 107 cells/ml in freezing medium consisting of Iscove modified Dulbecco medium, 40% fetal bovine serum, and 10% dimethyl sulfoxide. For analysis of unstimulated cells, cryopreserved primary samples were thawed and used immediately for isolation of RNA or protein.
Cell sorting and IκB expression.
Adenovirus vectors were constructed to express either green fluorescent protein (GFP) alone, or a combination of GFP with the NF-κB inhibitor IκB as previously described (12). The IκB allele that we used was mutated at serines 32 and 36 to generate a degradation-resistant form of the protein known as IκB super-repressor (IκB-SR) (2). Infection of primary cells were performed as previously described (13). Primary hematopoietic cells were labeled with CD34-phycoerythrin (PE) (Becton Dickinson) and sorted by using a FACSVantage flow cytometer (12). Isolated populations were at least 95% pure. Control populations of normal granulocytes and monocytes were obtained by labeling peripheral blood mononuclear cells with CD14-PE and CD15-fluorescein isothiocyanate (FITC) (Becton Dickinson). Cells were sorted by using appropriate forward-scatter versus side-scatter gates, and CD14+ CD15− (monocytes) and CD14− CD15+ (granulocytes) were isolated.
Overexpression studies of eIF4E.
The MSCV-pgk-GFP plasmid (kindly provided by Guy Sauvageau) was used to generate all retroviral vectors. eIF4E wild type and W56 and W73 mutants (as described previously [4]) were cloned into the EcoRI site to yield vectors expressing an eIF4E allele downstream of the retroviral long terminal repeat, followed by a pgk-GFP reporter cassette. Each plasmid was transiently transfected into the Phoenix-Ampho packaging line (kindly provided by Gary Nolan), and retroviral supernatants were used to infect human U937 cells (American Type Culture Collection). For experiments with adenovirus infection, purified populations of CD34+ GFP+ cells were isolated by using the FACSVantage flow cytometer. Dead cells were excluded by using propidium iodide, and sorted populations were at least 95% pure. U937 cells infected with retroviruses were sorted twice to obtain GFP+ populations that were at least 99% pure.
Western blot analysis and coimmunoprecipitation studies.
Cells were washed twice in 1× PBS (pH 7.2) and lysed in radioimmunoprecipitation assay buffer supplemented with Complete protease inhibitors on ice. Equal amounts of whole-cell protein lysates (20 μg) were loaded on sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. Membranes were probed with mouse monoclonal anti-eIF4E antibody (Ab; BD Transduction Laboratories), mouse monoclonal anti-cyclin D1 Ab (BD Pharmingen), mouse monoclonal anti-PML Ab (5E10) (38), mouse monoclonal anti-β-actin Ab (Sigma), mouse monoclonal anti-c-myc Ab (9E10; Covance), and affinity-purified rabbit polyclonal anti-PRH Ab (44). All primary antibodies were used at 1:2,000 except for monoclonal Ab 5E10, which was used at 1:100. Horseradish peroxidase-conjugated secondary antibodies were used at 1:20,000, and the signals were detected by chemiluminescence (Super Signal West Pico; Pierce).
Coimmunoprecipitations were carried out as described previously (3, 20). Briefly cells (∼108) were washed three times with PBS and lysed in immunoprecipitation buffer (150 mM NaCl, 20 mM Tris-Cl [pH 7.4], 1% NP-40, 100 μM phenylmethylsulfonyl fluoride) supplemented with protease inhibitors on ice. The appropriate Ab or immunoglobulin G (IgG; Calbiochem) previously cross-linked to protein A-Sepharose beads was added to precleared lysates, followed by incubation overnight at 4°C. Beads were washed five times with immunoprecipitation buffer, collected, and examined by Western blot analysis.
Cellular fractionation.
Cellular fractionation was performed as previously described (20, 43, 44). Briefly, cells were rinsed twice in ice-cold 1× PBS (pH 7.2) and then resuspended with slow pipetting in lysis buffer B (10 mM Tris [pH 8.4], 140 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40, 1 mM dithiothreitol, and RNasin [100 U/ml; Promega]). Nuclear suspensions were centrifuged at 1,000 × g for 3 min at 4°C, and the supernatant was saved as the cytoplasmic fraction. Nuclear pellets were resuspended in lysis buffer B. A 1/10 volume of the detergent (3.3% [wt/vol] sodium deoxycholate and 6.6% [vol/vol] Tween 40) was added with slow vortexing, and the nuclear suspension was incubated on ice for 5 min. Nuclei were pelleted by centrifugation at 1,000 × g for 3 min at 4°C, and the supernatant (postnuclear fraction) was saved and added to the cytoplasmic fraction. Together, these are considered the cytoplasmic fraction. The nuclei were rinsed once in lysis buffer B. This protocol yielded intact nuclei, as determined by light microscopy, with no significant cytoplasmic contamination, as determined from the tRNALys content (see Fig. 2). Further, no significant contamination of the cytoplasm by nuclei was observed, as determined by the amount of U6 snRNA (see Fig. 2).
FIG. 2.
Cyclin D1 levels are posttranscriptionally upregulated in M4 AML, M5 AML, and bcCML patients. (A) Whole-cell lysates were analyzed by Western blot (W.B.) as indicated. The levels of cyclin D1 are increased in all leukemia specimens (lanes 2, 3, and 5 to 8). β-Actin is shown as a control for protein loading. (B) Northern blot (N.B.) analysis of whole-cell lysates shows upregulation of cyclin D1 mRNA levels in cells derived from M1 AML, M2 AML, M2/AML-ETO AML, and ALL patients (lanes 4 to 7). Cyclin D1 mRNA levels were not altered in M4 AML, M5 AML, and bcCML specimens (lanes 2, 3, 9, and 10). (C) RNA, isolated from nuclear (n) and cytoplasmic (c) fractions, was analyzed by Northern blot as indicated. Nuclear export of cyclin D1 mRNA was increased in the cells derived from M5 AML (lanes 3, 4, 9, and 10) and bcCML (lanes 5 and 6) patients. tRNALys and U6snRNA were used as markers for the cytoplasmic and nuclear fractions, respectively. GAPDH is shown as a control for RNA loading. Samples are labeled as described in Fig. 1.
Northern blot analysis.
Total RNA and RNA from cytoplasmic and nuclear fractions were isolated by Trizol (Gibco) procedure according to the manufacturer's instruction. RNA from nuclear fractions was additionally treated with RNase-free DNase I (Promega). A total of 5 μg of total and fractionated RNA was loaded on 1% formaldehyde-agarose gel and subsequently transferred onto positively charged nylon membrane (Roche). Membranes were prehybridized in ULTRAhyb buffer (Ambion) for 1 h at 45°C and then probed with cyclin D1 cDNA probe (10 pM), eIF4E cDNA probe (5 pM), c-myc cDNA probe (5 pM), GAPDH cDNA probe (5 pM) (Ambion), tRNALys antisense oligoprobe (5′-7CTCATGCTCTACCGACTGAGCTAGCCGGGC-3′; 30 pM) and U6 antisense oligoprobe (5′-7GAATTTGCGTGTCATCCTTGCGCAGGGGCCATGCTAA-3′; 30 pM) in the same buffer for 16 h at 45°C. (Note that 7 indicates biotin.) cDNA probes were generated by PCR and labeled with BrightStar Psoralen-Biotin kit (Ambion), and signals were detected by using CDP Star Chemiluminescence (Ambion) according to the manufacturer's instructions.
Indirect immunofluorescence and laser-scanning confocal microscopy.
Cells were fixed and permeabilized as described previously (4, 43, 44) and then incubated, as indicated, with mouse monoclonal anti-SC35 Ab (1:100; BD Transduction Laboratories), mouse monoclonal anti-PML Ab (5E10; 1:10), rabbit polyclonal anti-Nopp140 Ab (RH-10; 1:100), or affinity-purified rabbit polyclonal anti-PRH Ab (1:100) in blocking buffer (10% fetal bovine serum-0.1% Tween 20 in 1× PBS [pH 7.2]) for 2 h at room temperature. After incubation with primary Ab, the cells were washed three times in 1× PBS (pH 7.2) and then further incubated with Texas red-conjugated donkey anti-rabbit Ab, Cy5-conjugated donkey anti-mouse Ab, Texas red-conjugated donkey anti-mouse Ab, or FITC-conjugated donkey anti-rabbit Ab (Jackson Immunoresearch Laboratories) for 45 min at room temperature. After incubation with the secondary Ab, cells were washed three times in 1× PBS (pH 7.4), mounted in Vectashield with DAPI (4′,6′-diamidino-2-phenylindole; Vector Laboratories, Inc.), and sealed. For triple staining, cells were additionally fixed with 3.7% paraformaldehyde for 10 min at room temperature, washed, and then incubated with a 1:20 dilution of FITC-conjugated mouse monoclonal anti-eIF4E Ab (BD Transduction Laboratories) at 4°C overnight. Fluorescence was observed by using 100× objective lens, further magnified by a zoom of 2 to 4 (as indicated), on a Leica inverted scanning confocal microscope at 488, 568, 633, or 351 and 364 nm. All channels were detected separately, and no cross talk between the channels was detected. Micrographs represent single sections with a thickness of ∼300 nm. Experiments were repeated three times with >500 cells in each sample.
Differentiation induction.
To induce differentiation, cells were seeded at 5 × 104 ml−1 and either all-trans-retinoic acid (ATRA; 5 μM) or 1,25(OH)2D3 (20 nM) were added. The appropriate carriers, dimethyl sulfoxide or ethanol, were added to control cultures. After 5 days in the presence of the differentiation agent, 106 cells were obtained for flow cytometry analysis. Each sample was washed in PBS and 1% bovine serum albumin (BSA) and then incubated in 100 μl of PBS with 1% BSA for 45 min on ice with either R-PE-conjugated IgG1 Ab, anti-CD11b-PE Ab, or anti-CD14-PE Ab (Caltag, Burlingame, Calif.). After incubation the cells were washed three times in ice-cold PBS with 1% BSA and resuspended in 0.5 ml of PBS with 1% BSA, and then flow cytometry carried out (FACScalibur; Becton Dickinson). For morphology assessment, 5 × 104 cells in 1% BSA and PBS were cytospun onto glass slides, air dried, and fixed in 100% methanol for 5 min. The cells were then Wright-Giemsa stained (Hematek 2000; Bayer, Pittsburgh, Pa.), and several fields from each condition examined.
RESULTS AND DISCUSSION
eIF4E and PRH are aberrantly regulated in a variety of primary human leukemia specimens.
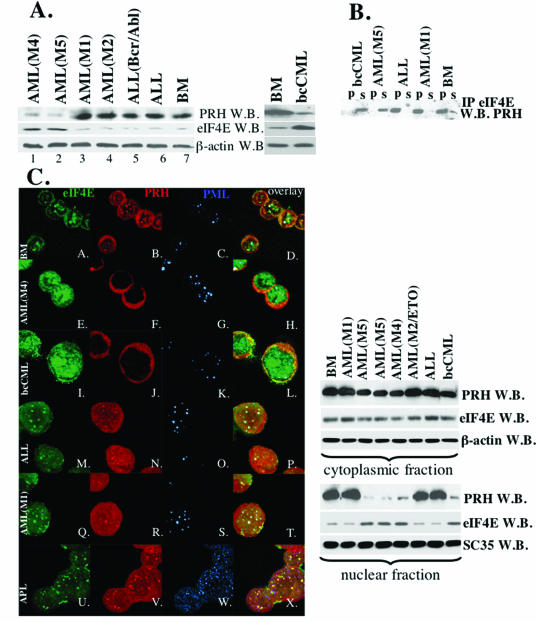
To investigate the potential role that eIF4E and PRH play in leukemia, we analyzed eIF4E and PRH protein levels in human primary leukemia specimens (Fig. 1 and Table 1). Western blot analysis demonstrated a substantial increase in eIF4E with concomitant repression of PRH in 13 of 13 primary AML (French American British FAB classification M4/M5) and 7 of 7 blast crisis CML (bcCML) specimens but not in 11 of 11 M1/M2, 7 of 8 acute lymphoid leukemia (ALL) specimens, or 2 of 2 chronic-phase CML specimens. In the M1/M2 AML and in the majority of the ALL specimens, the eIF4E and PRH levels appeared to be similar to those found in four of four normal specimens. Representative results are shown in Fig. 1A, with the total number of specimens analyzed and corresponding results summarized in Table 1. PRH levels were reduced in the M4 AML/M5 AML/bcCML patients, although there was some variability as to the level of reduction. Importantly, eIF4E levels were universally upregulated to apparently the same extent in these specimens (Table 1). Note that the levels and isoform distribution of PML are unchanged in any of these specimens versus normal controls (data not shown).
FIG. 1.
Levels, subcellular distribution, and interaction of eIF4E and PRH are altered in M4 AML, M5 AML, and bcCML specimens. (A) Western blot analysis of whole-cell extracts in cells derived from specimens as indicated. β-Actin is shown as a control for protein loading. (B) Whole-cell lysates were immunoprecipitated with anti-eIF4E Ab (IP eIF4E), and the resulting Western blot was probed for PRH. IP, immunoprecipitated fraction; s, supernatant after immunoprecipitation; W.B., Western blot. (C) In the left panel are shown confocal micrographs of cells stained with FITC-conjugated anti-eIF4E Ab (shown in green); anti-PRH Ab, followed by Texas red-conjugated anti-rabbit IgG Ab (shown in red); and anti-PML Ab (5E10), followed by Cy5-conjugated anti-mouse IgG Ab (shown in blue). The PML-eIF4E overlay is shown in light blue, the PML-PRH overlay is shown in pink, the PRH-eIF4E overlay is shown in yellow, and the triple eIF4E-PML-PRH overlay is shown in white. The objective was 100x with a further magnification of 2 (A-H) or 3 (I-X) fold. In the right panel, cells were fractionated into cytoplasmic and nuclear compartments and analyzed as indicated. β-Actin and SC 35 were used as a loading control for the cytoplasmic and nuclear fractions, respectively. BM, cells derived from the healthy individuals. Other specimens are as indicated. AML-ETO indicates that translocation was found in that M2 specimen (Table 1).
TABLE 1.
Characteristics of leukemia specimens examined herea
| Specimen | Subtype | eIF4E result
|
PRH result
|
Cyclin D1 result
|
c-Myc result (Western) (fold increase) | Cytogenetics | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Confocal | Western | Confocal | Western | Western | Northern (nuclear vs cytoplasmic) | Northern total | ||||
| UPN1 | M1 | Normal | Normal | NA | ||||||
| UPN2 | M1 | Normal | Normal | High | 46, XX | |||||
| UPN3 | M1 | Normal | Normal | Normal | Normal | High | Normal | High | 46, XY | |
| UPN4 | M1 | Normal | Normal | Normal | Normal | Normal | 46, XY | |||
| UPN5 | M1 | Normal | Normal | Normal | Normal | 46, XX | ||||
| UPN6 | M2 (t8;21) | Normal | Normal | Normal | Normal | Normal | 47, XY, +8, t(8;21) | |||
| UPN7 | M2 (t8;21) | Normal | Normal | Normal | 46, XY, t(8;21) | |||||
| UPN8 | M2 (t8;21) | Normal | Normal | Normal | Normal | High | Normal | High | 46, XY, t(8;21) | |
| UPN9 | M2 (t8;21) | Normal | 46, XY, t(8;21) | |||||||
| UPN10 | M2 | Normal | Normal | Normal | Normal | High | High | NA | ||
| UPN11 | M2 | Normal | Normal | Normal | NA | |||||
| UPN12 | M5 | Enlarged | High | Cytoplasmic | Low | High | Cytoplasmic | Normal | High (5) | 46, XX |
| UPN13 | M5 | Enlarged | High | Cytoplasmic | Low | High (5) | 46, XX | |||
| UPN14 | M4 | Enlarged | High | Cytoplasmic | Low | High | Cytoplasmic | Normal | High (5) | 47, XY, +9 |
| UPN15 | M4 | Enlarged | High | Cytoplasmic | Low | High (5) | 46, XX | |||
| UPN16 | M4 | High | Low | High | 45, XY, −7 | |||||
| UPN17 | M4/5 | High | Low | High | High (5) | 48, XY, +8, +13 | ||||
| UPN18 | M5 | Enlarged | High | Cytoplasmic | Low | High | 45, X, −Y | |||
| UPN19 | M4/5 | High | Low | High | NA | |||||
| UPN20 | M4/5 | High | Low | High | NA | |||||
| UPN21 | M5 | Enlarged | High | Cytoplasmic | Low | 47, XY, der(21),t(17;21) | ||||
| UPN22 | M4 | Enlarged | High | Cytoplasmic | Low | 46, XX | ||||
| UPN23 | MDS/AML | High | Low | High | NA | |||||
| UPN24 | MDS/AML | High | Low | High | 46, XY | |||||
| UPN25 | B-ALL | Normal | Normal | Normal | Normal | High | Normal | NA | ||
| UPN26 | B-ALL | Normal | Normal | High (2) | 46,XX,i(9),der(9)del(9), t(9;9;22), der(22), t(9;22) | |||||
| UPN27 | B-ALL | Normal | Normal | Normal | Normal | High | Normal | Complex | ||
| UPN28 | B-ALL | Normal | Normal | Normal | Normal | NA | ||||
| UPN29 | B-ALL | “Normal” | High | Disrupted | Normal | High | High (5) | 46, XY, t(4;11) | ||
| UPN30 | B-ALL | Normal | Normal | Normal | Normal | High (2) | 46, XY, t(9;22) | |||
| UPN31 | B-ALL | Normal | Normal | Normal | Normal | High | Normal | High | NA | |
| UPN32 | B-ALL | Normal | Normal | NA | ||||||
| UPN33 | BC-CML | High | Low | High (5) | BCR/ABL+ | |||||
| UPN34 | BC-CML | Enlarged | High | Cytoplasmic | Low | High | Cytoplasmic | Normal | BCR/ABL+ | |
| UPN35 | BC-CML | Enlarged | High | Cytoplasmic | Low | High (5) | BCR/ABL+ | |||
| UPN36 | BC-CML | High | Low | High | BCR/ABL+ | |||||
| UPN37 | BC-CML | Enlarged | High | Cytoplasmic | Low | High (5) | BCR/ABL+ | |||
| UPN38 | BC-CML | Enlarged | High | Cytoplasmic | Low | High | BCR/ABL+ | |||
| UPN39 | BC-CML | Enlarged | High | Cytoplasmic | Low | High | High (5) | BCR/ABL+ | ||
Abbreviations: UPN, unique patient number; M1 to M5, FAB subtype; MDS/AML, AML arising from myelodysplastic syndrome; B-ALL, B-cell ALL; BC-CML, blast crisis CML; confocal, microscopic image of eIF4E or PRH in nuclear bodies; Western, protein level for the indicated gene product as determined by Western blotting; northern (nuclear vs cytoplasmic), relative nuclear and cytoplasmic distribution of cyclin D1 mRNA; Northern total, relative abundance of total cyclin D1 mRNA; normal, eIF4E bodies were somewhat larger in this, the only t(4;11) specimen examined.
Controls for leukemic blast populations include both peripheral blood and marrow cells. We compared total white blood cell populations, as well as cells sorted to purify myeloid populations, including monocytes, granulocytes, and CD34+ cells from bone marrow. These studies indicate that differences between normal and leukemic specimens were due to the leukemic nature of cells and not due to differences in cell types (see supplemental Fig. 1 [http://atlas.physbio.mssm.edu/∼kbgroup/supplementary/MCB2]). Further, specimens included both wild-type and mutant Flt3, indicating that this common mutation in AML does not appear to correlate directly with the upregulation of eIF4E or downregulation of PRH (data not shown). The M4/M5 specimens represent a heterogeneous group of specimens. To our knowledge, there is no discernible pattern consistently observed in these samples. The bcCML specimens are all positive for the BCR/ABL translocation, but more detailed cytogenetics are not available. Given the typical pathogenesis of blast crisis CML, it is very likely that heterogeneous secondary mutations are present among our group of specimens. Characteristics of specimens are given in Table 1. Thus, eIF4E and PRH protein levels are altered in a distinct subset of primary leukemia specimens.
Nuclear architecture is disrupted in a subset of leukemias.
In cell culture, PRH is an important negative regulator of the transforming properties of eIF4E (44). Thus, we examined whether PRH was positioned to regulate eIF4E in primary specimens and whether this regulation was lost in the leukemic specimens. Immunoprecipitation studies indicate that eIF4E and PRH interact in normal specimens, in the M1/M2 AML and ALL specimens examined (Fig. 1B). However, no interaction is observed in the M4/M5/bcCML leukemic subtypes (Fig. 1B).
We examined the possibility that the interaction was lost due to alteration in the subcellular distribution of eIF4E and PRH (Fig. 1C). Confocal analysis of the M4/M5/bcCML specimens shows a dramatic reorganization of normal nuclear architecture versus normal specimens. In normal specimens, the majority of PML, PRH, and eIF4E colocalize to the same nuclear structures consistent with previous observations in cell lines (Fig. 1C, subpanels A to D [see also the supplementary figures at http://atlas.physbio.mssm.edu/∼kbgroup/supplementary/MCB2])(4, 20, 42, 44). In addition, eIF4E and PRH have a substantial cytoplasmic localization, as observed in a variety of cell lines (42, 44). In M4/M5/bcCML specimens, there are several striking changes (Fig. 1C, subpanels E to L). First, the majority of eIF4E is found in abnormally large nuclear bodies, with no significant increase in its cytoplasmic distribution. Second, there is a substantial loss of the nuclear fraction of PRH, where the remaining PRH is found diffusely throughout the cytoplasm. In these cases, PML appears nearly the same as in the normal specimens with the exception of some minor changes in body morphology in the CML patients, a finding consistent with the presence of activated Ras in these specimens (33). Protein fractionation studies confirmed the confocal results (Fig. 1C, right panel), where the majority of PRH is found in the cytoplasmic fraction in the M4/M5/bcCML specimens and the levels of eIF4E are not only upregulated, but a larger fraction of eIF4E is found in the nucleus in the M4/M5/bcCML specimens than others (Fig. 1C, right panel). Importantly, other nuclear structures such as splicing speckles and nucleoli are not altered in the leukemia specimens versus normal controls (supplementary Fig. 2, http://atlas.physbio.mssm.edu/∼kbgroup/supplementary/MCB2). Thus, alterations to the nuclear architecture are specific to PRH and eIF4E bodies.
There is no apparent difference in eIF4E and PRH nuclear architecture between normal and M1/M2/M3 AML specimens consistent with the observations that eIF4E and PRH protein levels were similar among these specimens to normals (Fig. 1C, subpanels M to X). The results in the M3 specimens (Fig. 1C, subpanels U to X), which express PML-RARα, are consistent with previous studies carried out in the APL patient derived cell line NB4, where we observed PRH and eIF4E bodies interact in the presence of PML-RARα (44). These findings suggest that dysregulation of eIF4E occurs because (i) eIF4E levels are upregulated and (ii) eIF4E cannot interact with its negative regulator, PRH, because PRH levels are depressed and the majority of PRH is in the cytoplasm. Previous studies in cell lines indicated that PRH only acts as a negative regulator of eIF4E-dependent transformation when found in the nuclear fraction (44), supporting the notion that its loss of interaction in the nucleus promotes dysregulation of eIF4E functions in these primary cells.
Regulation of cyclin D1 transport is disrupted in a subset of leukemia specimens.
In cell culture, the nuclear fraction of eIF4E mediates transport of a subset of transcripts, including cyclin D1, from the nucleus to the cytoplasm (35, 44). Thus, we hypothesized that cyclin D1 mRNA transport would be altered in leukemia specimens with dysregulated eIF4E and PRH (Fig. 2A). Consistent with the increased levels of eIF4E, we observe a substantial increase in cyclin D1 protein levels in M4/M5/bcCML specimens relative to normals (lanes 2, 3, and 5 versus lanes 1 and 4). Interestingly, there is also an increase in cyclin D1 protein levels observed in M1/M2 and ALL specimens (lanes 6 to 8). To establish the mechanism of cyclin D1 upregulation, we monitored levels of cyclin D1 transcripts by Northern analysis (Fig. 2B). There is substantially more cyclin D1 transcripts in M1/M2 AMLs and ALL than in normal specimens (lanes 4 to 7 versus lane 1). Importantly, the levels of cyclin D1 mRNA found in M4/M5/bcCML patients are the same as those found in samples from healthy subjects (compare lanes 1 to 3 with lanes 8 to 10). Thus, elevated cyclin D1 expression is common to the leukemic specimens examined; however, in the M4/M5/bcCML subset this was not due to the elevation of cyclin D1 transcript levels.
Since eIF4E overexpression promotes transport of cyclin D1 transcripts from the nucleus to the cytoplasm, we monitored transport of cyclin D1 by fractionation and Northern analysis (Fig. 2C). The subcellular distribution of tRNALys, a cytoplasmic RNA and U6 snRNA, a nuclear RNA, demonstrate the quality of the fractionation. Our results indicate that there is more cyclin D1 mRNA in the cytoplasmic fraction in the M4/M5/bcCML specimens with high eIF4E levels relative to normal marrow controls (lanes 3 to 6, 9, and 10 versus lanes 1, 2, 7, and 8). Thus, in this subset, cyclin D1 levels are elevated through increased transport of cyclin D1 transcripts to the cytoplasm. There is no increase in cyclin D1 transport in the M1/M2 AML or the ALL specimens compared to normals (lanes 11 to 16 versus lanes 7 and 8). Together, these data suggest that in the presence of these abnormally large eIF4E nuclear bodies and in the absence of PRH at these bodies, eIF4E inappropriately promotes transport of cyclin D1 transcripts to the cytoplasm. In cell culture systems, eIF4E-mediated upregulation of cyclin D1 transport is strongly correlated with its transforming activity (4, 44). Thus, it appears that the mRNA transport function of eIF4E contributes to the transformation process in a subset of primary leukemia cells.
NF-κB activity is correlated with disruption of eIF4E nuclear bodies.
NF-κB is a transcription factor commonly activated in primary leukemia specimens (8, 11, 19) and is implicated in pathways related to eIF4E regulation (36). We examined the effects of NF-κB activity by using an adenovirus vector encoding the IκB-SR that allows the complete repression of NF-κB activity within 12 h (12). AML or bcCML cells were transduced with Ad-GFP or Ad-IκB-SR-GFP, and CD34+ GFP+ cells were isolated by fluorescence-activated cell sorting (Fig. 3). In AML or CML CD34+ cells overexpressing GFP, the subcellular distribution of eIF4E and PRH proteins (as analyzed by confocal microscopy) is indistinguishable from their respective distributions in the untransduced patient cells. Here we observed abnormally large eIF4E nuclear bodies and, with the majority of PRH in the cytoplasm (compare Fig. 1E to L with Fig. 3A, subpanels A to D and I to L). Strikingly, introduction of IκB-SR-GFP leads to complete reorganization of eIF4E and PRH (Fig. 3A, subpanels E to H and M to P). PRH now colocalizes with eIF4E nuclear bodies. Further, the size and morphology of the eIF4E nuclear bodies now resemble those observed in normal specimens (compare with Fig. 1C, subpanels A to D; see also supplemental Fig. 1B [http://atlas.physbio.mssm.edu/∼kbgroup/supplementary/MCB2]). In addition, the introduction of IκB-SR resulted in downregulation of eIF4E protein levels and concomitant upregulation of PRH (Fig. 3B). Identical results were observed whether experiments were carried out in bcCML or M4/M5 AML specimens (Fig. 3 and supplemental Fig. 2 [http://atlas.physbio.mssm.edu/∼kbgroup/supplementary/MCB2]).
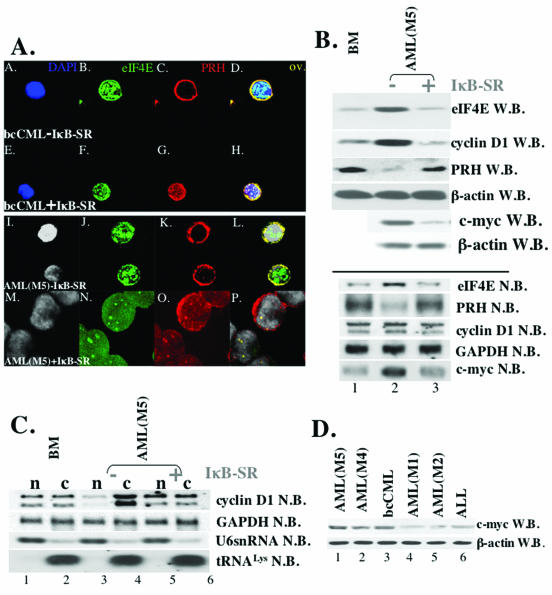
FIG. 3.
Expression of IκB-SR in CD34+ cells, derived from M5 AML and bcCML patients, correlates with the downregulation of the c-myc expression and the restoration of the expression and subcellular distribution of eIF4E and PRH proteins. (A) Cells were stained with anti-eIF4E Ab, followed by Texas red-conjugated anti-mouse IgG Ab (shown in green) and anti-PRH Ab, followed by Cy5-conjugated anti-rabbit IgG Ab (red). Nuclei were stained with DAPI (blue in panels A and E; gray in panels I and M). The PRH-eIF4E overlay (ov.) is shown in yellow. The objective was a 100× lens with a further magnification of two (A to L)- or three (G to R)-fold. (B) Western (W.B.) and Northern (N.B.) blot analysis of CD34+ cells derived from the healthy individuals (BM) and M5 AML patients [AML(M5)-IκB]. Blots were probed as indicated. (C) IκB-SR expression correlates with the restoration of cyclin D1 mRNA transport. Northern blot analysis of RNA isolated from nuclear (n) and cytoplasmic (c) fractions of CD34+ cells derived from healthy individuals (BM) and M5 AML patients. In contrast to Ad-GFP-transduced cells (−IκB-SR), CD34+ M5 AML cells that express IκB-SR (+IκB-SR) showed the same subcellular distribution of cyclin D1 mRNA (lanes 5 and 6) as CD34+ BM cells. tRNALys and U6snRNA were used as markers for cytoplasmic and nuclear fractions, respectively. −IκB-SR, cells transduced with Ad-GFP; +IκB-SR, cells transduced with Ad-GFP-IκB-SR. (D) Western blot analysis reveals upregulated levels of c-myc in M4 AML, M5 AML, and bcCML specimens (c-myc W.B.). β-Actin is shown as a control for protein loading.
We extended these studies to determine whether the reversion of nuclear architecture to that resembling a normal phenotype resulted in alterations in cyclin D1 mRNA transport. Strikingly, cells overexpressing IκB-SR-GFP had lower expression of cyclin D1 protein than the GFP-expressing cells. The total levels of cyclin D1 mRNA were nearly the same in the GFP- and IκB-SR-GFP-overexpressing systems (Fig. 3B; see also supplemental Fig. 2 [http://atlas.physbio.mssm.edu/∼kbgroup/supplementary/MCB2]), indicating that differences in protein expression were not due to decreased transcription of cyclin D1 mRNA in IκB-expressing cells. Thus, we fractionated cells into nuclear and cytoplasmic components and analyzed mRNA content by Northern methods (Fig. 3C). In normal specimens, the distribution of cyclin D1 mRNA is approximately equal in the nuclear and cytoplasmic compartments. In bcCML or AML M4/M5 expressing GFP, cyclin D1 mRNA is found mainly in the cytoplasmic fraction, as was found in the untransduced M4/M5 AML and bcCML cells (compare Fig. 3C with 2C). Importantly, in cells overexpressing IκB-SR-GFP the distribution of cyclin D1 mRNA is restored to that observed in normal specimens. Thus, the restoration of normal nuclear architecture is correlated with decreased cyclin D1 mRNA transport, and therefore decreased levels of cyclin D1 protein. Furthermore, these studies identify a previously undescribed function for NF-κB as a specific modulator of nuclear architecture.
Note that inhibition of NF-κB appears to result in at least partial sensitization of primary AML and CML cells to apoptosis (12; data not shown). Importantly, little to no cell death was evident within the first 12 h of culture. Thus, analysis of eIF4E and PRH in the present study was performed well before the onset of apoptosis.
IκB-SR reduces eIF4E and myc levels.
We extended these studies to determine how modulation of NF-κB activity alters eIF4E and PRH subcellular distribution and levels and thus affects cyclin D1 mRNA transport. Previous studies indicate that NF-κB activates myc (9). eIF4E is a myc-responsive gene, which contains two myc binding elements in its promoter (36). A total of 10 of 10 specimens examined that had high eIF4E protein levels also had elevated myc protein levels (ca. 5- to 6-fold) (Fig. 3D and Table 1), and 0 of 8 samples with normal eIF4E levels had significantly elevated (<2-fold) myc levels (Table 1). To ensure that elevated myc levels correlated with NF-κB activity, we monitored myc protein levels in the leukemia specimens in the presence or absence of IκB-SR (Fig. 3B, upper panel). These studies indicate that inhibition of NF-κB results in decreased myc protein levels, a finding consistent with the concomitant decrease in eIF4E levels observed above. Northern analysis shows that the levels of myc and eIF4E transcripts were reduced in the presence of IκB-SR (Fig. 3B, lower panel). In addition, these studies show that IκB-SR introduction is correlated with increased PRH transcript levels, a finding consistent with an increase in PRH protein levels. The mechanism by which NF-κB modulates PRH transcript levels is not known. Further, it is not known whether there is a connection between increasing eIF4E levels and decreasing PRH (see our conclusions below).
eIF4E-dependent mRNA transport contributes to a block in myeloid differentiation.
The consistent dysregulation of eIF4E-dependent mRNA transport in M4/M5 and bcCML specimens led us to investigate whether this activity of eIF4E contributes to a block in differentiation in addition to the loss of growth control associated with dysregulation of cyclin D1. U937 cells were retrovirally transduced with bicistronic constructs coding for GFP and wild-type or mutant forms of eIF4E. Subsequently, GFP-positive cells were isolated by using fluorescence-activated cell sorting. Two mutant forms of eIF4E were studied: W56A and W73A. The W56A mutation is in the m7G cap binding site of eIF4E and impedes both translation and mRNA transport functions (4, 18). The W73A mutation abolishes the binding of a number of eIF4E protein partners, including eIF4G, and thus formation of the translation initiation complex (10). Further, this mutant does not bind either of its reported nuclear regulators PRH or PML (4, 44). The W73A mutant cannot act in translation but is as efficient at upregulating mRNA transport as the wild-type protein (4, 10, 44). Wild-type and mutant constructs produced similar eIF4E protein levels (supplemental Fig. 1C [http://atlas.physbio.mssm.edu/∼kbgroup/supplementary/MCB2]). In addition, consistent with previous studies only wild-type or W73A, eIF4E increased cyclin D1 mRNA transport and subsequent protein levels (data not shown).
U937 cells overexpressing eIF4E or transduced with the vector controls were induced to differentiate along a granulocytic lineage with ATRA or along a monocytic lineage with 1,25(OH)2D3 (Fig. 4). The extent of differentiation induction was monitored by flow cytometric analysis. Here, the cell surface markers CD11b, a marker of ATRA-mediated granulocytic differentiation, and CD14, a marker of 1,25(OH)2D3-mediated monocytic differentiation, were monitored as described previously (46). In addition, cell morphology was examined by using Wright-Giemsa staining.
FIG. 4.
eIF4E blocks differentiation mediated by ATRA and vitamin D3. (A) Vector controls or cells expressing wild-type eIF4E, W56A mutant eIF4E, or W73A mutant eIF4E, as indicated, were treated for 5 days with either a vehicle control (untreated, white histogram), ATRA (red histogram), or 1,25(OH)2D3 (blue histogram) and then analyzed for cell surface marker expression. The median fluorescence intensity for each histogram (m) is given in each panel. The results are representative of three independent experiments. (B) Cells from panel A were stained with Wright-Giemsa, and changes in morphology were evaluated. Untreated cells shown are the vehicle control for vitamin D3; no effect on differentiation was seen with either carrier.
First, cells overexpressing wild-type or mutant forms of eIF4E were treated with ATRA. Upon ATRA treatment, vector control cells showed a striking (∼10-fold) increase in CD11b expression (Fig. 4A). Treatment of cells expressing wild-type or the W73A mutant did not lead to elevated CD11b levels. In fact, the levels of CD11b in wild-type and W73A mutant-expressing cells are similar to those in untreated controls. In contrast, cells expressing the W56A mutant differentiated in response to ATRA, as well as control cells (Fig. 4A). Furthermore, Wright-Giemsa staining of cell specimens showed that cells expressing the W56A mutant and vector controls showed clear morphological evidence of differentiation (Fig. 4B). Consistent with the flow cytometric data, wild-type and W73A-expressing cells showed no morphological evidence of differentiation upon ATRA treatment (Fig. 4B).
A similar pattern of results was observed with 1,25(OH)2D3. 1,25(OH)2D3 caused a striking increase in CD14 expression in the vector controls corresponding to an ∼250-fold increase in CD14 levels versus untreated controls (Fig. 4A). A similar level of CD14 upregulation was observed upon treatment of cells expressing the W56A mutant. In contrast, no alteration in CD14 levels upon 1,25(OH)2D3 treatment was observed in cells expressing wild-type or the W73A mutant. Here, the levels of CD14 are similar in untreated controls to those in treated cells. To be sure the CD14 upregulation reflected differentiation, treated cells were visually compared to untreated cells. After 1,25(OH)2D3 treatment, vector controls become adherent and develop morphological features characteristic of the monocytic lineage (Fig. 4B). Similar results were observed for cells expressing the W56A mutant. Consistently, 1,25(OH)2D3 treatment does not induce detectable morphological changes in cells overexpressing wild-type eIF4E or the W73A mutant. Thus, differentiation along two pathways is impeded by the expression of either wild-type eIF4E or the W73A mutant. Since the W73A mutant is as detrimental to differentiation in this context since the wild-type protein, these data strongly suggest that the mRNA transport function of eIF4E significantly contributes to its ability to impede differentiation. Furthermore, these are the first studies demonstrating that dysregulation of eIF4E expression leads to impaired differentiation of blood cell precursors.
Conclusions.
We demonstrated here that eIF4E-dependent mRNA transport of a model transcript, cyclin D1, is upregulated in a subset of primary leukemia specimens representing ∼35% of adult AML cases (http://www.myc-cancer-gene.org/site/mycTargetDB.asp) and also in blast crisis CML. In addition, the transport of ornithine decarboxylase transcripts (another mRNA subject to eIF4E-dependent mRNA transport [35]) is dysregulated specifically in the M4/M5/bcCML leukemia specimens (I. Topisisrovic and K. L. B. Borden, unpublished observations). Thus, the effects we observed on growth and differentiation are probably not due to cyclin D1 alone but likely attributable to dysregulation of the transport of a combination of eIF4E-sensitive mRNAs. Importantly, disruption of eIF4E-dependent mRNA transport promotes cell growth and impedes differentiation. Traditionally, eIF4E-dependent transformation both in cell culture and in vivo is thought to arise from its ability to inappropriately translate growth-promoting transcripts in the cytoplasm. Our findings extend this model, demonstrating that the nuclear function of eIF4E also mediates physiological effects that likely contribute to leukemogenesis in vivo.
Interestingly, cyclin D1 protein levels were upregulated in all of the leukemia specimens examined (Fig. 1). Thus, it appears that upregulation of cyclin D1 protein levels is an important event in a variety of leukemias and that this elevation is achieved through at least two distinct mechanisms. In M4/M5 and bcCML specimens, cyclin D1 expression is enhanced by increased mRNA transport, whereas in M1/M2 and ALL specimens expression is upregulated at the transcriptional level, either through increased transcription and/or increased transcript stability. Exactly why these two mechanisms are used in distinct leukemic subtypes is not yet clear.
Disruption of a wide variety of RNA processing events has been implicated in human diseases. One of the best-characterized examples is the disruption of splicing and alterations in transcript stability observed in thalassemias (32). Another example is found in CML primary specimens that contain the leukemogenic BCR/ABL fusion protein. Here, the production of the transcription factor C/EBPα, critical to normal granulocytic development, is repressed at the translational level through interactions with hnRNP E2 (16, 34). In addition, the mRNA transporter, hnRNP A1, is transcriptionally upregulated, potentially modifying general mRNA transport in these cells (16, 34). Both translational repression by hnRNP E2 of C/EBPα and upregulation of hnRNP A1 are recapitulated by expression of the BCR/ABL protein. The subsequent overexpression of hnRNP A1 resulted in early signs of differentiation, followed by cell death. These physiological consequences are quite different to those observed for eIF4E which promotes growth and impedes differentiation without inducing cell death. In fact, eIF4E overexpression rescues cells from apoptosis (39).
Importantly, eIF4E-dependent transport is upregulated in a variety of genetically distinct leukemias (Table 1), indicating that its dysregulation could be a wide-ranging phenomenon. In these leukemia specimens, dysregulation of eIF4E-dependent mRNA transport appears to arise from two distinct causes: the formation of very large eIF4E nuclear bodies and the loss of a key negative regulator, PRH, through both its downregulation and its near total exclusion from the nucleus. Our previous studies in cell culture indicated that PRH only represses eIF4E-dependent mRNA transport and transformation when in the nucleus (44). Consistently, the nuclear fraction of PRH is absent in thyroid carcinomas (7). In addition, PRH has been reported to act as a transcriptional repressor, and these activities could also be implicated in neoplastic transformation in certain tissues (40). The mechanism by which PRH is retained in the cytoplasm or excluded from the nucleus in these cells is not yet known. Thus, the loss of nuclear localization of PRH and therefore the loss of PRH mediated suppression of eIF4E transport activity could play a major role in the development of other human malignancies.
The use of IκB-SR proved to be a useful tool in our studies, since expression of this dominant-negative allele caused the nuclear architecture to be reconfigured to a state that resembled that found in normal marrow. These results are consistent with a model in which increased NF-κB activity leads to increased myc levels, leading in turn to elevated eIF4E levels, contributing to the transformed phenotypes observed (data not shown). Since we showed that the increased activity of NF-κB (11), in certain types of leukemia, correlates with upregulation of c-myc and eIF4E, we hypothesize that some of these effects could be mediated by upregulation and alteration of subcellular distribution of eIF4E. Increased activity of NF-κB was reported in other types of leukemia (11) that did not show alteration in eIF4E expression (Fig. 1), implicating the existence of other cellular factors, which could act in concert with NF-κB, that are specifically involved in leukemogenesis in M4/M5 AML and bcCML.
The growth and differentiation effects of both PRH and eIF4E are probably context dependent. Several recent studies show that PRH expression could promote or suppress cell proliferation during development of a variety of tissues, including liver, hematopoietic cell lines, vascular endothelium, and skin (26, 30, 31, 40). Therefore, depending on the cell type and possibly on the differentiation stage, both upregulation and downregulation of PRH could contribute to neoplastic transformation. In cases of PRH upregulation, other tissue-specific modulators of eIF4E could rescue and even promote eIF4E's transport activity. We showed that other homeodomain proteins, such as PRH, use conserved eIF4E-binding sites to directly interact with eIF4E (44). These homeodomains would be positioned to act as tissue-specific regulators of eIF4E in tissues which do not express PRH (44). Together, these results suggest that the mRNA transport function of eIF4E may contribute to transformation in other human cancers characterized by upregulation of eIF4E, such as breast cancer, head and neck squamous cell carcinomas, and non-Hodgkins lymphomas.
Acknowledgments
We are grateful for the kind gifts of antibodies and constructs from Paul Freemont, Gerd Maul, L. de Jong, Thomas Meier, Guy Sauvageau, Gary Nolan, David Grimwade, and Nahum Sonenberg. We thank Alex Kentsis for technical assistance and Liliana Ossowski for critical reading of the manuscript.
Confocal laser scanning microscopy was performed at the MSSM-LCSM core facility, supported with funding from NIH (1 S10 RR0 9145-01) and NSF (DBI-9724504). K.L.B.B. is a scholar of the Leukemia and Lymphoma Society. Financial support was provided by the NIH (CA 80728, CA88991, and CA90446) and by the Charlotte Geyer Cancer Foundation.
REFERENCES
- 1.Borden, K. L. 2002. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell. Biol. 22:5259-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brockman, J. A., D. C. Scherer, T. A. McKinsey, S. M. Hall, X. Qi, W. Y. Lee, and D. W. Ballard. 1995. Coupling of a signal response domain in IκBαa to multiple pathways for NF-κB activation. Mol. Cell. Biol. 15:2809-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlile, G. W., W. G. Tatton, and K. L. Borden. 1998. Demonstration of a RNA-dependent nuclear interaction between the promyelocytic leukaemia protein and glyceraldehyde-3-phosphate dehydrogenase. Biochem. J. 335:691-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen, N., M. Sharma, A. Kentsis, J. M. Perez, S. Strudwick, and K. L. Borden. 2001. PML RING suppresses oncogenic transformation by reducing the affinity of eIF4E for mRNA. EMBO J. 20:4547-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crompton, M. R., T. J. Bartlett, A. D. MacGregor, G. Manfioletti, E. Buratti, V. Giancotti, and G. H. Goodwin. 1992. Identification of a novel vertebrate homeobox gene expressed in haematopoietic cells. Nucleic Acids Res. 20:5661-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Benedetti, A., and A. L. Harris. 1999. eIF4E expression in tumors: its possible role in progression and malignacies. Int. J. Biochem. Cell Biol. 31:59-72. [DOI] [PubMed] [Google Scholar]
- 7.D'Elia, A. V., G. Tell, D. Russo, F. Arturi, F. Puglisi, G. Manfioletti, V. Gattei, D. L. Mack, P. Cataldi, S. Filetti, C. Di Loreto, and G. Damante. 2002. Expression and localization of the homeodomain-containing protein HEX in human thyroid tumors. J. Clin. Endocrinol. Metab. 87:1376-1383. [DOI] [PubMed] [Google Scholar]
- 8.Dokter, W. H., L. Tuyt, S. J. Sierdsema, M. T. Esselink, and E. Vellenga. 1995. The spontaneous expression of interleukin-1β and interleukin-6 is associated with spontaneous expression of AP-1 and NF-κB transcription factor in acute myeloblastic leukemia cells. Leukemia 9:425-432. [PubMed] [Google Scholar]
- 9.Duyao, M. P., D. J. Kessler, D. B. Spicer, and G. E. Sonenshein. 1992. Transactivation of the c-myc gene by HTLV-1 tax is mediated by NFκB. Curr. Top. Microbiol. Immunol. 182:421-424. [DOI] [PubMed] [Google Scholar]
- 10.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 11.Guzman, M. L., S. J. Neering, D. Upchurch, B. Grimes, D. S. Howard, D. A. Rizzieri, S. M. Luger, and C. T. Jordan. 2001. Nuclear factor-kappaB is constitutively activated in primitive human acute myelogenous leukemia cells. Blood 98:2301-2307. [DOI] [PubMed] [Google Scholar]
- 12.Guzman, M. L., C. F. Swiderski, D. S. Howard, B. A. Grimes, R. M. Rossi, S. J. Szilvassy, and C. T. Jordan. 2002. Preferential induction of apoptosis for primary human leukemic stem cells. Proc. Natl. Acad. Sci. USA 99:16220-16225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howard, D. S., D. A. Rizzierri, B. Grimes, D. Upchurch, G. L. Phillips, A. K. Stewart, J. R. Yannelli, and C. T. Jordan. 1999. Genetic manipulation of primitive leukemic and normal hematopoietic cells using a novel method of adenovirus-mediated gene transfer. Leukemia 13:1608-1616. [DOI] [PubMed] [Google Scholar]
- 14.Hromas, R., J. Radich, and S. Collins. 1993. PCR cloning of an orphan homeobox gene (PRH) preferentially expressed in myeloid and liver cells. Biochem. Biophys. Res. Commun. 195:976-983. [DOI] [PubMed] [Google Scholar]
- 15.Iborra, F. J., D. A. Jackson, and P. R. Cook. 2001. Coupled transcription and translation within nuclei of mammalian cells. Science 293:1139-1142. [DOI] [PubMed] [Google Scholar]
- 16.Iervolino, A., G. Santilli, R. Trotta, C. Guerzoni, V. Cesi, A. Bergamaschi, C. Gambacorti-Passerini, B. Calabretta, and D. Perrotti. 2002. hnRNP A1 nucleocytoplasmic shuttling activity is required for normal myelopoiesis and BCR/ABL leukemogenesis. Mol. Cell. Biol. 22:2255-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jordan, C. T., D. Upchurch, S. J. Szilvassy, M. L. Guzman, D. S. Howard, A. L. Pettigrew, T. Meyerrose, R. Rossi, B. Grimes, D. A. Rizzieri, S. M. Luger, and G. L. Phillips. 2000. The interleukin-3 receptor alpha chain is a unique marker for human acute myelogenous leukemia stem cells. Leukemia 14:1777-1784. [DOI] [PubMed] [Google Scholar]
- 18.Kentsis, A., E. C. Dwyer, J. M. Perez, M. Sharma, A. Chen, Z. Q. Pan, and K. L. Borden. 2001. The RING domains of the promyelocytic leukemia protein PML and the arenaviral protein Z repress translation by directly inhibiting translation initiation factor eIF4E. J. Mol. Biol. 312:609-623. [DOI] [PubMed] [Google Scholar]
- 19.Kordes, U., D. Krappmann, V. Heissmeyer, W. D. Ludwig, and C. Scheidereit. 2000. Transcription factor NF-κB is constitutively activated in acute lymphoblastic leukemia cells. Leukemia 14:399-402. [DOI] [PubMed] [Google Scholar]
- 20.Lai, H. K., and K. L. Borden. 2000. The promyelocytic leukemia (PML) protein suppresses cyclin D1 protein production by altering the nuclear cytoplasmic distribution of cyclin D1 mRNA. Oncogene 19:1623-1634. [DOI] [PubMed] [Google Scholar]
- 21.Lazaris-Karatzas, A., K. S. Montine, and N. Sonenberg. 1990. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 345:544-547. [DOI] [PubMed] [Google Scholar]
- 22.Lazaris-Karatzas, A., M. R. Smith, R. M. Frederickson, M. L. Jaramillo, Y. L. Liu, H. F. Kung, and N. Sonenberg. 1992. Ras mediates translation initiation factor 4E-induced malignant transformation. Genes Dev. 6:1631-1642. [DOI] [PubMed] [Google Scholar]
- 23.Lazaris-Karatzas, A., and N. Sonenberg. 1992. The mRNA 5′ cap-binding protein, eIF-4E, cooperates with v-myc or E1A in the transformation of primary rodent fibroblasts. Mol. Cell. Biol. 12:1234-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lejbkowicz, F., C. Goyer, A. Darveau, S. Neron, R. Lemieux, and N. Sonenberg. 1992. A fraction of the mRNA 5′ cap-binding protein, eukaryotic initiation factor 4E, localizes to the nucleus. Proc. Natl. Acad. Sci. USA 89:9612-9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao, W., C. Y. Ho, Y. L. Yan, J. Postlethwait, and D. Y. Stainier. 2000. Hhex and scl function in parallel to regulate early endothelial and blood differentiation in zebrafish. Development 127:4303-4313. [DOI] [PubMed] [Google Scholar]
- 26.Manfioletti, G., V. Gattei, E. Buratti, A. Rustighi, A. De Iuliis, D. Aldinucci, G. H. Goodwin, and A. Pinto. 1995. Differential expression of a novel proline-rich homeobox gene (Prh) in human hematolymphopoietic cells. Blood 85:1237-1245. [PubMed] [Google Scholar]
- 27.Martinez Barbera, J. P., M. Clements, P. Thomas, T. Rodriguez, D. Meloy, D. Kioussis, and R. S. Beddington. 2000. The homeobox gene Hex is required in definitive endodermal tissues for normal forebrain, liver and thyroid formation. Development 127:2433-2445. [DOI] [PubMed] [Google Scholar]
- 28.Nathan, C. A., P. Carter, L. Liu, B. D. Li, F. Abreo, A. Tudor, S. G. Zimmer, and A. De Benedetti. 1997. Elevated expression of eIF4E and FGF-2 isoforms during vascularization of breast carcinomas. Oncogene 15:1087-1094. [DOI] [PubMed] [Google Scholar]
- 29.Nathan, C. A., L. Liu, B. D. Li, F. W. Abreo, I. Nandy, and A. De Benedetti. 1997. Detection of the proto-oncogene eIF4E in surgical margins may predict recurrence in head and neck cancer. Oncogene 15:579-584. [DOI] [PubMed] [Google Scholar]
- 30.Newman, C. S., F. Chia, and P. A. Krieg. 1997. The XHex homeobox gene is expressed during development of the vascular endothelium: overexpression leads to an increase in vascular endothelial cell number. Mech. Dev. 66:83-93. [DOI] [PubMed] [Google Scholar]
- 31.Obinata, A., Y. Akimoto, Y. Omoto, and H. Hirano. 2002. Expression of Hex homeobox gene during skin development: increase in epidermal cell proliferation by transfecting the Hex to the dermis. Dev. Growth Differ. 44:281-292. [DOI] [PubMed] [Google Scholar]
- 32.Olivieri, N. F. 1999. The beta-thalassemias. N. Engl. J. Med. 341:99-109. [DOI] [PubMed] [Google Scholar]
- 33.Pearson, M., R. Carbone, C. Sebastiani, M. Cioce, M. Fagioli, S. Saito, Y. Higashimoto, E. Appella, S. Minucci, P. P. Pandolfi, and P. G. Pelicci. 2000. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 406:207-210. [DOI] [PubMed] [Google Scholar]
- 34.Perrotti, D., V. Cesi, R. Trotta, C. Guerzoni, G. Santilli, K. Campbell, A. Iervolino, F. Condorelli, C. Gambacorti-Passerini, M. A. Caligiuri, and B. Calabretta. 2002. BCR-ABL suppresses C/EBPα expression through inhibitory action of hnRNP E2. Nat. Genet. 30:48-58. [DOI] [PubMed] [Google Scholar]
- 35.Rousseau, D., R. Kaspar, I. Rosenwald, L. Gehrke, and N. Sonenberg. 1996. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc. Natl. Acad. Sci. USA 93:1065-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sonenberg, N., and A. C. Gingras. 1998. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell Biol. 10:268-275. [DOI] [PubMed] [Google Scholar]
- 37.Strudwick, S., and K. L. Borden. 2002. The emerging roles of translation factor eIF4E in the nucleus. Differentiation 70:10-22. [DOI] [PubMed] [Google Scholar]
- 38.Stuurman, N., A. de Graaf, A. Floore, A. Josso, B. Humbel, L. de Jong, and R. van Driel. 1992. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J. Cell Sci. 101:773-784. [DOI] [PubMed] [Google Scholar]
- 39.Tan, A., P. Bitterman, N. Sonenberg, M. Peterson, and V. Polunovsky. 2000. Inhibition of Myc-dependent apoptosis by eukaryotic translation initiation factor 4E requires cyclin D1. Oncogene 19:1437-1447. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka, T., T. Inazu, K. Yamada, Z. Myint, V. W. Keng, Y. Inoue, N. Taniguchi, and T. Noguchi. 1999. cDNA cloning and expression of rat homeobox gene, Hex, and functional characterization of the protein. Biochem. J. 339:111-117. [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas, P. Q., A. Brown, and R. S. Beddington. 1998. Hex: a homeobox gene revealing peri-implantation asymmetry in the mouse embryo and an early transient marker of endothelial cell precursors. Development 125:85-94. [DOI] [PubMed] [Google Scholar]
- 42.Topcu, Z., D. L. Mack, R. A. Hromas, and K. L. Borden. 1999. The promyelocytic leukemia protein PML interacts with the proline-rich homeodomain protein PRH: a RING may link hematopoiesis and growth control. Oncogene 18:7091-7100. [DOI] [PubMed] [Google Scholar]
- 43.Topisirovic, I., A. D. Capili, and K. L. Borden. 2002. Gamma interferon and cadmium treatments modulate eukaryotic initiation factor 4E-dependent mRNA transport of cyclin D1 in a PML-dependent manner. Mol. Cell. Biol. 22:6183-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Topisirovic, I., B. Culjkovic, N. Cohen, J. M. Perez, L. Skrabanek, and K. L. Borden. 2003. The proline-rich homeodomain protein, PRH, is a tissue-specific inhibitor of eIF4E-dependent cyclin D1 mRNA transport and growth. EMBO J. 22:689-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, S., I. B. Rosenwald, M. J. Hutzler, G. A. Pihan, L. Savas, J. J. Chen, and B. A. Woda. 1999. Expression of the eukaryotic translation initiation factors 4E and 2α in non-Hodgkin's lymphomas. Am. J. Pathol. 155:247-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ward, J. O., M. J. McConnell, G. W. Carlile, P. P. Pandolfi, J. D. Licht, and L. P. Freedman. 2001. The acute promyelocytic leukemia-associated protein, promyelocytic leukemia zinc finger, regulates 1,25-dihydroxyvitamin D3-induced monocytic differentiation of U937 cells through a physical interaction with vitamin D(3) receptor. Blood 98:3290-3300. [DOI] [PubMed] [Google Scholar]
- 47.Yatskievych, T. A., S. Pascoe, and P. B. Antin. 1999. Expression of the homebox gene Hex during early stages of chick embryo development. Mech. Dev. 80:107-109. [DOI] [PubMed] [Google Scholar]